Abstract
Nicotinamide phosphoribosyltransferase (NAMPT) is highly evolved to capture nicotinamide (NAM) and replenish the nicotinamide adenine dinucleotide (NAD+) pool during ADP-ribosylation and transferase reactions. ATP-phosphorylation of an active-site histidine causes catalytic activation, increasing NAM affinity by 160,000. Crystal structures of NAMPT with catalytic site ligands identify the phosphorylation site, establish its role in catalysis, demonstrate unique overlapping ATP and phosphoribosyltransferase sites, and establish reaction coordinate motion. NAMPT structures with beryllium fluoride indicate a covalent H247-BeF3− as the phosphohistidine mimic. Activation of NAMPT by H247-phosphorylation causes stabilization of the enzyme-phosphoribosylpyrophosphate complex, permitting efficient capture of NAM. Reactant and product structures establish reaction coordinate motion for NAMPT to be migration of the ribosyl anomeric carbon from the pyrophosphate leaving group to the nicotinamide-N1 while the 5-phosphoryl group, the pyrophosphate moiety, and the nicotinamide ring remain fixed in the catalytic site.
Keywords: nicotinamide, nicotinamide phosphoribosyltransferase, phosphohistidine, phosphoribosyltransferase, NAD+ regulation
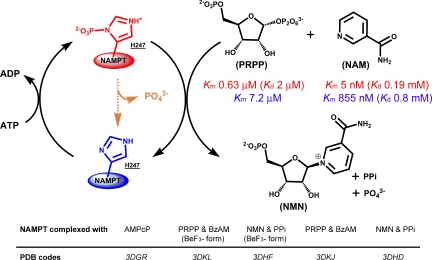
NAD+ is an essential cofactor in metabolic redox chemistry. It also functions in DNA repair reactions, poly- and mono-ADP-ribose polymerases, formation of cyclic ADP-ribose, and the Sirtuins (SIRT) (1–5). These reactions can deplete NAD+ by cleaving the N-ribosyl bond to generate free nicotinamide (NAM). Nicotinamide phosphoribosyltransferase (NAMPT; also known as pre-β cell colony enhancing factor, PBEF, and visfatin, an adipokine) is designed to efficiently recycle NAM (Km = 5 nM) by reaction with α-D-5-phosphoribosyl-1-pyrophosphate (PRPP, Fig. 1) to sustain the pool of NAD+ (6).
Fig. 1.
Human NAMPT exists in P-H247 (red) and H247 (blue) forms. P-H247 NAMPT is 1,125-fold more active (kcat/Km values) and requires continual hydrolysis of ATP. P-H247 NAMPT has low Km values for NAM and PRPP (red) while H247 NAMPT has higher Km values (blue). NAMPT slowly hydrolyzes ATP (orange) even in the absence of NAM and PRPP. In the presence of ATP, the stoichiometry of NMN:ADP formation is always 1.0 or less, indicating P-H247 hydrolysis on each NAM synthetic cycle. PRPP has a tighter affinity than NAM for P-H247, suggesting a preferential ordered binding of substrates to the enzyme, with PRPP binding first. Five crystallographic structures illustrate the main steps of this complex mechanism.
In mammals, NAMPT is the rate-limiting enzyme for NAD+ salvage from NAM and its overexpression increased cell lifespan (7) via activation of SIRT1 (8). Recently, NAMPT was also identified as the enzyme regulating mitochondrial NAD+ levels (9) and extending cell lifespan via the functions of SIR3 and SIR4. Because of its role in NAD+ maintenance, NAMPT is a target in cancer research (10). Its inhibition by FK866 causes depletion of NAD+ and a decrease in SIRT1 activity (8) resulting in cell senescence.
Bacterial nicotinic acid phosphoribosyltransferase (NAPT) is mechanistically and structurally similar to human NAMPT but is specific for nicotinic acid rather than NAM (11). Both transferases have an ATPase activity that nonstoichiometrically couples with the transferase reaction to shift the thermodynamic equilibrium toward mononucleotide formation (6, 12). For Salmonella typhimurium NAPT (13, 14) the ATPase reaction involves phosphorylation of H219. Conservation of this His in Saccharomyces cerevisae NAPT (1VLP, H232) and human NAMPT [2GVG (11), H247] suggests autophosphorylation of H247 in human NAMPT. This assignment is supported by mutagenesis studies [H247E (15) and H247A (11, 15)] where mutants exhibited a lower enzymatic activity. Recent ADP/ATP isotope exchange and thermodynamic experiments with human NAMPT established a high-energy, unstable phosphorylated enzyme intermediate (ΔG°hydrolysis = −9.2 kcal mol−1) consistent with a phosphohistidine at H257, but structural evidence has been lacking (6).
At physiological MgATP concentrations (6), 80% of human NAMPT is phosphorylated, but none of the reported NAMPT crystal structures (11, 15, 16) have described the phosphorylated enzyme or characterized the ATP/ADP binding site. The NAMPT inhibitor, FK866 (10) has validated the enzyme as a target for the development of new anticancer agents. Although it binds with high picomolar affinity, the Km/Ki is only 33, because the enzyme has an extraordinary affinity for NAM (KmNAM of 5 nM) when it is in its phosphorylated state (6). The unusual catalytic site of human NAMPT (11) [supporting information (SI) Fig. S1] may explain the difficulty encountered with FK866 in clinical trials (17). Considering the elaborate reaction catalyzed by NAMPT (3 substrates and 4 products, including the ATPase system, Fig. 1), the previously reported binary complexes have characterized only a small part of this complicated molecular complex (11, 15, 16).
Here we define the molecular mechanism by which the ATPase activity of NAMPT is coupled to increase affinity for NMA. We also define the atomic excursions in both ATP and non-ATP-coupled NMN synthesis catalyzed by NAMPT. Two crystal structures of human NAMPT in ternary complexes with reactants and products are used to establish reaction coordinate motion for ribosyl transfer. An ATP binding site is characterized with bound α,β-methylene-adenosine-diphosphate (AMPcP). With beryllium fluoride, we report a unique mimic of the active phosphorylated NAMPT. The BeF3− ion appears in a covalent interaction with (Nδ1)H247. Two data sets with resolution to 1.8 Å and 1.9 Å define the molecular interactions for ATP-coupled NMN synthesis. Those data define the mechanism of activation by ATP and support a reaction mechanism of ribosyl group migration, similar to that recognized in other N-ribosyltransferases.
Results
Overall Fold of Human NAMPT.
Crystal structures of H247-unphosphorylated human NAMPT, in complex with AMPcP, PRPP·BzAM, and NMN·PPi, were determined at resolutions of 2.1, 2.0, and 2.0 Å, respectively. Two BeF3−-containing crystal structures were determined at resolutions ranging from 1.8 to 1.9 Å and provided models of phosphorylated NAMPT bound to reactants or products in the active site. All models contain residues 9–41 and 54–484 (residues 42–53 are disordered and not represented by electron density) and display good stereochemistry (Table S1).
NAMPT forms a dimer with an extensive intermolecular interface that buries 4,064 Å2 of surface area. Each monomer is composed of 16 α-helices and 18 β-strands (Fig. S2a) and can be divided into 3 domains (11, 15, 16). The dimer interface includes numerous salts bridges, hydrogen bonds, and van der Waals interactions. Each active site lies at the entrance of a tunnel at the dimer interface (Fig. S2b). Residues contributed from 5 internal β-strands (F193-β6, 383-GGG-385-β12, β11, 311-RPD-313-β8, D279-β7) and 7 external α-helices (α8, α6, α15, α14, α12, α11, H247-α10) interact with the 5-phosphoryl group and the ribosyl group of the substrates. Additional interactions are provided by the second monomer (Y18*-α1*, K415*/K423*-loop between β14* and β15*) and consist primarily of hydrogen bonds with the PPi moiety of the substrates. This arrangement is proposed to sequester the active site from solvent as discussed for other phosphoribosyltransferases (18–21).
Substrate and Product Complexes with Nonphosphorylated NAMPT.
In ternary complexes of NAMPT, the catalytic site is filled to provide complete characterization of protein and divalent cation contacts to NMN and Mg2PPi. The base of the NMN mononucleotide is sandwiched between F193 of the β6 strand and that of Y18* from the α1* helix (Fig. S3a), exhibiting classic π-stacking interactions. The amide function of NMN is located between R311 (β8) and D219, with the later sharing a hydrogen bond with the NMN amido group, to provide NAM specificity (11, 15). Near the solvent interface, the 5-phosphoryl group is surrounded by several water molecules and is partially neutralized by an oxyanion hole (G355, 383-GGG-385, Fig. S3b). Beneath the NMN, an oxygen of the incipient PPi nucleophile, OPα (Ο5), is 3.5 Å from C-1 of the ribose ring (Fig. S3b) and PPi forms polar interactions with cationic residues from the loop between β14* and β15*, R40*-β1 and R392*. The PPi is also partially neutralized by 2 Mg2+ ions with octahedral coordination. The first Mg2+ makes contact with O2 and O5 (each at 2.1 Å, Fig. S3b) and 4 water molecules while the second Mg2+ has (Oδ1)D313, O2R, and O3R at 2.4, 2.2, and 2.4 Å, respectively, in the coordination sphere (Fig. S3a). The second cation is proposed to play the more important role in catalysis as D313 is part of a His-Asp-Asp triad conserved in all annotated NAMPT and NAPT enzymes (H247, D279, and D313).
The ternary complex formed by NAMPT, PRPP, and BzAM differs from the NMN·Mg2PPi·NAMPT complex in that the PRPP-containing structure does not exhibit bound Mg2+, despite its presence in the crystallization mixture (Fig. S3 c and d). In this case, pyrophosphate charge stabilization is achieved by replacing the Mg2+ with arginine and lysine groups (Fig. S3d). The BzAM ring, a chemically unreactive mimic of NAM, is tilted 8° with respect to the NAM of NMN and is positioned 3.8 Å from C-1 of the PRPP between the F193 and Y18* residues (Fig. S3c).
AMPcP Binding Mode.
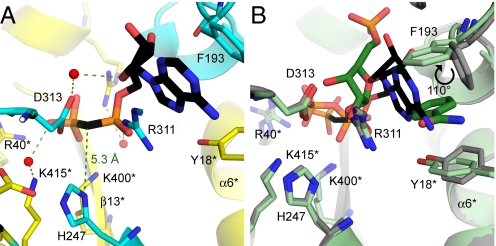
The stable ADP analogue, AMPcP, involves an unexpected π-stacking interaction of the adenine ring with F193 (β6) and R311 (β8, Fig. 2A). Stabilization of the negatively charged diphosphate moiety is provided by arginine and lysine residues (R40*, R392*, K400*, Fig. 2A). The 2′ and 3′-OHs of the ribosyl group make favorable hydrogen bonds with the backbone carbonyl oxygens (G353 at 2.7 Å and F193 at 2.6 Å, respectively). The β-phosphate points away from the Nδ1 phosphorylation site of H247 with a distance of 5.3 Å between H247 atom and the methylene group of AMPcP (Fig. 2A). A rotation of ≈130° of the β-phosphate around the CH2/α-phosphate bond, would be needed to align a reactive γ-phosphate group for favorable phosphorylation of H247.
Fig. 2.
The AMPcP-binding site of human NAMPT. (A) Ribbon diagram showing the AMPcP binding mode with H247-unmodified human NAMPT. The 2 monomers are colored in yellow and cyan, respectively. AMPcP is shown in black and water molecules are depicted as red spheres with hydrogen bonds represented as olive dashed lines. The 5.3 Å green dashed line represents the distance from (Nδ1)H247 to the methylene α,β-bridge of AMPcP. (B) The structural overlap for PRPP·BzAM (substrates shown as green sticks with an overall representation of the structure in light green) and AMPcP complexes in nonphosphorylated human NAMPT (black stick representation of AMPcP with the corresponding structure in gray) is demonstrated. Produced with PyMol v 1.1.
Comparison of PRPP and AMPcP positions in the catalytic site demonstrates considerable steric overlap, precluding simultaneous ATPase and transferase reactions. Most of the amino acids in the active site have similar conformations in both AMPcP and PRPP·BzAM complexes (Fig. 2B). The diphosphate group of AMPcP and pyrophosphate group of PRPP superimpose, as do the amino acids in this region. When PRPP binds to His-BeF3−-NAMPT, the side chain of F193, which is involved in π-stacking with BzAM and adenine rings, undergoes a 30° tilt of its side chain toward the active site and a 110° rotation of the phenyl group around the CβCγ axis (Fig. 2B).
Structures of Substrates and Products Bound to the Phosphorylated Mimic of NAMPT.
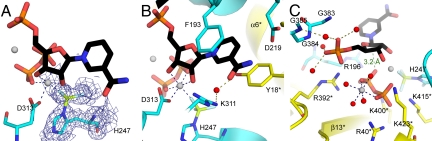
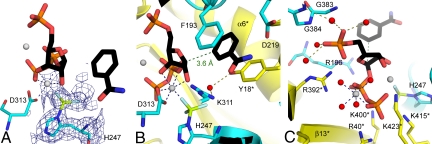
NAMPT in a complex with NMN, Mg2PPi, and BeF3− revealed a well-defined and continuous electron density around (Nδ1)H247 that was modeled as a BeF3− (Fig. 3A). A similar geometry was found for the H247-beryllium complex with Mg2PRPP·BzAM (Fig. 4A). The (Nδ1)H247 and 3 fluoride atoms adopt a tetrahedral geometry around the beryllium atom, with a 1.7 to 1.8 Å beryllium-nitrogen bond. This geometry closely resembles the N-PO3 geometry of N-phosphohistidine. The pyridyl and benzamide rings are sandwiched between F193 (β6) and Y18* (α1*), similar to that observed in the BeF3−-free structures (Figs. 3B and 4B). Both pyrophosphate and 5′-phosphoryl groups contribute to catalytic site organization in the complex with BeF3−. Pyrophosphate interacts with 2 magnesium ions and with arginine and lysine residues from the loop separating the β14* and β15* strands (Figs. 3C and 4C). The 5′-phosphate in these complexes is bound to an oxyanion hole (383-GGG-385, 353-GDGV-356, Figs. 3C and 4C).
Fig. 3.
H247-BeF3− human NAMPT in complex with NMN and Mg2PPi. (A) 1.8 Å 2Fo–Fc electron density map contoured at 1σ level around H247-BeF3− (blue); Mg2+ atoms are depicted as gray spheres. (B) Interactions for the NAM, ribose and BeF3− moieties, with 2 monomers presented in yellow and cyan. NMN is colored in black, Be is in lemon, and F in light blue. Blue dashed lines represent Mg2+ coordination. Red spheres represent water molecules and the olive dashed lines represent the corresponding hydrogen bonds involved. (C) The main interactions for the diphosphate moiety. The green dashed line represents distance d2 as summarized in Table 1. Produced with PyMol v 1.1.
Fig. 4.
H247-BeF3− human NAMPT in complex with Mg2PRPP and BzAM. (A) Final 2Fo−Fc electron density map for H247-BeF3− at 1.9 Å. The contour level (blue) is at 1σ; gray spheres correspond to Mg2+ atoms. (B) Interactions for the BzAM, ribose and BeF3− moieties. The 2 monomers are depicted in yellow and cyan, while PRPP·BzAM is colored in black, Be is in lemon, and F represented in light blue. Blue dashed lines represent the Mg2+ coordination while the green dashed line represents distance d1 as summarized in Table 1. Water molecules and the corresponding hydrogen bonds are depicted as red spheres and olive dashed lines, respectively. (C) Interactions for the diphosphate moiety. Produced with PyMol v 1.1.
Although the overall binding contacts of NMN·Mg2PPi to NAMPT with and without BeF3− are similar, there are significant differences. First, the distances in the reaction coordinate [N(C)/C-1; Fig. S3c and S4b and OPα(O5)/C-1 Fig. S3b; Fig. 3C] shorten by 0.3 Å (d1 and d2 of Table 1). Second, coordination for one of the magnesium ions differs. The water molecule in the first coordination sphere [(Oδ1)D313, O2R, O3R, OPα(O5), OPβ(O2), and W; Fig. S3a], which makes a contact with Mg2+ at 2.0 Å, is replaced by a 1.8 Å, fluoride contact, F1 (Fig. 3B). Finally, the Mg2PRPP·BzAM complex binds to the BeF3− form of NAMPT with 2 Mg2+ ions (Fig. 4 B and C) where, in comparison, no Mg2+ is found in the enzyme without BeF3− (Fig. S3 c and d). Two of the fluorine atoms are positioned to form hydrogen bonds to (Nδ1)H247 (F2, 2.9 Å) and to (Nζ)K400* and (Nζ)K415* (F3, 2.6 and 2.8 Å, respectively, Fig. 5). As a consequence, the loop separating the β14* and β15* strands is in a position to provide increased stability of PRPP in the active site. The interactions between NAMPT and PRPP are stronger when the enzyme is activated by BeF3− and thermodynamic and kinetic experiments confirm those structural observations (Fig. 1; Figs. S6 and S7).
Table 1.
Atomic excursions observed in the four NAMPT complexes
| Human NAMPT |
||||||||
|---|---|---|---|---|---|---|---|---|
| N(C) | d1* | C-1 | d2* | OPα(O5) | 5PO | d3* | d4* | |
| NMN·PPi | 1.5 | 3.5 | 3.5 | 3.0 | ||||
| Reaction (I) | 0.4†; −8‡ | 2.2 | 0.7; −25 | 0.3 | ||||
| PRPP·BzAM | 3.8 | 1.5 | 2.4 | 2.7 | ||||
| PRPP·BzAM (Be) | 3.6 | 1.5 | 2.3 | 2.7 | ||||
| Reaction (II) | 0.7; 12 | 1.8 | 0.3; 11 | 0.2 | ||||
| NMN·PPi (Be) | 1.5 | 3.2 | 3.7 | 2.9 | ||||
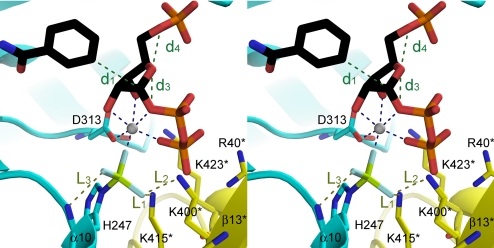
*Distances d1, d2, d3, and d4 are linear atomic distances in angstroms as depicted in Fig. 5.
†Distances are linear interatomic distances changes in angstroms.
‡Angles are tilts in degrees of the corresponding atom; a positive value reflects a tilt inside the active site.
Fig. 5.
Interaction between (Nδ1)H247-BeF3− and the neighboring residues (cross-eye stereo representation). The structure of one monomer is in yellow, the other is in cyan, PRPP·BzAM is in black, Be is in lemon, and F is in light blue. Blue dashed lines represent Mg2+ interactions with PRPP and BeF3−, olive dashed lines represent interatomic distances L1, L2, and L3 and green dashed lines represent distances d1, d2, d3, and d4 as summarized in Table 1. Produced with PyMol v 1.1.
Substrate Affinity.
Direct binding studies of PRPP with the phosphorylated enzyme gave a Kd of 2.0 ± 0.2 μM, (Fig. 1), similar to its kinetic constant for interaction with the enzyme·NAM complex (KmPRPP = 0.63 ± 0.03 μM, Fig. 1). The direct binding of NAM to NAMPT or phospho-NAMPT is near mM in affinity. However, the Km for NAM decreases dramatically to the nanomolar range for phosphorylated NAMPT saturated with PRPP (6) (Fig. 1). Thus, PRPP binding precedes binding of NAM to create a high-affinity interaction (Fig. 1). The crystal structures reveal that phosphorylation of NAMPT on His-247 enhances affinity between the enzyme and PRPP and as a result, lowers the KmNAM for the E-P·PRPP·NAM Michaelis complex (Figs. 2, 4, and 5).
Kinetic Properties of the NAMPT-BeF3− Complex.
Formation of (Nδ1)H247-BeF3− under crystallographic conditions raised the question of whether BeF3− might be a significant inhibitor by preventing phosphorylation of H247 by ATP. ADP/ATP isotope exchange experiments (6) in the presence of BeF3− would be inhibited if the complex of enzyme and BeF3− is kinetically stable. Under conditions that varied BeF3− from 0 to 500 μM, no significant inhibition of the isotope exchange was observed. Under conditions for crystallography the beryllium complex mimics the unstable, transient nature of the phosphohistidine intermediate on the normal reaction path, but does not form an inhibitory complex under exchange conditions.
Ribosyl Migration.
Ternary reactant and product complexes with NAMPT reveal atomic positions as the reaction progresses. This approach has been informative in defining reaction coordinate motion in several N-ribosyltransferases [purine nucleosidase phoshorylase, hypoxanthine-guanine phosphoribosyltransferase, and adenine phosphoribosyltransferase (20, 22, 23)].
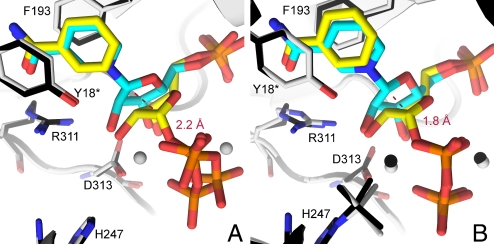
Molecular overlays of the NMN·Mg2PPi and PRPP·BzAM complexes from unmodified H247 human NAMPT establish that the major atomic motion of catalytic site reactants involves translation of the C-1 portion of the ribosyl ring. In contrast, the base (NAM, BzAM), the 5-monophosphate and the PPi moiety remain relatively fixed in position (Fig. 6A, Table 1).
Fig. 6.
Ribosyl migration during NMN synthesis catalyzed by human NAMPT. (A) Superimposition of PRPP·BzAM (yellow, with structure in black) and NMN·Mg2PPi (cyan, with structure in gray) complexes in the active site of the H247-unmodified human NAMPT. Mg2+ atoms are depicted as gray spheres. The red dashed line represents the interatomic distance between C-1 in reactants and C-1 in products. (B) Superimposition of the Mg2PRPP·BzAM (yellow, with structure in black) and NMN·Mg2PPi (cyan, with structure in gray) complexes in the active site of the H247-BeF3− complex of human NAMPT. Mg2+ atoms are depicted as black and gray spheres (with respect to the corresponding structure) and the red dashed line represents the interatomic distance between C-1 in reactants and C-1 in products. Produced with PyMol v 1.1.
A comparison of reaction coordinate motion can also be made between unmodified and the (Nδ1)H247-BeF3− forms of NAMPT. With H247 unmodified NAMPT, the atomic positions reveal that the ribosyl C-1 excursion from substrates to the products is 2.2 Å. In the BeF3− form of the enzyme the C-1 anomeric carbon migrates 1.8 Å upon conversion of reactants to products (Fig. 6B, Table 1). While PRPP is bound in the same conformation in both structures, the sugar pucker of bound NMN differs, revealing a 24° tilt of the ribosyl ring oxygen C-1/O4R/C-4 toward the ribosyl 2′- and 3′-Hs in BeF3−-modified NAMPT. The ribosyl ring oxygen of NMN (O4R) in unmodified NAMPT is sandwiched between the oxygens of the incipient PPi nucleophile [OPα(O5)] and of the 5′-phosphate (O5R) with distances of 3.5 and 3.0 Å, respectively, for these electron-rich centers (d3 and d4, Table 1). In the BeF3− form of NAMPT the distance from C-1 to the PPi moiety is marginally longer (d3 = 3.7 Å) but the interaction between the ring O4R oxygen and the lone pairs of O5R are not significantly changed from the unmodified enzyme (3.0 and 2.9 Å, respectively).
Discussion
Significance of NAMPT.
The NAMPT protein has been reported to have several distinct biological roles. As visfatin, it is proposed to interact with insulin receptors as an adipokine. As PBEF (pre-B cell enhancing factor), it acts as a novel cytokine to stimulate early B-lineage precursor cells (24). As a nicotinamide phosphoribosyltransferase, it recycles nicotinamide to NAD+ for its essential roles in metabolism and cell regulatory pathways. NAMPT inhibition is reported to have antiproliferative activity and FK866 is a nanomolar inhibitor in clinical trials as an anticancer agent. Despite its biological significance, the mechanistic link to structure was not previously defined. The structure of the covalent high-energy phosphohistidine species, disposition of the ATP and 5-phosphoribosyl 1-pyrophosphate binding sites, and mechanism of activation by formation of the phosphohistidine were unknown at the outset of this study.
Prokaryotes and lower eukaryotes (3, 25, 26), deamidate NAM to NA for recycling, but in mammals, NAMPT is the only path for NAM salvage. Reactions including ADP-ribosylations and NAD+-linked deacetylations can deplete the NAD+ pool by cleaving the N-ribosyl bound to NAM (1–4). NAMPT, when phosphorylated at H247, is a highly efficient scavenger of free NAM by virtue of its 5 nM Km and its catalytic efficiency of 1.8 × 106 M−1 s−1 (6). These kinetic properties ensure that nanomolar concentrations of this vitamin will be recycled to constantly restore the NAD+ pool. Inhibition of NAMPT activity (10, 17) causes low levels of cellular NAD+, a decrease of SIRT1 activity and induction of cell senescence (8). The link of NAMPT to the NAD+ pool, cell lifespan, and cellular senescence make this enzyme an important target for cell lifespan and cancer research.
ATPase and Phosphotransferase Functions Share a Common Site.
The activation of NAMPT by ATP hydrolysis has suggested that phosphorylation on a conserved histidine is a common catalytic feature for the pyridine phosphoribosyltransferases (6, 11, 15). In the presence of Mg2+ and ATP, NAMPT undergoes a fast, near quantitative phosphorylation (6). Phospho-NAMPT is approximately 3 orders of magnitude more active than unphosphorylated enzyme. Structures of the NAMPT·AMPcP complex show the binding of adenine nucleotides in proximity to H247. Occupation of the same ribosyl binding site by both AMPcP and PRPP requires sequential H247 phosphorylation and phosphoribosyltransferase activities (Figs. 1 and 2B). Kinetically, ADP/ATP isotope exchange occurs without the other reactants (6). This structural arrangement explains the inhibition of ADP/ATP exchange reactions in the presence of PRPP and/or PPi (6). Likewise, the ribosyl site overlap explains the inhibition of NMN synthesis observed at high ATP concentrations, even though ATP is required to activate the enzyme.
The conformation of AMPcP bound to NAMPT would place the γ-phosphate of ATP in a position to phosphorylate H247 with a 130° rotation of the β-phosphate group around the CH2/α-phosphate bond. Subsequent release of ADP is then required to permit binding of PRPP. The reaction scheme for NAMPT is sequential with (i) ATP binding, (ii) H247 phosphorylation, (iii) ADP release, (iv) PRPP binding, (v) NAM binding, and (vi) the chemical step. The stoichiometry of ATP hydrolyzed to NMN formed is always >1.0 in vitro, a consequence of some phosphorylated enzyme being hydrolyzed before NMN is formed (Fig. 1).
BeF3− as a Mimic Phospho-H247.
Crystallization of NAMPT with substrates or products in the presence of beryllium fluoride provided a structural mimic of the active enzyme with phospho-H247. Phospho-H247 is a high-energy complex (−9.2 kcal mol−1 for hydrolysis) with a short lifetime (6). Two X-ray data sets with BeF3− exhibit continuous electron density that is readily modeled as a tetrahedral species formed between the (Nδ1)H247 and BeF3− (Figs. 3A and 4A). New protein interactions are observed in the vicinity of the H247-BeF3− moiety (Fig. 5). Although this complex of BeF3− with H247 is not a chemically stable inhibitory species, it provides a structural model for the phosphorylated NAMPT (Fig. 5).
Structural Mechanism of Phospho-H247 Activation.
By coupling ATP hydrolysis to NMN synthesis, the catalytic efficiency of the system is increased by ≈103 and the chemical equilibrium shifted toward NMN as long as ATP is being hydrolyzed (6). Although the kinetic and thermodynamic consequences of the ATPase activity are known, the structural mechanisms underlying the coupling of phosphohistidine formation to the phosphoribosyltransferase rate and the chemical equilibrium were unknown. Interactions provided by (Nδ1)H247-BeF3− together with the pyrophosphate group stabilize the K375, R385 loop that links β14* and β15* in human NAMPT. This loop is disorganized in the apo enzyme and when organized, is integrated into the active site. Phosphorylated histidine provides a new structural element that contributes to higher affinity between PRPP and the phosphorylated enzyme (Fig. 5). The presence of (Nδ1)H247-BeF3− (and by extrapolation, the histidyl phosphate), is not essential for loop closure as the loop is also closed for NMN synthesis when H247 is not modified (Fig. S3). However, the phosphorylated enzyme strengthens the interactions between the two monomers forming the catalytic site (hydrogen bonds between fluoride atoms, from H247-BeF3−, and K400* and K415*, Fig. 5) and provides stronger affinity for PRPP. While NAM alone binds weakly to the enzyme, a Kd/Km change of 160,000 results from the structural modifications between the complexes E·PRPP and E-P·PRPP where E-P·PRPP allows efficient synthesis of NMN (Fig. 1). Formation of a ribocationic transition state to permit ribosyl migration during the reaction coordinate would also be favored by the additional negative charge arising from the vicinal phosphohistidine (15). However, the F to C-1 distances of 4.0 and 4.7 Å would provide only weak electrostatic stabilization of an oxacarbenium by the histidyl phosphate, until these distances were reduced by reaction coordinate motion.
Disposition of the Ribosyl Group.
The ribose moieties of bound NMN and PRPP show considerable differences in sugar pucker near C-1. The BeF3−-NAMPT structures suggest that PRPP binds in a half-chair conformation and is converted to an envelope conformation in the NMN product (Fig. 6B). The reaction coordinate motion for BeF3−-NAMPT involves a 1.8 Å excursion of the ribosyl anomeric carbon and is consistent with the enzymatic mechanism of nucleophilic substitution by electrophile migration (23). The transferase reaction catalyzed by human NAMPT, like other phosphoribosyltransferase reactions, involves a nucleophilic substitution in which NAM and PPi are held tightly to the active site, and the C-1 carbon translocates between those 2 relatively immobile nucleophiles during catalysis. This mechanism requires the formation of a ribocationic transition state at some point in the reaction coordinate, supporting an SN1 mechanism. The mechanism for physiological activation as a consequence of forming phospho-His-247 permits the enzyme system to be saturated at low concentrations of NAM. Small changes in distances in the reaction coordinate also have large effects in catalysis, and may account for the activation of NAMPT when phosphorylated at H247. Thus, in His-unmodified NAMPT the C-1 excursion from PRPP to NAM is 2.2 Å while this reaction coordinate distance is reduced to 1.8 Å in the activated enzyme.
Binding of the Pyrophosphate.
Like other phosphoribosyltransferases (20, 27), arginine and lysine residues form ion pairs to pyrophosphate at the catalytic site. One of the Mg2+ ions reduces the negative charge and links pyrophosphate to the active site and to the ribosyl group by interactions with (Oδ1)D313, (F)BeF3−, O2R, O3R, OPα(O5), and OPβ(O2). The octahedral coordination includes participation of the 2′-OH in the coordination. An important role for this interaction is suggested by the weak inhibition of NAMPT by a 2′-deoxyribocationic analogue of NMN [5P-DADMe-NMN (6)].
Summary of Catalytic Site Features.
Both ATPase and the NAM-PRPP transferase substrates for NAMPT sequentially share the same active site. The chemical instability of phospho-H247 leads to some premature hydrolysis of the complex and explains the inherent excess stoichiometry of ATP hydrolysis to NMN synthesis. This structural overlap also explains inhibition of ATPase by excess NMN and inhibition of NMN formation by excess ATP. Despite the “musical chairs” nature of the catalytic site, the energetic coupling between ATPase and transferase reactions permits NAMPT to catalyze a thermodynamically favorable capture of NAM and efficient NMN synthesis. The enzyme couples only a small portion of the energy provided by chemical hydrolysis of ATP toward the chemical equilibrium. Thus, H247 phosphorylation contributes more than the primary chemical energy. Phosphorylated H247 serves as a hydrogen-bonding link where the modified residue alters the monomers to create the catalytic site into a preferential arrangement to provide higher affinity between the modified enzyme and PRPP. The resulting E-P·PRPP complex provides a more efficient synthesis of NMN from NAM. Crystallization of the enzyme with beryllium fluoride provided unprecedented information on the tetrahedral complex of BeF3−-H247 as a mimic of histidyl phosphate. Finally, the structures permit description of nucleophilic displacement by electrophile migration to define the reaction coordinate motion.
Materials and Methods
Protein Expression and Purification.
Protein Crystallization.
Crystals of human NAMPT were grown by sitting drop vapor diffusion at 18 °C. The initial crystallization condition (25% PEG 3350, 100 mM Tris pH 8.5 and 200 mM sodium chloride) was identified using Hampton Research Index HT screening. The final optimized condition used a 2:1 ratio of protein (5 mg mL−1) to the crystallization buffer (15% PEG 3350, 100 mM Tris pH 8.5, 200 mM sodium chloride, and 20% glycerol). Condensed clusters of thin plate-shaped crystals were obtained from the optimized crystallization condition. Single crystals were obtained using microseeding and were frozen in liquid nitrogen.
Crystals of human NAMPT in complex with AMPcP were obtained by co-crystallizing the enzyme in the presence of 30 mM AMPcP and using the same conditions described above.
H247 unmodified and BeF3−-H247 human NAMPT in complex with PRPP and BzAM were obtained at 18 °C by soaking methods. Crystals for PRPP and BzAM bound to human NAMPT were soaked in the crystallization buffer supplied with PRPP, BzAM, and MgCl2 (5 mM each) before flash-freezing. In a similar way, crystals of the BeF3− complex were obtained by soaking for at least 5 hours, native human NAMPT crystals in the crystallization buffer containing: 5 mM BeCl2, 30 mM NaF, 5 mM MgCl2, 5 mM PRPP, and 5 mM BzAM.
H247 unmodified and H247-BeF3− human NAMPT in complex with NMN and PPi were obtained at 18 °C by soaking methods. The crystals used to characterize NMN and Mg2PPi bound to human NAMPT were soaked in crystallization buffer containing NMN, PPi, and MgCl2 (5 mM each) before flash-freezing. Likewise, crystals of H247-BeF3− enzyme were obtained by soaking, for at least 5 hours, native human NAMPT crystals in the crystallization buffer supplied with 5 mM BeCl2, 30 mM NaF, 5 mM MgCl2, 5 mM NMN, and 5 mM PPi.
Data Collection and Processing.
X-Ray diffraction data were collected at the X29A beamline of Brookhaven National Laboratory on an ADSC Q315 detector at 100 K and processed with the HKL2000 program suite (28). The data processing statistics are summarized in Table S1.
Structure Determination and Refinement.
The structures of all liganded human NAMPT were determined by molecular replacement in Molrep (29) using the published structure of human NAMPT [2GVL (11)] as the search model. The models without any ligand were first rebuilt in COOT (30) and refined in Refmac5 (31). The ligands were built in COOT using the Fo−Fc map and refined in Refmac5. Mg2+ positions were confirmed by the anomalous difference Fourier map calculated from Mn2+ substituted crystal (Fig. S5). Apo crystals of NAMPT were soaked with 5 mM BeCl2, 30 mM NaF, 5 mM PPi, 5 mM NMM, and 5 mM MnCl2 for 5 h. Diffraction data for a crystal of the resulting complex were collected at an X-ray wavelength of 1.743 Å to a resolution of 2.2 Å. Sigma weighted 2mFo−DFc phases were generated by minimization of the NAMPT coordinates (PDB code 3DHF) against this data set. Anomalous difference Fourier synthesis at 3 Å revealed 2 peaks of each NAMPT subunit (14.4σ and 15σ of subunit A and 15.5σ and 16.5σ of subunit B, Fig. S5) in the immediate vicinity of the Mg sites modeled in the original structure, consistent with replacement of Mg by Mn. All of the final models were validated by Procheck (32). The refinement statistics are summarized in Table S1.
ADP/ATP Isotope Exchange Assay.
The protocol is similar to the one previously described (6) (SI Text).
Substrates Affinities with NAMPT and Phosphorylated NAMPT.
Dissociation constants (Kd) for NAM were determined by ultrafiltration. Nicotinamide (120 to 3800 μM) was incubated in buffer (50 mM Hepes, 100 mM KCl, 5 mM MgCl2, 1 mM THP, pH 7.5, supplied with 200,000 cpm [CONH2-14C]NAM, with or without 3.6 mM MgATP, at 25 °C) inside ultrafiltration units (Millipore, Biomax UltraFree 0.5 mL centrifugal filter, 10 KD NMWL). NAMPT was added to a 300-μM final concentration. After mixing, 25 μL of solution (upper sample) was collected and added to a scintillation vial containing 1 mL of water. The remaining 75 μL of solution from each ultrafiltration unit were spun down (4 min, 10,000 rpm). After mixing, 25 μL of filtrate (lower sample) was collected and added to a scintillation vial containing 1 mL of water. Scintillation liquid was added to each vial (10 mL) and radioactivity was assayed for 10 min. Data analysis from the upper and lower parts allows determination of the Kd (Fig. S6). The affinity for PRPP and the phosphorylated NAMPT was assayed by fluorescence (λEx = 288 nm, λEm = 330 nm with SlitEx = 2.5, SlitEm = 2.5). The enzyme (2 μM) was dissolved in buffer (50 mM Hepes, 100 mM KCl, 5 mM MgCl2, 2.5 mM MgATP, 1 mM THP, pH 7.5, 25 °C). Successive additions of PRPP (final concentration 130 μM) allow titration of NAMPT followed by increase of fluorescence emission. Data analysis provides the corresponding Kd value (Fig. S7).
Supplementary Material
Acknowledgments.
We are grateful to Dr. Udupi Ramagopal for his helpful comments on this manuscript. This work was supported by National Institutes of Health (NIH) research Grant GM-41916. Data for this study were measured at beamline X29A of the National Synchrotron Light Source. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy, and from the National Center for Research Resources of the NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factor data have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3DGR (human NAMPT·AMPcP complex), 3DHD (human NAMPT·NMN·Mg2PPi complex), 3DKJ (human NAMPT·PRPP·BzAM complex), 3DKL (human BeF3−-NAMPT·Mg2PRPP·BzAM complex), and 3DHF (human BeF3−-NAMPT·NMN·Mg2PPi complex)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0903898106/DCSupplemental.
References
- 1.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 2.Guarente L, Picard F. Calorie restriction–the SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 3.Magni G, et al. Enzymology of NAD+ homeostasis in man. Cell Mol Life Sci. 2004;61:19–34. doi: 10.1007/s00018-003-3161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marmorstein R. Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases. Biochem Soc Trans. 2004;32:904–909. doi: 10.1042/BST0320904. [DOI] [PubMed] [Google Scholar]
- 5.Ziegler M. New functions of a long-known molecule. Emerging roles of NAD in cellular signaling. Eur J Biochem. 2000;267:1550–1564. doi: 10.1046/j.1432-1327.2000.01187.x. [DOI] [PubMed] [Google Scholar]
- 6.Burgos ES, Schramm VL. Weak coupling of ATP hydrolysis to the chemical equilibrium of human nicotinamide phosphoribosyltransferase. Biochemistry. 2008;47:11086–11096. doi: 10.1021/bi801198m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 8.van der Veer E, et al. Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J Biol Chem. 2007;282:10841–10845. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63:7436–7442. [PubMed] [Google Scholar]
- 11.Khan JA, Tao X, Tong L. Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents. Nat Struct Mol Biol. 2006;13:582–588. doi: 10.1038/nsmb1105. [DOI] [PubMed] [Google Scholar]
- 12.Vinitsky A, Grubmeyer C. A new paradigm for biochemical energy coupling. Salmonella typhimurium nicotinate phosphoribosyltransferase. J Biol Chem. 1993;268:26004–26010. [PubMed] [Google Scholar]
- 13.Gross J, Rajavel M, Segura E, Grubmeyer C. Energy coupling in Salmonella typhimurium nicotinic acid phosphoribosyltransferase: Identification of His-219 as site of phosphorylation. Biochemistry. 1996;35:3917–3924. doi: 10.1021/bi9517906. [DOI] [PubMed] [Google Scholar]
- 14.Grubmeyer CT, Gross JW, Rajavel M. Energy coupling through molecular discrimination: Nicotinate phosphoribosyltransferase. Methods Enzymol. 1999;308:28–48. doi: 10.1016/s0076-6879(99)08004-0. [DOI] [PubMed] [Google Scholar]
- 15.Wang T, et al. Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme. Nat Struct Mol Biol. 2006;13:661–662. doi: 10.1038/nsmb1114. [DOI] [PubMed] [Google Scholar]
- 16.Kim MK, et al. Crystal structure of visfatin/pre-B cell colony-enhancing factor 1/nicotinamide phosphoribosyltransferase, free and in complex with the anti-cancer agent FK-866. J Mol Biol. 2006;362:66–77. doi: 10.1016/j.jmb.2006.06.082. [DOI] [PubMed] [Google Scholar]
- 17.Holen K, Saltz LB, Hollywood E, Burk K, Hanauske AR. The pharmacokinetics, toxicities, and biologic effects of FK866, a nicotinamide adenine dinucleotide biosynthesis inhibitor. Invest New Drugs. 2008;26:45–51. doi: 10.1007/s10637-007-9083-2. [DOI] [PubMed] [Google Scholar]
- 18.Shi W, et al. The 2.0 A structure of human hypoxanthine-guanine phosphoribosyltransferase in complex with a transition-state analog inhibitor. Nat Struct Biol. 1999;6:588–593. doi: 10.1038/9376. [DOI] [PubMed] [Google Scholar]
- 19.Shi W, et al. Crystal structures of Giardia lamblia guanine phosphoribosyltransferase at 1.75 A(,) Biochemistry. 2000;39:6781–6790. doi: 10.1021/bi000128t. [DOI] [PubMed] [Google Scholar]
- 20.Shi W, et al. Closed site complexes of adenine phosphoribosyltransferase from Giardia lamblia reveal a mechanism of ribosyl migration. J Biol Chem. 2002;277:39981–39988. doi: 10.1074/jbc.M205596200. [DOI] [PubMed] [Google Scholar]
- 21.Gross JW, Rajavel M, Grubmeyer C. Kinetic mechanism of nicotinic acid phosphoribosyltransferase: Implications for energy coupling. Biochemistry. 1998;37:4189–4199. doi: 10.1021/bi972014w. [DOI] [PubMed] [Google Scholar]
- 22.Fedorov A, et al. Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis. Biochemistry. 2001;40:853–860. doi: 10.1021/bi002499f. [DOI] [PubMed] [Google Scholar]
- 23.Schramm VL, Shi W. Atomic motion in enzymatic reaction coordinates. Curr Opin Struct Biol. 2001;11:657–665. doi: 10.1016/s0959-440x(01)00269-x. [DOI] [PubMed] [Google Scholar]
- 24.Samal B, et al. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol. 1994;14:1431–1437. doi: 10.1128/mcb.14.2.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 26.Preiss J, Handler P. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J Biol Chem. 1958;233:488–492. [PubMed] [Google Scholar]
- 27.Gonzalez-Segura L, Witte JF, McClard RW, Hurley TD. Ternary complex formation and induced asymmetry in orotate phosphoribosyltransferase. Biochemistry. 2007;46:14075–14086. doi: 10.1021/bi701023z. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Vagin AA, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 30.Emsley P, Cowtan K. Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski RA, McArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;265:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.