Abstract
Lipid homeostasis in vertebrates is regulated by 3 sterol regulatory element binding protein (SREBP) isoforms. Here, we identify targets of SREBP-1 in mammalian liver using chromatin immunoprecipitation–high-throughput DNA sequencing. Antisera to SREBP-1 were used with liver chromatin from mice fed a high-carbohydrate diet after a fast, which leads to superinduction of hepatic SREBP-1c expression. SREBP-1–DNA complexes were subjected to massive parallel DNA sequencing using the Illumina Genome Analyzer II, resulting in 5.7 million sequence reads. Mapping these reads to the mouse reference genome identified 426 peaks of SREBP-1 binding vs. a control antibody. These binding peaks show a striking enrichment in proximal promoter regions, with 52% located within 1 kb upstream of a transcription start site. A previously undescribed sequence motif (5′-ACTACANNTCCC-3′) was present in 76% of the total peaks, and we show that it is a functional SREBP-1 response element. Our analysis also reveals that an Sp1 consensus site is present as a “coregulatory” motif in 50% of the SREBP-1 binding peaks, consistent with previous functional studies. SREBP-1 bound not only to many well-characterized SREBP-1 target genes but to several other previously unknown targets in lipid and carbohydrate metabolism as well as many putative target genes in other diverse biological pathways.
Keywords: ChIP-seq, fasting/refeeding, Sp1, Kolmogorov–Smirnov test
All living organisms have evolved strategic regulatory systems to coordinate metabolic flux into potentially competing biochemical pathways to manage metabolite and end-product pools for optimal cellular fitness. This is particularly important in mammalian lipid homeostasis because cholesterol and fatty acids, the 2 major classes of mammalian lipids, are key to the structure and function of all cells, and unbalanced flux into these pathways can be both energy-inefficient and toxic. In vertebrates, lipid homeostasis is maintained in part by sterol regulatory element binding protein (SREBP) transcription factors, which are synthesized as ≈125-kDa precursors containing 2 membrane-spanning domains that localize the immature proteins to the endoplasmic reticulum membrane (1).
Overlapping transcripts from 1 gene encode SREBP-1a and SREBP-1c isoforms, and the singular SREBP-2 is encoded from a distinct unlinked gene (2). There is an intricate system of nutrient sensing that relates cellular levels of cholesterol and other key lipid components to the expression and multistep membrane trafficking/protease pathway that converts the membrane-bound SREBPs into the much smaller nuclear targeted transcription factors.
SREBPs activate genes encoding most of the key enzymes in cholesterol and fatty acid metabolism (3); thus, coupling SREBP processing to pathway flux provides a direct link between the activity of the regulatory factors and metabolic demand. Overexpression and targeted elimination studies in mice have produced crucial information on the roles of SREBPs in lipid regulation (3). The available evidence indicates that SREBP-1 and SREBP-2 are mainly involved in fatty acid and cholesterol metabolism, respectively (3). However, there is still much to be learned about individual roles and physiological targets of the 3 mammalian SREBPs. Additionally, SREBPs activate genes not directly related to core reactions of lipid biosynthesis (4, 5); thus, additional studies are required to provide a more comprehensive understanding of SREBP function in physiology and metabolism.
We have performed a genome-wide analysis of SREBP-1 binding in liver using chromatin immunoprecipitation–high-throughput DNA sequencing (ChIP-seq) (6). We identified 426 SREBP-1 binding sites in the mouse liver genome, and, interestingly, 52% are located within 1 kb of the 5′-end of a mapped gene (the null expectation is 1%). We also performed an expression microarray analysis for genes regulated by the fasting/refeeding transition, in which SREBP-1c expression is dramatically increased. When we compared the fasting/refeeding differentially expressed genes with genes located close to SREBP-1 binding peaks, there was a high degree of correlation, indicating that our ChIP-seq data set contains a functional collection of SREBP-1c binding target genes.
The data also reveal a previously unrecognized DNA binding motif for SREBP-1, which matches an orphan motif that is highly conserved in promoters across several mammalian species, but a corresponding transcription factor was not known (7). We show that this previously undescribed motif is a functional SREBP-1 target using electrophoretic mobility-shift and transfection assays, further supporting the identity of SREBP-1 as the previously unidentified transcription factor.
Results
We prepared an antibody to a fragment of mouse SREBP-1 that shares no homology with SREBP-2 and used it to detect SREBP-1 in liver chromatin from a control group of mice, a fasted group, or a group that was fasted followed by refeeding a high-carbohydrate diet. The mature SREBP-1 was significantly decreased by fasting and increased to a level several-fold higher in the refed sample (Fig. S1) than in the control group. Because this antibody is highly specific and robust, we used it to enrich SREBP-1 containing DNA fragments from the refed chromatin using ChIP. In gene-specific ChIP analyses (Fig. S1), SREBP-1 association with 3 known SREBP target genes was confirmed; thus, we were encouraged to perform a genome-wide analysis of SREBP-1 binding. Chromatin from the SREBP-1 or IgG-enriched samples was used as a starting sample for high-throughput sequencing analysis (6).
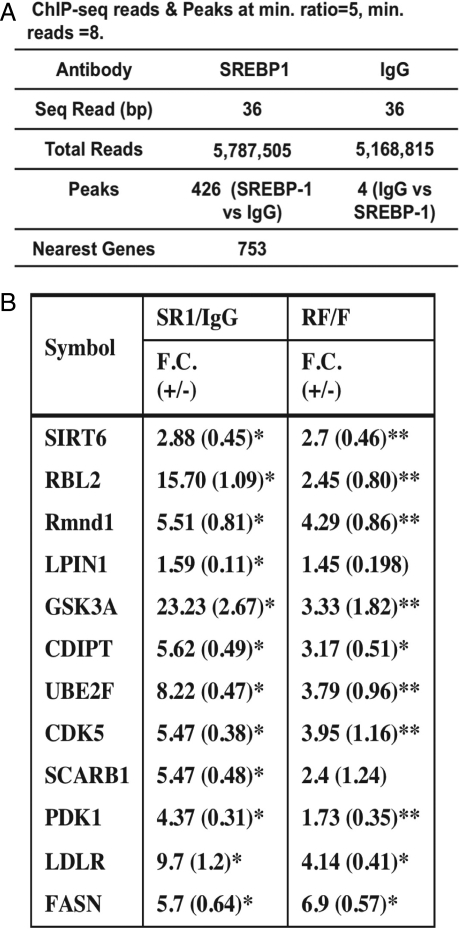
Fig. 1A shows a summary of the sequencing analysis, and Fig. S2 shows the data for 1 peak with the sequence reads mapped onto a window in the UCSC Genome Browser Database. For defining SREBP-1 binding peaks, we set the minimum sequence reads to 8 and the minimum read ratio comparing SREBP-1 with IgG to 5. Using this stringent cutoff, we identified a total of 426 SREBP-1 binding sites. When the samples were reversed, there were only 4 peaks detected. Thus, the background is low and the specificity is high. We searched for the peak proximal genes (on both sides) and identified 753 total genes. Additional information on the peak locations is provided in Table S1, and the nearest gene list is provided in Table S2.
Fig. 1.
ChIPSeq analysis for SREBP-1–DNA binding in hepatic chromatin. (A) Summary of peak analysis. (B) Peak validation. Ten randomly selected peaks were chosen for validation by gene-specific ChIP. The fold change (FC) is the fold increase for the signal from SREBP-1 antibody–enriched chromatin relative to a control IgG. SREBP-1 binding to these 10 peaks as well as to the LDL receptor and FAS promoters was also compared in hepatic chromatin prepared from fasting vs. refed mice as indicated. The negative control L32 showed no enrichment (31). *P < 0.01, **P < 0.05
To validate the accuracy of the genome-wide binding data, we randomly picked 10 regions and performed ChIP analysis with peak specific primers. SREBP-1 binding to all 10 regions was enriched between 1.6- and 23-fold in the SREBP-1 chromatin relative to IgG (Fig. 1B). There was also a significant increase in SREBP-1 binding to most of the tested promoters in refed vs. fasted hepatic chromatin (Fig. 1B).
To find any commonalities among the SREBP-1 target genes, we searched the Database for Annotation, Visualization, and Integrated Discovery (DAVID) gene ontology (GO) database (8) for enrichment and found gene clusters in lipid (P = 7e-4) and carbohydrate metabolism (P = 5.6e-3) as expected (Table S3). Many of the genes in these categories are known SREBP targets, and others are previously unknown. Additionally, genes in other GO categories associated with intracellular protein movement and targeting (P = 3.8e-7), cell proliferation and differentiation (P = 5.3e-4), and apoptosis (P = 3.6e-5) showed significant enrichment. This is interesting and likely significant, because SREBPs undergo a multi-intracellular membrane itinerary during their life span (9) and are involved in cell proliferation (10) and apoptosis (11–13).
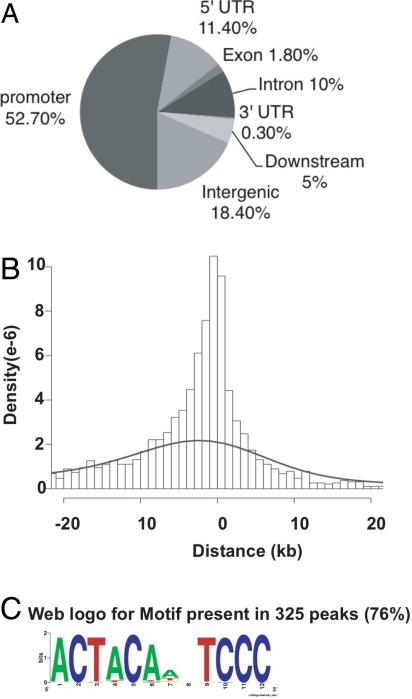
Interestingly, a peak distribution analysis revealed that 52% mapped to within 1 kb 5′ of a gene transcription start site (TSS) and 11.4% were in 5′ UTR regions. Altogether, this accounts for over 63% of the total SREBP-1 binding events, suggesting a strong preference for promoter proximal binding by SREBP-1. The remaining sites were distributed mostly within intergenic regions and introns, with very few located in 3′ UTRs and exons (Fig. 2A). Overall, 95% of the peaks are located within 20 kb of a known TSS (Fig. 2B; the null expectation is 15%).
Fig. 2.
Genome-wide distribution of ChIP-seq peaks. (A) Location of SREBP-1 binding peaks relative to known genes. The promoter and downstream regions are defined as 2 kb of 5′ or 3′ flanking DNA. (B) Peak distance relative to TSS of the closest gene. The dashed line shows the distribution of a random sequence of DNA the same size as the average peak. (C) The 426 peak regions were analyzed for overrepresented motifs using MEME (14). The top-scoring motif is shown.
The sequences around the peaks were then analyzed by MEME to search for enriched sequence motifs (14). The web logo derived from the MEME motif with the top score is shown in Fig. 2C. This motif was present in 325 peaks (76% with a position-specific frequency matrix model and z > 4.5; the number of motifs per peak is provided in Fig. S3). The motif is split into 2 halves, with 1 resembling part of the SREBP motif we proposed based on functional comparisons and sequence alignments of a small set of promoters (15). However, overall, this represents a putative previously undescribed recognition site for SREBP-1. Interestingly, this motif is preferentially located close to the center of the peaks, consistent with it corresponding to the recognition site for SREBP-1 (z = 4.8, P = 1e-6; Fig. S4). This is expected because of the high resolution provided by ChIP-seq (6).
SREBPs are very inefficient transcription factors by themselves, and they stimulate gene expression robustly only when close to a binding site for a coregulatory protein like Sp1, NF-Y/CBF, and CREB (16). A search for additional motifs that might be enriched close to the SREBP peaks revealed a match to the Sp1 consensus site within 150 bp of 215 (50%) of the SREBP-1 binding peaks. No other significantly enriched elements were identified by this analysis.
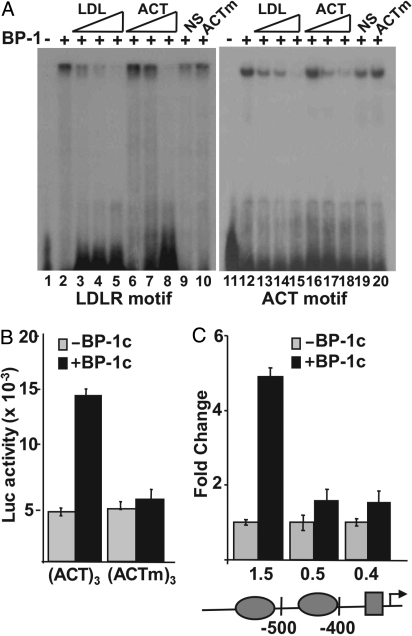
To determine whether this previously undescribed motif is indeed functional, we sought additional evidence. We note that the motif (5′-ACTACAnnTCCC-3′) is split into 2 halves, with 1 resembling part of the SREBP motif we proposed previously based on functional comparisons and sequence alignments of a small set of promoters (15). Although the motif did not match any known motifs deposited in the Transfac database, it is almost identical to a computationally identified motif [M4 motif (7): 5′-ACTAYRnnnCCC-3′] based on detecting sequence elements exhibiting strong evolutionary conservation across several mammalian genomes (7). Indeed, M4 was ranked as the most highly conserved motif with an unidentified DNA binding protein. Then, to analyze experimentally if this previously undescribed motif was a bona fide site of SREBP-1 binding and activation, we prepared an oligonucleotide probe containing the unique motif for the EMSA, and we also prepared a synthetic promoter containing 3 tandem copies next to an Sp1 site in a luciferase reporter vector for functional studies. The EMSA revealed that the previously undescribed motif probe bound specifically to recombinant SREBP-1 (Fig. 3A), and the competition studies suggest that the LDL receptor SREBP response element (SRE) has a higher relative affinity for SREBP-1 binding. Transient transfection analysis showed that the synthetic reporter was activated by cotransfected SREBP-1c, whereas an identical construct in which 6 bases in the previously undescribed motif were mutated was not activated by cotransfected SREBP-1c (Fig. 3B). Additionally, we showed that SREBP-1 activated expression of a natural promoter identified in our study (GSK-3a) and stimulation was blunted when 1 copy of the previously undescribed motif was deleted (Fig. 3C). This is similar to other natural SREBP-responsive promoters in which mutation of 1 element where another binding site remains intact results in a loss of SREBP responsiveness (2, 15, 17).
Fig. 3.
Functional analysis of the previously undescribed SREBP-1 binding motif. (A)32-P–labeled probes representing the well-characterized SREBP site from the human LDL receptor promoter (lanes 1–10) or the previously undescribed SREBP-1 motif (lanes 11–20) were incubated with buffer (lanes 1 and 10) or with recombinant SREBP-1 [BP-1 (2 ng), lanes 2–10 and 12–20] and analyzed by EMSA. Increasing concentrations of unlabeled probe (30-, 100-, or 300-fold molar excess) for either LDL receptor (LDL, lanes 3–5 and 13–15) or for the previously undescribed motif (ACT, lanes 6–8 and 16–18) were included in the binding reactions as indicated. In lanes 9 and 19, or 10 and 20, 300-fold molar excess of a nonspecific (NS) DNA fragment or a mutant version of the previously undescribed motif probe (ACTm) was included in the binding reaction. DNA sequences for probes and competitors are provided in Table S1. (B) Reporter plasmids containing 3 copies of the previously undescribed motif upstream of an Sp1 site (ACT)3 or a similar plasmid containing 3 copies of a mutated version (ACTm)3 were fused to the luciferase coding sequence and analyzed by transient transfection in 293T cells. The reporters were cotransfected with the pcDNA3.1 empty vector (gray bars) or an SREBP-1c expression vector (black bars). (C) Luciferase vectors for the WT and deleted versions of the GSK3a promoter were also prepared and analyzed for SREBP-1 responsiveness as above. The 3 promoter constructs contain a 1.5-, 0.5-, or 0.4-kb 5′ flanking sequence as indicated. The ovals on the promoter diagram denote positions of the 2 copies of the 5′-ACTACANNTCCC-3′, and the rectangle marks the position of an Sp1 consensus site.
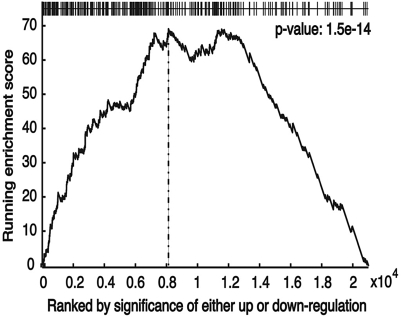
Because SREBP-1c levels are repressed by fasting and increased above control levels by refeeding a high-carbohydrate diet following a fast (Fig. S1), genes that are regulated by SREBP-1c in the liver should be differentially expressed by this protocol. Therefore, we compared expression profiles for genes from RNA isolated from livers of fasted and fasted/refed mice (Dataset S1). This analysis revealed that expression of 1,063 and 803 genes, respectively, was increased or decreased by refeeding. A GO analysis revealed that, as expected, genes in lipogenesis and biosynthetic metabolism were increased and genes involved in fatty acid oxidation and catabolic metabolism were decreased. We ranked the list of genes by significance of differential regulation by refeeding and then asked how the genes containing an SREBP-1c binding site were distributed over the expression list using the Kolmogorov–Smirnov (KS) test (18). The analysis showed that SREBP-1c target genes are highly enriched with genes showing differential expression (Fig. 4; P = 1.5e-14). The high degree of correlation indicates that the majority of the SREBP-1 binding sites identified by the ChIP-seq analysis are functionally important.
Fig. 4.
Gene set enrichment analysis. The ChIP-seq peaks were analyzed for their representation within an expression array data set from refed vs. fasting liver as described in the text (18). Expression values for all genes in the microarray are ranked by fold regulation (from fasting to refed) on the x axis. The graph plots a running enrichment (KS) score on the ordinate across the complete differential expression data set on the abscissa. The location of each gene from the ChIP-seq data set within the ranked differential expression list is given as a thin vertical line on the horizontal axis displayed at the top of the graph. Here, the clustering of the ChIP-seq genes within the top portion of the differentially ranked genes is clearly visible (P = 1.5 e-14).
We also performed an unbiased search of the 5′-flanking regions of the genes that were differentially expressed by fasting/refeeding for a conserved motif that might emerge. Interestingly, the SREBP-1c motif described previously again emerged as the top motif using this unbiased data set, lending additional support for the pivotal role of SREBP-1c in mediating systematic gene expression changes during fasting/refeeding.
Discussion
We identified 426 SREBP-1 binding sites in chromatin from livers of mice treated to a fasting/high-carbohydrate feeding protocol that is known to increase hepatic levels of nuclear SREBP-1c, the major SREBP-1 isoform in liver, significantly. It is interesting that 52% of the binding sites are located in proximity to TSSs for mapped genes (Fig. 2). This is unusual because most genome-wide reports of transcription factor binding sites have shown no particular clustering or association relative to positions of known genes (19–22).
We also showed that there is a high correlation between the identity of genes located nearest to the SREBP-1 binding peaks and differentially expressed genes from RNA isolated from livers of fasted vs. fasted/refed mice. Because nuclear SREBP-1c levels are very low during fasting and dramatically induced by high-carbohydrate feeding, it is likely that the binding sites identified here are functionally important.
SREBPs are members of the basic helix-loop-helix (bHLHL) leucine zipper class of transcription factors, and most bHLH proteins dimerize and bind to an inverted repeat “E-box” element (23). The core E-box reads 5′-CANNTG-3′, and individual family members exhibit unique preferences for specific bases in the center and flanking residues. All 3 SREBPs bind avidly to the E-box 5′-CACGTG-3′ in vitro (24); however, in all the promoters carefully analyzed to date, none has been shown to be dependent on an E-box for activation by SREBP (2). The first well-characterized functional SRE was from the human LDL receptor promoter (25), which was used for affinity purification and subsequent cloning of SREBP cDNAs (26–28). This SRE element, 5′-ATCACCCCAC-3′, is a direct repeat as opposed to an inverted repeat E-box. The DNA binding flexibility that allows SREBPs to recognize E-boxes and direct repeat elements in vitro is attributable to a signature tyrosine residue that is unique to the SREBPs (24). All other E-box binding bHLH proteins contain an arginine at this position, which makes DNA contacts critical for specific recognition of E-box elements (29).
The crucial tyrosine allows the SREBP DNA binding domain to adopt an altered conformation to interact specifically with the direct repeat SRE element (29). We showed that the tyrosine residue was crucial to the nutritional regulatory response of SRE-containing promoters (30). Because this tyrosine was crucial for SRE element binding and sterol-dependent regulation, in vivo regulated SREs likely do not contain E-box motifs but, rather, some form of the direct repeat SRE similar to the LDL receptor promoter element. Consistent with this prediction, our ChIP-seq results identified a functional variant of the direct repeat SRE 5′-ACTACANNTCCC-3′ as an in vivo recognition site in 76% of the SREBP-1 binding peaks.
In promoter activation studies, SREBPs are weak activators of transcription by themselves and require a neighboring coregulatory site for efficient activation and nutritional regulation (16). Transcription factors Sp1, NF-Y/CBF, and CREB have all been shown to function as coregulators in different SREBP-responsive promoters (2, 17). Consistent with these earlier studies, our genome-wide analysis identified an Sp1 site located close to 215 of the 426 peaks of SREBP-1 binding. However, predicted binding sites for NF-Y and CREB were not significantly enriched when analyzed on a genome-wide scale.
When the nearest genes to the SREBP-1 peaks were analyzed by GO software, clusters for lipid and carbohydrate metabolism were enriched as expected. Additionally, GO categories associated with intracellular membrane targeting and movement, apoptosis, cell proliferation, and proteolysis were significantly enriched (Table S3). These categories are interesting because SREBPs are known to undergo a regulated itinerary from endoplasmic reticulum to Golgi, followed by proteolytic maturation (9), and SREBPs are also associated with cell proliferation and apoptosis signaling (11–13).
Our analysis likely represents the binding pattern for SREBP-1c because it is the major hepatic isoform and it is specifically induced by the high-carbohydrate refeeding regimen used here (ref. 31; Fig. S1. However, because SREBPs function as dimers and 1c monomers can freely dimerize with both SREBP-1a and SREBP-2 (32), we cannot rule out the possibility that some of the binding sites identified here result from different monomeric combinations. It will be interesting to compare these results with future experiments under conditions in which only a single SREBP isoform is expressed.
The SREBP-1c binding motif identified by our studies corresponds to a motif identified in over 500 human promoters and is conserved in 317 promoters across diverse mammalian species (7). Although conservation of the motif was revealed by this earlier report (7), a corresponding transcription factor was not known. Our studies suggest that the unidentified transcription factor is SREBP-1c.
A genome-wide analysis of SREBP-1 binding in cultured human hepatoma HepG2 cells using ChIP on chip hybridization technology identified an E-box as a preferred site for SREBP-1 binding (33). The differences with our study may be that a cultured line of cells was used and SREBP-1a is more abundant than SREBP-1c in all cultured cell lines examined, including HepG2 (34). Additional studies are required to evaluate this further.
Materials and Methods
Animals.
Six-week-old male C57BL/6 mice were obtained from Taconic and maintained on a chow diet (2020X; Harlan Teklad Global) for 1 week with a 12-h light/12-h dark cycle for acclimatization. The fasting/refeeding experiments were performed as described elsewhere (31). Livers were removed, and ≈5% slices were snap-frozen for later. The remaining tissue was immediately used for ChIP as described elsewhere in this article. Animal experiments were approved by the University of California, Irvine Institutional Animal Care and Use Committee (protocol 97–1545).
Preparation of Samples for ChIP Assays.
Samples for ChIP assays using mouse tissues were prepared as described previously (31). Final DNA samples were analyzed for gene-specific ChIP (31) or ChIP-seq. The quantitative PCR oligonucleotide pairs for the mouse promoters are provided in Table S4. The library preparation protocol from Illumina was used to prepare samples for analysis with the Illumina/Solexa Genome Analyzer (6). Peak identification was performed using the software developed by the Wold laboratory at the California Institute of Technology (6). We used the default settings of a minimal number of 8 reads with a difference ratio of 5 between SREBP-1 antibody and control IgG to define peaks. At this cutoff, a false discovery rate of <0.2% was estimated by randomly generating a set of sequence reads corresponding to the size and number generated by the SREBP-1 antibody–enriched sample. Also, we did perform more stringent and less stringent analyses. These conditions resulted in fewer or more peaks as expected, but the most significantly enriched motif was the same as in Fig. 2.
EMSA.
The sequences for the oligonucleotides used for the EMSA are provided in Table S4. The EMSA was performed with recombinant SREBP-1 protein (amino acids 1–490) as described elsewhere (16). Briefly, labeled probe and unlabeled competitor DNAs (as indicated in Fig. 3) were combined together on ice at the indicated molar equivalents before recombinant SREBP-1 protein was added. After a further 30-min incubation on ice, free DNA and DNA-protein complexes were resolved by native PAGE (16).
Transient Transfection.
293T cells were transfected with luciferase reporter constructs and a CMV–β-galactosidase internal control plasmid as described previously (32). Oligonucleotides used for plasmid cloning are provided in Table S4.
Motif Analysis.
We applied the motif-finding program MEME (14) to SREBP-1 target peak regions to search for statistically overrepresented consensus SREBP-1 binding sites in the peak regions. Motifs were presented as position-dependent letter-probability matrices.
To find the binding sites for coregulators near the SREBP-1 binding sites, we masked all the SREBP-1 motif sites (z > 3) in the peak sequences with “N” and scanned the masked region ±150 bp for coregulator binding sites using MEME. We also applied an enumeration-based method, k-mer analysis, to the sequences to search for coregulator motifs. For the k-mer analysis, we examined each motif length separately, from 6 to 15 bp. The following 11-mers were enriched in the extended peak regions (z > 3) and are matches to the Sp1 consensus site: CCCAGCCCCAG, CCCCAGCCCCA, CCAGCCCCAGC, GCCCCAGCCCC, AGCCCCAGCCC, and CAGCCCCAGCC. The 6-mer CCAGCC is common to all these sites and is present in 215 of 426 extended peaks (±150 bp; z = 7.4).
RNA Isolation, Quantitative PCR, and Gene Expression Profiling.
Mouse liver total RNAs for refeeding and fasting were isolated using TRIzol(Invitrogen) and the RNeasy Mini Kit (Qiagen), followed by reverse transcription of 0.5 μg of RNA with iScript Reverse Transcriptase (Bio-Rad) following the manufacturer's instructions. Analysis was performed using the standard curve method and normalization of all genes of interest to the housekeeping control L32. Gene expression profiling was carried out using the Mouse Gene 1.0 OST (Affymetrix) by hybridizing RNA from refeeding and fasting livers in triplicate. Differential expression was then assessed using the cyberT analysis program (35).
KS Analysis.
The obtained ChIP-seq data were compared with expression microarray data by using a KS plot, a modified method of gene set enrichment analysis (18). The KS plot was obtained by calculating the running sum statistics for our SREBP-1 ChIP-seq gene set to observe enrichment in the ranked gene list from genes differentially expressed comparing the fast/refeeding expression microarray data.
Supplementary Material
Acknowledgments.
We thank Dr. Bing Zhu and Mary Bennett for their contributions to the early portions of this work. This study was supported in part by National Institute of Health Grant HL48044 (to T.F.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904246106/DCSupplemental.
References
- 1.Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 2.Osborne TF. Sterol regulatory element binding proteins (SREBPs): Key regulators of nutritional homeostasis and insulin action. J Biol Chem. 2000;275:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- 3.Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeon TI, Zhu B, Larson JL, Osborne TF. SREBP-2 regulates gut peptide secretion through intestinal bitter taste receptor signaling in mice. J Clin Invest. 2008;118:3693–3700. doi: 10.1172/JCI36461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park HJ, et al. Parasympathetic response of the heart is controlled by SREBP. J Clin Invest. 2008;118:259–271. doi: 10.1172/JCI32011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 7.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 9.Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakakuki M, et al. A transcription factor of lipid synthesis, sterol regulatory element-binding protein (SREBP)-1a causes G(1) cell-cycle arrest after accumulation of cyclin-dependent kinase (cdk) inhibitors. FEBS J. 2007;274:4440–4452. doi: 10.1111/j.1742-4658.2007.05973.x. [DOI] [PubMed] [Google Scholar]
- 11.Gibot L, et al. Human caspase-7 is positively controlled by SREBP-1 and SREBP-2. Biochem J. 2009;420(3):473–483. doi: 10.1042/BJ20082057. [DOI] [PubMed] [Google Scholar]
- 12.Logette E, Solary E, Corcos L. Identification of a functional DNA binding site for the SREBP-1c transcription factor in the first intron of the human caspase-2 gene. Biochim Biophys Acta. 2005;1738(1–3):1–5. doi: 10.1016/j.bbalip.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, et al. Cleavage of sterol regulatory element binding proteins (SREBPs) by CPP32 during apoptosis. EMBO J. 1996;15:1012–1020. [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey TL. Discovering novel sequence motifs with MEME. Current Protocol Bioinformatics. 2002 doi: 10.1002/0471250953.bi0204s00. [DOI] [PubMed] [Google Scholar]
- 15.Magaña MM, Lin SS, Dooley KA, Osborne TF. Sterol regulation of acetyl coenzyme A carboxylase promoter requires two interdependent binding sites for sterol regulatory element binding proteins. J Lipid Res. 1997;38:1630–1638. [PubMed] [Google Scholar]
- 16.Sanchez HB, Yieh L, Osborne TF. Cooperation by sterol regulatory element-binding protein and Sp1 in sterol regulation of low density lipoprotein receptor gene. J Biol Chem. 1995;270:1161–1169. doi: 10.1074/jbc.270.3.1161. [DOI] [PubMed] [Google Scholar]
- 17.Osborne TF. Transcriptional control mechanisms in the regulation of cholesterol balance. Crit Rev Eukaryotic Gene Expression. 1995;5(3):317–335. doi: 10.1615/critreveukargeneexpr.v5.i3-4.50. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefterova MI, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Bolton EC, et al. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murre C, et al. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 24.Kim JB, et al. Dual DNA binding specificity of ADD1/SREBP-1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol Cell Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawson PA, et al. Sterol-dependent repression of low density lipoprotein receptor promoter mediated by 16-basepair sequence adjacent to binding site for transcription factor Sp1. J Biol Chem. 1988;263:3372–3379. [PubMed] [Google Scholar]
- 26.Briggs MR, Yokoyama C, Wang X, Brown MS, Goldstein JL. Nuclear protein that binds sterol regulatory element of low density receptor promoter I: Identification of the protein and delineation of its target nucleotide sequence. J Biol Chem. 1993;268:14490–14496. [PubMed] [Google Scholar]
- 27.Wang X, et al. Nuclear protein that binds sterol regulatory element of low density receptor promoter II: Purification and characterization. J Biol Chem. 1993;268:14497–14504. [PubMed] [Google Scholar]
- 28.Yokoyama C, et al. SREBP-1, a basic-helix-loop-helix-zipper protein that controls transcription of the low density receptor gene. Cell. 1993;75:185–197. [PubMed] [Google Scholar]
- 29.Parraga A, Bellsolell L, Ferre-D'Amare AR, Burley SK. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3Å resolution. Structure. 1998;6:661–672. doi: 10.1016/s0969-2126(98)00067-7. [DOI] [PubMed] [Google Scholar]
- 30.Athanikar JN, Osborne TF. Specificity in cholesterol regulation of gene expression by coevolution of sterol regulatory DNA element and its binding protein. Proc Natl Acad Sci USA. 1998;95:4935–4940. doi: 10.1073/pnas.95.9.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett MK, Seo Y-K, Datta S, Shin D-J, Osborne TF. Selective binding of SREBP isoforms and co-regulatory proteins to promoters for lipid metabolic genes in liver. J Biol Chem. 2008;283:15628–15637. doi: 10.1074/jbc.M800391200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datta S, Osborne TF. Activation domains from both monomers contribute to transcriptional stimulation by SREBP dimers. J Biol Chem. 2005;280:3338–3345. doi: 10.1074/jbc.M411222200. [DOI] [PubMed] [Google Scholar]
- 33.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4(7):e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long AD, et al. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem. 2001;276:19937–19944. doi: 10.1074/jbc.M010192200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.