Abstract
Box C/D guide RNAs are abundant noncoding RNAs that primarily function to direct the 2′-O-methylation of specific nucleotides by base-pairing with substrate RNAs. In archaea, a bipartite C/D RNA assembles with L7Ae, Nop5, and the methyltransferase fibrillarin into a modification enzyme with unique substrate specificity. Here, we determined the crystal structure of an archaeal C/D RNA–protein complex (RNP) composed of all 3 core proteins and an engineered half-guide RNA at 4 Å resolution, as well as 2 protein substructures at higher resolution. The RNP structure reveals that the C-terminal domains of Nop5 in the dimeric complex provide symmetric anchoring sites for 2 L7Ae-associated kink-turn motifs of the C/D RNA. A prominent protrusion in Nop5 seems to be important for guide RNA organization and function and for discriminating the structurally related U4 snRNA. Multiple conformations of the N-terminal domain of Nop5 and its associated fibrillarin in different structures indicate the inherent flexibility of the catalytic module, suggesting that a swinging motion of the catalytic module is part of the enzyme mechanism. We also built a model of a native C/D RNP with substrate and fibrillarin in an active conformation. Our results provide insight into the overall organization and mechanism of action of C/D RNA–guided RNA methyltransferases.
Keywords: crystal structure, methylation, RNA–protein complex, non-coding RNA
More than 100 types of chemical modification are introduced at specific sites of cellular RNAs after their transcription (1). These modifications, which greatly increase the chemical diversity of RNA, are generally beneficial and sometimes even critical for the structure and function of host RNAs. RNA modifications are normally carried out by individual protein enzymes. However, an exception is the large number of pseudouridine and 2′-O-methylated nucleotides in rRNAs, snRNAs, and tRNAs that are synthesized by H/ACA and C/D RNA–protein complexes (RNPs), respectively (2–6). In each such complex, a distinct guide RNA belonging to the H/ACA or C/D RNA family determines the substrate specificity by base-pairing with the substrate around the modification site. These 2 classes of RNA-guided RNA modification enzymes are ubiquitous in eukaryotic and archaeal organisms but are not found in bacteria.
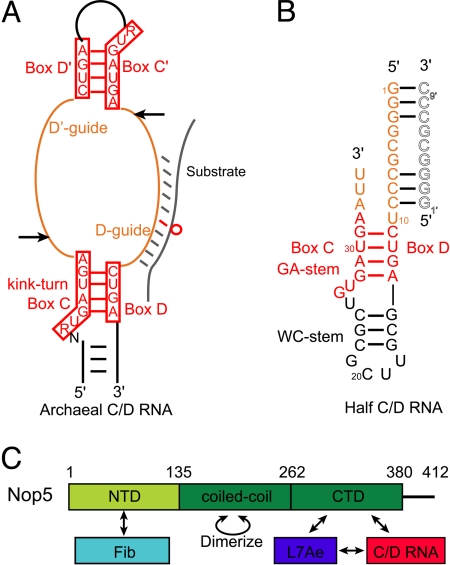
The methylation guide C/D RNAs contain a C box (RUGAUAG, R is purine) near the 5′ end and a D box (CUGA) near the 3′ end, and related boxes C′ and D′ at the internal region. One or 2 antisense sequences are present upstream of boxes D and D′ and form a 10–21-bp duplex with the substrate, thereby selecting the nucleotide paired to the fifth nucleotide upstream of box D/D′ for modification (7–9). Boxes C and D combine and fold into a kink-turn (K-turn) structural motif (10–13). A canonical K-turn is composed of 2 bent stems linked by a 3-nt bulge with an extruded nucleotide. One stem contains tandem sheared G·A pairs (hereafter called the GA-stem, also known as stem II or noncanonical stem), and the other contains Watson–Crick pairs (WC-stem, also known as stem I or canonical stem). In archaeal C/D RNAs (14), the internal C′ and D′ boxes often form a related K-loop structure, in which a loop replaces the WC-stem in the canonical K-turn (15, 16).
In archaea, C/D RNA associates with 3 proteins—fibrillarin, Nop5 (also known as Nop56/58), and L7Ae—to form an enzyme capable of catalyzing site-specific modification (17). In eukaryotic complex, paralogs Nop56 and Nop58 replace the single Nop5 protein, and a 15.5-kDa protein (15.5K, Snu13 in yeast) is equivalent to L7Ae. Fibrillarin is the methyltransferase, and it catalyzes the transfer of a methyl group from bound S-adenosyl-methionine (SAM) to the 2′-hydroxyl group of the target ribose (17–19). L7Ae/15.5K is a primary RNA-binding protein that recognizes the K-turn (10, 11, 13). L7Ae also binds well to the K-loop (20, 21), but eukaryotic 15.5K does not (21–23). Nop5 is a scaffold protein composed of an N-terminal domain (NTD) that binds fibrillarin, a coiled-coil domain that mediates self-dimerization, and a C-terminal domain (CTD, also known as the Nop domain) that likely binds the L7Ae–RNA complex (24). C/D RNP assembly proceeds in a stepwise manner (17, 20, 24, 25) and begins with the formation of the L7Ae–C/D RNA complex. Nop5 associates with the preassembled L7Ae–C/D RNP but cannot stably bind C/D RNA or L7Ae alone. Fibrillarin is recruited through its interaction with Nop5.
The structure of archaeal C/D RNP has a 2-fold symmetry, with structurally equivalent C/D and C′/D′ motifs in RNA, 2 copies of each protein, and self-association of Nop5. The integrity of the symmetric structure is important for the activity (20, 26–28), particularly for the substrate specificity (29). By contrast, the structure of the eukaryotic C/D RNP is less symmetric at both the RNA and protein levels. The eukaryotic C′ and D′ boxes are frequently divergent from the consensus sequence required for K-turn formation and cannot bind 15.5K in isolation (21–23). Nop58 and Nop56 preferentially bind box C/D and box C′/D′, respectively (30).
Crystal structures have been solved for fibrillarin alone (19), 2 Nop5–fibrillarin complexes [from Archaeoglobus fulgidus (AF) and Pyrococcus furiosus (PF)] (24, 31), and a number of L7Ae/15.5K and K-turn/K-loop RNA complexes (11, 13, 16, 32). The spliceosomal U4 small nuclear RNP (snRNP) shares 3 homologous components with the C/D RNP: a K-turn in the 5′ stem loop (5′SL) of the U4 snRNA, 15.5K, and the Nop5 homolog Prp31. The crystal structure of a ternary U4 snRNP complex revealed that the CTD of Prp31 is an RNP binding domain recognizing a composite surface formed by the K-turn and 15.5K (33). The ternary U4 snRNP structure provides a model for understanding C/D RNP assembly, yet is insufficient to explain the unique guide function of the C/D RNA. Despite these advances, the structure of a fully assembled C/D RNP is not yet available. To understand the structural organization, assembly specificity, and mechanism of action of C/D RNP, we have studied its structure by x-ray crystallography and modeling.
Results
Crystallization and Structure Determination.
Archaeal C/D RNAs have a bipartite circular structure with 2-fold symmetry (Fig. 1A). To simplify RNP assembly and crystallization, we engineered a half-C/D RNA that contains a single copy of boxes C and D and a 10-nt guide region upstream of box D (Fig. 1B). The WC-stem is sealed by a UUCG tetra-loop, whereas the GA-stem is open-ended. We assembled the half-C/D RNA with Nop5, which contains residues 1–380 without the C-terminal tail, fibrillarin, and L7Ae from Sulfolobus solfataricus (SS) (17), and obtained crystals that diffracted x-ray to 4.0 Å. The crystal contains 2 copies of each component in the asymmetric unit or a dimeric RNP when the complex of Nop5, L7Ae, fibrillarin, and the half-C/D RNA is regarded as a unit. The structure was initially solved by the method of single isomorphous replacement with anomalous scattering (SIRAS) based on a mercury derivative. However, de novo model building is formidable at 4.0 Å without homologous structure (34). To assist the model building, 2 substructures were determined at higher resolution. Given the modular organization of Nop5 (Fig. 1C) (24), 2 Nop5 fragments were constructed, 1 lacking the CTD (residues 1–262, Nop5ΔCTD) and 1 lacking the NTD (residues 135–380, Nop5ΔNTD). We determined the structure of Nop5ΔCTD in complex with fibrillarin to 2.6 Å and the structure of Nop5ΔNTD alone to 3.1 Å [Table S1]. These 2 substructures were solved by molecular replacement using the structure of the PF Nop5–Fib complex as a search model (31). The 2 substructures and available other homologous structures were used to build a nearly complete model for the C/D RNP, which has been refined to a free R value of 0.304 and exhibits excellent stereochemistry (Table S1).
Fig. 1.
Natural and half-C/D RNAs. (A) Schematic of the bipartite structure of an archaeal C/D RNA. The terminal boxes C and D form a canonical K-turn, whereas the internal boxes C′ and D′ fold into a K-loop. Boxes C/C′ and D/D′ are represented by their consensus sequences surrounded by red boxes. The guide upstream of box D (D-guide) is shown to associate with the substrate (gray), whereas the guide upstream of box D′ (D′-guide) is unbound. The hollow red circle strands for the modification target that is paired with the 5th nucleotide upstream of box D. Arrows indicate the cutting sites for generating the half-C/D RNA. (B) The sequence and secondary structure of the half-C/D RNA used in crystallization. The guide region pairs with the same sequence (gray hollow letters) from a crystallographic neighboring RNP. (C) Domain organization and interaction network of Nop5. The NTD, the coiled-coil domain, and the CTD are indicated. Numbers indicate the domain boundaries. Arrows indicate interaction relationships among RNP components.
Structures of Nop5 and Fibrillarin.
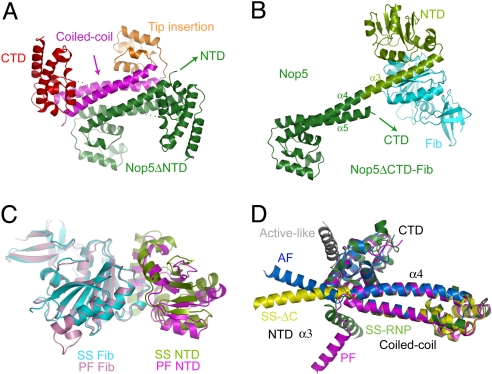
The structure of Nop5ΔNTD adopts an all-helical fold composed of an oval-shaped CTD, a coiled-coil domain, and a prominent globular domain inserting at the tip of the coiled-coil domain (Fig. 2A). The coiled-coil domain, comprising the antiparallel α4 and α5 helices, dimerizes into a left-handed 4-helix bundle with a total burial of 3091 Å2 of solvent-accessible area. The dimer interface, as well as the relative orientation between the coiled-coil domain and the CTD, are identical in the Nop5ΔCTD–Fib and the C/D RNP complex structures and are highly similar to those in the AF and PF Nop5 structures (24, 31), confirming the conservation of these features. By contrast, the tip insertion is a variable element, which is also present in PF Nop5 but not in AF Nop5.
Fig. 2.
The structures of Nop5 and fibrillarin. (A) The structure of dimeric Nop5ΔNTD. One subunit is colored deep blue, and the other is colored according to individual domains, with the CTD in red, the coiled-coil domain in magenta, and the tip insertion domain in orange. Dots strand for disordered loops. (B) Structure of the Nop5ΔCTD–Fib complex. Only a single copy is shown. The coiled-coil domain has the same orientation as in Fig. 2A. The CTD and NTD of Nop5 are colored dark and light green, respectively, and fibrillarin (Fib) is indicated in cyan. (C) The SS and PF Nop5 NTD–Fib complex structures aligned by fibrillarin. (D) Variable orientation between the NTD and the coiled-coil domain of Nop5. Nop5 structures in the SS Nop5ΔCTD–Fib (SS-ΔC, yellow), SS C/D RNP (SS-RNP, green), PF Nop5–Fib (PF, magenta), and AF Nop5–Fib (AF, blue) complexes, as well as an active model (active-like, gray), are shown. Only the α3 helix of the Nop5 NTD is shown for clarity. These structures are aligned on the basis of their CTDs and coiled-coil domains.
The structure of the Nop5ΔCTD–Fib complex shows that fibrillarin associates with the Nop5 NTD, leading to the burial of a total of 2739 Å2 of solvent-accessible area (Fig. 2B). The SS fibrillarin structure closely resembles other fibrillarin structures (Fig. 2C), with an rmsd in the range of 0.7–1.2 Å (19, 26, 31). The SAM binding pocket of fibrillarin shows significant residual density, which was plausibly modeled as a SAM molecule, despite only weak density around the methionine (Fig. S1). The methyl donor was not added during protein purification or crystallization and must have copurified with the Nop5–Fib complex, as shown previously for the PF Nop5–Fib complex (31). The SS Nop5 NTD has the same fold and similar fibrillarin-binding surface as the PF Nop5 NTD (31), but otherwise the 2 structures are quite divergent, with an rmsd of 2.84 Å over 100 Cα atoms (Fig. 2C). The NTD is a highly variable region in the Nop5 family, being considerably smaller in AF Nop5 (24).
Structure of the C/D RNP.
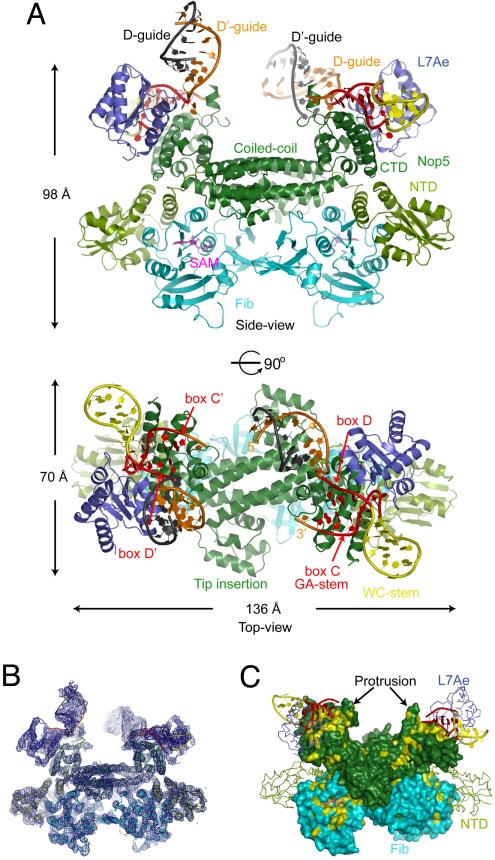
The dimeric C/D RNP structure has an overall dimension of 136 × 98 × 70 Å and resembles a butterfly when viewed from the side (Fig. 3A). The unbiased experimental map shows clear electron density for most protein and RNA structural elements (Fig. 3B). The top wing pairs are the RNA-binding modules composed of L7Ae, the Nop5 CTD, and the half-C/D RNA, each of which interacts with the other 2. The bottom wing pairs are the catalytic modules, which are composed of fibrillarin in association with the Nop5 NTD. The coiled-coil domain and the tip insertion domain of Nop5 dimerize to form a flat platform that supports the top and bottom wings (Fig. 3A, Top). There are 2 half-C/D RNAs associated with the dimeric RNP; to distinguish them, 1 is labeled as box C and D, and the other is denoted by primes, by analogy with the natural C/D RNA designation.
Fig. 3.
Overview of the C/D RNP structure. (A) Ribbon representation of the C/D RNP structure shown in side (Top) and top (Bottom) views. Boxes C, D, C′, and D′ are red, the guides are orange, the rest of the RNA is yellow, and the pairing guides from a crystallographic molecule are indicated in gray. The coiled-coil domain and CTD of Nop5 are dark green, the NTD of Nop5 is light green, L7Ae is blue, and fibrillarin is cyan. SAM is represented by magenta sticks. (B) Experimental electron density surrounding the entire C/D RNP structure. The map is contoured at 1.5σ, and the C/D RNP structure is shown as Ca or P traces in the side view. (C) Conserved surfaces in Nop5 and fibrillarin. Residues conserved in >90% of 56 archaeal Nop5 and fibrillarin sequences are colored yellow. None of the residues in the Nop5 NTD meet this conservation cutoff. The 2 copies of Nop5 and fibrillarin in the dimeric structure display nearly opposite surfaces. This view is slightly tilted from the side view to maximize visualization of the conserved region. The Nop5 NTD and L7Ae are shown as Cα traces. The guide regions of the RNA are not shown for clarity.
Each half-C/D RNA folds into a standard K-turn structure. The GA-stems are orientated toward the inside of the structure center, whereas the WC-stems point outward. The 2 opposite half-C/D RNAs bound in the dimeric RNP are not covalently linked, but the orientation of the GA-stem is compatible with the covalent linkage between them in natural C/D RNAs. The termini of boxes C and D are separated from the opposite ends of boxes D′ and C′ by ≈50 Å (A33 P to C11′ P), and are raised by ≈18 Å above the coiled-coil platform. The large space between the 2 opposite upper wings is suitable to accommodate guides and guide–substrate duplexes (see below). By comparison, an A-form RNA helix has a 23-Å diameter and the 5′ to 3′ phosphate distance spans 31 Å for every 11-bp turn.
Surprisingly, the 2 guide regions (G1–C9), which were not designed to be self-complementary, pair with neighboring equivalent regions in a crystallographic dimeric RNP. The guides form 9-bp duplexes that include 6 Watson–Crick pairs and 3 mismatches (Fig. 1B). This guide self-association results in a nonphysiologic tetrameric organization of RNPs in the crystal. The 2 guide self-duplexes adopt different orientations relative to the coiled-coil protein platform. One duplex formed by D-guide has a ≈45° inclination to the platform, and the other formed by D′-guide is nearly perpendicular to the platform. The guide self-duplexes are analogous with the guide–substrate duplex formed upon substrate loading. However, the 2 conformations are unlikely to represent an active state of the substrate–guide duplex because the end of D or D′-guide is too far away from the end of the opposite C′ or C box (23 or 59 Å). The covalent connection that is present in native C/D RNAs would bring the duplex closer and more parallel to the protein platform.
The CTD of Nop5 is highly conserved (Fig. 3C), reflecting its critical role in binding L7Ae and RNA. The most conserved surface residues of fibrillarin are located at the Nop5 NTD binding interface and near the SAM binding pocket. The latter region is likely to be associated with the substrate–guide RNA duplex.
Recognition of the C/D RNA.
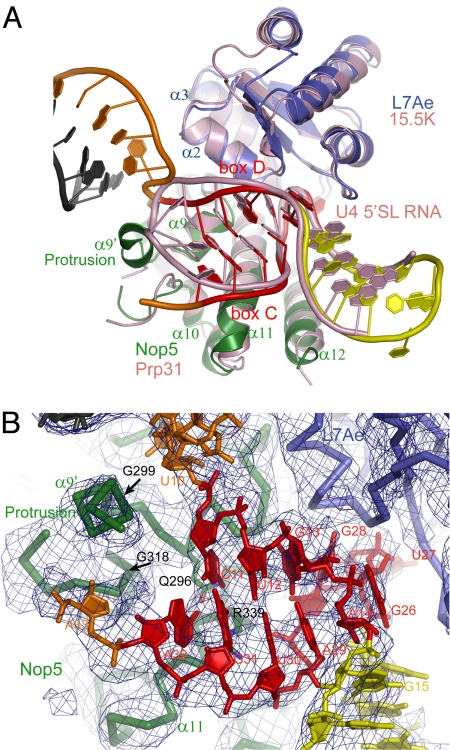
The mutual interactions between L7Ae, the Nop5 CTD, and the RNA closely resemble those present in the structure of the 15.5K–Prp31–U4 5′SL RNA ternary complex (33). L7Ae and the Nop5 CTD can be collectively aligned with their counterparts in the U4 snRNP structure, yielding an rmsd of 1.18 Å over 185 Cα atoms (Fig. 4A). The Nop5 CTD uses helix α8 and the C-terminal half of helix α11 to associate with helices α2 and α3 of L7Ae. The Nop5 CTD and L7Ae jointly contact the major groove of the GA-stem of the K-turn. By contrast, the WC-stem of the K-turn is minimally contacted, suggesting that a K-loop lacking the WC-stem frequently formed by boxes C′ and D′ would similarly assemble in the archaeal RNP (15, 20). However, the presence of a WC-stem in the K-turn seems to be important for eukaryotic C/D RNP assembly (21–23). The structure of L7Ae and the K-turn in the C/D RNP can be well aligned with the binary AF L7Ae–C/D RNA complex structure (Fig. S2) (13), suggesting that the Nop5 CTD recognizes a preformed L7Ae–C/D RNA complex. Nop5 cannot stably associate with L7Ae or RNA alone (17), apparently owing to the incomplete binding surface.
Fig. 4.
Recognition of the C/D RNA. (A) Structural superposition of the C/D RNP and the 15.5K–Prp31–U4 5′SL snRNP complex. The C/D RNP is color-coded as in Fig. 3A, and the U4 snRNP is pink. (B) Interaction between the Nop5 CTD and the half C/D RNA. The σA-weighted 2Fo-Fc map is shown at the 1.0σ contour level. RNAs and candidate residues for specific RNA recognition are shown as sticks, and proteins are shown as Cα traces. The protrusion boundaries (G299 and G318) are indicated by arrows.
The Nop5 CTD structure contains a prominent protrusion that docks at the end of the GA-stem and separates the 2 following guide strands. The protrusion (residues 300–317) is located between helices α9 and α10 and consists of a short helix named α9′ and a partially disordered loop, which were modeled as polyalanine chain on the basis of the experimental map and difference map (Fig. S3). The protrusion is apparently stabilized by RNA interaction, because this region is unstructured in all RNA-free Nop5 structures currently determined (Fig. 2D).
The opposite α9′ helices interact in a different way with the guide self-duplexes in 2 orientations (Fig. S4). At the C/D side, the α9′ helix contacts at the minor groove of the guide self-duplex. At the C′/D′ side, some unmodeled density connects the α9′ helix and the 5′-end of the guide RNA from a crystallographic molecule, suggesting that there are interactions between them. However, the functional relevance of these interactions with guide self-duplexes is unclear because the conformation of substrate–guide duplex in natural C/D RNP is unknown.
The GA-stem is composed of 4 bp, including 2 tandem G·A pairs followed by a U30·U12 noncanonical pair and a G31·C11 Watson–Crick pair. A32 stacks over G31 and, contrary to prediction, does not pair with U10. The 5 residues distal to the G·A pairs (U30, G31, A32, C11, and U12), although not required for K-turn formation, are conserved in C/D RNAs and are functionally essential (35). The Nop5 CTD could recognize the base edges of these residues via hydrogen bonding. Residue Q296 probably contributes to sequence recognition, because its side chain has the potential to contact the base edges of U12 and A32 (Fig. 4B). Consistent with its proposed role, Q296 is invariant among Nop5 and its eukaryotic counterparts Nop56 and Nop58. The corresponding residue is variable in Prp31 proteins, which does not recognize these sequences in U4 RNA. Residue R339 may also mediate sequence recognition owing to its proximity to G31. However, the equivalent residue in Prp31 interacts with the phosphate backbone of U4 RNA (33), suggesting that R339 of Nop5 might mediate a similar interaction. R339 is invariant in the Nop5 family but is occasionally replaced by lysine and other residues in the Prp31 family. Although the moderate resolution of our structure prevents the determination of hydrogen bonding, residues R339 and Q296 have been shown to be critical for binding L7Ae-C/D RNA complex (24, 36).
Flexibility of the Catalytic Module.
The catalytic module comprising fibrillarin and the NTD of Nop5 occupies markedly different positions in our C/D RNP and Nop5ΔCTD–Fib complex structures. This dissimilarity is a result of the different orientations of the α3 and α4 helices of Nop5 (Fig. 2D). The 2 helices fuse into a single continuous long helix in the Nop5ΔCTD–Fib complex, whereas they meet at nearly right angles in the C/D RNP complex. Variable orientation of the catalytic module has been observed between the AF and PF Nop5 structures. However, these previous 2 Nop5 proteins are from different species and demonstrate significant differences in the NTD and tip insertion domain structures. The possibility that different orientations of the catalytic module reflect a species-dependent effect cannot be excluded (24, 31). However, we observed multiple conformations for the same Nop5 protein, providing strong evidence that the linker connecting the Nop5 NTD and the coiled-coil domain is inherently flexible. The wildly varied conformations of the Nop5 catalytic module are primarily due to the different crystallographic environments, rather than to species-dependent effects. All of the observed positions of fibrillarin are too far away from the predicted substrate position to be an active conformation, and large movement should occur for fibrillarin to access the loaded substrate.
Structural Model of Substrate-Loaded Active C/D RNP.
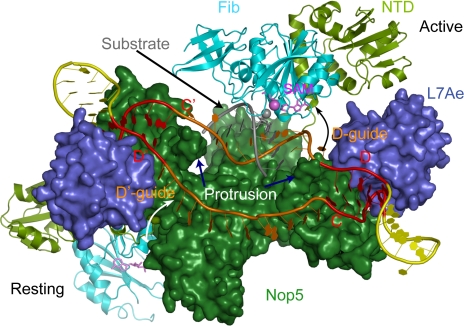
To provide further insight into substrate recruitment and potential conformational transitions of fibrillarin upon substrate loading, we modeled a structure bound with a natural bipartite C/D RNA (Fig. 5). One half of the complex bound by box C/D is modeled in the active state, with loaded substrate and fibrillarin approaching the modification site. The other half bound by box C′/D′ is in a resting state with empty guide. The guides are set to 12 nt, which is the most common length in archaeal C/D RNAs (37). The space between boxes C and D′ (≈50 Å) is large enough to accommodate a 12-nt single-stranded empty guide. With regard to substrate binding, the substrate is set to be paired with the 2nd to 10th residues upstream of box D, forming a 9-bp guide–substrate duplex. This region corresponds to the substrate–guide pairing core that is predicted by statistical analysis (38) and is experimentally important for methylation to occur (39, 40). The 9-bp guide–substrate duplex could reasonably be placed over the coiled-coil platform, in which case the guide strand of the duplex would be linked to boxes D and C′ via 1-nt and 2-nt single-stranded linkers, respectively. The distance between the 5′ and 3′ phosphates of a 9-nt A-form duplex strand is ≈27 Å, and the C and D′ boxes opposite are ≈50 Å away. Therefore, the 2 short linkers need to be significantly stretched to fill the gaps between the guide–substrate duplex and their respective ends, given that a single nucleotide spans, at most, ≈7.4 Å (5′ P to 3′ P distance). The guide–substrate duplex was orientated to allow the methyl-accepting 2′-OH group to face the intruding fibrillarin. The catalytic module composed of the Nop5 NTD and fibrillarin was rotated around the junction between helices α3 and α4 of Nop5, so that the SAM donor methyl group is juxtaposed to the target 2′-OH group. The path of rotation is such that the catalytic module can pass the valley formed between the tip insertion domain and L7Ae without steric hindrance. The resulting model illustrates a potential conformation of the guide–substrate duplex that allows fibrillarin to access the substrate via a swing movement. Nevertheless, the precise conformation of the active C/D RNP requires further structural characterization of substrate-bound native C/D RNP.
Fig. 5.
Model of the native C/D RNP in resting and active states. The C/D half of the RNP complex is in the active state, with the D-guide forming a 9-bp duplex with the substrate and fibrillarin approaching the substrate. The 2′-OH group of the target nucleotide paired to the 5th nt upstream of box D is shown as a gray sphere, and the SAM methyl group to be transferred is shown as a violet sphere. The other half of the complex is in a resting state, with boxes C and D′ connected by a linear 12-nt guide. The curved arrows indicate the transition path for the catalytic module from the resting state to the active state.
Discussion
In this study, we present the structure of a C/D RNP that includes its full complement of 3 proteins assembled with a split half-C/D RNA. Although the RNA is unnatural, to our knowledge this is the most comprehensive C/D RNP structure yet solved. This RNP structure also formed the basis for a model of the natural RNP in its substrate-loaded active state. Together, the structure and the model provide insight into the overall organization and mechanism of action of C/D RNA–guided RNA methyltransferases.
Structural Organization of C/D RNP.
The opposing CTDs in the dimeric C/D RNP provide 2 anchoring sites for the dual K-turns in the C/D RNA, thereby bridging the symmetry in the protein and RNA structure. The positioning of the CTDs is organized by the self-associated coiled-coil domains at the center of the RNP complex. The fact that the structures and the sequences of both the coiled-coil and the CTD domains are highly conserved suggests that the 2 CTDs are separated by a fixed distance in all C/D RNPs, and accordingly the 2 bound C/D motifs are also separated by a fixed distance (≈50 Å). This fixed inter-motif spacing predicts that the proper assembly of the C/D motifs into Nop5's CTD requires the guide connecting opposite K-turns to be longer than approximately 7 nt, by our estimation. However, for a guide to form a duplex of sufficient length with the substrate, the guide needs to be longer than the minimal length, because a strand in a duplex has a significantly shorter end-to-end distance. In our modeling process, a length of 12 nt seems to be the lower limit for the formation of a 9-bp substrate–guide duplex. Guides shorter than 12 nt would allow RNA assembly but would interfere with substrate interaction. Indeed, it has been shown for an archaeal RNP that reducing the length of a guide to 10 nt had no effect on RNP assembly but significantly blocked the modification activity mediated by the specific guide (37). Interestingly, increasing the guide length to 14 nt or longer also blocked activity (37). This suggests that the modifying enzyme might have little tolerance for excess unpaired guide residues in the active conformation of the RNP. This upper limit on the length of the guide apparently does not to apply to eukaryotic C/D RNAs, which are highly variable in terms of the spacing of the C/D motifs.
Our C/D RNP structure shows that the protrusion in the Nop5 CTD is a key structural element in the organization of the guide RNA conformation. The protrusion wedges between the 2 guides and separates them into different paths, which may prevent accidental pairing between 2 guides and liberate them for substrate association. In support of this role, a C/D RNA with paired guides was shown not to be able to bind the target unless it assembled with Nop5 (41). In addition, the 2 protrusions at opposite ends of the dimeric RNP are close to the guide–substrate duplex in the substrate-loaded model, implying their potential role in substrate loading (Fig. 5). The residues in the protrusion are highly conserved in Nop5 proteins (Fig. 3C), underscoring their importance.
Previous studies showed that RNPs assembled with split half-C/D RNAs lost site specificity, leading to promiscuous modification around the target ribose (29). The flexibility in the guide–substrate duplex in the absence of fixation at one end as shown in our C/D RNP structure, as well as the inherent mobility of the catalytic module, may contribute to the nonspecific modification.
Structural Determinants of C/D and U4 RNP Assembly Specificity.
The assembly mode of C/D RNP closely resembles that of U4 snRNP, confirming their evolutionary relationship. Nevertheless, Nop5 and Prp31 assemble specifically into C/D and U4 RNP, respectively. What is the structural determinant for this assembly specificity? It was previously proposed that the length of the GA-stem distinguishes the 2 RNP assemblies, because the GA-stem of the C/D RNA was thought to be longer by 1 bp than the U4 RNA stem, and Prp31 cannot bind a GA-stem with 1 additional base pair (33, 42). However, our structure shows that the U10·A32 pair in our C/D RNA is not formed. Therefore, its GA-stem is the same length as that of U4 RNA. Rather than being guided by the length of the GA-stem, the assembly specificity seems to derive from different terminal structures of the GA-stem. The GA-stem of U4 snRNA is closed by a pentaloop, whereas that of C/D RNA is an open-ended structure that leads to 2 single-stranded guides. The CTD protrusions seem to be a key factor for discriminating the different terminal structures of GA-stems. The α9′ helix of Nop5 would sterically interfere with the pentaloop of U4 RNA (Fig. 4A), whereas the corresponding protrusion in Prp31 is tailored to interact with the pentaloop of U4 snRNA (33). The protein sequences of the protrusions are highly divergent between Nop5 and Prp31, consistent with their different roles in RNP assembly and function. Moreover, the Nop5 CTD may specifically recognize the conserved residues distal to the G·A pairs in the GA-stem of C/D RNA, which further distinguishes C/D from U4 RNA.
Methods
For detailed information regarding the cloning, protein purification, crystallization, and structure determination, see SI Text.
Supplementary Material
Acknowledgments.
We thank staff at SPring-8 and the Beijing Synchrotron Radiation Facility for help in data collection. This research was supported by the Chinese Ministry of Science and Technology and Beijing Municipal Government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3ID5 (C/D RNP structure), 3ICX (Nop5ΔNTD structure), and 3ID6 (Nop5ΔCTD-Fib complex structure)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0905128106/DCSupplemental.
References
- 1.Czerwoniec A, et al. MODOMICS: A database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 3.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 4.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Omer AD, et al. RNA-modifying machines in archaea. Mol Microbiol. 2003;48:617–629. doi: 10.1046/j.1365-2958.2003.03483.x. [DOI] [PubMed] [Google Scholar]
- 6.Reichow SL, Hamma T, Ferre-D'Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiss-Laszlo Z, et al. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 8.Tycowski KT, Smith CM, Shu MD, Steitz JA. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc Natl Acad Sci USA. 1996;93:14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavaille J, Nicoloso M, Bachellerie JP. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 10.Watkins NJ, et al. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 11.Vidovic I, et al. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 12.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 14.Omer AD, et al. Homologs of small nucleolar RNAs in Archaea. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 15.Nolivos S, Carpousis AJ, Clouet-d'Orval B. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res. 2005;33:6507–6514. doi: 10.1093/nar/gki962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 17.Omer AD, Ziesche S, Ebhardt H, Dennis PP. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc Natl Acad Sci USA. 2002;99:5289–5294. doi: 10.1073/pnas.082101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tollervey D, et al. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, et al. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution. EMBO J. 2000;19:317–323. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran EJ, Zhang X, Maxwell ES. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J. 2003;22:3930–3940. doi: 10.1093/emboj/cdg368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charron C, et al. The archaeal sRNA binding protein L7Ae has a 3D structure very similar to that of its eukaryal counterpart while having a broader RNA-binding specificity. J Mol Biol. 2004;342:757–773. doi: 10.1016/j.jmb.2004.07.046. [DOI] [PubMed] [Google Scholar]
- 22.Szewczak LB, DeGregorio SJ, Strobel SA, Steitz JA. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem Biol. 2002;9:1095–1107. doi: 10.1016/s1074-5521(02)00239-9. [DOI] [PubMed] [Google Scholar]
- 23.Szewczak LB, et al. Molecular basis for RNA kink-turn recognition by the h15.5K small RNP protein. RNA. 2005;11:1407–1419. doi: 10.1261/rna.2830905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aittaleb M, et al. Structure and function of archaeal box C/D sRNP core proteins. Nat Struct Biol. 2003;10:256–263. doi: 10.1038/nsb905. [DOI] [PubMed] [Google Scholar]
- 25.Bortolin ML, Bachellerie JP, Clouet-d'Orval B. In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNA(Trp) Nucleic Acids Res. 2003;31:6524–6535. doi: 10.1093/nar/gkg860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rashid R, et al. Functional requirement for symmetric assembly of archaeal box C/D small ribonucleoprotein particles. J Mol Biol. 2003;333:295–306. doi: 10.1016/j.jmb.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Omer AD, Zago M, Chang A, Dennis PP. Probing the structure and function of an archaeal C/D-box methylation guide sRNA. RNA. 2006;12:1708–1720. doi: 10.1261/rna.31506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, et al. The coiled-coil domain of the Nop56/58 core protein is dispensable for sRNP assembly but is critical for archaeal box C/D sRNP-guided nucleotide methylation. RNA. 2006;12:1092–1103. doi: 10.1261/rna.2230106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardin JW, Batey RT. The bipartite architecture of the sRNA in an archaeal box C/D complex is a primary determinant of specificity. Nucleic Acids Res. 2006;34:5039–5051. doi: 10.1093/nar/gkl644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cahill NM, et al. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 2002;21:3816–3828. doi: 10.1093/emboj/cdf376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oruganti S, et al. Alternative conformations of the archaeal Nop56/58-fibrillarin complex imply flexibility in box C/D RNPs. J Mol Biol. 2007;371:1141–1150. doi: 10.1016/j.jmb.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 32.Hamma T, Ferre-D'Amare AR. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 A resolution. Structure. 2004;12:893–903. doi: 10.1016/j.str.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 33.Liu S, et al. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science. 2007;316:115–120. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 34.Brunger AT, DeLaBarre B, Davies JM, Weis WI. X-ray structure determination at low resolution. Acta Crystallogr D Biol Crystallogr. 2009;65:128–133. doi: 10.1107/S0907444908043795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watkins NJ, Dickmanns A, Luhrmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol Cell Biol. 2002;22:8342–8352. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardin JW, Reyes FE, Batey RT. Analysis of a critical interaction within the archaeal box C/D small ribonucleoprotein complex. J Biol Chem. 2009;284:15317–15324. doi: 10.1074/jbc.M901368200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran E, Zhang X, Lackey L, Maxwell ES. Conserved spacing between the box C/D and C′/D′ RNPs of the archaeal box C/D sRNP complex is required for efficient 2′-O-methylation of target RNAs. RNA. 2005;11:285–293. doi: 10.1261/rna.7223405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen CL, Perasso R, Qu LH, Amar L. Exploration of pairing constraints identifies a 9 base-pair core within box C/D snoRNA-rRNA duplexes. J Mol Biol. 2007;369:771–783. doi: 10.1016/j.jmb.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 39.Cavaille J, Bachellerie JP. SnoRNA-guided ribose methylation of rRNA: Structural features of the guide RNA duplex influencing the extent of the reaction. Nucleic Acids Res. 1998;26:1576–1587. doi: 10.1093/nar/26.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Appel CD, Maxwell ES. Structural features of the guide:target RNA duplex required for archaeal box C/D sRNA-guided nucleotide 2′-O-methylation. RNA. 2007;13:899–911. doi: 10.1261/rna.517307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh SK, Gurha P, Gupta R. Dynamic guide-target interactions contribute to sequential 2′-O-methylation by a unique archaeal dual guide box C/D sRNP. RNA. 2008;14:1411–1423. doi: 10.1261/rna.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz A, Nottrott S, Hartmuth K, Luhrmann R. RNA structural requirements for the association of the spliceosomal hPrp31 protein with the U4 and U4atac small nuclear ribonucleoproteins. J Biol Chem. 2006;281:28278–28286. doi: 10.1074/jbc.M603350200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.