Abstract
Gene silencing via heterochromatin formation plays a major role in cell differentiation and maintenance of homeostasis. Here we report the identification and characterization of a novel heterochromatinization factor in vertebrates, bromo adjacent homology domain–containing protein 1 (BAHD1). This nuclear protein interacts with HP1, MBD1, HDAC5, and several transcription factors. Through electron and immunofluorescence microscopy studies, we show that BAHD1 overexpression directs HP1 to specific nuclear sites and promotes the formation of large heterochromatic domains, which lack acetyl histone H4 and are enriched in H3 trimethylated at lysine 27 (H3K27me3). Furthermore, ectopically expressed BAHD1 colocalizes with the heterochromatic inactive X chromosome (Xi). The BAH domain is required for BAHD1 colocalization with H3K27me3, but not with the Xi chromosome. As highlighted by whole genome microarray analysis of BAHD1 knockdown cells, BAHD1 represses several proliferation and survival genes, in particular the insulin-like growth factor II gene (IGF2). When overexpressed, BAHD1 specifically binds the CpG-rich P3 promoter of IGF2, which increases MBD1 and HDAC5 targeting at this locus. This region contains DNA-binding sequences for the transcription factor SP1, with which BAHD1 coimmunoprecipitates. Collectively, these findings provide evidence that BAHD1 acts as a silencer by recruiting at specific promoters a set of proteins that coordinate heterochromatin assembly.
Keywords: BAH domain, epigenetics, heterochromatin, HP1, polycomb
Gene repression in eukaryotes in response to developmental or environmental signals is a complex multistep process requiring the concerted action of many cellular factors, including DNA-binding transcription factors and cofactors. It also involves chromatin components and chromatin-binding/-modifying proteins, which are implicated in the establishment and maintenance of a condensed chromatin state, also referred to as facultative heterochromatin (1). Deregulation of these chromatin regulatory factors and aberrant chromatin modifications play important roles in various human diseases, including cancers, developmental abnormalities, and neurologic disorders (2–6). Moreover, several recent reports have linked histone modifications and infectious diseases (7). During a screen for human proteins that interact with microbial factors and that might play a role in bacterial pathogenicity, we identified a putative chromatin-associated protein, bromo adjacent homology domain–containing protein 1 (BAHD1), which remains uncharacterized. One study correlated repression of the BAHD1 gene to poor prognosis in lung cancer (8), raising the possibility that BAHD1 might act as a tumor suppressor. This connection between BAHD1 and pathological processes prompted us to characterize the function of this protein.
BAHD1 is so named because it contains a C-terminal BAH domain, which is found in several chromatin-binding proteins involved in transcriptional repression (9), including the yeast Sir3 protein (10). We investigated whether BAHD1 could be involved in heterochromatin formation using a combination of approaches, including two-hybrid screen, knockdown, and gain-of-function experiments.
Results
BAHD1 Is a Nuclear Protein.
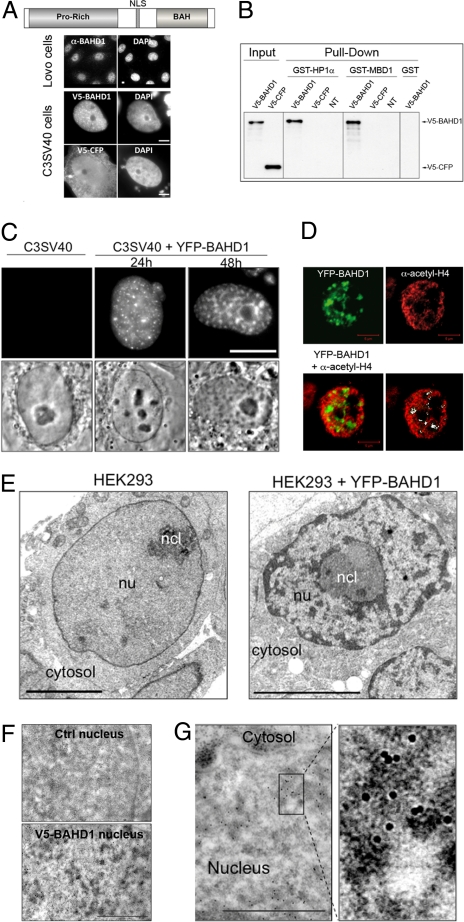
A single gene encodes BAHD1 in vertebrates, and no ortholog is found in invertebrates or plants, suggesting that BAHD1 is involved in vertebrate-specific functions. In addition to its C-terminal BAH domain, the BAHD1 protein contains a N-terminal proline-rich region and a central nuclear localization signal (Fig. 1A). Immunofluorescence labeling with an antibody generated against the last 17 residues of the protein indicated that BAHD1 localizes to interphase nuclei in human cells (Fig. 1A). But the staining was relatively weak, in agreement with the reportedly low BAHD1 gene expression in a broad range of human tissues (11, 12). Consequently, we cloned BAHD1 cDNA into an expression vector. One day after transfection, BAHD1 tagged with the V5 epitope localized exclusively to the nucleus, whereas V5-tagged cyan fluorescent protein (V5-CFP), used as a control, localized to both the nucleus and the cytoplasm (Fig. 1A), confirming that BAHD1 is a nuclear protein.
Fig. 1.
BAHD1 promotes heterochromatin formation. (A) Endogenous BAHD1 localizes predominantly to nuclei, labeled with DAPI, when detected with an anti-BAHD1 antibody in Lovo epithelial cells. V5-BAHD1 expressed in C3SV40 fibroblasts localizes only to the nucleus, whereas control V5-CFP localizes to both the cytosol and the nucleus. (Scale bar: 5 μm.) (B) BAHD1 binds HP1α and MBD1. Nuclear extracts from HEK293 cells either untransfected (NT) or transfected with V5-BAHD1 or V5-CFP constructs were incubated with purified GST-HP1α, GST-MBD1, or GST. Bound BAHD1 was detected in immunoblots using anti-V5 antibody; 10% of nuclear extracts were analyzed in parallel (input). (C–E) Increasing the amount of BAHD1 in cells stimulates chromatin condensation. (C) In living C3SV40 cells, YFP-BAHD1 forms nuclear foci that increase in size with increasing transfection time (24–48 h) and appear as dense material in phase contrast (Movie S1). Large dark bodies are nucleoli. (Scale bar: 10 μm.) (D) Confocal image of a YFP-BAHD1–transfected C3SV40 cell labeled with acetyl-H4 antibody. The white regions on the last image are positive for YFP-BAHD1 and negative for acetyl-H4. (Scale bar: 5 μm.) (E) Transmission electron microscopy reveals dense patches of condensed chromatin formed in nucleus of YFP-BAHD1 expressing HEK293 cells. (Scale bar: 5 μm.) (F) Anti-V5 immunogold cytochemistry demonstrates localization of V5-BAHD1 at de novo formed heterochromatin. V5-BAHD1, detected with 10-nm colloidal gold particles, specifically localizes to electron-dense areas in the nucleus that are not present in untransfected cells (Ctrl). (Scale bar: 1 μm.) (G) High magnification highlights localization of V5-BAHD particles within electron-dense heterochromatin areas.
BAHD1 Interacts With Chromatin Repressors and Transcription Factors.
Given that BAH domain–containing proteins, such as the yeast Sir3 and mammalian MTA proteins, are components of multiprotein silencing complexes (10, 13), we hypothesized that BAHD1 also belongs to a chromatin-modifying complex, and searched for its partners using a large-scale yeast two-hybrid screen. This screen identified a series of nuclear BAHD1 interactors [supporting information (SI) Table S1], several of which are well-characterized chromatin-associated repressors, including the 3 isoforms of heterochromatin protein 1 (HP1α, β, and γ), the methyl-CpG–binding protein MBD1, the histone H3 methyltransferase KMT1E, the histone deacetylase HDAC5, and the ATP-dependent chromatin remodeling enzyme CHD1. This screen also revealed several transcription and RNA-processing factors. We validated the ability of BAHD1 to associate with at least 2 of its partners, HP1α and MBD1, through pull-down assays. In contrast to GST alone, GST-HP1α or GST-MBD1 purified proteins added to nuclear extracts of HEK293 cells expressing V5-BAHD1 or the control V5-CFP were able to pull down V5-BAHD1, but not the control V5-CFP (Fig. 1B), demonstrating that BAHD1 associates with HP1α and MBD1.
BAHD1 Represses Basal Transcription.
The chromatin-associated factors HP1α and MBD1 play key roles in the formation of repressive chromatin by binding methylated histone H3 and methylated DNA, respectively (14–16). Consequently, we expected that BAHD1 would induce transcriptional repression. To test this hypothesis, BAHD1 was fused to the DNA-binding domain (DBD) of the GAL4 protein and coexpressed in HEK293 cells with reporters containing GAL4 binding sites linked to either TK or MMTV promoters upstream of the luciferase gene. GAL4-BAHD1 inhibited luciferase activity in a dose-dependent manner, with a repressive effect comparable to that of GAL4-HP1α and GAL4-MBD1 (Fig. S1). These results demonstrate that BAHD1 acts as a repressor when targeted to a promoter. Because BAHD1 can interact with heterochromatin factors, it may promote gene silencing through an heterochromatinization process.
Increasing BAHD1 Expression Levels in Cells Triggers Heterochromatin Formation.
An important feature of heterochromatin is its ability to spread from a specific nucleation site (14). We reasoned that if BAHD1 promotes the assembly of a heterochromatinization machinery at genomic microdomains, then increasing its amount in cells should stimulate the spread of heterochromatin throughout large genomic regions, a phenomenon visible by microscopy techniques. To address this question, we expressed YFP-BAHD1 in human C3SV40 fibroblasts for 24 h or 48 h. YFP-BAHD1 localized in the nucleus as dispersed foci that increase in size with increasing YFP-BAHD1 expression to form larger structures (Fig. 1C). Importantly, these structures were visible in living cells as patches of phase-dense material (Movie S1), which did not result from unspecific aggregation, because they were not observed on expression of either YFP or another tagged nuclear protein, such as GFP-PML (data not shown). To examine whether YFP-BAHD1–associated foci were heterochromatin areas, we first examined whether they correlated with histone hypoacetylation, a specific feature of heterochromatin (1). Accordingly, acetyl-histone H4 was largely excluded from YFP-BAHD1 foci (Fig. 1D). We then confirmed by electron microscopy that these nuclear structures were domains of condensed chromatin. In HEK293 cells expressing YFP-BAHD1, electron-dense patches of heterochromatin were visible throughout the nucleus, but were rarely detected in nuclei of untransfected cells (Fig. 1E). Immunogold labeling further demonstrated that BAHD1 localized to this de novo–formed heterochromatin. In cells expressing V5-BAHD1, the V5 antibody reacted with electron-dense regions of condensed chromatin, but did not label nuclei of untransfected cells (Fig. 1 F and G).
BAHD1 Directs HP1α at Specific Nuclear Sites.
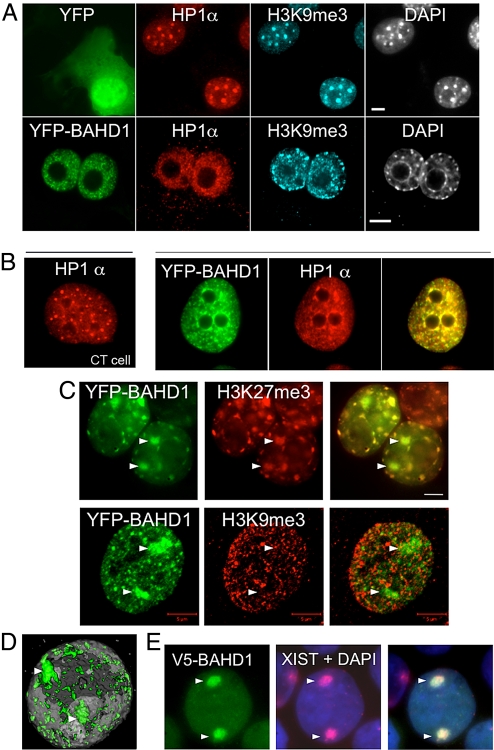
We next addressed whether BAHD1-mediated assembly of heterochromatin occurs after recruitment of chromatin modifiers by BAHD1 at specific nuclear sites, or vice versa. For this, we focused on HP1α, whose localization within murine nuclei is well established. In interphase nuclei of murine cells, HP1α bound to histone H3 trimethylated at lysine 9 (H3K9me3) accumulates mainly at chromocenters (i.e., pericentromeric heterochromatin), which appear as distinct large heterochromatic bodies after DAPI staining or labeling with a specific H3K9me3 antibody (17). In contrast, in murine nuclei expressing YFP-BAHD1, HP1α was displaced from chromocenters to YFP-BAHD1 foci (Figs. 2A and S2A). In human cells—C3SV40 fibroblasts (Fig. 2B) and JEG3 epithelial cells (Fig. S2B)—in which HP1α localizes at dispersed small nuclear foci, expression of YFP-BAHD1 also triggered important changes in HP1α localization, with its recruitment to BAHD1 foci. This HP1α relocalization to YFP-BAHD1–associated heterochromatin sites establishes that BAHD1 can displace and recruit HP1α in vivo, either directly or indirectly.
Fig. 2.
BAHD1 recruits HP1α and colocalizes with H3K27me3 in vivo. (A) BAHD1 delocalizes HP1α from chromocenters in mouse fibroblasts. In 3T3 fibroblasts expressing YFP, HP1α localizes to chromocenter nuclear bodies labeled with H3K9me3 antibodies and DAPI. In YFP-BAHD1–transfected cells, HP1α no longer accumulates at chromocenters, but instead localizes mainly at YFP-BAHD1 discrete foci (Fig. S2). (Scale bar: 5 μm.) (B) HP1α is recruited to BAHD1-associated heterochromatin in human fibroblasts. In C3SV40 cells expressing control YFP (CT cell), HP1α localizes at dispersed nuclear foci. In YFP-BAHD1–expressing cells, HP1α colocalizes with BAHD1, as shown by yellow regions in the merge image. (C and D) YFP-BAHD1 heterochromatic foci have features of facultative heterochromatin. (C) YFP-BAHD1 expressed in HEK293 cells colocalizes with H3K27me3 at several nuclear territories, including Xi (indicated by triangles). YFP-BAHD1 does not colocalize with H3K9me3. (Scale bar: 5 μm.) (D) Three-dimensional reconstruction of confocal images shows localization of V5-BAHD1 at the 2 Xi chromosomes (indicated by triangles), as well as several other domains dispersed in a HEK293 cell nucleus (Movie S2). (E) Exogenous V5-BAHD1 is recruited to the Xi chromosomes when expressed for 24 h in HEK293 cells. The Xi chromosomes were detected by RNA FISH using a Xist probe (in red) in the nucleus labeled with DAPI (in blue).
Ectopically Expressed BAHD1 Colocalizes With Histone H3K27me3, Including at the inactive X chromosome (Xi).
Our initial two-hybrid screen suggested that BAHD1 could link chromatin condensation activities to DNA-binding transcription factors (Table S1), thereby inducing the formation of facultative heterochromatin rather than constitutive heterochromatin at euchromatic loci. To test this hypothesis, we examined specific histone modifications that can distinguish between the 2 forms of heterochromatin (1). We found that the YFP-BAHD1 heterochromatic foci overlapped mainly with H3K27me3, a specific feature of facultative heterochromatin, and overlapped only rarely with H3K9me3, found in constitutive heterochromatin (Fig. 2C). In addition, as noted earlier, YFP-BAHD1 was not recruited at constitutive pericentromeric heterochromatin (Fig. 2A). Strikingly, when expressed in HEK293 cells, YFP-BAHD1 or V5-BAHD1 colocalized with H3K27me3 not only at heterochromatic foci dispersed throughout the nucleus, but also at 2 large bodies located at the nuclear periphery corresponding to 2 Xi (Movie S2; Fig. 2D). Indeed, HEK293 are female cells with an atypical karyotype, usually containing 3 X chromosomes, 2 of which are inactive and visible as large heterochromatic bodies (18). Through FISH, using as a probe the X-inactive specific transcript (XIST), an untranslated RNA that coats the Xi (18, 19) we confirmed that exogenous BAHD1 was recruited at these 2 Xi . V5- or YFP-BAHD1 localized at the 2 Xi in HEK293 cells (Figs. 2E and S2C), as well as at the unique Xi in diploid fibroblasts IMR90 and WI38 (data not shown). These findings indicate that BAHD1 is associated with facultative heterochromatin. They also suggest that it may target genes modified with H3K27me3, thus acting upstream or downstream of the histone H3 (K27) methyltransferase EZH2 of the polycomb (PcG) repressive complex PRC2, which catalyzes this histone modification (20).
The BAH Domain Does Not Bind H3K27me3 In Vitro but Is Required for BAHD1 Colocalization With H3K27me3 In Vivo.
The BAH domain has been well characterized in the yeast Sir3 protein as a nucleosome and histone tail–binding domain (10). To investigate whether the BAH domain of BAHD1 could directly bind H3K27me3, we used purified GST-BAH in binding assays with biotin-labeled histone H3 peptides (i.e., H3K27me3, H3K9me3, or unmodified H3 peptides) and histone H2B or H4 peptides. In contrast to GST-HP1α, which strongly binds H3K9me3 through its chromodomain, GST-BAH did not specifically bind H3K27me3 (Fig. S3A).
We next investigated whether the BAH domain could play a role in BAHD1 localization to H3K27me3 in vivo, by generating a truncated form of BAHD1 lacking the last 188 amino acids encompassing the BAH domain (BAHD1-ΔBAH). Because high expression of this YFP-tagged truncated protein in cells led to the formation of abnormal large nuclear bodies (data not shown), we further analyzed only those cells that moderately expressed BAHD1-ΔBAH. Strikingly, BAHD1-ΔBAH failed to localize with H3K27me3 at nuclear foci in C3SV40 male cells (Fig. S3B) and HEK293 female cells, except at the Xi chromosome (Fig. S3C).
Taken together, our in vitro and in vivo results suggest that the BAH domain is necessary but not sufficient for BAHD1 colocalization with H3K27me3. In addition, recruitment of exogenous BAHD1 at the Xi chromosome does not require the BAH domain and may occur through interaction with Xi-specific components other than H3K27me3.
BAHD1 Depletion With siRNA Induces Expression of PRC2 Targets Such as IGF2.
To identify genes targeted and repressed by BAHD1, we searched for genes induced in HEK293 cells depleted of endogenous BAHD1 with small interfering RNA (siRNA). Our analysis on whole human genome microarrays identified 192 transcripts induced by at least 1.4-fold in BAHD1 knockdown (KD) cells (Table S2). Differences in expression between control and BAHD1 KD cells were consistent with gene expression changes described in PcG-deficient cells (21). The expression array data were then validated by quantitative real-time (qRT)-PCR analysis in independent experiments for a selection of several genes (Fig. S4). Of note, various proliferation and survival genes were induced on BAHD1 depletion; in particular, half of the genes associated with our data set and analyzed with the Ingenuity software were related to cell death, cellular growth and proliferation, and cancers (Table S3). These data suggest that BAHD1 may directly silence several cancer-related genes.
According to the colocalization studies shown above, BAHD1 may target genes modified with H3K27me3. Thus, we compared the set of genes induced in BAHD1 KD cells with PcG targets previously identified in human embryonic fibroblasts (21). We found that 35 genes induced on BAHD1 depletion overlapped with PcG targets (Table S3). Interestingly, the gene most induced by BAHD1 depletion was the IGF2 gene, found as a PRC2 target in several genome-wide mapping studies (21–23). Like BAHD1, IGF2 is specific to vertebrates. It encodes a growth factor that plays a key role in fetal growth and influences body mass in adults (24). Moreover, IGF2 overexpression correlates with carcinogenesis (3, 24, 25). We thus addressed whether BAHD1 directly regulates IGF2.
BAHD1 Binds the IGF2 P3 Promoter Region and Represses IGF2 and IGF2AS.
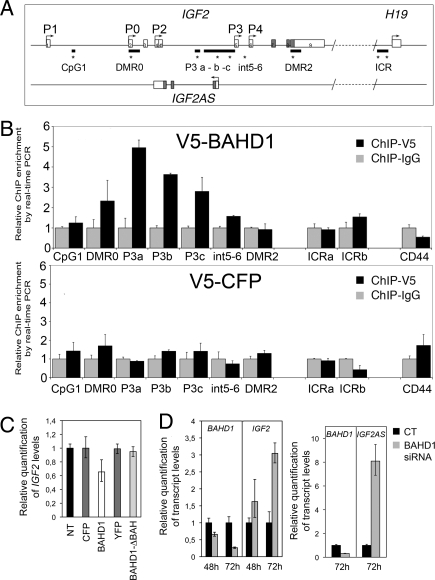
In humans, IGF2 regulation is a complex process involving 5 different promoters (Fig. 3A) in a tissue- and development-specific manner (26). IGF2 is also submitted to genomic imprinting via transcriptional repression of the maternal allele (3, 24, 26). IGF2 imprinting involves differential methylation of CpG islands between the 2 parental alleles (DMR), 1 of which—the imprinted control region (ICR)—is an insulator located upstream of the adjacent imprinted gene H19 (Fig. 3A) (3, 24, 26). Because BAHD1 interacts with the methyl-CpG binding protein MBD1, and because of the importance of CpG methylation in IGF2 expression, we investigated whether BAHD1 directly binds IGF2 at CpG islands in HEK293 cells. In these cells, IGF2 expression is governed mainly by promoter 3 (P3), the most active promoter in fetal and nonhepatic adult tissues (27) and the major up-regulated promoter in many tumor tissues (25, 28). We performed chromatin immunoprecipitation (ChIP) from cells expressing V5-BAHD1 or the control V5-CFP, using the V5 antibody (which is efficient in ChIP assays) or mouse IgG to control nonspecific binding. Three DNA fragments present in the 2 CpG islands located upstream of P3 (P3a, P3b, and P3c) were selectively enriched in ChIP samples from V5-BAHD1–transfected cells, whereas no specific binding of BAHD1 was observed elsewhere in the IGF2/H19 region (Fig. 3B). BAHD1 binding to the P3 region was correlated with repression of IGF2, as demonstrated by a 40% decrease in IGF2 transcript level in BAHD1-overexpressing cells. In contrast, overexpression of the truncated BAHD1-ΔBAH did not repress IGF2, suggesting that the BAH domain is required for BAHD1 silencing function (Fig. 3C). In cells depleted of endogenous BAHD1, the increased amount of IGF2 transcript was correlated with decreased expression of BAHD1 (Fig. 3D). Taken together, our results strongly suggest that BAHD1 promotes heterochromatinization at the P3 proximal region, thereby repressing transcription of IGF2.
Fig. 3.
BAHD1 targets IGF2. (A) Organization of the human IGF2/IGF2AS/H19 genomic region. Shown are the positions of IGF2 promoters (P0→P4), CpG islands (black lines), exons (open boxes), and ORF (gray boxes) (adapted from ref. 26). Stars indicate the position of primers used in ChIP. The position of IGF2AS on the opposite strand is shown. (B) BAHD1 binds IGF2 in the P3 proximal region. ChIP assays with V5 antibodies or control IgG using chromatin prepared from V5-BAHD1– or control V5-CFP–expressing HEK293 cells. DNA fragments were quantified by qPCR with the primer sets indicated in (A). The CD44 promoter was used as a negative control because it shares features with IGF2 P3 (i.e., a CpG island and localization on chromosome 11). The amount of DNA precipitated with V5 antibody was normalized to the amount precipitated with IgG control. The data are averages (± SD) of qPCR replicates of 1 of 3 independent ChIP experiments. (C and D) BAHD1 represses IGF2 and IGF2AS. (C) HEK293 cells were transfected for 24 h with V5-CFP, V5-BAHD1, YFP, or BAHD1-BAH. The levels of IGF2 transcript quantified by qRT-PCR decreased with BAHD1 overexpression, but not with BAHD1-BAH overexpression. (D) HEK293 cells were transfected with BAHD1 siRNA or control siRNA (CT). BAHD1, IGF2, or IGF2AS transcript levels were quantified by qRT-PCR after 48 h or 72 h.
The CpG-rich region upstream of P3 contains several potential binding sites for transcription factors regulating IGF2, as well as regulatory elements for IGF2AS, an antisense transcript of IGF2 also expressed on the paternal allele and targeted by PRC2 (Fig. 3A) (21–23, 29). We reasoned that if BAHD1 promotes heterochromatinization at P3, then it also should repress IGF2AS expression. In agreement with this hypothesis, we found that IGF2AS transcript level significantly increased on BAHD1 depletion with siRNA (Fig. 3D). Together, these results demonstrate that BAHD1 physically interacts with the CpG-rich P3 regulatory region and represses IGF2 and IGF2AS transcription.
BAHD1 Coimmunoprecipitates With MBD1, HDAC5, and SP1.
We explored whether transcription factors known or predicted to regulate P3, including AP2, SP1, EGR1, MEF2, TEF1, and HAND1 (Fig. S5), are connected to BAHD1 through protein–protein interactions. We extended the network of interactions obtained in our two-hybrid screen to known interactors of BAHD1 partners, reported in Ingenuity and HPRD knowledge databases. Strikingly, this global analysis demonstrated a network in which BAHD1 couples chromatin modifiers to transcription factors able to target promoter P3 (Fig. S5; Table S1). This network also connects BAHD1 to EZH2 and to the PRC1 ubiquitin ligase RING1b (20), supporting a link between BAHD1- and PcG-mediated silencing (Fig. S5; Table S1).
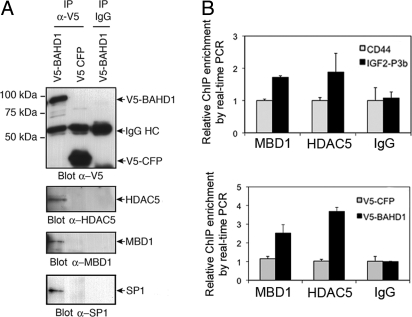
We then used coimmunoprecipitation to address whether BAHD1 could interact in vivo with some of these proteins. Nuclear extracts from HEK293 cells expressing either V5-BAHD1 or control V5-CFP were immunoprecipitated with the anti-V5 antibody or with control IgG, and the immunoprecipitates were probed for the presence of HDAC5, MBD1, SP1, and AP2. HDAC5, MBD1, and SP1 clearly coimmunoprecipitated with V5-BAHD1 but not with the control V5-CFP (Fig. 4A). In contrast, no signal of AP2 copurifying with V5-BAHD1 was detected (data not shown). These results indicate that ectopically expressed BAHD1 forms a complex with chromatin regulatory proteins and at least one transcription factor (SP1).
Fig. 4.
(A) Exogenous BAHD1 coimmunoprecipitates with endogenous HDAC5, MBD1, and SP1. Nuclear extracts from V5-BAHD1– or V5-CFP–expressing HEK293 cells were analyzed after IP with anti-V5 mAbs (IP α-V5) or with mouse IgG (IP IgG). Western blots were probed with antibodies against V5, HDAC5, MBD1, or SP1. IgG HC, heavy chains of immunoglobulins. (B) MBD1 and HDAC5 are recruited at IGF2 P3 in BAHD1-overexpressing cells. Chromatin from V5-BAHD1– or V5-CFP–expressing HEK293 cells was used in ChIP with MBD1 or HDAC5 antibodies or control IgG. P3b and CD44 DNA fragments were quantified by qPCR, as in Fig. 3B. The amount of DNA precipitated with the different antibodies was normalized to the amount precipitated with IgG. In cells expressing V5-BAHD1, MBD1 and HDAC5 recruitment at IGF2 P3b increased compared with that at the CD44 promoter (top) and was greater than in control cells expressing V5-CFP (bottom).
Increasing BAHD1 Expression Induces MBD1 and HDAC5 Recruitment at IGF2 P3.
To provide evidence that BAHD1 mediates gene silencing by recruiting heterochromatin factors to specific loci, we examined MBD1 and HDAC5 recruitment at the IGF2 promoter P3 in BAHD1-overexpressing cells. In these cells, exogenous BAHD1 is recruited at P3 (Fig. 3B), represses IGF2 expression (Fig. 3C) and coimmunoprecipitates with MBD1 and HDAC5 (Fig. 4A). As demonstrated by ChIP assays, increased MBD1 and HDAC5 recruitment at the P3b region of IGF2 occurred on BAHD1 ectopic expression (Fig. 4B). These results strongly suggest that targeting BAHD1 to a specific locus triggers the recruitment of heterochromatin factors.
Discussion
In this work, we have demonstrated through multiple approaches that BAHD1 is a new factor involved in heterochromatin formation in vertebrates. We propose that BAHD1 acts as a silencer that connects heterochromatin factors, such as HP1, MBD1, and HDAC5, to DNA-bound transcription factors, such as SP1, to repress transcription at specific euchromatic loci. Our data strongly suggest that BAHD1 targets include several genes previously associated with the H3K27me3 mark, including IGF2 (21–23). Our findings also provide evidence for a role of the conserved BAH domain in subnuclear localization of BAHD1 and silencing. We also found that when ectopically expressed, BAHD1 is targeted to the Xi in female cells independent of the presence of the BAH domain, an important issue to be addressed in future work.
The massive chromatin compaction triggered by BAHD1 overexpression in cells suggests that BAHD1 may spread along the chromatin fiber, as reported for the yeast BAH-containing protein Sir3 (10). Like Sir3, BAHD1 may interact directly with nucleosomes through its conserved BAH domain; however, the BAH domain does not specifically bind H3K27me3, at least in vitro, suggesting that it does not function as the chromodomain of HP1 or PcG proteins in recognizing methylated histone H3. Nevertheless, the BAH domain is important for BAHD1 colocalization with H3K27me3 in vivo, as well as down-regulation of IGF2 levels. These findings generalize the role of BAH domains in silent chromatin assembly.
Based on the reported functions of chromatin proteins found in the BAHD1 interactome (Fig. S5; Table S1), as well as on the fact that BAHD1 ectopic expression is sufficient to induce massive heterochromatinization and endogenous BAHD1 depletion induces expression of IGF2 and IGF2AS, we propose a model that explains how the BAHD1-containing complex triggers IGF2 silencing. BAHD1 is first targeted to the IGF2–IGF2AS overlapping regulatory region through selective binding to cofactors of transcription factors (Fig. 5). BAHD1 then orchestrates the assembly of a chromatin condensation machine by sequential recruitment of at least HP1 (which binds methyl-H3K9), MBD1 (which binds methyl-CpG), and the histone deacetylase HDAC5, to stimulate heterochromatin nucleation and spreading, as detailed in Fig. 5. More work is needed to validate the presence in the BAHD1 multiprotein complex of several key regulators found in our two-hybrid screen, such as the histone methyltransferase KMT1E and the nucleosome remodeler CHD1, as well as to search for a possible link to DNA methylation and RNA processing. Another important issue is the connection between BAHD1 and PcG repressive complexes (20). Whether BAHD1 acts upstream or downstream of EZH2 that forms H3K27me3, and whether components of PRC1, which bind this mark, are associated with BAHD1 heterochromatic foci remain to be addressed (Fig. S5).
Fig. 5.
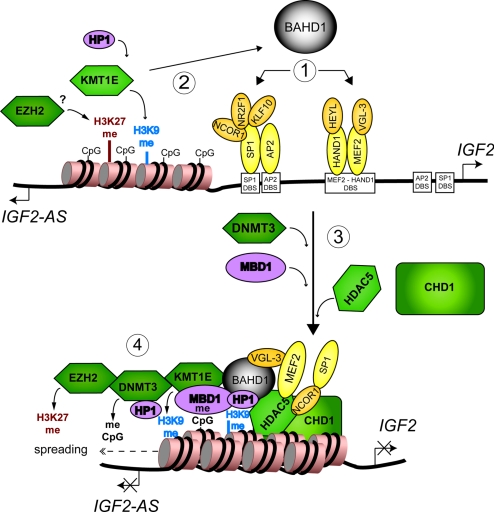
Hypothetical model for BAHD1-mediated heterochromatic silencing at the IGF2 P3 region. 1, BAHD1 targets IGF2 P3 by interacting with cofactors of DNA-bound transcription factors. 2, BAHD1 recruits KMT1E, which methylates K9 of histone H3 (H3K9me). HP1 is tethered to chromatin by binding H3K9me and BAHD1. EZH2 is recruited at this locus, before or after BAHD1, by an unknown mechanism and methylates K27 of H3 (H3K27me). 3, KMT1E and HP1 interact with DNMT3, which methylates cytosines (meCpG), creating binding sites for MBD1. BAHD1 recruits HDAC5 and CHD1, both of which bind NCOR1, which interacts with SP1. HDAC5 also binds MEF2. The complex is stabilized by cross-interactions between BAHD1, MBD1 and HDAC5 (Fig. S5). HDAC5 and CHD1 activities lead to histone deacetylation and nucleosome compaction. 4, Heterochromatin spreads to surrounding sequences after successive loading of BAHD1, histone, and DNA methylation marks, preventing access of the transcription machinery to the promoter and resulting in transcriptional repression of IGF2 and IGF2AS. Abnormal down-regulation of BAHD1 would reactivate IGF2 expression.
We propose that BAHD1 belongs to a growing family of scaffold proteins, including Sin3, Kap1, MTA, and MTG proteins (13, 30–32), that tether chromatin-modifying enzymes to sequence-specific transcription factors, thus enabling local chromatin regulation at specific gene targets. Pathological deregulation of such scaffolds is likely to have deleterious effects. Of note, BAHD1 expression is down-regulated in invasive lung cancer cells (8), and a recent study indicated reduced levels of BAHD1 transcript in RNA in blood samples from patients with double tumors compared with patients with single tumors (http://www.nextbio.com/b/home/home.nb?q = BAHD1). Our findings support the hypothesis that BAHD1 acts as a tumor suppressor through the silencing of cancer-related genes, such as IGF2, in adult tissues. Further studies are needed to address this possibility, and also to explore a putative relationship between BAHD1-induced silencing and IGF2 imprinting.
Materials and Methods
Cell Lines, Plasmids, Antibodies, siRNA, and Oligonucleotides.
The cell lines, plasmids, antibodies, siRNA, oligonucleotides, and transfection procedures used in this study are described in SI Materials and Methods, Table S4, and Table S5.
Identification of BAHD1 Interactors by Yeast Two-Hybrid Screening and Construction of an Interaction Network.
The Y2H screen was performed on a whole human genome cDNA placental library, as described in SI Materials and Methods. To generate the BAHD1 interactome, BAHD1 interactors were uploaded in the Ingenuity Pathways Analysis software (Ingenuity Systems; www.ingenuity.com) and also checked for interactors in the Human Protein Reference Database (http://www.hprd.org).
Transmission Electron Microscopy, Immunofluorescence Analysis, RNA FISH, and Live-Cell Imaging.
The light and electron microscopy studies and immunostaining procedures performed are described in SI Materials and Methods. For RNA FISH, the DNA probe detected Xist RNA from the fourth intron to the 3′ end of the human Xist gene.
Protein Purification, Preparation of Nuclear Extracts, GST Pull-Down and Histone Peptide-Binding Assays, Immunoprecipitation, and Immunoblotting.
GST-tagged proteins were produced and purified following standard procedures (Novagen or Amersham Pharmacia Biotech). Biotinylated histone peptides were from Millipore. The nuclear extraction, pull-down and histone peptide-binding assays, and immunoprecipitation (IP) studies performed are described in SI Materials and Methods.
Microarray Analysis.
Total RNA from cells transfected for 72 h with siRNA BAHD1 or siRNA control was extracted and purified using the Qiagen RNeasy Kit. The quality of RNAs and cRNAs was monitored on Agilent RNA Nano LabChips. cRNA was obtained according to Affymetrix standard protocols and hybridized on GeneChip Human Genome U133Plus 2.0. For each condition, 3 biological replicates were hybridized. The procedures, data, and statistical analyses are described in detail in SI Materials and Methods.
RNA Isolation, ChIP, and RT-PCR Analysis.
RNA was extracted using the Qiagen RNeasy Kit and treated with TURBO DNA-free DNase (Applied Biosystems). Chromatin was prepared from HEK293 cells transfected for 24 h with V5-BAHD1 or V5-CFP expression vectors. Three independent chromatin preparations per vector were made, each corresponding to 107 cells. The procedures are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank M. Yaniv for helpful discussions and A. Lebreton, M. Hamon, and S. Bruck for their critical reading of the manuscript. We are grateful to E. Gouin for generating the BAHD1 antiserum, S. Dupuis for providing C3SV40 cells, and A. Dejean for facilitating the work in her laboratory. We thank the Institut Pasteur Technopole, especially J.Y. Copée, M.C. Prevost, P. Roux, S. Shorte, and H. Kiefer-Biasizzo, for the use of their technological facilities. Work in P.C.'s laboratory was supported by the Pasteur Institute, INRA (AIP291), INSERM, ANR (ERANET Spatelis), and ERC (Advanced Grant 233348). Work in J.F.'s laboratory was supported by ARC and the Canceropole Ile-de-France. P.C. is an international research scholar of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE16097).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901259106/DCSupplemental.
References
- 1.Trojer P, Reinberg D. Facultative heterochromatin: Is there a distinctive molecular signature? Mol Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Moss TJ, Wallrath LL. Connections between epigenetic gene silencing and human disease. Mutat Res. 2007;618:163–174. doi: 10.1016/j.mrfmmm.2006.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 4.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, part II: ATP-dependent chromatin remodeling. Trends Mol Med. 2007;13:373–380. doi: 10.1016/j.molmed.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, part I: Covalent histone modifications. Trends Mol Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Bird A. The methyl-CpG–binding protein MeCP2 and neurological disease. Biochem Soc Trans. 2008;36(Pt 4):575–583. doi: 10.1042/BST0360575. [DOI] [PubMed] [Google Scholar]
- 7.Hamon MA, Cossart P. Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe. 2008;4:100–109. doi: 10.1016/j.chom.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Brena RM, et al. Aberrant DNA methylation of OLIG1, a novel prognostic factor in non–small cell lung cancer. PLoS Med. 2007;4:e108. doi: 10.1371/journal.pmed.0040108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callebaut I, Courvalin JC, Mornon JP. The BAH (bromo-adjacent homology) domain: A link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999;446:189–193. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- 10.Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Nagase T, et al. Prediction of the coding sequences of unidentified human genes, XIII: The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999;6:63–70. doi: 10.1093/dnares/6.1.63. [DOI] [PubMed] [Google Scholar]
- 12.Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 14.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 15.Smallwood A, Esteve PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klose RJ, Bird AP. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Rice JC, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert SL, Pehrson JR, Sharp PA. XIST RNA associates with specific regions of the inactive X chromatin. J Biol Chem. 2000;275:36491–36494. doi: 10.1074/jbc.C000409200. [DOI] [PubMed] [Google Scholar]
- 19.Masui O, Heard E. RNA and protein actors in X-chromosome inactivation. Cold Spring Harb Symp Quant Biol. 2006;71:419–428. doi: 10.1101/sqb.2006.71.058. [DOI] [PubMed] [Google Scholar]
- 20.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 23.Pan G, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Chao W, D'Amore PA. IGF2: Epigenetic regulation and role in development and disease. Cytokine Growth Factor Rev. 2008;19:111–120. doi: 10.1016/j.cytogfr.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eriksson T, et al. Methylation changes in the human IGF2 p3 promoter parallel IGF2 expression in the primary tumor, established cell line, and xenograft of a human hepatoblastoma. Exp Cell Res. 2001;270:88–95. doi: 10.1006/excr.2001.5336. [DOI] [PubMed] [Google Scholar]
- 26.Monk D, et al. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum Mol Genet. 2006;15:1259–1269. doi: 10.1093/hmg/ddl041. [DOI] [PubMed] [Google Scholar]
- 27.Jouvenot Y, et al. Targeted regulation of imprinted genes by synthetic zinc-finger transcription factors. Gene Ther. 2003;10:513–522. doi: 10.1038/sj.gt.3301930. [DOI] [PubMed] [Google Scholar]
- 28.Rietveld LE, Koonen-Reemst AM, Sussenbach JS, Holthuizen PE. Dual role for transcription factor AP-2 in the regulation of the major fetal promoter P3 of the gene for human insulin-like growth factor II. Biochem J. 1999;338(Pt 3):799–806. [PMC free article] [PubMed] [Google Scholar]
- 29.Okutsu T, et al. Expression and imprinting status of human PEG8/IGF2AS, a paternally expressed antisense transcript from the IGF2 locus, in Wilms' tumors. J Biochem. 2000;127:475–483. doi: 10.1093/oxfordjournals.jbchem.a022630. [DOI] [PubMed] [Google Scholar]
- 30.van Oevelen C, et al. A role for mammalian Sin3 in permanent gene silencing. Mol Cell. 2008;32:359–370. doi: 10.1016/j.molcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein–mediated transcriptional repression. Mol Cell Biol. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossetti S, Hoogeveen AT, Sacchi N. The MTG proteins: Chromatin repression players with a passion for networking. Genomics. 2004;84:1–9. doi: 10.1016/j.ygeno.2004.02.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.