Abstract
Changes in the expression of Hox genes have been widely linked to the evolution of animal body plans, but functional demonstrations of this relationship have been impeded by the lack of suitable model organisms. A classic case study involves the repeated evolution of specialized feeding appendages, called maxillipeds, from anterior thoracic legs, in many crustacean lineages. These leg-to-maxilliped transformations correlate with the loss of Ultrabithorax (Ubx) expression from corresponding segments, which is proposed to be the underlying genetic cause. To functionally test this hypothesis, we establish tools for conditional misexpression and use these to misexpress Ubx in the crustacean Parhyale hawaiensis. Ectopic Ubx leads to homeotic transformations of anterior appendages toward more posterior thoracic fates, including maxilliped-to-leg transformations, confirming the capacity of Ubx to control thoracic (leg) versus gnathal (feeding) segmental identities. We find that maxillipeds not only are specified in the absence of Ubx, but also can develop in the presence of low/transient Ubx expression. Our findings suggest a path for the gradual evolutionary transition from thoracic legs to maxillipeds, in which stepwise changes in Hox gene expression have brought about this striking morphological and functional transformation.
Keywords: functional studies, maxillipeds, morphological evolution, Parhyale, Ultrabithorax (Ubx)
Hox genes play a key role in assigning region-specific identities in the body of diverse animals (1), and changes in Hox gene function have been linked to the evolution of animal body plans. For example, changes in Hox gene regulation (2–6), in Hox coding sequences (7, 8), and in the interactions of Hox genes with their downstream targets (9, 10) have all been implicated in the evolution of segment diversity in arthropods.
Crustaceans exhibit impressive diversity and specialization in their appendages, more extensive than any other animal group (11). One of the most illuminating case studies, suggesting an association between changes in Hox gene expression and evolutionary changes in morphology, involves the Hox gene Ubx in the specification of distinct thoracic appendage identities in crustaceans (4). Ubx is typically expressed in the thoracic region of crustaceans, but in many lineages it is excluded from anterior thoracic segments that develop specialized feeding appendages, called maxillipeds (Mxp). Thus, in crustaceans without maxillipeds Ubx is expressed throughout the thorax, but in crustaceans with 1, 2, or 3 pairs of maxillipeds Ubx is excluded from the maxilliped-bearing segments. Maxillipeds display characteristics of both thoracic and head feeding appendages—for instance, they develop a main limb branch (endopod) like thoracic legs, but they also have prominent proximal endites that are used for the manipulation of food, similar to the maxillary appendages Mx1 and Mx2 (Fig. 1). Maxillipeds are thought to be specified by expression of the Hox genes Sex combs reduced (Scr; also expressed in Mx1 and Mx2) and/or Antennapedia (Antp; expressed throughout the thorax and Mx2) and by absence of Ubx expression (4, 6, 12, 13).
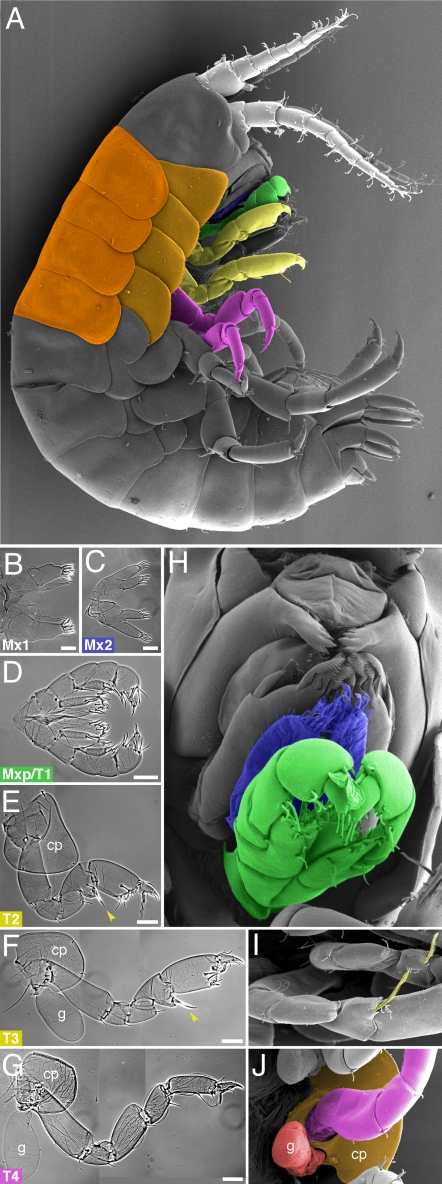
Fig. 1.
Appendage diversity in the head and anterior thorax of Parhyale hatchlings. (A) Scanning electron micrograph (SEM) of a Parhyale hatchling, highlighting the two antennal (white), Mx2 (blue), Mxp/T1 (green), T2–T3 (yellow), and T4–T5 (magenta) appendages on the right side of the animal and the tergal and coxal plates of T2–T5 segments (dark and light orange). (B–G) Cuticle preparations of wild-type dissected appendages: bilateral pairs of Mx1 (B), Mx2 (C), and Mxp/T1 (D) and individual T2 (E), T3 (F) and T4 (G) appendages, marking the presence of coxal plates (cp), and gills (g). (Scale bars, 50 μm.) T2/3-type appendages are distinguished from T4/5-type by the shape and size of their distal podomeres and by the presence of a characteristic bristle (yellow arrowhead), also shown in I. (H) SEM of the gnathal region, highlighting the Mx2 (blue) and Mxp/T1 (green) appendages, which lie just posterior to the Mx1 appendages and the mandibles. (I) SEM of characteristic T2 and T3 bristles. (J) SEM of the coxal plate (orange) and gill (red) at the base of a thoracic appendage.
Such expression studies have furnished persuasive evidence implicating shifts in Ubx expression in crustacean morphological evolution, but these conclusions are based entirely on correlations rather than direct tests of gene function. To overcome this limitation, we have focused on an emerging crustacean model, the amphipod Parhyale hawaiensis. Parhyale represents an attractive model for comparative developmental research because of its phylogenetic position, its rapid life cycle and ease of use in the laboratory, and the suitability of its embryos for embryological and genetic manipulations (14, 15). Moreover, the early cell lineage and fate map of Parhyale embryos have been determined (16), and considerable effort is being invested to study the expression patterns and functions of developmental genes in this species (17–21). Parhyale develop a pair of maxillipeds in the first thoracic segment (Mxp/T1; Fig. 1) that lack Ubx expression (21). Further modulation of Ubx expression in the anterior thorax correlates with thoracic appendage (leg) modifications: the second and third segments (T2–T3) develop subchelate grasping appendages in the presence of low levels of Ubx, while more posterior segments (T4–T8) develop locomotory appendages with high levels of Ubx (Fig. 1) (21).
The establishment of transgenesis in Parhyale (22) has provided the basis for developing functional genetic approaches in this species. Here, we exploit the opportunities offered by transgenesis to establish a method for conditional (heat-inducible) misexpression of genes in Parhyale. By altering Ubx expression experimentally, we are able to functionally test the capacity of Ubx to control thoracic appendage identity in this species.
Results
Establishing Tools for Gene Misexpression in Parhyale.
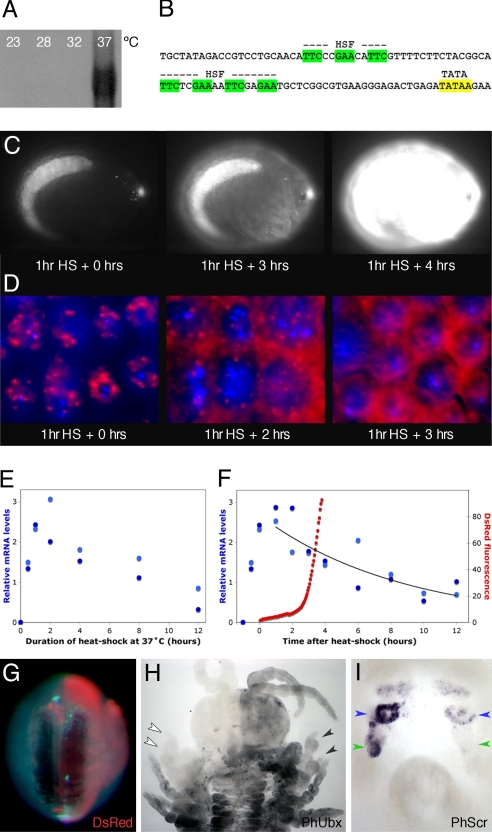
To establish a method for conditional misexpression, we have characterized a heat-inducible cis-regulatory element. We used PCR to obtain the coding sequence of hsp70 family genes from Parhyale. Northern analysis shows that these genes are not expressed at detectable levels when Parhyale are kept at 23°–32 °C, but become strongly induced after a 1-h incubation at 37 °C (Fig. 2A), defining the conditions for a robust heat-shock response in this species. Using inverse PCR, we isolated upstream regulatory regions of these genes and tested their activity with DsRed reporter constructs introduced into Parhyale using the Minos transformation vector (22). Through these studies, we defined a 2.5-kb fragment, named PhHS, that gives heat-inducible expression of DsRed in independent transgenic lines (Fig. 2C). Consistent with heat inducibility, 2 clusters of putative binding sites for the heat-shock factor (HSF) are found in PhHS, upstream of a putative basal promoter (Fig. 2B). The on/off kinetics of PhHS were examined at the transcriptional level, using quantitative RT-PCR and in situ hybridization on transgenic embryos. Transcription is strongly induced within 30 min from the start of heat shock at 37 °C, peaks at 1–2 h, and ceases 2–3 h after return to 25 °C (Fig. 2 D–F). The onset of DsRed fluorescence lags by ≈3 h, reflecting the time required for maturation of the DsRed protein [Fig. 2 C and F; supporting information (SI) Movie S1]. Expression can be induced at any time after embryonic stage 11 (staging according to ref. 14) and appears uniform in all tissues.
Fig. 2.
Heat-inducible expression in Parhyale. (A) Northern blot showing expression of Parhyale hsp70 transcripts in wild-type animals incubated at 23°, 28°, 32°, or 37 °C for 1 h before collection; transcripts become strongly induced only at 37 °C. (B) Part of the 2.5-kb PhHS sequence, containing clusters of HSF binding sites upstream of a putative TATA promoter. (C) Induction of DsRed fluorescence in a live transgenic embryo carrying the PhHS-DsRed reporter, after a 1-h heat shock at 37 °C. The 3 images were taken at 0, 3, and 4 h after heat shock (full time-lapse movie in Movie S1). Fluorescence at 0 h is because of expression of the 3xP3-DsRed transformation marker (anterior spots) and because of autofluorescence of the yolk (dorsal crescent). (D) Time course of heat-inducible transcription following a 1-h heat shock at 37 °C, observed by in situ hybridization for PhUbx on transgenic embryos carrying multiple insertions of the PhHS-Ubx-I construct. Transcripts are shown in red and cell nuclei stained with DAPI in blue. Nuclear dots, representing nascent transcripts from multiple loci (transgenes) within each nucleus, appear immediately after the heat shock and are clearly visible up to 2 h post-heat shock. Active transcription ceases 2–3 h post-heat shock, when most of the transcripts are cytoplasmic. (E and F) Quantification of heat-inducible expression in 2 independent PhHS-DsRed transgenic lines (light and dark blue datapoints), using quantitative real-time RT-PCR. (E) DsRed transcript levels during prolonged heat shock. Maximum levels are observed after 1–2 h of heat shock at 37 °C; longer heat shocks do not induce a stronger response. (F) On/off kinetics of the heat-shock response, upon 1 h heat shock at 37 °C. Transcript levels reach maximum levels ≈1 h after the heat shock and drop off with a half-life of ≈6 h after return to 25 °C (trend line). The onset of DsRed fluorescence (red data points) lags by ≈3 h. (G–I) Embryos injected with PhHS constructs and Minos transposase mRNA at the 2-cell stage, giving rise to unilaterally transformed embryos. (G) Dorsal view of embryo injected with a PhHS-DsRed construct, showing unilateral DsRed fluorescence several hours after a 1-h heat shock at 37 °C. (H) Ventral view of an embryo injected with PhHS-PhUbx-II, subjected to a 4-h heat shock at 37 °C, and stained with the FP6.87 antibody for Ubx. Ectopic PhUbx is seen unilaterally, superimposed on the normal pattern. The levels of expression at T2 and T3 (black arrowheads) approach the wild-type levels of T4/5 (compare to T2–T3 on the contralateral side, indicated by white arrowheads). (I) Ventral view of an embryo injected with PhHS-PhUbx-II, subjected to a 4-h heat shock at 37 °C, and hybridized with a probe for PhScr. Downregulation of PhScr is observed unilaterally in the Mx2 and Mxp (blue and green arrowheads, respectively).
Ubx Misexpression Causes Homeotic Transformations Toward Thoracic Identities.
To directly test the capacity of Ubx to control thoracic appendage morphology, we used PhHS to misexpress each of the 2 identified splice variants of Parhyale Ubx (PhUbx isoforms I and II), which differ in their first N-terminal amino acids (21). Misexpression in embryos from stable transgenic lines that express uniform low levels of PhUbx-I or PhUbx-II upon heat shock resulted in homeotic transformations toward thoracic identities, including ectopic coxal and tergal plates in the head and antennal-to-leg and Mx2-to-Mxp transformations (Fig. 3E). However, misexpression in transgenic lines that express near wild-type levels of PhUbx-I or II resulted in embryonic lethality before appendage diversification, associated with secondary phenotypes, most notably abnormal morphogenesis of the gut (Fig. S1). To overcome this problem, we generated genetic mosaics in which only part of the developing embryo is transformed (22) (Fig. 2 G and H). These mosaic embryos can express near wild-type levels of PhUbx in the affected tissues and yet survive to hatching, allowing us to score the full spectrum of homeotic transformations. Specifically, we injected PhHS-PhUbx constructs (carrying isoforms I or II) together with transposase mRNA into 1- or 2-cell stage Parhyale embryos, generating hundreds of mosaic embryos that were heat-shocked for 1 h daily over the entire period of appendage development (stages 12–27; ref. 14). In situ hybridizations and antibody stainings showed that 20–60% of these embryos express ectopic PhUbx in different parts of their body for several hours after each heat shock (Fig. 2H and Fig. S2).
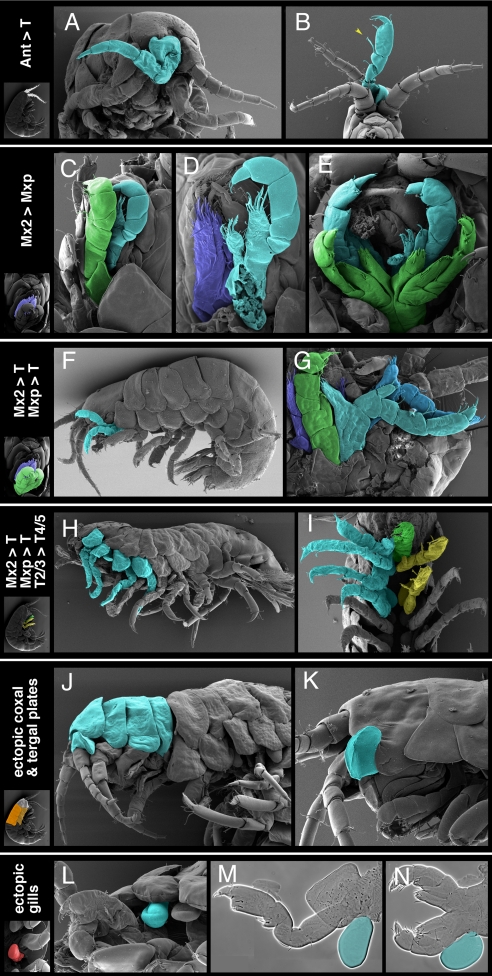
Fig. 3.
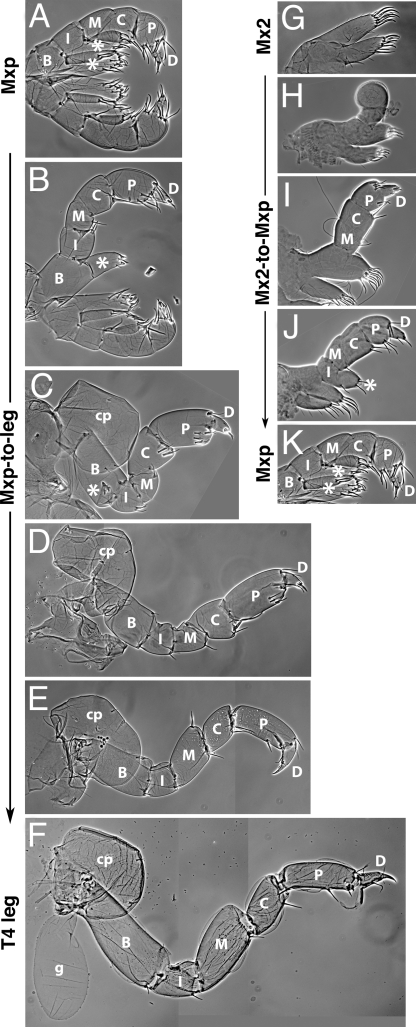
Homeotic phenotypes resulting from PhUbx misexpression in Parhyale. Phenotypes are highlighted in cyan, in scanning electron micrographs (A–L) or cuticle preparations of dissected appendages (M and N). (A) Partial antennal-to-leg transformation, seen in characteristic shape and flexure of proximal leg podomeres. (B) Complete antennal-to-T2/3 transformation (arrowhead on characteristic T2/3 bristle). (C) Near complete transformation of Mx2 (cyan) to Mxp; normal Mxp is shown in green for comparison. (D) Same specimen after Mxp have been dissected away to reveal the structure of the transformed Mx2 (cyan), compared to the unaffected Mx2 on the contralateral side (blue). (E) Bilateral transformation of Mx2 (cyan) to Mxp, in a stable transgenic line; Mxp/T1 appendages are shown in green for comparison. (F) Mx2 and Mxp (cyan) overgrown and transformed to thoracic-like appendages. (G) Overgrowth of Mx2 and Mxp (dark and light cyan) and partial transformation toward thoracic leg, compared to unaffected Mx2 (blue) and Mxp (green) on the contralateral side. (H) More extensive transformations of Mx2 and Mxp toward thoracic legs, combined with partial transformations of T2 and T3 toward T4/5-like legs. (I) Extensive Mx2-to-leg, Mxp-to-leg, and T2/3-to-T4/5 transformations, compared to unaffected Mxp (green) and T2/3 (yellow) appendages on the contralateral side. (J) Ectopic tergal plates and small coxal plate in the head region (compare to the unsegmented head shield in the wild type, Fig. 1A). (K) Ectopic coxal plate in the head region. (L and M) Ectopic gill associated with the T2 appendage. (N) Ectopic gill associated unilaterally with the Mxp. The phenotypes shown resulted from misexpression of PhUbx-I (C, D, G, and K–N) or PhUbx-II (A, B, F, and H–J) in mosaic embryos or uniform misexpression of PhUbx-II in a stable transgenic line (E).
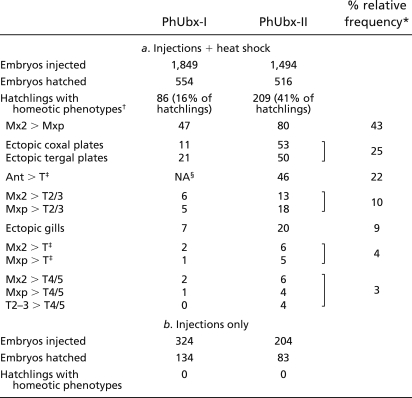
Table 1 summarizes the homeotic phenotypes that we scored in these hatchlings, typically transformations of anterior thoracic and head structures toward more posterior thoracic fates. These transformations include the appearance of ectopic coxal and tergal plates (normally present from T2 to T8) on head segments and on T1; the appearance of ectopic gills (normally from T3 to T7) on T1 and T2; the transformation of Mx2 appendages into Mxp; and the transformation of antennae, Mx1, Mx2, and Mxp/T1 appendages into elongated thoracic-type legs (with characteristics of T2/3 or T4/5 morphology) (Figs. 3 and 4). A similar range of phenotypes was obtained by misexpressing either of the 2 PhUbx isoforms, but the penetrance and severity of these transformations was significantly higher when using isoform PhUbx-II (Table 1). These phenotypes are a consequence of PhUbx misexpression, as no transformations were observed among hundreds of heat-shocked wild-type and PhHS-DsRed embryos, or among PhHS-PhUbx-I and -II injected embryos not subjected to heat shock (Table 1). The specificity of these phenotypes is also evident in hatchlings where only one side of the body is affected (left or right side) (16, 22), while the other side exhibits wild-type morphology that can serve as an internal control (e.g., Fig. 3 A, B, D, G, and I).
Table 1.
Homeotic phenotypes resulting from misexpression of PhUbx isoforms I and II
*Frequency of individuals with at least 1 transformation of that category, among hatchlings with homeotic phenotypes.
†Scores include both complete and partial homeotic transformations; mandibular and maxilla 1 transformations were not scored.
‡Scored as thoracic-like appendage, making no distinction between T2/3- and T4/5-type legs.
§NA, not applicable. Antennal-to-leg transformations were observed but not quantified.
Fig. 4.
Mxp-to-leg and Mx2-to-Mxp transformations resulting from PhUbx misexpression. (A) Bilateral pair of wild-type Mxp/T1 appendages, with characteristic segmented endopod, including ischium and basis with characteristic endites (asterisks). (B–E) T1 appendages with transformations of increasing severity, resulting from misexpression of PhUbx in mosaic embryos. (B) Bilateral pair of Mxp/T1 appendages with unilateral phenotype, including loss of the endite on the ischium, and elongation of endopod segments (especially basis and propodus). (C) More severe transformation of Mxp/T1 toward a T2/3-type leg, including loss of the endite on the ischium, reduction of the endite on the basis (folded on the basis, asterisk), the appearance of a coxal plate, and the shapes of the leg segments, which are characteristic of T2/3 legs. (D) Almost complete transformation of Mxp to T2/3-type appendage. Compared to the appendages in previous panels, this appendage was positioned more laterally and separated from the wild-type Mxp on the contralateral side, a characteristic of T2–T8 appendages. (E) Transformation of Mxp to T4/5-type appendage. T4 characteristics include the narrower propodus and characteristic shapes of the merus and carpus. (F) Wild-type T4 leg. (G) Wild-type Mx2 appendage. (H–J) Mx2 appendages with transformations of increasing severity, resulting from misexpression of PhUbx in mosaic embryos. (H) Mx2 with lateral outgrowth. (I) Mx2 with segmented Mxp-like endopod, comprising dactyl, propodus, carpus, and merus. Proximal portions of the appendage retain the characteristics of Mx2. (J) Mx2 with segmented endopod, including ischium with Mxp-like endite (asterisk). The proximal podomere retains the characteristics of Mx2, suggesting that the normal Mx2 appendages represent protopodal endites. (K) Wild-type Mxp/T1 appendage, as in A. All panels show cuticle preparations of dissected hatchling appendages at the same magnification. B, basis; I, ischium; M, merus; C, carpus; P, propodus; D, dactyl; cp, coxal plate; g, gill.
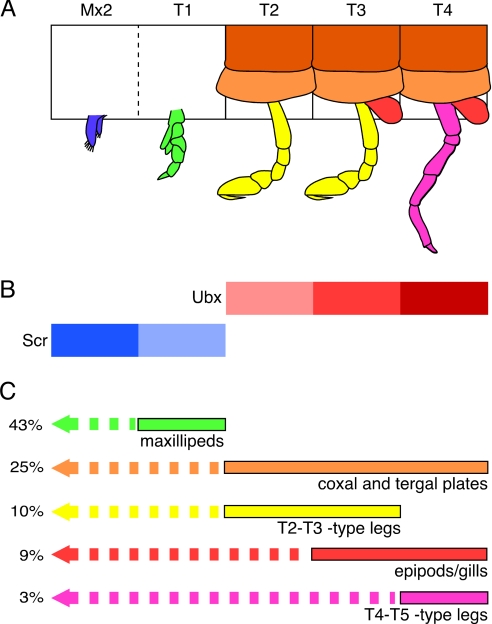
With the exception of Mx2-to-Mxp transformations, to which we return below, the homeotic transformations recovered in these experiments are fully consistent with the wild-type expression pattern of PhUbx (21) and the posterior prevalence model of Hox gene function (23): Misexpression of PhUbx induces anterior segments that do not normally express the gene to acquire characteristics of segments that do, or segments that express low levels to acquire characteristics of segments that express higher levels (Fig. 5). Furthermore, the intensity of PhUbx expression normally associated with each thoracic character is reflected in the frequency with which this character is induced ectopically by misexpression of PhUbx (Fig. 5C); e.g., coxal/tergal plates, which develop starting from T2 (low expression of PhUbx), are more frequently induced than gills, which develop starting from T3 (slightly higher expression), and these in turn are more frequently induced than T4/5-type legs (high/persistent expression). Thus, the effects of PhUbx misexpression appear to capture some of the quantitative requirements for PhUbx function in each of these structures.
Fig. 5.
Distribution of segmental characters, Hox gene expression, and homeotic transformations caused by PhUbx misexpression. (A) Illustration of morphological characters associated with Mx2, T1/Mxp, T2, T3, and T4 segmental identities in Parhyale, including second maxillary appendage (blue), maxilliped (green), T2/3-type leg (yellow), T4/5-type leg (magenta), gill (red), and coxal and tergal plates (light and dark orange). (B) Segmental domains of wild-type Scr (blue) and Ubx (red) expression (12, 21); levels of expression are illustrated by lighter or darker shades. (C) Segmental distribution of each morphological character in wild type (solid bars, color-coded as in A) and upon PhUbx misexpression (dashed bars). Percentage values represent the frequency with which each of these characters was observed ectopically in a more anterior segment, among PhUbx misexpressing hatchlings with homeotic transformations (Table 1). There is a clear correlation between these frequencies and the intensity of PhUbx expression associated with each of these characters in the wild type.
To assess the extent of mosaicism and the strength of PhUbx misexpression in these experiments, we carried out immunostainings for PhUbx on stage 23–24 embryos that had been injected with PhHS-PhUbx-II and heat-shocked daily, as in previous experiments. We were able to detect ectopic PhUbx in 30% of screened embryos: 12% that displayed ectopic PhUbx exclusively in mesodermal or endodermal tissues and 18% that showed various levels and patterns of expression in the ectoderm, including 7% with low levels of ectopic PhUbx (Fig. S2 A and B), 2% with intermediate levels (Fig. S2C), 2% with high levels (Fig. S2D), and 7% with varying levels of expression in different groups of cells (Fig. S2E). These frequencies are broadly consistent with the frequencies of homeotic transformations documented earlier (Fig. S2F). Among these embryos, we observed several cases of Mx2-to-Mxp transformations in embryos that expressed low levels of ectopic PhUbx (some at the limit of detection; Fig. S2A′) and examples of ectopic gills, tergal and coxal plates, and antennal-to-leg, Mx2-to-leg, and Mxp-to-leg transformations in embryos with higher or variable levels of expression (Fig. S2 B′, C′, and E′).
Ubx Can Induce Mx2-to-Mxp Transformations by Downregulating Scr.
A puzzling outcome of both PhUbx-I and -II misexpression was the frequent transformation of Mx2 into Mxp (Table 1 and Figs. 3 C–E and 4 G–K). This is unexpected because Mxp normally develop in the absence of detectable PhUbx expression (21), and their evolution has been associated with a retraction of Ubx expression from Mxp-bearing segments in a wide range of species (4). One possible explanation is that there may be weak or transient PhUbx expression in Mxp/T1 that was previously overlooked. Careful examination of PhUbx expression in the anterior thorax found no evidence for such expression, beyond expression in posterior T1 in the neuroectoderm (21). In addition, this explanation is unlikely because knockdown of PhUbx does not appear to have any effect on Mxp (21).
Alternatively, this phenotype could be an indirect consequence of PhUbx misexpression, through cross-regulation of another Hox gene. Scr is the prime candidate, as it is the Hox gene predominantly associated with Mxp/T1 and Mx2 development (12) (Fig. 5B). Scr has also been shown to be repressed by Ubx in Drosophila (24, 25). To address this possibility, we examined the expression of the Parhyale orthologue of Scr (PhScr) in embryos that had been injected with the PhHS-PhUbx-II construct at the 2-cell stage and subjected once to heat shock: 18% (32/182) of these embryos showed unilateral partial or complete downregulation of PhScr 4–12 h after the onset of heat shock (Fig. 2I) and lower levels of PhScr could be observed up to 1 day later (3/22 embryos). This downregulation provides an explanation for the transformation of Mx2 (which normally expresses high levels of PhScr) to Mxp (which expresses lower levels) in ≈16% of surviving embryos that had been injected with the PhHS-PhUbx-II construct (80/516, see Table 1). The fact that this transformation is among the most frequent outcomes of PhUbx misexpression (Table 1, Fig. 5C) suggests that PhScr is a sensitive PhUbx target.
Discussion
During the course of evolution, several crustacean lineages diversified their body plans by transforming locomotory appendages (legs) into feeding appendages (maxillipeds) in anterior thoracic segments. The establishment of tools for gene misexpression has allowed us to test the role of Ubx directly in Parhyale. The homeotic transformations reported here demonstrate that Ubx has the capacity to direct locomotory appendage identities when expressed in anterior thoracic and head segments. The frequency of these transformations suggests that the strength and/or duration of Ubx expression may be important in determining different subtypes of thoracic locomotory appendages (T2/3 versus T4/5, presence of gills). Considering the transient and mosaic nature of the misexpression system used, it is not possible yet to demonstrate a direct association between levels and timing of ectopic PhUbx and specific appendage morphology in a reproducible manner—each injected animal has a unique history of spatial and temporal misexpression. Despite this caveat, we have noted a broad agreement in the frequencies of experimentally induced homeotic transformations with the levels of ectopic PhUbx observed. Thus, as suggested by expression studies (4), changes in Ubx expression are likely to have been a major driving force in the evolution of segmental specialization in the anterior thorax of crustaceans.
This study has also allowed us to uncover an aspect of Ubx function that could not be deduced from expression studies or extrapolated from established experimental models—the observation that PhUbx misexpression can downregulate PhScr and induce Mx2-to-Mxp transformations. This has interesting evolutionary implications. First, it indicates that maxillipeds can develop in the presence of low and/or transient Ubx expression. This is corroborated by loss-of-function studies in the accompanying paper by Liubicich et al., where lowering PhUbx levels induces maxilliped-like features in T2 and T3 appendages (21). Second, this observation reinforces the idea that maxillipeds represent a hybrid segmental identity—part gnathal and part thoracic—that may be realized through different Hox codes. Expression studies have also indicated that spatial and temporal modulation of Hox gene expression within a segment can generate appendages with intermediate morphologies (4, 12). Together, these findings suggest an evolutionary path where successive changes with small effects on morphology—starting from lowered or mosaic Ubx expression to a stepwise loss of Ubx and expansion of Scr—can drive a gradual evolutionary transition from thoracic legs to maxillipeds. This gradualist scenario, which postulates no sudden morphological transformations, addresses concerns over the viability of intermediate forms during body plan evolution (26).
Materials and Methods
Cloning of PhHS and Transgenesis Constructs.
A 240-bp fragment of the Parhyale hsp70 coding sequence was amplified from genomic DNA, using degenerate primers Hsp70F1 (5′-ACIACITAYTCITGYGTIGG-3′) and Hsp70R1 (5′-AAIGGCCARTGYTTCAT-3′). To test the conditions for heat inducibility, this fragment was used as a probe in a Northern blot on total RNA extracted from wild-type animals. The 240-bp sequence was extended upstream by 3 rounds of inverse PCR and the entire contig (EMBL/GenBank accession no. FM991730) was amplified from genomic DNA using primers Phhsp70F (5′-TTACTGTAACCGCAGGGGCAAAAGA-3′) and Phhsp70R (5′-ACAGCATCCTTCACGTCTCCTCCAA-3′) and cloned into pGEM-T-Easy (Promega). The sequence upstream of the start codon (5′-UTR and promoter/regulatory region) was cloned in front of a DsRedT1/SV40polyA reporter cassette to generate plasmid pSL(PhHS-DsRed). The coding sequence of PhUbx-I was obtained as a PmeI-NotI fragment from plasmid pCR4TOPOPhUbx (21) and cloned downstream of PhHS in BstXI (blunted)/NotI-digested pSL(PhHS-DsRed), replacing DsRed. Similarly, the coding sequence of PhUbx-II was amplified from cDNA clone pPhUbxII#5 using primers PhUbxII_BspHI_F (5′-TTAGTCATGAACTCCTACTTTGAAC-3′) and PhUbx_NotI_R (5′-TATTGCGGCCGCTTAGTTTTGTCCGGGGTT-3′), digested with BspHI-NotI, and cloned downstream of PhHS in NcoI/NotI-digested pSL(PhHS-DsRed). The PhHS-DsRed, PhHS-Ubx-I, and PhHS-Ubx-II constructs were then cloned as AscI fragments into the Minos{3xP3-DsRed} and Minos{3xP3-EGFP} vectors (22, 27), generating the plasmids pMi{3xP3-DsRed; PhHS-DsRed}, pMi{3xP3-EGFP; PhHS-PhUbx-I}, and pMi{3xP3-EGFP; PhHS-PhUbx-II}, respectively, which were used in microinjections as donor plasmids.
Parhyale Husbandry, Microinjections, and Heat Shock.
Parhyale rearing, embryo collection, and staging were carried out as described previously (14, 17, 22). Microinjections were carried out by co-injecting donor plasmids with in vitro synthesized capped mRNA encoding the Minos transposase (27) at 300 and 100 ng/μL, respectively, as described previously (22) (detailed microinjection protocols are available on request). Injections at the 1- and 2-cell stage produced a significant proportion of mosaic embryos with unilaterally transformed thoracic and head segments, as noted before (22). Injected embryos were kept at 25 °C in filtered artificial seawater (FASW) supplemented with penicillin/streptomycin and amphotericin B (Gibco). To heat-shock, embryos were transferred to dishes with prewarmed FASW in a 37 °C incubator. After this incubation, the dishes with the embryos were transferred back to 25 °C.
Quantification of PhHS Heat-Shock Response.
The analysis was carried out on late embryos from 2 independent transgenic lines carrying a single copy of the PhHS-DsRed construct. Clutches of 6–10 transgenic embryos were collected immediately after heat shock of varying durations at 37 °C (Fig. 2E) or at various time points after a 1-h heat shock at 37 °C (Fig. 2F). Total RNA from each clutch was extracted with TRIzol (Invitrogen), treated with DNAseI (amplification grade, Invitrogen), reextracted with TRIzol, quantified on a Nanodrop spectrophotometer, and reverse transcribed with SuperScript III (Invitrogen). Quantification of DsRed transcripts was carried out relative to a control (calibrator) sample on a Roche 480 LightCycler 480 Real-Time PCR instrument, using SYBR Green I. For each sample, the expression levels of reporter transcripts were measured with primers that hybridized on SV40polyA (SV40F 5′-CCACATTTGTAGAGGTTTTACTTGC-3′ and SV40R 5′-TGAGTTTGGACAAACCACAACTA-3′) and normalized against 2 reference Parhyale ribosomal genes, PhRpL21 (PhRpL21F 5′-CCGAGGCTTCAAGAAGAATG-3′ and PhRpL21R 5′-AAAATCCGGCCTCGTACTCT-3′) and PhRpL32 (PhRpL32F 5′-CCAGCATTGGTTATGGTTCA-3′ and PhRpL32R 5′-TTGAGCTTAGCCTTGCCATT-3′). All reactions were carried out in triplicate, fluorescence crossing points were calculated with the second derivative maximum method using the Lightcycler 1.5 software, and relative quantifications were carried out with amplification efficiency correction using the geNorm freeware (28). Relative DsRed fluorescence levels (Fig. 2F) were calculated from average pixel intensities in Movie S1, excluding regions with 3xP3-driven fluorescence and yolk autofluorescence, using ImageJ 1.38x.
Scanning Electron Microscopy (SEM) and Cuticle Preparations.
Hatchlings were scored for phenotypes under a dissecting stereoscope, and affected specimens were examined in more detail using SEM or cuticle preparations. For SEM analysis, hatchlings were fixed for 1 h in 1% glutaraldehyde in FASW and for 1 h in 1% osmium tetroxide in FASW, washed in FASW, dehydrated through an ethanol series, and stored in 90% ethanol. Subsequently they were washed 3 times in absolute ethanol, subjected to critical point drying, coated with gold or platinum in a Polaron SC7640 Sputter Coater, and observed under a JEOL JSM-6700F scanning electron microscope. For cuticle preparations, hatchlings were fixed for 1 h in 3.7% formaldehyde in FASW (or in 1% glutaraldehyde in FASW), washed and dissected in PBS with 0.1% Triton X-100 (PTx), dehydrated through an ethanol series, mounted in Hoyer's medium/lactic acid (1:1), and cleared overnight on a 60 °C plate.
Immunohistochemistry and in Situ Hybridization.
For antibody stainings, embryos at stages 23–24 were immersed in a drop of fixative (3.7% formaldehyde in FASW) on a silicone rubber plate, the egg membranes were removed with sharp tungsten needles within 15 min, and the embryos were washed and processed using standard protocols (14, 17). We used antibody FP6.87 against Ubx and AbdA (4, 29) (kindly provided by Rob White) at 1:5 and goat anti-mouse horseradish peroxidase (Jackson ImmunoLabs) at 1:500. In situ hybridizations were carried out following previously published protocols (15), except that hybridizations were carried out for approximately 40 h at 65 °C.
Image Capture and Analysis.
Live embryos and hatchlings were observed on a Leica MZFLIII fluorescence stereomicroscope and photographed with a Leica DFC500 camera. Stained embryos and cuticle preparations were viewed on Zeiss Axiophot or Zeiss Axioskop2 compound microscopes and images were captured with a Zeiss AxioCam MRm. For thick whole-mount preparations, images captured at multiple focal planes were combined into a single focused image using the Helicon Focus software. Brightfield, fluorescence, and SEM images were adjusted for contrast, merged, and color overlaid using Photoshop CS. The time-lapse movie (Movie S1) was captured using Zeiss Axiovision 4.6 software and assembled using Volocity (Improvision).
Supplementary Material
Acknowledgments.
We are grateful to Franco and Gennaro Iamunno for scanning electron microscopy at the Stazione Zoologica Anton Dohrn in Naples and to Ina Arnone for hosting us during visits to the facility. We also thank Evangelia Stamataki, Judith Gallagher, Cassandra Extavour, Carsten Wolff, and Maura Strigini for their practical help, advice, and comments on the manuscript. This work was supported by the Marie Curie Research Training Network program ZOONET (European Union), by an EPAN collaborative program (General Secretariat for Research and Technology, Greece), and by European Molecular Biology Organization and Marie Curie Intra-European fellowships (to A.P.).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902804106/DCSupplemental.
References
- 1.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 2.Warren RW, Nagy L, Selegue J, Gates J, Carroll S. Evolution of homeotic gene regulation and function in flies and butterflies. Nature. 1994;372:458–461. doi: 10.1038/372458a0. [DOI] [PubMed] [Google Scholar]
- 3.Averof M, Akam M. Hox genes and the diversification of insect and crustacean body plans. Nature. 1995;376:420–423. doi: 10.1038/376420a0. [DOI] [PubMed] [Google Scholar]
- 4.Averof M, Patel NH. Crustacean appendage evolution associated with changes in Hox gene expression. Nature. 1997;388:682–686. doi: 10.1038/41786. [DOI] [PubMed] [Google Scholar]
- 5.Stern DL. A role of Ultrabithorax in morphological differences between Drosophila species. Nature. 1998;396:463–466. doi: 10.1038/24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abzhanov A, Kaufman TC. Crustacean (malacostracan) Hox genes and the evolution of the arthropod trunk. Development. 2000;127:2239–2249. doi: 10.1242/dev.127.11.2239. [DOI] [PubMed] [Google Scholar]
- 7.Ronshaugen M, McGinnis N, McGinnis W. Hox protein mutation and macroevolution of the insect body plan. Nature. 2002;415:914–917. doi: 10.1038/nature716. [DOI] [PubMed] [Google Scholar]
- 8.Galant R, Carroll SB. Evolution of a transcriptional repression domain in an insect Hox protein. Nature. 2002;415:910–913. doi: 10.1038/nature717. [DOI] [PubMed] [Google Scholar]
- 9.Weatherbee SD, et al. Ultrabithorax function in butterfly wings and the evolution of insect wing patterns. Curr Biol. 1999;9:109–115. doi: 10.1016/s0960-9822(99)80064-5. [DOI] [PubMed] [Google Scholar]
- 10.Jeong S, Rokas A, Carroll SB. Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell. 2006;125:1387–1399. doi: 10.1016/j.cell.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 11.Brusca RC, Brusca GJ. Invertebrates. Sunderland, MA: Sinauer Associates; 1990. [Google Scholar]
- 12.Abzhanov A, Kaufman TC. Novel regulation of the homeotic gene Scr associated with a crustacean leg-to-maxilliped appendage transformation. Development. 1999;126:1121–1128. doi: 10.1242/dev.126.6.1121. [DOI] [PubMed] [Google Scholar]
- 13.Abzhanov A, Kaufman TC. Embryonic expression patterns of the Hox genes of the crayfish Procambarus clarkii (Crustacea, Decapoda) Evol Dev. 2000;2:271–283. doi: 10.1046/j.1525-142x.2000.00066.x. [DOI] [PubMed] [Google Scholar]
- 14.Browne WE, Price AL, Gerberding M, Patel NH. Stages of embryonic development in the amphipod crustacean, Parhyale hawaiensis. Genesis. 2005;42:124–149. doi: 10.1002/gene.20145. [DOI] [PubMed] [Google Scholar]
- 15.Rehm EJ, Hannibal RL, Chaw RC, Vargas-Vila MA, Patel NH. Emerging Model Organisms: A Laboratory Manual. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2009. p. 592. [Google Scholar]
- 16.Gerberding M, Browne WE, Patel NH. Cell lineage analysis of the amphipod crustacean Parhyale hawaiensis reveals an early restriction of cell fates. Development. 2002;129:5789–5801. doi: 10.1242/dev.00155. [DOI] [PubMed] [Google Scholar]
- 17.Extavour CG. The fate of isolated blastomeres with respect to germ cell formation in the amphipod crustacean Parhyale hawaiensis. Dev Biol. 2005;277:387–402. doi: 10.1016/j.ydbio.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 18.Browne WE, Schmid BG, Wimmer EA, Martindale MQ. Expression of otd orthologs in the amphipod crustacean, Parhyale hawaiensis. Dev Genes Evol. 2006;216:581–595. doi: 10.1007/s00427-006-0074-7. [DOI] [PubMed] [Google Scholar]
- 19.Price AL, Patel NH. Investigating divergent mechanisms of mesoderm development in arthropods: The expression of Ph-twist and Ph-mef2 in Parhyale hawaiensis. J Exp Zoolog B Mol Dev Evol. 2008;310:24–40. doi: 10.1002/jez.b.21135. [DOI] [PubMed] [Google Scholar]
- 20.Kizil GO, Havemann J, Gerberding M. Germ cells in the crustacean Parhyale hawaiensis depend on Vasa protein for their maintenance but not for their formation. Dev Biol. 2008;218:333–339. doi: 10.1016/j.ydbio.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 21.Liubicich DM, et al. Knockdown of Parhyale Ultrabithorax recapitulates evolutionary changes in crustacean appendage morphology. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0903105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pavlopoulos A, Averof M. Establishing genetic transformation for comparative developmental studies in the crustacean Parhyale hawaiensis. Proc Natl Acad Sci USA. 2005;102:7888–7893. doi: 10.1073/pnas.0501101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 24.Struhl G. Genes controlling segmental specification in the Drosophila thorax. Proc Natl Acad Sci USA. 1982;79:7380–7384. doi: 10.1073/pnas.79.23.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller DF, et al. Cross-regulation of Hox genes in the Drosophila melanogaster embryo. Mech Dev. 2001;102:3–16. doi: 10.1016/s0925-4773(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 26.Akam M. Hox genes, homeosis and the evolution of segment identity: No need for hopeless monsters. Int J Dev Biol. 1998;42:445–451. [PubMed] [Google Scholar]
- 27.Pavlopoulos A, Berghammer AJ, Averof M, Klingler M. Efficient transformation of the beetle Tribolium castaneum using the Minos transposable element: Quantitative and qualitative analysis of genomic integration events. Genetics. 2004;167:737–746. doi: 10.1534/genetics.103.023085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelsh R, Weinzierl RO, White RA, Akam M. Homeotic gene expression in the locust Schistocerca: An antibody that detects conserved epitopes in Ultrabithorax and abdominal-A proteins. Dev Genet. 1994;15:19–31. doi: 10.1002/dvg.1020150104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.