Abstract
Myotonic dystrophy type 1 (DM1) is caused by toxicity of an expanded, noncoding (CUG)n tract in DM protein kinase (DMPK) transcripts. According to current evidence the long (CUG)n segment is involved in entrapment of muscleblind (Mbnl) proteins in ribonuclear aggregates and stabilized expression of CUG binding protein 1 (CUGBP1), causing aberrant premRNA splicing and associated pathogenesis in DM1 patients. Here, we report on the use of antisense oligonucleotides (AONs) in a therapeutic strategy for reversal of RNA-gain-of-function toxicity. Using a previously undescribed mouse DM1 myoblast−myotube cell model and DM1 patient cells as screening tools, we have identified a fully 2′-O-methyl-phosphorothioate-modified (CAG)7 AON that silences mutant DMPK RNA expression and reduces the number of ribonuclear aggregates in a selective and (CUG)n-length-dependent manner. Direct administration of this AON in muscle of DM1 mouse models in vivo caused a significant reduction in the level of toxic (CUG)n RNA and a normalizing effect on aberrant premRNA splicing. Our data demonstrate proof of principle for therapeutic use of simple sequence AONs in DM1 and potentially other unstable microsatellite diseases.
Keywords: antisense oligonucleotide, microsatellite, muscle, pathogenesis, RNA silencing
Myotonic dystrophy type 1 (DM1) is the most common neuromuscular dystrophy in adults and the first triplet-repeat RNA-mediated disorder described (1, 2). Its complex, multisystemic pathology involves the production of toxic (CUG)n transcripts after expansion of a mutant (CTG)n tract in the DMPK gene. In DM1 patients with expansions ranging from 50 to >2,000 triplets in their blood DNA severity of disease correlates to (CTG)n tract length.
Currently, the weight of evidence argues in favor of a model in which the harmful effect of a mutation in the DMPK gene occurs at the gene product level and involves protein binding to the long (CUG)n segment. Other cell stress effects of expression of the mutant DMPK gene cannot be excluded, however (1, 2). Members of the Mbnl and CELF families of RNA-binding proteins are most likely implicated in DM1 pathogenesis (3, 4). Mbnl1 is sequestered together with expanded (CUG)n transcripts in ribonucleoprotein (RNP) aggregates, resulting in reduced free Mbnl1 levels in the cell. Conversely, steady-state levels of CUGBP1 are increased in cells or tissues with a high (CUG)n dose. The ensuing imbalance in splicing factors causes missplicing of several transcripts, usually resulting in expression of fetal proteins inappropriate for adult tissue (2).
Use of DM1 mouse models has pointed out that harmful effects of toxic (CUG)n RNA are largely reversible. For the HSAlr model (5), characterized by high expression of skeletal α-actin transcripts bearing a (CUG)250 tract, it was demonstrated that viral overexpression of Mbnl1 reverses splicing defects and alleviates myotonia (6). In 5–313 mice, which inducibly overexpress the human DMPK (hDMPK) 3′-UTR including a (CUG)5 repeat tract as part of a GFP transcript, typical DM1 myotonia and cardiac defects disappeared when the transgene was silenced (7). These observations underscore the idea that reducing (CUG)n RNA dosage is beneficial to patients and forms an attractive therapeutic goal for improvement of DM1 features.
Here, we investigated the feasibility of antisense oligonucleotide (AON) technology, which has shown a range of promising applications (8–11). Among a series of chemically modified AONs designed to induce posttranscriptional gene silencing of mutant hDMPK transcripts a (CAG)7 AON was identified that causes specific reduction of expanded (CUG)n transcripts and reverses DM1 features in cell and mouse models. We demonstrate that an effective, therapeutic strategy based on an AON consisting of simple repeats may become feasible, which may also have bearing for other microsatellite disorders.
Results
A Novel Myogenic Cell Model for DM1.
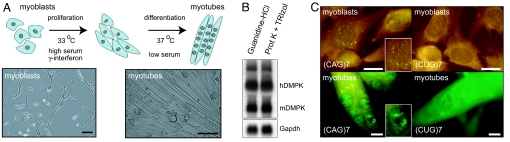
As a model for testing DM1-specific AONs in vitro, we derived conditionally immortalized myoblast–myotubes from skeletal muscle of the DM300 lineage (Fig. 1A) (12). DM300 mice were originally generated as carriers of a hDMPK (CTG)300 transgene, but ongoing expansion was observed in this lineage. The DM1 repeat locus in our cell model at the time of derivation comprised around 500 triplets. Hence, we will refer to these cells as DM500 myoblasts/myotubes.
Fig. 1.
Novel cell model for testing DM1 therapy. (A) Conditionally immortalized myoblasts derived from DM500 mice differentiate to myotubes under low serum conditions. Bars, 30 μm. (B) Northern blot analysis of RNA isolated by 2 different methods from DM500 myoblasts demonstrated expression of transgenic hDMPK mRNA bearing a (CUG)500 tract next to endogenous mDMPK mRNA. (C) FISH using a (CAG)7 probe revealed ribonuclear (CUG)n foci, mainly in cell nuclei. A (CUG)7 probe was used as negative control. Insets show nuclei at higher magnification. Bars, 10 μm.
DM500 myoblasts expressed hDMPK (CUG)500 mRNA at levels comparable to those of endogenous mouse DMPK (mDMPK) mRNA (Fig. 1B). Ribonuclear foci, a hallmark of DM1 pathogenesis (13), were observed in proliferating myoblasts and in differentiated myotubes (Fig. 1C). FISH using a (CAG)7 probe confirmed the presence of 5 to 10 foci per nucleus, not seen with a (CUG)7 control. Altogether, our DM500 cells displayed molecular characteristics of DM1, which makes them a useful test system for AON effects on cell stress caused by mutant hDMPK transcripts.
Silencing of hDMPK (CUG)500 RNA by PS58, a Chemically Modified (CAG)7 Oligo.
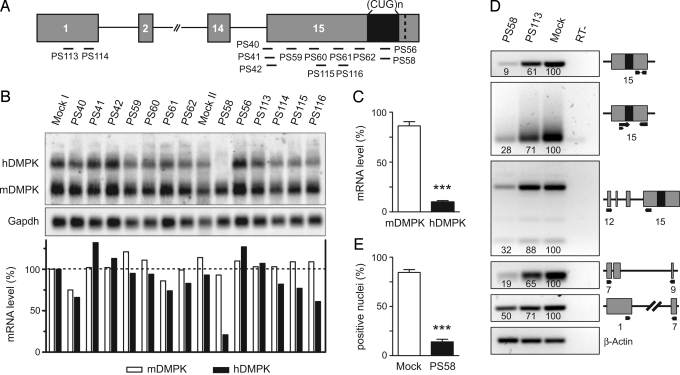
A series of modified RNA- and DNA-based AONs was designed with a target sequence for a particular region within the hDMPK primary transcript [Fig. 2A and supporting information (SI) Table S1]. Thirteen AONs were tested for their ability to suppress hDMPK (CUG)500 RNA expression in DM500 myotubes. Northern blotting revealed that silencing capacity varied between AONs (Fig. 2B). PS58, a 2′-O-methyl (2′-OMe) phosphorothioate (PT) modified (CAG)7 oligo designed to target the (CUG)n segment was most effective, causing a ≈90% reduction in hDMPK (CUG)500 mRNA (Fig. 2C). Not surprisingly, endogenous mDMPK mRNA bearing a (CUG)2(CAG)2(CUG) sequence instead of an uninterrupted (CUG)n segment was left intact. To ensure that our appraisal of PS58's silencing capacity was correct and that the disappearance of hDMPK (CUG)500 mRNA signal was not the result of events that occurred during the RNA isolation procedure, we tested various methods (Fig. S1) and also carried out RT-PCR analysis across the hDMPK transcript (Fig. 2D). Data obtained are consistent with a breakdown of hDMPK (CUG)500 transcripts by PS58. For reasons unknown, the 5′ region (exons 1–7) of the mRNA appeared relatively resistant to degradation compared to 3′ regions (Fig. 2D). In this respect, it is important to note that no short DMPK mRNA products of discrete size were detected on Northern blot or by RT-PCR.
Fig. 2.
PS58-mediated silencing of expanded hDMPK transcripts in DM500 cells. (A) Location of target sites of AONs (black bars) along the hDMPK premRNA. (B) Northern blot analysis to detect ability of AONs to silence hDMPK mRNA in DM500 myotubes. Gapdh was used as loading control. Oligos were tested at least twice; a representative blot is shown. (C) PS58, directed at the (CUG)n segment, was the most successful AON (P < 0.001, n = 19). (D) PS58 activity was corroborated by semiquantitative RT-PCR analysis of individual segments of the hDMPK transcript (primers indicated with black arrows). Numerals in lanes indicate relative abundance to mock samples (set at 100), using β-actin for normalization. A representative result from 2 experiments is shown. (E) A reduction in the number of nuclei containing (CUG)n foci was observed after PS58 treatment of DM500 myoblasts (P < 0.001, n = 4).
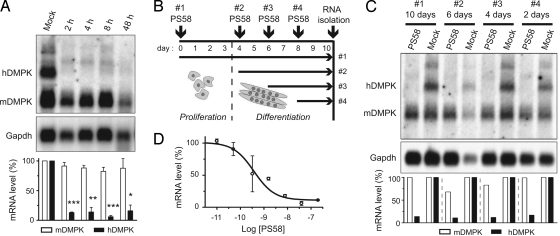
Examination of time and concentration effects revealed that maximal efficacy of PS58 in our DM500 cell model was already reached within 2 hours after transfection (Fig. 3A). hDMPK (CUG)500 mRNA levels remained virtually undetectable for up to 10 days after a single oligo dose (Fig. 3 B and C). From concentration response analyses, we deduced an IC50 of 0.40 ± 0.15 nM (Fig. 3D). Combined, our studies demonstrated that PS58 silenced expanded hDMPK transcripts quickly and with high affinity.
Fig. 3.
PS58-induced silencing is rapid, persistent, and occurs with high efficacy. (A) DM500 myotubes were mock-treated or treated with PS58. RNA was isolated 2–48 h after start of transfection. Northern blot analysis indicated that expanded hDMPK was degraded within 2 hours. A representative blot is shown. The means of 3 experiments are summarized in the Bottom panel (P < 0.05). (B and C) DM500 myoblasts and myotubes in different stages of myogenesis were treated with a single 200-nM PS58 dose or mock-treated and then cultured for up to 10 days. Northern blot analysis demonstrated that hDMPK RNA remained undetectable for up to 10 days. (D) Concentration-response curve in DM500 myotubes. Data points are the mean of at least 3 measurements.
Because PS58 is a (CAG)7 triplet repeat oligonucleotide, (CUG)n-bearing transcripts other than DMPK could potentially serve as targets too. We searched databases for (CUG)n tracts in the mouse transcriptome and identified 9 candidate transcripts that contain at least 6 CUG triplets (Table S2). RT-PCR-mediated quantification of 4 candidates revealed that PS58 treatment only marginally affected expression; for Mapkap1 (CUG)26 transcripts, a more pronounced effect was noted at high PS58 concentration (Fig. S2).
In Human Cells, PS58 Preferentially Silences DMPK Transcripts with an Expanded (CUG)n Repeat.
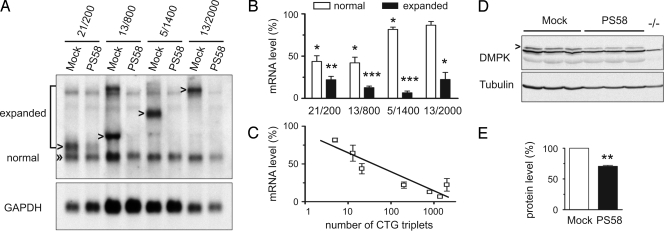
Symptoms in DM1 strongly correlate with the length of the (CTG)n segment in the mutated DMPK allele (1). To be functional in patients, PS58 should ideally silence transcripts from the expanded allele and not be active toward transcripts from the normal-sized allele, which may carry up to 37 triplets (14). To test this requirement, we analyzed PS58 efficacy in 4 patient myoblast cultures (15). These myoblasts expressed transcripts from expanded alleles that contained 200, 800, 1,400, or 2,000 CTG triplets, representing different severities of disease, and normal-sized alleles bearing 5, 13, or 21 triplets. Northern blot analysis demonstrated that PS58 silenced expanded DMPK transcripts in these human cells to the same degree as in DM500 mouse myotubes (Fig. 4 A and B). PS58 efficacy clearly correlated with the number of CUG triplets in the hDMPK transcript since normal-sized transcripts were only mildly reduced (Fig. 4 A–C). Effects on nonmutant transcripts appeared variable, as (CUG)13 transcripts were reduced in 13/800 cells, but unchanged in 13/2,000 cells. Western blot analysis confirmed that undesired silencing of normal-sized transcripts did not result in great loss of DMPK protein, as 71% of control level was still present at 72 h after PS58 treatment in 21/200 myoblasts (Fig. 4 D and E).
Fig. 4.
PS58 silences preferentially expanded DMPK transcripts in patient cells. (A) DM1 myoblasts expressing different expanded and normal-sized DMPK alleles were treated with PS58 or mock treated. Northern blot analysis indicated that expanded mRNA was strongly reduced whereas normal-sized DMPK transcripts were less sensitive to breakdown. A representative blot is shown. (B) Quantification of 3 experiments as described in A (P < 0.05). (C) A clear correlation was demonstrated between the number of CTG triplets and PS58 efficacy (P < 0.05, Spearman correlation, r = −0.79). (D and E) Myoblasts (21/200) were analyzed by Western blotting 72 h after mock or PS58 treatment. A muscle sample from a DMPK KO mouse (−/−) served as negative control. A 29% reduction in DMPK protein (arrowhead) was observed (P < 0.01, n = 3).
Secondary Effects of PS58 Activity in DM500 Cells.
Next, we used our DM500 cell model to examine whether PS58 treatment had beneficial effects on the DM1-typical formation of nuclear RNA foci or occurrence of embryonic modes of premRNA splicing. Muscleblind proteins and CUGBP1 are prominently involved in these events (2). FISH analysis demonstrated that PS58 treatment caused a 4-fold reduction in the percentage of cells that contained ribonuclear foci (Fig. 2E). The presence of PS58 had no effect on CUGBP1 level, but it should be noted that the CUGBP1 level in DM500 myotubes was not increased compared to controls (similar to DM500 mice). Also the pattern of alternative splicing of a representative subset of DM1-specific transcripts was not altered in the presence of PS58 in DM500 myotubes or WT myotubes (Fig. S3). This excludes anomalous effects of PS58 on RNA processing. Effects of PS58 activity on splicing required study in vivo, as the terminal end stage of myogenic differentiation and adult programming of RNA processing cannot be achieved in cells in culture (Fig. S3).
PS58 Is Active in DM1 Mouse Models in Vivo.
As a prelude to in vivo experiments, we investigated the distribution of fluorescently labeled PS58 after injection into the GPS (gastrocnemius-plantaris-soleus) complex of DM500 mice. A nonhomogeneous staining was observed throughout the tissue (Fig. S4A). Virtually no signal was found in the tibialis anterior (TA) demonstrating that intertissue spreading was minimal. Analysis of muscle sections showed that PS58 molecules concentrated in cell nuclei (Fig. S4B).
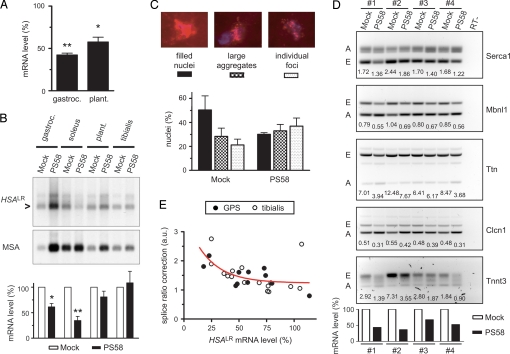
Next, we assessed efficacy of PS58 in 2 different mouse models. In DM500 mice, injection in the GPS complex followed by RT-PCR analysis of RNA content confirmed silencing of hDMPK (CUG)500 mRNA in the gastrocnemius and plantaris, but not the TA control (Fig. 5A). Again, the 5′ end of the hDMPK transcript appeared relatively resistant to degradation (Fig. S5). PS58 injection in the GPS complex of HSAlr mice (5) resulted in significant silencing of toxic (CUG)250 transcripts, predominantly in soleus and to a lesser extent in gastrocnemius (Fig. 5B).
Fig. 5.
PS58 reverses molecular features in DM1 mouse models in vivo. (A) PS58 was injected in the GPS complex of DM500 mice, followed by RNA isolation at day 15. Semiquantitative RT-PCR analysis demonstrated hDMPK mRNA silencing in gastrocnemius (P < 0.01, n = 3) and plantaris (P < 0.05, n = 3) (see Fig. S5). (B) A similar protocol was used in HSAlr mice. TA muscle served as uninjected control. HSAlr transcripts were quantified on Northern blot, using mouse α-actin (MSA) for normalization. The blot represents 1 of 4 independent experiments. The histogram shows average values: lower HSAlr mRNA levels were observed in soleus (35 ± 8%; P < 0.01) and gastrocnemius (61 ± 7%; P = 0.01), but not in plantaris (81 ± 11%; P = 0.19). (C) FISH analysis was performed on sections of PS58- or mock-injected HSAlr gastrocnemius muscle. (CUG)n foci were found in every nucleus. Nuclei were categorized in 3 groups according to their (CUG)n foci appearance (Top panels each show 1 representative nucleus). A significant shift toward smaller and fewer foci was observed after PS58 treatment (P < 0.01, n = 2). (D) TA muscle in HSAlr mice was electroporated with PS58 or mock treated. RNA was isolated after 7 days. Northern blotting showed significantly lower HSAlr mRNA levels after PS58 treatment (Bottom panel; 51 ± 7%; P < 0.01, n = 4). Splice modes were investigated by RT-PCR. Numerals indicate embryonic (E):adult (A) splice ratio in each lane. Lower ratios in PS58-treated samples, compared to their mock-treated samples, demonstrate positive effects on alternative splicing (see also Fig. S7). (E) Correlation between splicing correction and HSAlr mRNA levels in PS58-treated muscles. Each symbol represents a TA muscle or 1 muscle from the GPS complex. Splice ratio correction illustrates the mean effect on alternative splicing of 5 transcripts (see D) (P < 0.01; Spearman correlation, r = −0.64; n = 27).
The very high expression of transgenic transcripts in the HSAlr mouse culminates in formation of large RNP foci in every cell nucleus in skeletal muscle, which makes it a particular attractive model for examining effects of PS58 treatment on this feature. We categorized appearance of RNP foci according to FISH signal intensity and pattern after PS58 treatment and observed a significant shift from nuclei packed with foci toward nuclei containing smaller, discrete foci (Fig. 5C). No nuclei void of foci were detected. Simultaneous study of nuclear aggregation behavior of Mbnl1 in this model revealed a substantially more diffuse distribution of Mbnl1 (Fig. S6) (6).
Many embryonic splice modes in skeletal muscle observed in DM1 patients are replicated in HSAlr mice (16)—in contrast to DM500 mice, which display hardly any DM1-typical splice modes in skeletal muscle and also RNP foci much less overtly (21). We measured splice modes of 5 different transcripts—Serca1, Mbnl1, Ttn, Clcn1, and Tnnt3—upon PS58 treatment in TA and GPS muscles. In electroporated TA muscle, a reduction of ≈50% of HSAlr mRNA was achieved after 7 days, which was paralleled by a significant shift in splicing pattern from an embryonic-like (E) to normal-adult (A) mode for 4 out of 5 transcripts tested (Fig. 5D and Fig. S7).
Finally, we combined all data obtained with HSAlr mice, irrespective of the mode of PS58 administration, muscle type, or timing of muscle isolation. Collective evaluation of the results revealed a clear correlation between PS58-induced reduction of HSAlr (CUG)250 mRNA levels and the accompanying effects on splicing (i.e., splice-ratio correction, defined as the mock-treated embryonic:adult splice ratio divided by the PS58-treated embryonic:adult splice ratio; Fig. 5E). This demonstrated that PS58-induced suppression of abnormally expanded (CUG)n RNA mitigates toxicity effects in vivo.
Discussion
DM1 was the first RNA toxic-gain-of-function disorder described (2). Here, we explored a therapeutic approach that directly deals with its root cause, i.e., expression of expanded (CUG)n RNA. Only few experimental studies were dedicated in the past to the breakdown of normal or mutant hDMPK RNA, all restricted to use of cell models and mostly viral-based vectors for the delivery of the active component (17–20). To circumvent problems associated with viral-based therapy in humans, we chose AONs to degrade toxic (CUG)n RNA. The (CAG)7 AON PS58 identified in this paper was successfully delivered into cells with the help of transfection reagents or locally via injection-electroporation protocols in mouse muscle, to demonstrate proof of principle. The chemical nature of PS58 precludes efficient uptake in tissues. We will therefore need to concentrate on the design of more sophisticated delivery techniques to promote uptake in vivo, before we can proceed with a study of systemic treatment of DM1 animal models or patients. Clearly, this remains a challenge for future efforts.
PS58 silencing capacity for expanded (CUG)n RNA ranged from up to 90% in cells to between 30 and 80% in vivo, with differences between muscle types (soleus > gastrocnemius ∼ tibialis anterior > plantaris). The HSAlr and DM500 models clearly differ in the degree of muscle pathology, presumably because the level of expanded (CUG)n transcripts in muscle from the HSAlr model is much higher than in DM500 muscle. Still, a similar range of silencing was observed for muscles of both models. On the basis of these observations, we think that membrane integrity or physiological state of individual muscle types, rather than the initial cellular concentration of target RNA forms the most important parameter for AON uptake and breakdown capacity. Also size of the (CUG)n segment or differences in sequence context or subcellular localization of target RNAs are considered parameters of lesser importance.
Studies of Mbnl1 protein overexpression in HSAlr mice (6), expression inhibition of a (CUG)5 RNA-encoding transgene in an inducible mouse model (7) or correction of Clcn1 splicing (11) have illustrated that particular DM1 features are reversible. Our findings demonstrate that also nongenetic modalities, e.g., treatment with a simple repeat AON, can reverse DM1 pathogenesis in vivo. Our findings on the PS58-induced reduction of nuclear RNP foci are in line with the notion that foci containing (CUG)n RNA, Mbnl1 protein, and potentially other unknown factors are dynamic structures (2). We therefore may assume that upon degradation of the (CUG)n RNA component, less Mbnl1 will be captured in the aggregated state and more Mbnl1 will be available for regulation of splicing. Our findings in the HSAlr model confirm this idea. PS58 treatment significantly improved splicing of 5 different transcripts, in a manner clearly correlating to the degree of (CUG)n RNA reduction in this model.
Whether the (CUG)n repeat dose, splice abnormalities, and phenotypic features in HSAlr mice accurately reflect the situation in patients is a hotly debated question in the DM field. The disbalance introduced by transgene expression in the currently available models may be considered either too extreme (e.g., the HSAlr model) or too mild (e.g., DM500 model). Further progress in knowledge on this topic is needed before we can productively proceed with studies toward the development of PS58-based therapy. Analysis on the reversal of phenotype in DM1 mouse models and its value for prediction of possible effectiveness in clinical trials in DM1 patients therefore needs further work.
Our AONs were originally designed to bind and degrade expanded (CUG)n RNA or induce exon skipping (compare ref. 22) or abortive splicing (23). The unique effectiveness of PS58 can best be explained by induction of selective breakdown, presumably starting at the 3′ target segment with the (CUG)n tract, as we noticed relatively stable behavior of the 5′ part of the hDMPK transcripts. Currently, we do not know much about the molecular mechanism involved in this breakdown, but do not expect that PS58, being an AON with a 2′ O-methyl-based backbone, will work via recruitment of RNase H. Selective effects of PS58 on premRNA processing events, like the use of premature polyadenylation sites or complete skipping of exon 15 or segments thereof, can also not be involved as no smaller DMPK mRNA products of discrete size were identified using RT-PCR or Northern blot analyses. PS58-induced breakdown of repeat expanded transcripts in DM500 myotubes was fast and went nearly to completion. So we assume that both the cytoplasmic mature mRNA pool (constituting probably only a minor fraction) and the nuclear pool of primary and mature expanded (CUG)n transcripts served as targets. Moreover, in patient cells we observed effects on nonmutant DMPK transcripts residing in the cytoplasm. Thus, an exclusive effect of PS58 in the nucleus seems unlikely.
It is known that 2′-OMe PT modifications, as in PS58, protect against enzymatic breakdown in the cell, resulting in higher AON efficacy (9). The PT moiety is probably also responsible for the dominant nuclear localization that we observed for PS58 (24). A fully modified 2′-OMe PT AON has to our knowledge not been used successfully before in antisense silencing strategies (9, 25, 26). Our data suggest that the structural composition of PS58 determines entry in novel RNA degradation mechanisms, but this will require further testing.
Regardless of the mechanism operating, we have observed that PS58 provides selectivity between expanded and normal-sized hDMPK transcripts. Patients carrying a normal-sized allele in the (CTG)18–37 range are only present in ≈10% of the DM1 population (14) and also other transcripts with a (CUG)n ≥ 10 segment are scarce in the human transcriptome (Table S3). From a kinetic point of view, it is important to note that PS58 has multiple binding sites on a long (CUG)n segment but only 1 or a few on a normal (CUG)n segment. Assuming that binding of 1 oligo to a (CUG)n transcript is sufficient for degradation, a lower dose may thus increase selectivity and reduce the chance of undesired side effects. Changing the number of CAG triplets in the AON could also positively influence selectivity. Although at this point, unfamiliar with the working mechanism of PS58, it is impossible to predict whether a shorter or longer (CAG)n AON will be equally effective. Experiments with (CAG)n AONs of different lengths in cells expressing DMPK transcripts with diverse (CUG)n segments should provide answers.
Successful development of RNA-directed therapy for DM1 will also create new opportunities for treatment of other microsatellite diseases caused by RNA toxicity, like DM2, SCA8, SCA10, FRAXA, and HDL2, but maybe also disorders like Huntington's or oculopharyngeal muscular dystrophy. Each of these diseases progressively affects muscle, heart, and brain and functioning of various body systems. For none of these disorders is a cure or therapy available.
Materials and Methods
For details see SI Materials and Methods.
Animals.
DM500 mice, derived from the DM300–328 line (12), and HSAlr20b mice have been described (5). All animal experiments were approved by the Institutional Animal Care and Use Committees of the Radboud University Nijmegen and the University of Rochester Medical Center.
Cell Culture.
An immortal mouse myoblast cell culture expressing hDMPK (CUG)500 transcripts was derived from GPS tissue isolated from DM500 mice additionally expressing 1 copy of the H-2Kb-tsA58 allele. The generation of human DM1 myoblast lines has been described (15).
Oligos.
All AONs used in this study are listed in Fig. 2A and Table S1. PS58 is a synthetic RNA oligonucleotide with sequence 5′-CAGCAGCAGCAGCAGCAGCAG-3′, which carries full-length 2′-OMe-substituted ribose molecules and PT internucleotide linkages.
Oligo Transfection.
Cells were transfected with AONs complexed with polyethyleneimine (PEI; ExGen500, Fermentas) according to the manufacturer's instructions.
RNA Isolation.
RNA from cultured cells was isolated using the Aurum Total RNA mini kit (Bio-Rad) according to the manufacturer's protocol. RNA from muscle tissue was isolated using TRIzol reagent (Invitrogen). Alternative methods are described in SI Materials and Methods.
Northern Blotting and RT-PCR.
Northern blotting and semiquantitative RT-PCR analysis were done following standard procedures. For analysis of alternative splicing, embryonic (E):adult (A) splice ratio was defined as embryonic form signal divided by adult form signal in each sample. Splice ratio correction illustrates the effect of PS58 treatment on alternative splicing. Splice ratio correction is defined as the mock-treated embryonic:adult splice ratio divided by the PS58-treated embryonic:adult splice ratio for a given transcript per mock/PS58-treated muscle pair (i.e., per mouse). Hence, a splice ratio correction of 1.0 means no change in alternative splicing, while a correction >1.0 implies an increase in adult splice forms.
Western Blotting.
Western blotting was done by standard methods described in SI Materials and Methods.
In Vivo Treatment.
The GPS complex of DM500 mice was pretreated with PEI-5% glucose injections on days 1 and 2. This pretreatment was followed by 2 injections with 2 nmoles of PS58 in a PEI-5% glucose solution on days 3 and 4. Mock-treated mice received the same treatment except that oligo PS58 was left out. Mice were killed and muscles were isolated on day 15.
Diverse treatment protocols were tested in HSAlr mice. In each case, 1 leg of a HSAlr mouse was treated with 1 or 2 nmoles PS58, while the contralateral leg served as mock control. (i) HSAlr mice were injected twice in the GPS complex as described above, without PEI pretreatment. Injections took place at days 1 and 8, and tissues were isolated on day 22. Noninjected TA muscle served as negative control. (ii) TA muscle was injected once as described above, without PEI pretreatment, and muscle tissue was isolated 7 days later. (iii) TA muscle was injected with or without PEI, followed by electroporation (11) and analyzed on day 8 or 15.
Immunofluorescence and FISH.
Immunofluorescence and FISH were performed by standard methods. Images were collected on a Zeiss Axioplan fluorescence microscope (Carl Zeiss). Pictures were captured by a Zeiss AxioCam MR camera with Axiovision 3.1 software.
Intratissue Imaging.
The distribution of FAM-labeled PS58 was imaged in the GPS complex of DM500 mice with the Olympus OV110 whole mouse imaging system (Olympus).
Statistical Analysis.
See details in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank J. van Deutekom (Prosensa) for discussions and support; M. Swanson (University of Florida, Gainesville, FL) for antibody A2764 against Mbnl1; R. Donders (Department of Epidemiology, Biostatistics and HTA, Nijmegen, The Netherlands) for help with statistics; G. Bouw (Olympus) for help with intratissue imaging; M. Huynen (Centre for Molecular and Biomolecular Informatics) for advice in bioinformatics. We thank members of the Department of Cell Biology for discussions; S. Snellenberg, D. Nijholt, and J. Hooijer for initial experiments; and J. Fransen for microscopical advice. This work was supported by grants from SenterNovem (a Dutch agency in the Ministry of Economic Affairs), the Prinses Beatrix Fonds, the Stichting Spieren voor Spieren, the Association Française contre les Myopathies, and National Institutes of Health AR046806.
Footnotes
Conflict of interest: P.v.K.-R., S.J.d.K., and G.J.P. report being employed by or having an equity interest in Prosensa B.V. The method described in this paper is the subject of a patent application (inventors S.J.d.K, G.J.P., and D.G.W.).
This article contains supporting information online at www.pnas.org/cgi/content/full/0905780106/DCSupplemental.
References
- 1.Groenen P, Wieringa B. Expanding complexity in myotonic dystrophy. BioEssays. 1998;20:901–912. doi: 10.1002/(SICI)1521-1878(199811)20:11<901::AID-BIES5>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 2.Wheeler TM, Thornton CA. Myotonic dystrophy: RNA-mediated muscle disease. Curr Opin Neurol. 2007;20:572–576. doi: 10.1097/WCO.0b013e3282ef6064. [DOI] [PubMed] [Google Scholar]
- 3.Miller JW, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mankodi A, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1772. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 6.Kanadia RN, et al. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahadevan MS, et al. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Deutekom JC, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 9.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur J Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 10.Krützfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs.’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 11.Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seznec H, et al. Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum Mol Genet. 2000;9:1185–1194. doi: 10.1093/hmg/9.8.1185. [DOI] [PubMed] [Google Scholar]
- 13.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zerylnick C, Torroni A, Sherman SL, Warren ST. Normal variation at the myotonic dystrophy locus in global human populations. Am J Hum Genet. 1995;56:123–130. [PMC free article] [PubMed] [Google Scholar]
- 15.Furling D, Lemieux D, Taneja K, Puymirat J. Decreased levels of myotonic dystrophy protein kinase (DMPK) and delayed differentiation in human myotonic dystrophy myoblasts. Neuromuscul Disord. 2001;11:728–735. doi: 10.1016/s0960-8966(01)00226-7. [DOI] [PubMed] [Google Scholar]
- 16.Lin X, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 17.Furling D, et al. Viral vector producing antisense RNA restores myotonic dystrophy myoblast functions. Gene Ther. 2003;10:795–802. doi: 10.1038/sj.gt.3301955. [DOI] [PubMed] [Google Scholar]
- 18.Langlois MA, Lee NS, Rossi JJ, Puymirat J. Hammerhead ribozyme-mediated destruction of nuclear foci in myotonic dystrophy myoblasts. Mol Ther. 2003;7:670–680. doi: 10.1016/s1525-0016(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 19.Langlois MA, et al. Cytoplasmic and nuclear retained DMPK mRNAs are targets for RNA interference in myotonic dystrophy cells. J Biol Chem. 2005;280:16949–16954. doi: 10.1074/jbc.M501591200. [DOI] [PubMed] [Google Scholar]
- 20.Krol J, et al. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol Cell. 2007;25:575–586. doi: 10.1016/j.molcel.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 21.Guiraud-Dogan C, et al. DM1 CTG expansions affect insulin receptor isoforms expression in various tissues of transgenic mice. Biochim Biophys Acta. 2007;1772:1183–1191. doi: 10.1016/j.bbadis.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Tiscornia G, Mahadevan MS. Myotonic dystrophy: The role of the CUG triplet repeats in splicing of a novel DMPK exon and altered cytoplasmic DMPK mRNA isoform ratios. Mol Cell. 2000;5:959–967. doi: 10.1016/s1097-2765(00)80261-0. [DOI] [PubMed] [Google Scholar]
- 23.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 24.Lorenz P, Misteli T, Baker BF, Bennett CF, Spector DL. Nucleocytoplasmic shuttling: A novel in vivo property of antisense phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 2000;28:582–592. doi: 10.1093/nar/28.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holen T, Amarzguioui M, Babaie E, Prydz H. Similar behaviour of single-strand and double-strand siRNAs suggests they act through a common RNAi pathway. Nucleic Acids Res. 2003;31:2401–2407. doi: 10.1093/nar/gkg338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krützfeldt J, et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.