Abstract
Yeast and mammalian genomes are replete with nearly identical copies of long dispersed repeats in the form of retrotransposons. Mechanisms clearly exist to maintain genome structure in the face of potential rearrangement between the dispersed repeats, but the nature of this machinery is poorly understood. Here we describe a series of distinct “retrotransposon overdose” (RO) lineages in which the number of Ty1 elements in the Saccharomyces cerevisiae genome has been increased by as much as 10 fold. Although these RO strains are remarkably normal in growth rate, they demonstrate an intrinsic supersensitivity to DNA-damaging agents. We describe the identification of mutants in the DNA replication pathway that enhance this RO-specific DNA damage supersensitivity by promoting ectopic recombination between Ty1 elements. Abrogation of normal DNA replication leads to rampant genome instability primarily in the form of chromosomal aberrations and confirms the central role of DNA replication accuracy in the stabilization of repetitive DNA.
Keywords: chromosome rearrangement, genome instability, Ty1 elements
The transmission of genetic information requires multiple distinct mechanisms to ensure its fidelity. DNA replication and repair proteins can be grouped into modules, pathways, and, more broadly, a global network based upon the congruence of their synthetic fitness and lethality profiles (1–3). However, the DNA sequence and structure itself also determines the fidelity of genome propagation, with each class of repeat DNA (i.e., ribosomal DNA, telomeres, trinucleotide repeats, and transposons) presenting unique challenges to the genome. There are pathways that predominate to ensure control of each repeat type [e.g., chromatin cohesion and transcriptional silencing are needed to stabilize ribosomal DNA repeat copy number whereas trinucleotide repeat array integrity depends on accurate DNA replication and mismatch repair (4, 5)], but the precise interactions of repetitive DNA with the various DNA repair pathways remain unidentified.
Retrotransposons present a unique threat to stability as a result of their dispersed nature, with 32 copies of the Ty1 element per haploid yeast genome (6). Ty1 retrotransposons replicate through an RNA intermediate that is reverse-transcribed following encapsidation into a virus-like particle, and then integrated into the host cell genome (7, 8). These integrated Ty1 copies provide two challenges to genome stability: when Ty1 repeats are present as inverted pairs, they function to stall replication forks; and when recombination machinery is subsequently recruited to these stalled forks, the dispersed Ty1 elements provide numerous templates for ectopic repair (9–11). Abundant evidence exists for the association of Ty1 sequences with chromosomal translocations that may result from ectopic recombination in industrial, laboratory, and evolutionary settings (9, 12–15). Most notably, strains evolved under nutrient limitation contained chromosomal translocation breakpoints largely coincident with Ty1 elements (16, 17).
Although Ty1 elements are certainly capable of contributing to the formation of chromosome aberrations, the relative importance of this phenomenon to the maintenance of genome instability remains unclear because (i) repair is highly efficient, (ii) ectopic recombination between Ty1 repeats in yeast is apparently limited (18), and (iii) there are relatively few elements in the genome as a result of transcriptional and post-transcriptional copy number control that regulates both transposition rates and Ty1 loss rates (19, 20). Although increasing Ty1 copy number generally leads to modest defects on cell growth (20), occasional strains containing specific Ty1 insertions enjoy a significant selective advantage over the WT (21). Yet, these studies were limited by modest twofold increases in Ty1 copy number, and much higher levels of Ty1 copy number must be achieved to determine whether Ty1 sequences contribute significantly to overall ectopic inter-repeat recombination rates. We describe a set of yeast strains with Ty1 copy numbers as many as 10 times that of the WT strain; this increased Ty1 content not only promotes genome instability, but further enables the identification of genes and pathways that contribute to the suppression of retrotransposon-mediated instability.
Results
To determine the contribution of Ty1 elements to genome integrity, we sought to reduce the efficiency of host cell mechanisms that prevent Ty1-Ty1 recombination by increasing Ty1 element abundance. Multiple cycles of galactose-induced transposition, followed by curing of the donor plasmid produced a series of “retrotransposon overdose” (RO) strains, constituting a set of independent isogenic lineages loaded with new Ty1 retrotransposon copies (named L26–10C through L31–10C). Control strains (lineage L48) were also cycled on galactose, but in the presence of an empty vector.
Introduction of Ty1 Transposons to the Genome.
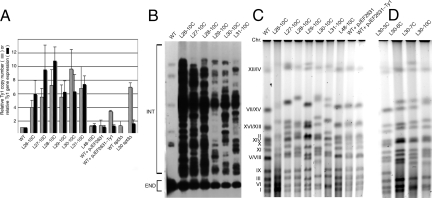
We previously doubled Ty1 copy number, which was stably maintained (20), but the new strains have 3- to 10-fold higher levels of Ty1 accumulation (Fig. 1A). For unknown reasons, the number of Ty1 elements varied considerably among the strains, with lineages 26 and 27 accumulating far fewer elements than lineages 28, 30, and 31. The addition of Ty1 elements in RO strains increases the overall DNA content of the cell by as much as 15%. To control for the effects of replicating an increased amount of DNA, we used a super high-copy 2 μm plasmid (pJEF2631), and a derivative with the WT Ty1 sequence that maintains Ty1 levels in the cell at a copy number similar to that of many RO strains (pJEF2631-Ty1, Fig. 1A), thereby allowing a distinction between phenotypes resulting from increased overall DNA load, from an increase in overall Ty1 copy number, or from an increased number of integrated, dispersed Ty1 elements.
Fig. 1.
DNA content of high-copy Ty1 strains. (A) Real-time PCR (hatched bars) and RT-PCR (solid bars) analysis of Ty1 copy number and gene expression in RO strains (L26–10C, L27–10C, L28–10C, L29–10C, L30–10C, and L31–10C), control strain (L48–10C), and plasmid-bearing strains (WT + pJEF2631 and WT + pJEF2631-Ty1) relative to the parental strain (GRF167). (B) Southern blot of Ty1 elements in RO strains, distinguishing endogenous Ty1 elements that contain 2 restriction sites (END) from introduced Ty1 elements that contain 1 site (INT; including some native Ty1 elements lacking an AvaI/XhoI site in one LTR) in RO strains. (C) Pulsed-field gel electrophoresis of yeast chromosomes, indicating the increased relative mobility of chromosomes of RO strains. The migration of chromosomes in the WT strain is indicated (Left). (D) Pulsed-field gel electrophoresis of strains from the L30 lineage after 3, 6, 7, and 10 cycles of Ty1 transposition.
Although WT yeast cells express abundant levels of Ty1 RNA, there is an increase in the abundance of full-length Ty1 RNA in RO strains that roughly parallels the increased Ty1 DNA content [Fig. 1A and supporting information (SI) Fig. S1], and increased Ty1 Gag protein abundance (Fig. S1). Ty1 RNA and protein levels in both WT and RO strains were markedly reduced upon deletion of Ty1 transcription factor gene SPT3 (Fig. 1A and Fig S1), indicating that transcription of the introduced Ty1 elements remains under its control.
Each RO strain shows a distinct pattern of Ty1 insertions that is readily visualized by Southern blot analysis of genomic DNA with a Ty1 probe (Fig. 1B); although these strains contain certain insertion sites in common (i.e., those that pre-existed in the parental strain), the overall pattern of targeting in each lineage is unique, reflecting its independent origin. The magnitude of the increased Ty1 load can also be visualized by pulsed-field gel electrophoresis; the RO lineages show dramatic mobility alterations in many chromosomes (Fig. 1C). In contrast, no alterations in electrophoretic karyotype can be seen in the control strain L48–10C or in WT strains containing the pJEF2631 and pJEF2631-Ty1 plasmids. Looking more closely at L30 after 3, 6, 7, and 10 cycles on galactose (Fig. 1D) reveals bands progressively shifted in subsequent cycles, consistent with the prediction that most alterations result from sequential accumulation of Ty1 copies throughout the genome. This extreme number of new insertions present in some RO Ty1 strains would likely saturate the ability of Ty1 elements to insert singly at loci that serve as hotspots for Ty1 integration. Integration of pairs of Ty1 elements and Ty1 arrays have been correlated with susceptibility to chromosome breaks (9). PCR analysis of RO Ty1 strains confirms that they contain a greater number of such Ty1 element arrays, and that, even though these insertions occur preferentially in the head-to-tail orientation, arrays of head-to-head and tail-to-tail elements are also detected in each of the RO strains (Fig. S2).
Although Ty1 elements preferentially integrate into genomic regions largely devoid of ORFs (22), we did not know whether the targeting mechanism remained intact in the face of the RO state. RO Ty1 strains displayed no gross phenotypic defects, with only a modest decrease in overall growth rate (Fig. S3 a and b), and were therefore screened for auxotrophy, respiratory deficiency, and growth defects at high and low temperatures that could result from novel insertions. Only a few phenotypes were noted. Whereas strains L26–10C, L27–10C, and L28–10C displayed no defects, strain L29–10C showed a strong tendency to flocculation, strain L30–10C became slightly temperature-sensitive at 37 °C after the tenth cycle on galactose, and strain L31–10C contained an insertion into MET15 during the ninth cycle that rendered the strain a methionine auxotroph (23). Interestingly, 2 of 3 RO Ty1 strains were able to sporulate when back-crossed to either the WT or L26–10C strains (Table S1), with the exception being L31–10C, which became diploid during the eighth round of cycling on galactose, and showed a deletion between the MAT and HMR loci that allowed the strain to mate as MATα (24; Fig. S3c).
Damage Sensitivity Depends on Repeat Dispersion and Copy Number.
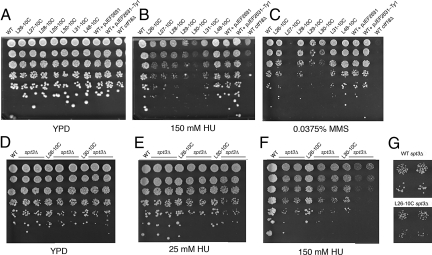
We predicted that increasing Ty1 copy number would increase the propensity to ectopic recombination. Indeed, RO Ty1 strains show increased transformation efficiency with marked linear Ty1 DNA, reflecting the increased copy number of Ty1 elements that can serve as targets for recombination of the incoming Ty1 into the genome (Table S2). This increase in ectopic recombination suggests that RO Ty1 strains should be less able to cope with DNA damage. WT and RO Ty1 strains were therefore challenged with a panel of DNA- or microtubule-damaging drugs: the alkylating agent methyl methanesulfonate (MMS), the ribonucleotide reductase inhibitor hydroxyurea (HU), the microtubule depolymerizing agent benomyl, and the topoisomerase inhibitor camptothecin (Fig. 2 A–C and Fig. S4 a–c). Both the control strain L48–10C and the strains containing the pJEF2631 and pJEF2631-Ty1 plasmids were as resistant as parental strains to both drugs, confirming that a simple increase in DNA or Ty1 content does not enhance genome instability. In marked contrast, all RO Ty1 strains displayed hypersensitivity to both HU and MMS, with a reduction in colony size and, notably in strains L29–10C and L30–10C, a reduction in the efficiency of plating as well (Fig. 2 B and C), suggesting a correlation between high Ty1 copy number and damaging agent sensitivity.
Fig. 2.
Sensitivity of high-copy Ty1 strains to DNA-damaging agents. Serial dilutions of yeast strains were plated onto YPD for 2 d (A), YPD + 150 mM HU for 4 d (B), or YPD + 0.0375% MMS for 2 d (C). Serial dilutions of high-copy strains with and without deletion of the SPT3 gene were plated onto YPD for 2 d (D), YPD + 25 mM HU for 3 d (E), or YPD + 150 mM HU for 4 d (F). (G) Higher magnification of WT and L26–10C spt3Δ colonies grown on 150 mM HU reveals that the L26–10C strain forms smaller colonies even in the absence of ongoing retrotransposition.
The probability of hitting one gene leading to HU or MMS hypersensitivity by transposition in this collection of strains is reasonably high (0.587; see Methods), and consistent with this, strain L30–10C is much more sensitive than the WT to HU and L27–10C is much more sensitive than the WT to MMS. Although it is formally possible that the haploid strains each contain a different insertion that leads to damage sensitivity, the probability of independently obtaining MMS hypersensitivity as a result of Ty1 transposition in all 6 strains assayed is exceedingly low (6.69 × 10−6; see Methods). To rule out this possibility, we created heterozygous diploid strains by crossing strains of the L30 lineage to either the WT strain or another RO Ty1 strain, L29–10C. Although less sensitive to HU than their haploid counterparts, the heterozygous diploid strains remain much more sensitive to HU than the WT diploid (Fig. S4 j and k), confirming that increasing Ty1 copy number increases HU sensitivity independently of Ty1-induced recessive mutations. Although all RO Ty1 strains are hypersensitive to HU and MMS, none are sensitive to benomyl or camptothecin (Fig. S4 a–c), confirming hypersensitivity of RO strains to very specific types of damage.
The sensitivity of RO strains to DNA-damaging agents could result from increased ectopic recombination or from ongoing retrotransposition of the numerous newly dispersed Ty1 copies. If the sensitivity of RO Ty1 lineages to HU and MMS is the result of nonallelic recombination, sensitivity should increase with retrotransposon copy number, and indeed HU sensitivity increases with each cycle of Ty1 transposition for lineages L26 and L30, with decreases in both the number and size of colonies on HU, as is visible in spot assays and by measuring the median area of colonies grown on 150 mM HU (Fig. S4 d–i). In addition to this dose-dependent increase in HU sensitivity, lineage L30 contains 2 dramatic stepwise increases in HU sensitivity, observable in strains L30–7C and L30–10C, that are likely caused by specific insertions of Ty1 elements that affect adjacent repair genes. In contrast, there seems to be little role for ongoing Ty1 transposition in the hypersensitivity of RO Ty1 strains, as deletion of the transcriptional transactivator SPT3, which profoundly reduces Ty1 transposition efficiency, does not diminish the differential sensitivity of the RO strains; whereas deletion of SPT3 increases the intrinsic sensitivity of the control and RO strains to HU treatment, the RO spt3 strains retain HU hypersensitivity relative to their parental counterparts (Fig. 2 D–F), with the L26–10C spt3 strain producing much smaller colonies than the WT spt3 strain (Fig. 2G). We conclude that sensitivity to DNA-damaging drugs results from the dispersed copies of Ty1 DNA throughout the genome.
Enhanced RO Damage Sensitivity in Replication Mutants.
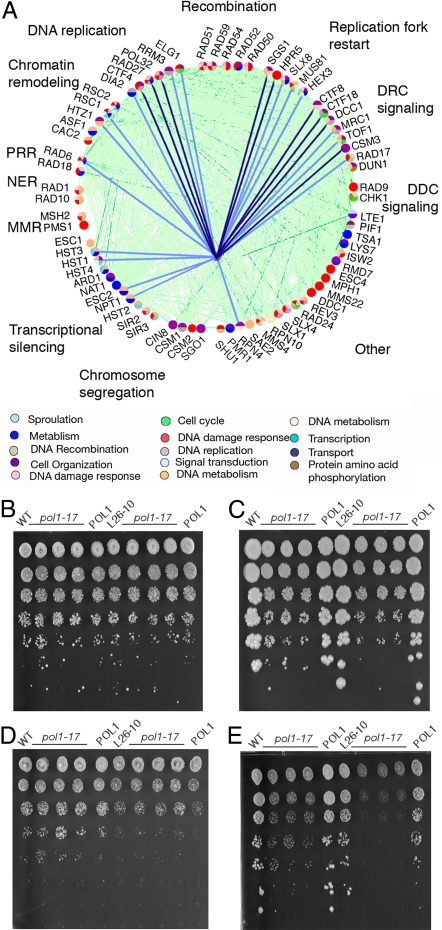
Because RO strains are sensitive to only a subset of DNA-damaging agents, it suggests that specific pathways may be required to cope with the genomic stress of high Ty1 copy number. To test whether interruption of these pathways would preferentially affect RO Ty1 strains, we screened for interactions between the RO state and mutations in specific DNA repair genes. Query gene deletion cassettes were transformed into the parental and 2 RO Ty1 strains (L26–10C and L30–10C) and screened for those that led to a specific growth defect of the 2 RO Ty1 strains relative to the WT on transformation. Despite the high degree of genetic interaction and the extensive network of synthetic lethality and fitness defects between members of these pathways, their differential effects on WT and RO strain growth were remarkably specific (Fig. 3A and Tables S3 and S4). Most notably, disruption of the genes that participate in DNA replication and DNA replication checkpoint signaling led to a specific reduction in the colony size and/or efficiency of plating of the RO strains. In contrast, other DNA repair pathways, including mismatch repair and DNA damage checkpoint signaling, were dispensable for stability of RO strains. Mutations affecting chromatin remodeling and transcriptional silencing genes had varied effects, with only a subset of gene deletions producing RO-specific growth defects. Importantly, deletion of genes required for homologous recombination had no effect on the RO strains, presumably because while DNA damage may be elevated, ectopic recombination frequency is reduced. It is possible that these mutations actually suppress the genome instability phenotype caused by the RO state, but we were unable to test this because the intrinsic damage sensitivity phenotypes of rad52 and rad51 strains are much more severe than the RO-induced sensitivity. Further, in many cases, the deletions also exacerbate the sensitivity of RO strains to DNA-damaging drugs, as would be expected from an elevation in the incidence of ectopic Ty1 recombination (Fig. 3A and Tables S3 and S4). For example, although deletion of DCC1, CTF8, or CTF18 affects the growth of the WT strain on HU, these deletions display much more dramatic effects on growth of L26–10C and L30–10C strains than the WT on HU, with no significant growth following extensive incubation (Fig. S5). Numerous host cell factors have been identified that repress Ty1 transposition, with deletion of particular DNA replication and repair proteins leading to increased retrotransposition, likely as a result of DNA damage-induced activation of the S-phase checkpoint (Fig. 3) (25). It is possible that deletion of the identified genes gives rise to RO-specific growth defects as a result of an increase in the transposition rate and the ectopic Ty1 recombination rate.
Fig. 3.
RO synthetic fitness interactions. (A) Nodes in the network diagram represent all the DNA replication and repair genes we assayed for genetic interaction with the RO Ty1 state and are colored according to GO biological process as annotated in the legend. Light green and dark green lines represent synthetic growth defects and synthetic lethality interactions, respectively, between cellular genes (from Yeast GRID database); light blue lines represent synthetic fitness interactions generated by deletion of cellular genes from RO strains (present study) whereas dark blue lines represent synthetic fitness interactions generated by deletion of cellular genes from RO strains that further result in HU supersensitivity. (B–E) Sensitivity of RO strains to a mutant allele of POL1. WT and an RO strain each carrying a WT or mutant pol1–17 allele were serially diluted and plated onto YPD (B and C) or YPD + 150 mM HU (D and E) and incubated at 22 °C for 3 d (B and D) or 30 °C for 5 d (C and E). Strains designated WT and L26–10C contain the WT POL1 allele, those designated pol1–17 contain the mutant allele, and those labeled POL1 contain the mutant allele as well as the pRS413-POL1 plasmid.
Polymerase Mutant Enhances RO Damage Sensitivity and Genome Rearrangement.
Because many of the genes we identified are involved in DNA replication, and because those yielding the most dramatic effects function in replication fork progression, we reasoned that slowing of the replication fork is a common mechanistic thread underlying the instability of the RO strains. This hypothesis is bolstered by the finding that cells with limited polymerase concentration suffer breaks at inverted Ty1 pairs (9). To directly test this hypothesis, we introduced a mutant allele of DNA polymerase-α (pol1–17) that has been demonstrated to affect the fidelity of replication of short repeats (26, 27) into both the WT and RO strains. There is little increased sensitivity of the L26–10C pol1–17 strain relative to the WT pol1–17 strain at the semi-permissive temperature of 30 °C (Fig. 3C). However, when these strains are also challenged with HU, the hypersensitivity of the RO strain relative to the WT is quite obvious (Fig. 3E), with reduced titers and colony size of L26–10C pol1–17 strains. These results confirm the increased susceptibility of RO Ty1 strains to replication stress.
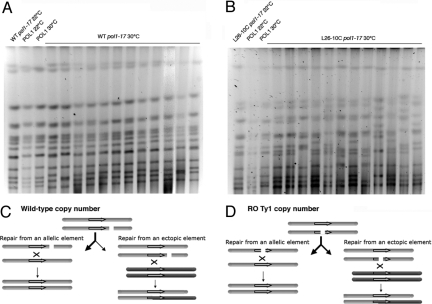
To directly demonstrate that repetitive DNA causes the formation of genomic rearrangements, the WT and an RO strain were simultaneously submitted to 2 transient replication stresses (i.e., reduced POL1 activity and HU treatment) and assayed for chromosomal rearrangements. For the parental pol1–17 strain, modest alterations in chromosome electrophoretic mobility are seen in clones obtained from 5 of 12 independent cultures, whereas in the RO strain L26–10C, there is evidence of massive chromosome instability in all strains following 2.5 h of replication stress (Fig. 4 A and B), with as many as 5 chromosomes migrating at novel positions in a single isolate (Fig. 4B, lane 12). By simply counting the number of chromosome bands that migrate in an altered position compared with the untreated control, we observe that there are two fold more changes in the RO strain than in the native strain, increasing to 3.7-fold when we except chromosome III, which commonly undergoes spontaneous length variations in conventional strains (28). The broad spectrum of changes to genome structure in RO strains confirms that dispersed retrotransposon DNA causes genome instability. The RO state increases the relative frequency of ectopic/allelic DNA recombination between Ty1s, presumably by increasing the frequency of breaks within Ty1 elements as a simple consequence of their increased copy number (Fig. 4 C and D) and by providing more potential recombination partners.
Fig. 4.
Chromosomal alterations in RO pol1–17 strains. (A and B) WT and RO strain L26–10C containing WT or mutant alleles of POL1 were held at 22 °C or shifted to 30 °C for 2.5 h in the presence of 200 mM HU; recovered isolates were examined for changes in chromosome structure by pulsed-field gel electrophoresis. Strains are labeled as in Fig. 3. (C and D) Model for generation of chromosomal translocations in RO strains. DNA lesions within chromatids (gray and red bars) are more likely to occur within Ty1 elements (purple arrows) in RO Ty1 strains (D) relative to a WT strain (C) as they comprise a greater fraction of the RO strain genome. These lesions can either be repaired from an allelic element (Left), regenerating the original chromosome structure, or from an ectopic Ty1 sequence (Right) located intra- or inter-chromosomally and resulting in the formation of deletions, inversions, and translocations.
Discussion
We have previously reported that doubling the copy number of Ty1 retrotransposons has no significant deleterious effects to cells, suggesting that the maximum sustainable burden of Ty1 elements had not yet been reached (20). However, increasing the number of Ty1 elements to as many as 10 times the WT copy number is achievable, with no gross defects in colony size or morphology after 10 cycles of transposition. Despite the ability to load the genome with Ty1 elements, this increase in copy number leads to a defect when strains are challenged with DNA-damaging agents, and all RO strains—although to somewhat variable degrees—are hypersensitive to certain DNA-damaging compounds. The sensitivity of RO strains to a subset of DNA-damaging agents pointed to specific pathways that may be necessary to maintain chromosomes replete with repetitive DNA, including the DNA replication pathway (Fig. 3) and the MRC1, CSM3, TOF1, CTF18, CTF8, DCC1, RAD27, and POL32 genes that promote replication fork progression (29, 30). The identification of these proteins is consistent with the sensitivity of RO strains specifically to MMS, an alkylating agent that produces DNA lesions that slow replication rates (31), and to HU, which depletes intracellular dNTP pools, leading to replication fork collapse (11).
Our results therefore support the assertion that repetitive DNA becomes frankly deleterious to chromosome integrity when cells are faced with the abrogation of normal DNA replication and the associated double strand break repair pathways (Fig. 4 A and B), with several possible mechanisms. It seems unlikely that a replication delay resulting from an increase in the total amount of DNA would account for the DNA damage sensitivity, as a strain carrying the high-copy pJEF2631-Ty1 plasmid displays no effects (Fig. 2). The most parsimonious explanation is that increasing the number of Ty1 elements simply increases the likelihood that stalling or collapse of the replication fork will occur within the Ty1 sequence as a result of its predominance within the genome (RO strains increase Ty1 load from 1.6% of the genome as full-length elements to as much as 15%). This is consistent with our observation that the response to DNA damage roughly parallels the copy number of Ty1 elements (Fig. S4).
As an alternative to this model, the dispersed retrotransposon copies may themselves affect the rate of replication fork progression. Indeed, regions of the genome that slow replication fork progression are apparent sites of double strand breaks, replication fork collapse, and chromosomal rearrangements in cells with a temperature-sensitive mec1 allele (32, 33). Repetitive DNA itself may serve as an impediment to fork progression, as both the ribosomal DNA (29, 34) and inverted pairs of Ty1 elements are prone to recombinational instability (9, 35, 36). We have demonstrated the presence of an elevated number of tandem Ty1 integrations in our high-copy strains (Fig. S2), and it is therefore reasonable to assume that the increased number of these DNA structures may specifically affect the orderly progression of DNA replication. Further, a subset of tRNA genes—by far the predominant integration sites of Ty1 retrotransposons, even in RO strains (37)—contain a unidirectional block to fork progression (38) at which DNA replication stress leads to chromosomal instability and non-allelic recombination at the closely linked Ty or LTR elements (10).
Regardless of the mechanism by which lesions are generated during DNA replication, cells that contain high levels of repetitive DNA, such as the RO strains, can amplify the effect by increasing the number of genetic loci available to participate in an ectopic event, thereby further increasing the chance of genome alteration, potentially exponentially (Fig. 4 C and D). In our experimental system, the Ty1 elements are all of nearly identical sequence, further enhancing this effect of increased inter-element recombination more than would be apparent in an evolved genome with more Ty element sequence diversity.
We have shown that simply increasing retrotransposon abundance is itself detrimental to the fitness and stability of the yeast genome, even in the absence of ongoing retrotransposition. The ability of higher eukaryotic cells to maintain levels of repetitive DNA at much higher levels than in yeast cells underscores the importance of highly efficient and precisely controlled DNA replication and error prevention machinery to safeguard against the deleterious potential of such expansion. Yet the persistence of retrotransposon DNA, despite the inherent risk posed by its maintenance, suggests that the resulting genome alterations confer an important and ongoing underlying evolutionary benefit to cells, driving adaptation and evolution of the genome.
Methods
Media, Strains, and Plasmids.
Yeast strains were propagated at 30 °C in YPD medium, prepared as described (39). Minimal (SD) medium (39) was supplemented with 0.2 mM uracil, 0.3 mM histidine, and 1.2 mM methionine. To assay sensitivity to DNA-damaging drugs, strains were grown to saturation in either YPD or SC–His (to select for the pJEF2631 or pJEF2631-Ty1 plasmids). Cell densities were normalized by OD600, and fivefold serial dilutions were plated onto freshly prepared YPD or minimal plates containing either the drug or DMSO as a control and incubated at 30 °C for 2 to 5 d.
RO strains were derivatives of GRF167 (MATα, ura3–52, his3Δ200) and were constructed as previously described with either plasmid unmarked pGTy1-H3 (pJEF724) or control empty vector pGal1-XhoI (pCGE329); induction was performed on SD plates containing 2% (wt/vol) casamino acids and 2% (wt/vol) galactose, a formulation that produced a greater number of transposition events than previously described (20). By using the casamino acid medium in conjunction with an unmarked Ty1, we boosted retrotranspositions per cycle by approximately three fold. Following each cycle of induction, strains were colony purified both on SC–Ura to retain the plasmid for the next cycle of galactose induction and on YPD to allow plasmid loss. Colony purified isolates from each round that were confirmed to have lost the plasmid were used for all experiments and were frozen in 15% glycerol at −80 °C so that L26–5, for example, represents the isolate of lineage 26 that went through 5 cycles of galactose induction and L26–5C represents that same isolate that was cured of the donor plasmid (Table S5). Well isolated colonies were picked from the center of the plate to minimize bias toward the selection of large colonies and were screened on YPGE medium to ensure that they were not petite. The pol1–17 allele (26, 27) was introduced by transforming WT and RO strains with an XhoI linearized pJH1138 plasmid (26), colony purification on SC–Ura, patching onto YPD for 2 d at 22 °C and then onto SC + 5-FOA for 5 d at 22 °C, and colony purification of papillae on YPD medium. Temperature-sensitive isolates were transformed with the pRS413-POL1 plasmid carrying the WT sequence to verify that growth was restored at the restrictive temperature.
Plasmid pJEF2631 will be described in detail elsewhere; a plasmid map is available on request. Its structure is similar to a plasmid previously described by Ludwig and Bruschi (40) called pBH-2L in that it contains an intact copy of 2 μm plasmid; it consists of the HIS3 vector pRS403 inserted within the unique HpaI site of native 2 μm circle. The derivative pJEF2631-Ty1 was constructed by subcloning a BamHI fragment of pGN821 into pJEF2631. Plasmid pRS413-POL1 was created by PCR amplification of the POL1 locus with primers JB9854 (catttgaccgcggTTTGAGAAGGTTCAGAAAGAATAAAAT) and JB9855 (catttgacccgggGTCACCTCGAAAGCAAGAGC), digestion with SacII and SmaI, and ligation into pRS413.
Supplementary Material
Acknowledgments.
We thank members of the Boeke laboratory and Douglas Koshland for valuable comments and advice, Maitreya Dunham for array-CGH analysis, and David Pai for technical support. We thank James Haber for sharing yeast strains and plasmid constructs. Supported in part by National Institutes of Health Grant GM36481 (to J.D.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906552106/DCSupplemental.
References
- 1.Pan X, et al. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 2.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 3.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi T. Strategies to maintain the stability of the ribosomal RNA gene repeats-collaboration of recombination, cohesion and condensation. Genes Genet Syst. 2006;81:155–161. doi: 10.1266/ggs.81.155. [DOI] [PubMed] [Google Scholar]
- 5.Lenzmeier BA, Freundereich CH. Trinucleotide repeat instability: a hairpin curve at the crossroads of replication, recombination, and repair. Cytogenet Genome Res. 2003;100:7–24. doi: 10.1159/000072836. [DOI] [PubMed] [Google Scholar]
- 6.Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 7.Boeke JD, Sandmeyer SB. Yeast transposable elements. In: Broach J, Jones E, Pringle J, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1991. pp. 193–261. [Google Scholar]
- 8.Lesage P, Todeschini AL. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet Genome Res. 2005;110:70–90. doi: 10.1159/000084940. [DOI] [PubMed] [Google Scholar]
- 9.Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 10.Admire A, et al. Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes Dev. 2006;20:159–173. doi: 10.1101/gad.1392506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 12.Rachidi N, Barre P, Blondin B. Multiple Ty-mediated chromosomal translocations lead to karyotype changes in a wine strain of Saccharomyces cerevisiae. Mol Gen Genet. 1999;261:841–850. doi: 10.1007/s004380050028. [DOI] [PubMed] [Google Scholar]
- 13.Fischer G, James SA, Roberts IN, Oliver SG, Louis EJ. Chromosomal evolution in Saccharomyces. Nature. 2000;405:451–454. doi: 10.1038/35013058. [DOI] [PubMed] [Google Scholar]
- 14.Umezu K, Hiraoka M, Mori M, Maki H. Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics. 2002;160:97–110. doi: 10.1093/genetics/160.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koszul R, Caburet S, Dujon B, Fischer G. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004;23:234–243. doi: 10.1038/sj.emboj.7600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams J, Oeller PW. Structure of evolving populations of Saccharomyces cerevisiae: adaptive changes are frequently associated with sequence alterations involving mobile elements belonging to the Ty family. Proc Natl Acad Sci USA. 1986;83:7124–7127. doi: 10.1073/pnas.83.18.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunham MJ, et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kupiec M, Petes TD. Allelic and ectopic recombination between Ty elements in yeast. Genetics. 1988;119:549–559. doi: 10.1093/genetics/119.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garfinkel DJ, Nyswaner KM, Stefanisko KM, Chang C, Moore SP. Ty1 copy number dynamics in Saccharomyces. Genetics. 2005;169:1845–1857. doi: 10.1534/genetics.104.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boeke JD, Eichinger DJ, Natsoulis G. Doubling Ty1 element copy number in Saccharomyces cerevisiae: host genome stability and phenotypic effects. Genetics. 1991b;129:1043–1052. doi: 10.1093/genetics/129.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilke CM, Adams J. Fitness effects of Ty transposition in Saccharomyces cerevisiae. Genetics. 1992;131:31–42. doi: 10.1093/genetics/131.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji H, et al. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- 23.Cost GJ, Boeke JD. A useful colony colour phenotype associated with the yeast selectable/counter-selectable marker MET15. Yeast. 1996;12:939–941. doi: 10.1002/(SICI)1097-0061(199608)12:10%3C939::AID-YEA988%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 24.Hicks J, Strathern J, Klar A, Ismail S, Broach J. Structure of the SAD mutation and the location of control sites at silent mating type genes in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1278–1285. doi: 10.1128/mcb.4.7.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes AM, Haber JE. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 27.Gutierrez PJ, Wang TS. Genomic instability induced by mutations in Saccharomyces cerevisiae POL1. Genetics. 2003;165:65–81. doi: 10.1093/genetics/165.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wicksteed BL, et al. A physical comparison of chromosome III in six strains of Saccharomyces cerevisiae. Yeast. 1994;10:39–57. doi: 10.1002/yea.320100105. [DOI] [PubMed] [Google Scholar]
- 29.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bermudez VP, et al. The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc Natl Acad Sci USA. 2003;100:10237–10242. doi: 10.1073/pnas.1434308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 32.Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- 33.Cobb JA, et al. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 2005;19:3055–3069. doi: 10.1101/gad.361805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brewer BJ, Fangman WL. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 35.McMurray MA, Gottschling DE. An age-induced switch to a hyper-recombinational state. Science. 2003;301:1908–1911. doi: 10.1126/science.1087706. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wheelan SJ, Scheifele LZ, Martinez-Murillo F, Irizarry RA, Boeke JD. Transposon insertion site profiling chip (TIP-chip) Proc Natl Acad Sci USA. 2006;103:17632–17637. doi: 10.1073/pnas.0605450103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- 39.Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1986. [Google Scholar]
- 40.Ludwig DL, Bruschi CV. The 2-micron plasmid as a nonselectable, stable, high copy number yeast vector. Plasmid. 1991;25:81–95. doi: 10.1016/0147-619x(91)90019-s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.