Abstract
Gene profiling techniques allow the assay of transcripts from organs, tissues, and cells with an unprecedented level of coverage. However, most of these approaches are still limited by the fact that organs and tissues are composed of multiple cell types that are each unique in their patterns of gene expression. To identify the transcriptome from a single cell type in a complex tissue, investigators have relied upon physical methods to separate cell types or in situ hybridization and immunohistochemistry. Here, we describe a strategy to rapidly and efficiently isolate ribosome-associated mRNA transcripts from any cell type in vivo. We have created a mouse line, called RiboTag, which carries an Rpl22 allele with a floxed wild-type C-terminal exon followed by an identical C-terminal exon that has three copies of the hemagglutinin (HA) epitope inserted before the stop codon. When the RiboTag mouse is crossed to a cell-type-specific Cre recombinase-expressing mouse, Cre recombinase activates the expression of epitope-tagged ribosomal protein RPL22ha, which is incorporated into actively translating polyribosomes. Immunoprecipitation of polysomes with a monoclonal antibody against HA yields ribosome-associated mRNA transcripts from specific cell types. We demonstrate the application of this technique in brain using neuron-specific Cre recombinase-expressing mice and in testis using a Sertoli cell Cre recombinase-expressing mouse.
Keywords: gene profiling, immunopreciptation, mouse genetics
Quantitative genomic and proteomic approaches in mammals are challenging due to the fact that mammalian tissues are made up of multiple unique cell types with widely divergent RNA and protein expression profiles. Some of the difficulty in analyzing transcriptomes from complex tissues has been met by taking advantage of transgenic techniques to drive expression of epitope tags and/or reporter genes such as green fluorescent protein (GFP) in specific cell types in complex tissues, thus allowing investigators to identify cells of interest. These marked cells can be isolated from a complex organ such as brain using manual dissection, enzymatic dissociation combined with fluorescence-activated cell sorting (FACS), or laser capture microdissection (LCM) (1–7).
Although these technologies provide a mechanism for isolating specific cell types from a complex mixture, they present a number of obstacles to successful isolation of cell-type-specific transcripts. With manual dissection or enzymatic dissociation combined with FACS, the three-dimensional relationship of multiple cell types present in a tissue must be completely disrupted, often via enzymatic dissociation, and this may lead to changes in gene and protein expression (5–7). In the case of LCM, cells are removed from their in situ environment, but the tissue must be sectioned, processed, and dehydrated; furthermore, in cells with very complex anatomy such as astrocytes and neurons in the brain, only the cell bodies are captured, leaving dendritic and axonal mRNAs behind.
We have created a line of mice carrying an epitope-tagged ribosomal protein allele that can be activated in specific cell types by mating to a Cre recombinase-expressing mouse. Conditional activation of this allele leads to epitope-tagged polyribosomes that can be purified from the target cell population via immunoprecipitation. Messenger RNA transcripts found in the target cell population can be isolated from the immunoprecipitated polyribosomes and analyzed using standard genomic profiling technologies such as quantitative RT-PCR (qRT-PCR) or microarray. The advantages of this approach are the widespread availability of well-characterized Cre recombinase-expressing mouse lines and the robust expression of the epitope-tagged ribosomal protein subunit from the endogenous allele.
Results
Selection of Ribosomal Proteins for Epitope Tagging and Design of the Targeting Vector.
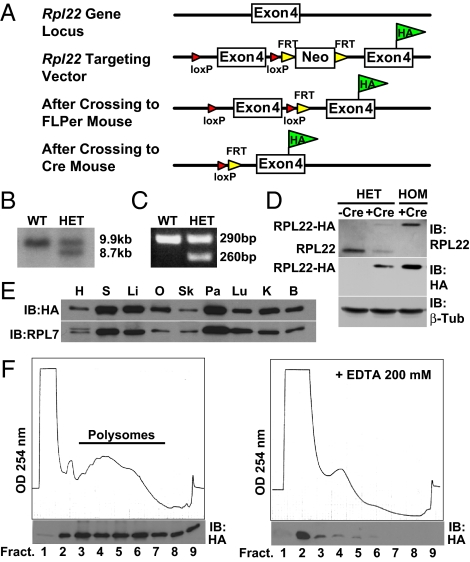
Approximately 80 proteins form the ribosome particle (8). Based on examination of large-scale yeast and mammalian protein tagging studies (9, 10), four core ribosome protein subunits (RPS17, RPS25, RPL22, and RPL36) that localized to the appropriate cellular compartments (nucleolus and cytoplasm) when C-terminally tagged with GFP were identified. Pilot studies expressing mouse cDNAs in HeLa cells established that all four proteins, when tagged with HA, were incorporated into polyribosomes. From this group we chose to target the Rpl22 gene. A 9.8-kb genomic fragment was isolated from a bacterial artificial chromosome (BAC) containing the Rpl22 gene and used to construct the targeting vector as described in SI Materials and Methods. The targeted allele consists of a duplicated HA-tagged exon 4 that is preceded by the wild-type exon 4, which is flanked by loxP sites as shown in Fig. 1A and Fig. S1. Southern blotting and PCR analysis was used to identify correctly targeted embryonic stem (ES) cell clones and genotype offspring from chimeras (Fig. 1 B and C). The targeting vector is designed to express wild-type RPL22 protein in the absence of Cre recombination and after Cre recombination to express the RPL22ha protein (Fig. 1D).
Fig. 1.
Targeting the Rpl22 genomic locus. (A) Targeting strategy for the genomic locus of Rpl22. A loxP site was inserted 5′ to the wild-type exon 4. A loxP-FRT-neomycin resistance-FRT cassette was inserted 3′ to the wild-type exon 4 followed by a modified Rpl22 exon 4 containing the HA epitope tag inserted before the Rpl22 stop codon. F1 heterozygous offspring were bred to FLPeR mice to remove the neomycin cassette used for selection in the ES cells. Crossing the RiboTag mouse to a Cre recombinase-expressing mouse results in deletion of the wild-type exon 4 in the target cell population and replacement with the Rpl22HA exon 4. (B) Southern blot strategy used to identify correctly targeted ES cells. The wild-type HindIII/SfiI DNA fragment is 9,908 bp, while the correctly targeted HindIII/SfiI fragment is 8,749 bp. (C) PCR products using oligonucleotides that amplify the loxP-containing intron sequence 5′ to the wild-type exon 4 of the Rpl22 gene. The wild-type PCR product is 260 bp, while the mutant PCR product is 290 bp. (D) Western analysis of wild-type RPL22 or RPL22ha in a control Meox2+/+:Rpl22ha/+ mouse, a double heterozygote Meox2Cre/+:Rpl22ha/+ mouse, and a Rpl22ha-expressing homozygous mouse. Blots were probed with anti-RPL22 and anti-HA antibodies. RPL22ha results in a 23-kDa protein, while the native RPL22 is a 15-kDa protein. (E) Western blot using tissue homogenates from a Rpl22ha-expressing homozygous mouse. The blot was probed with anti-HA antibody and then reprobed with anti-RPL7 antibody. H, heart; S, spleen; Li, liver; O, ovary; Sk, skeletal muscle; Pa, pancreas; Lu, lung; K, kidney; B, brain. (F) A254 absorbance profile of 15% to 50% sucrose density gradients from a Rpl22ha-expressing homozygous mouse brain in the absence (left) or presence (right) of 200 mM EDTA. Western blot analysis (below each profile) of trichloroacetic acid-precipitated fractions probed with anti-HA antibody.
Expression of RPL22ha Protein in Mouse Tissues.
To test the functionality of the Rpl22ha allele, we first crossed the RiboTag mouse to a mesenchyme homeobox 2 (Mox2, Meox2)-Cre recombinase-expressing mouse to activate the Rpl22ha allele in all adult tissues. Mox2-Cre:RiboTag offspring were identified via PCR using primers to Cre recombinase and primers flanking the loxP site located 5′ to the wild-type exon 4 (see SI Materials and Methods). Offspring from Mox2-Cre:RiboTag mice that expressed the floxed Rpl22ha allele in epiblast-derived tissue, including germ cells, were bred with each other to produce RPL22ha mice that express only HA-tagged RPL22 protein in all tissues. Using antibodies against the wild-type RPL22 protein or the HA epitope, we demonstrate the expected expression of wild-type RPL22 protein, which runs at 15 kDa, or RPL22ha protein, which runs at 23 kDa, in brain homogenates from control mice and mice expressing one or both Rpl22ha alleles (Fig. 1D). Western blotting across a panel of tissues from Rpl22ha-expressing homozygous mice demonstrates that RPL22ha protein is stably expressed in all tissues. Differences in RPL22ha protein levels between tissues appear to be related to varying ribosome content in each tissue as assessed by western analysis of another large-subunit ribosomal protein, RPL7 (Fig. 1E). Rpl22ha-expressing homozygous mice, which express only the RPL22ha protein, appear normal and are fertile.
RPL22ha Protein Is Incorporated into Polyribosomes.
Having demonstrated that the RPL22ha protein is stable, it was critical to demonstrate incorporation of RPL22ha into polyribosomes in vivo. Thus, sucrose density gradients were performed using brain homogenates from Rpl22ha-expressing homozygous mice followed by Western blot analysis of the fractions. As shown in Fig. 1F, RPL22ha protein is primarily found in the polysome fractions, with very little protein found in the first fraction of the gradient (cytoplasmic). Treatment of brain polysome homogenates with EDTA, which disassembles the polyribosomes, results in a shift of the RPL22ha protein into the lighter fractions (Fig. 1F), as expected for a polysome-associated protein. Brain polysome homogenates were also treated with low concentrations of RNase (10 ng/mL), which cleaves ribosome-associated mRNA transcripts between ribosomes while leaving the ribosome particle intact (11), and this also resulted in a majority of RPL22ha protein moving to the monoribosome peak.
The RPL22ha C-Terminal Hemagglutinin Tag Can Be Used To Immunoprecipitate Polysomes.
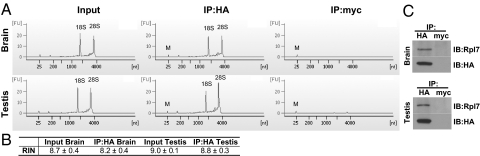
Fig. 2 is a cartoon that outlines the RiboTag methodology. Crossing the RiboTag animal to a Cre recombinase-expressing mouse line results in deletion of the wild-type exon 4 and replacement with the HA-tagged exon 4 only in cells that express Cre recombinase. The tissue containing cells that express RPL22ha-tagged ribosomes is homogenized, and HA antibody-coupled magnetic beads are added to the cleared homogenate. After an overnight incubation at 4 °C, the magnetic beads containing immunoadsorbed polysomes are washed with a high salt buffer, and the RNA extracted. To preserve intact RNA during the prolonged manipulations, RNasin and heparin are present to control RNase activity. An Agilent Technologies 2100 Bioanalyzer was used to assess the integrity of purified RNA samples obtained after overnight incubation of Rpl22ha-expressing homozygote brain or testis homogenates with anti-HA or control anti-Myc antibodies. The electropherograms in Fig. 3A demonstrate intact 18S and 28S ribosomal RNA using antibodies to the HA epitope. Immunoprecipitation with control Myc antibodies resulted in undetectable levels of 18S and 28S ribosomal RNA. The RNA integrity number (RIN) values (Fig. 3B), which reflect the quality of the rRNA in the analyzed samples (12), range from 8.0 to 9.2 for testis and brain samples, suggesting that the RNA obtained from the immunoprecipitation procedure is of sufficient quality for all standard genomic approaches, including microarray, qRT-PCR, and RNA-seq analysis. Western blot analysis of proteins immunoprecipitated with anti-HA- or anti-Myc-coated beads showed that the RPL22ha was only detectable in the HA-IP and that another ribosomal protein (RPL7) was also co-immunoprecipitated as expected (Fig. 3C).
Fig. 2.
A cartoon that outlines the RiboTag methodology. The RiboTag mouse is crossed to any available Cre recombinase-expressing mouse line, which results in the deletion of wild-type exon 4 and replacement with the HA-tagged exon 4 only in cells that express Cre recombinase. The cells within the tissue or organ that now express RPL22ha-tagged ribosomes are homogenized, and HA antibody-coupled magnetic beads are added to the cleared homogenate. After an overnight incubation at 4 °C, the magnetic beads that have immunoadsorbed polysomes are washed with a high salt buffer before extraction of the mRNA transcripts, which can then be analyzed by qRT-PCR, microarray, or RNA-seq.
Fig. 3.
Immunoprecipitation of polysomes from Rpl22ha-expressing homozygous mouse brain and testis. (A) Agilent Technologies 2100 Bioanalyzer electropherogram analysis of total RNA from brain and testis immunoprecipitates. (B) RIN values from Agilent Technologies 2100 Bioanalyzer analysis demonstrating recovery of high quality RNA from the pellets (RIN values >8.0). (C) Western blots using anti-HA antibody and anti-RPL7 antibody demonstrating the presence of RPL7 (a component of the large subunit of the ribosome) specifically in anti-HA versus anti-Myc pellets.
Enrichment of Transcripts from Specific Cell Populations.
The RiboTag mouse was crossed to a variety of Cre recombinase-expressing mouse lines, including knock-in mice expressing Cre recombinase under the control of the dopamine transporter (DAT, Slc6a3) promoter, or transgenic mice expressing Cre under the control of the dopamine- and cAMP-regulated phosphoprotein (DARPP-32, Ppp1r1b) or the anti-mullerian hormone (Amh) promoters. In each case, we observed robust expression of the RPL22ha protein in specific cell types, including DAT-expressing dopaminergic neurons, DARPP32-expressing medium spiny neurons of the striatum, and AMH-expressing Sertoli cells in the testis (Fig. 4A). As expected, the level of RPL22ha protein expression depends on the abundance of the cell type targeted (Fig. 4B); after loading equivalent amounts of input protein, RPL22ha protein was detected at low levels in DAT-Cre:RiboTag mouse brain homogenates, intermediate levels in DARPP32-Cre:RiboTag mouse brain homogenates, and higher levels in AMH-Cre:RiboTag mouse testis homogenates. Messenger RNA transcripts associated with tagged polysomes were immunoprecipitated using extracts from these HA-expressing tissues and the enrichment of cell-specific transcripts was determined. Although it would be possible to enrich significantly for the DAT-Cre or DARPP32-Cre expressing neurons by manually dissecting out the appropriate brain regions, total brain was used for these test examples. In Fig. 4C, the relative efficiency of RPL22ha protein immunoprecipitation using the anti-HA antibody in homogenates from brain and testis is demonstrated. In all three cases, the efficiency of immunoprecipitation did not change significantly whether the HA-expressing cells make up a substantial fraction of the total (AMH-Cre:RiboTag and DARPP32-Cre:RiboTag) or only a very small fraction (DAT-Cre:RiboTag). The overall efficiency of immunoprecipitating the RPL22ha was about 25% in each case. The control Myc antibody did not precipitate any detectable RPL22ha, indicating a low background.
Fig. 4.
Enrichment of transcripts expressed in dopaminergic neurons (DAT-Cre:RiboTag), medium spiny neurons of the striatum (DARPP32-Cre:RiboTag), and Sertoli cells of the testis (AMH-Cre:RiboTag) using the RiboTag technology. (A) Immunohistochemistry on 25-μm brain and testis sections using anti-HA antibody and marker antibodies to proteins known to be expressed in the cell type or area under study. (Top panels) Immunohistochemistry on DAT-Cre:RiboTag mouse brain. Anti-HA staining is shown in green and anti-tyrosine hydroxylase (TH) staining is shown in red. Merged image shows the presence of both TH and HA staining in the same cell types. (Middle panels) Immunohistochemistry on DARPP32-Cre:RiboTag mouse brain. Anti-HA staining is shown in green and anti-TH staining is shown in red. TH was used as a striatal marker. Merged image shows striatal localization of the HA staining. (Bottom panels) Immunohistochemistry on AMH-Cre:RiboTag mouse testis. Anti-HA staining is shown in green and TO-PRO-3 (Invitrogen) nuclear staining is shown in blue. (B) Anti-HA Western blots using 10% weight per volume homogenates of DAT-Cre:RiboTag brain, DARPP32-Cre:RiboTag brain, and AMH-Cre:RiboTag testis demonstrating varying levels of RPL22ha protein depending on the Cre recombinase-expressing mouse line used. (C) Western blots after immunoprecipitation of brain and testis homogenates using anti-HA or control anti-Myc antibodies and demonstrating specific immunoprecipitation of RPL22ha protein in HA versus Myc pellets. Two percent of input (I) and supernatant (S) samples, and 4% of pellet (P) samples were loaded on the gel. (D) qRT-PCR analysis of transcripts expressed in dopaminergic neurons, medium spiny neurons of the striatum, and Sertoli cells of the testis after immunoprecipitation of polysomes from DAT-Cre:RiboTag brain, DARPP32-Cre:RiboTag brain, and AMH-Cre:RiboTag testis. Total RNA was isolated from input and from anti-HA or control anti-Myc pellets after overnight immunoprecipitation of polysomes from 10% weight per volume homogenates of brain or testis. All cell-specific marker genes and control genes not expressed in the target cells were normalized to beta-actin (Actb) levels using the ΔΔCt method. The immunoprecipitated RNA samples were compared to the input sample in each case. The transcripts analyzed by qRT-PCR were: Tyrosine hydroxylase (Th); dopamine transporter (DAT, Slc6a3); 2′,3′-cyclic nucleotide 3′ phosphodiesterase (CNPase, Cnp); dopamine- and cAMP-regulated phosphoprotein (DARPP-32, Ppp1r1b); prodynorphin (Pdyn); proenkephalin (Penk1); follicle-stimulating hormone receptor (Fshr); transferrin (Trf); inhibin beta-B (Inhbb); protamine 2 (Prm2). ***, P < 0.001; **, P < 0.01. Values are the mean ± SEM of three independent experiments.
Fig. 4D shows the enrichment of cell-type-specific mRNAs in the immunoprecipitate compared to the input RNA. In the DAT-Cre:RiboTag mice, a 10- to 15- fold enrichment for transcripts expressed in dopaminergic neurons of the midbrain, including tyrosine hydroxylase (Th) and the dopamine transporter (DAT, Slc6a3), was observed. Conversely, a statistically significant depletion of transcripts such as 2′,3′-cyclic nucleotide 3′ phosphodiesterase (CNPase, Cnp), a marker of oligodendrocytes (13), was not seen. We hypothesize that the lack of depletion of CNPase in DAT-Cre immunoprecipitates has to do with the ratio of signal-to-noise. Because dopaminergic neurons represent a much smaller fraction of total cells in the brain compared to striatal medium spiny neurons, the relative signal-to-noise (background) is much lower in DAT-Cre versus DARPP32-Cre whole brain; this is reflected in the lack of depletion of beta-actin-normalized CNPase compared to the input sample. As the percentage of mRNA transcripts specific to HA-tagged cells increases (as is the case for DARPP32-Cre), there is an increase in the signal-to-noise ratio, which results in de-enrichment for mRNA transcripts that are not expressed in the HA-tagged cells, including CNPase, which is specific to oligodendrocytes. In DARPP-32-Cre:RiboTag mice, a 5- to 6-fold enrichment of transcripts expressed in medium spiny neurons of the striatum, including DARPP32 (Ppp1r1b) itself, prodynorphin (Pdyn, expressed in the striatonigral neurons), and proenkephalin (Penk1, expressed in the striatopallidal neurons) was observed. In this case, a depletion of the transcript for CNPase, which is not expressed in medium spiny neurons, was observed. In the AMH-Cre:RiboTag mice, a 5-fold enrichment for transcripts expressed in Sertoli cells, including follicle stimulating hormone receptor (Fshr), transferrin (Trf), and inhibin beta-B (Inhbb) was observed. In this case, a 10-fold depletion of protamine 2 (Prm2) transcripts that are not expressed in Sertoli cells and are specific to round and elongating spermatids (14) was also observed, indicating that 90% of the immunoprecipitated mRNA is from Sertoli cells.
Distinguishing Between Ribosome and Non-Ribosome Associated mRNAs.
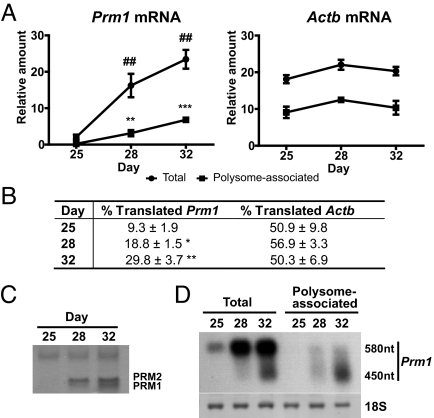
The RiboTag methodology allows us to capture mRNA transcripts associated physically with the ribosome; however, pools of translationally repressed mRNA transcripts are present in many cell types (15, 16) and would most likely be sequestered away from ribosomes. The expression of proteins involved in sperm DNA compaction during sperm development is a well-established example of translational control (14). During the first wave of spermatogenesis, protamine 1 (Prm1) mRNA is transcribed but kept translationally repressed by Y-box binding proteins until several days later in development. Protamines are specific to developing spermatids and Prm1 mRNA is transcribed in round spermatids and then first translated in elongating spermatids. To determine whether the RiboTag approach can distinguish between translationally repressed versus translationally active mRNA transcripts, the levels of total and polysome-associated Prm1 mRNA were measured during the first wave of spermatogenesis. Testes from Rpl22ha-expressing homozygous mice were isolated at postnatal day 25, 28, and 32, then frozen for later analysis; frozen testis were homogenized, and polysomes immunoprecipitated using anti-HA-coupled magnetic beads to obtain the polysome-associated transcripts at these different time points. qRT-PCR analysis using a Taqman probe specific to the murine Prm1 mRNA (Fig. 5A) revealed that Prm1 mRNA begins to increase at day 25 and is near maximal at day 28. However, polysome-associated Prm1 mRNA as determined by assay of the anti-HA immunoprecipitated transcripts is only 9% of total Prm1 mRNA at day 25 and 18.8% of total at day 28. At day 32, the percent of Prm1 mRNA in polysomes increases to 30% (Fig. 5B). In contrast, no significant change in the levels of polysome-associated beta-actin (Actb) mRNA was observed at the different time points (Fig. 5 B and C). Additionally, PRM1 protein content in nuclear basic protein extracts was determined for the three developmental time points (Fig. 5C). Coomassie brilliant blue staining of acid-urea gels revealed increasing amounts of PRM1 protein starting at postnatal day 28; no significant levels of PRM1 protein were detected at postnatal day 25. Partial deadenylation of Prm1 mRNA has been observed when Prm1 mRNA moves from the Y-box-protein-bound translationally repressed state to the translationally active state in polysomes (17). Thus, the 580 nt Prm 1 mRNAs are found in the nonpolysomal compartment, while the 450 nt Prm1 mRNAs are enriched in polysomes. To confirm the presence of these partially deadenylated transcripts in the polysome-associated fraction, Northern blot analysis was performed using both total and anti-HA immunoprecipitated RNA. Northern blot analysis revealed the presence of nondeadenylated and deadenylated forms in the total RNA samples, while deadenylated forms were preferentially found in the polysome-associated samples (Fig. 5D).
Fig. 5.
Immunoprecipitation of actively translated Prm1 mRNA from Rpl22ha-expressing homozygous mouse testis. (A) qRT-PCR Taqman assay using a probe specific for Prm1 mRNA comparing input (total RNA) and HA-immunoprecipitated (polysome-associated RNA) from P25, P28, and P32 Rpl22ha-expressing homozygous mouse testis homogenates. qRT-PCR analysis of beta-actin (Actb) transcripts was performed as a control. Values are the mean ± SEM of three independent experiments. (B) Table showing the percentage of polysome-associated transcripts compared to total transcripts (% Translated) of Prm1 and Actb at each time point. (C) Coomassie Brilliant Blue staining showing basic proteins isolated from nuclear pellets of P25, P28, and P32 Rpl22ha-expressing homozygous mouse testis. Samples were resolved on 15% acrylamide acid-urea gels. (D) Northern blot analysis revealing the predominance of partially deadenylated Prm1 transcripts in the polysome-associated samples.
Discussion
To address the ongoing challenge in defining transcriptional and translational changes in specific cell populations in complex tissues, we developed a method to tag ribosomes in specific cell types using Cre-loxP-dependent recombination. The usefulness of the RiboTag approach was demonstrated through isolation of cell-specific transcripts from dopaminergic and striatal neurons starting with whole brain homogenates. We also specifically isolated Sertoli cell mRNA from whole testis using the RiboTag mouse.
The Heintz laboratory has recently reported a similar approach (18, 19). These authors created a series of bacTRAP transgenic mice to drive expression of an EGFP-tagged ribosomal protein in specific neuronal cell types, which then allows the use of immunoprecipitation techniques to isolate polysome-bound mRNA. There are several advantages to the approach we have developed that make it more generally applicable. We have modified the endogenous Rpl22 gene, and therefore the expression of the ribosomal protein is high and proportional to the ribosomal content of the cell, whereas the bacTRAP technique depends on expression levels from cell-type-specific promoters that are quite variable in transcriptional activity and generally much lower than competing ribosomal protein promoters. A second major advantage is that the RiboTag methodology takes advantage of the full range of Cre recombinase-expressing mouse lines that have already been created, including knock-in Cre-expressing mice that may more accurately replicate the endogenous cell-type specificity of particular genes. By using the RiboTag technology, we also avoid concerns about random insertion of the BAC construct into the mouse genome and the observation that BACs often contain additional intact genes that may produce phenotypes and complicate interpretation of the data (19).
We designed the RiboTag mouse such that wild-type RPL22 would be synthesized in the absence of Cre recombination, ensuring that the animals would not have a deficiency in RPL22 that could lead to phenotypes. However, recently an Rpl22 knockout mouse was made that is viable and fertile with only a minor phenotype in the development of a subset of T cells (20). The viability of these mice is likely due to the existence of a homolog, Rpl22-Like 1, which is expressed in all tissues examined together with Rpl22 (B. Kennedy and M. Stanfel, personal communication). Because a deficiency in T cell development in Rpl22ha-expressing homozygous mice was not observed (M. Stanfel and B. Kennedy, personal communication), we conclude that tagging the RPL22 protein does not affect its function in vivo. It is possible that some cell types may express higher levels of RPL22-like-1 compared to RPL22, which could affect our ability to universally apply the RiboTag technology to these cell types. Because no RPl22-like-1 antibody exists, we cannot make a direct comparison of RPL22 and RPL22-Like 1 across tissues. However to date, we have crossed the RiboTag mouse to 12 different Cre recombinase-expressing lines and have always observed robust expression of tagged RPL22ha in the expected target cell population.
Although substantial enrichments for cell-type-specific mRNAs have been obtained, there remains a low level of background absorption of untagged ribosomes to the magnetic beads, as measured by control immunoprecipitations with Myc antibody and by the continued presence of transcripts that are not expressed in the Cre-dependent cell population. We are optimistic that methods can be developed to reduce nonspecific polysome immunoprecipitation even further. A simple approach to improve the signal-to-noise for rare neuronal populations is to dissect the appropriate brain region to increase the proportion of ribosome-tagged cells in the homogenate.
The translational control of specific mRNAs has been extensively studied and shown to play a critical role in development as well as neuronal function (15, 16, 21). The RiboTag approach described here and the bacTRAP approach (18, 19) isolate mRNAs by virtue of their association with polysomes. It should be noted that trafficking neuronal RNA granules in neurons have been demonstrated to contain both small and large ribosomal subunits (15). Therefore, depending upon the particular transcript and the tissue or cell type under study, a fraction of the immunoprecipitated mRNA transcripts may be translationally repressed but still associated with ribosomes. Nonetheless, in the well-understood example of Prm1 mRNA translational control in male germ cells (14), RiboTag isolation clearly discriminates between translationally repressed and active Prm1 mRNA.
These techniques provide a unique opportunity to measure ribosome-associated mRNA populations in genetically identified cell types within the intact physiological system. Coupling the RiboTag approach with additional strategies, such as the recently described ribosome footprinting technique (22), will facilitate an even more complete analysis of cell-type specific translational regulation. The RiboTag approach can be easily combined with genetic mutations, hormonal, and metabolic manipulations, and pharmacological treatments that perturb normal physiological function to examine changes in ribosome-associated mRNA transcripts from unique cell types in vivo in complex tissues. In addition, immunoprecipitation of polysomes from specific cell types should allow investigators to detect cell-type specific modifications of ribosomal proteins and the presence of nonribosomal regulatory proteins that may be involved in processes such as localization, repression, or transport.
Materials and Methods
Animal Maintenance.
Mice were housed in a temperature- and humidity-controlled facility with a 12-h light/dark cycle. All animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee at the University of Washington. The Mox2-Cre (23) and DAT-Cre mice (24) lines have been described previously. The DARPP32-Cre mice were provided by Dr. Michelle Ehrlich (25) and the AMH-Cre mice were provided by Dr. Robert Braun at the Jackson Labs, Bar Harbor, Maine (26). The RiboTag mouse was bred to each Cre recombinase-expressing mouse line to obtain double heterozygotes for immunoprecipitation experiments. Once Rpl22ha-expressing homozygous mice were obtained, we maintained the colony as a separate line.
Targeting the Rpl22 Gene and Generation of the RiboTag Mouse.
A detailed description of the targeting vector along with the generation and genotyping of the RiboTag mouse can be found in the SI Materials and Methods.
Western Blot Analysis.
Western blots were performed as described (27). We used the following antibodies: Mouse monoclonal anti-HA (purified and ascites, 1:1,000 dilution; Covance); rabbit polyclonal anti-RPL7 (1:1,000 dilution; Novus Biologicals); mouse monoclonal anti-RPL22 (1:200 dilution; BD Transduction Laboratories).
Sucrose Density Gradient Analysis.
Sucrose density gradients were performed as described (28).
Immunohistochemistry.
Immunohistochemistry was performed as described (29). The following primary antibodies were used: Alexa Fluor 488-conjugated mouse monoclonal anti-HA (1:500 dilution; Covance), mouse monoclonal anti-HA (ascites fluid, 1:500 dilution; Covance), and rabbit anti-tyrosine hydroxylase (1:500 dilution; Chemicon).
Immunoprecipitation of Polysomes.
A detailed description of the immunoprecipitation procedure can be found in the SI Materials and Methods.
RNA Integrity Analysis.
RNA quality of input, HA pellet, and control Myc pellet samples were assessed on the Center for Array Technology (CAT) Agilent 2100 Bioanalyzer (Agilent Technologies). For input and HA pellet samples, 150 ng total RNA were used. For Myc pellet samples, a volume equivalent to the HA pellet samples was used.
qRT-PCR Analysis.
Total RNA from input, anti-HA, and anti-Myc pellet samples was quantified using a NanoDrop 1,000 spectrophotometer (Thermo Scientific) and the RiboGreen RNA quantitation kit (Molecular Probes). Equal amounts (for input and HA pellet samples) or equal volumes (for myc samples) were used for assays. Transcript levels were assessed by one-step qRT-PCR assays using Brilliant II SYBR green qRT-PCR master mix (Stratagene) in a MX3000P PCR machine (Stratagene). Fold-enrichment was calculated using the ΔΔCt method with normalization to Actb. Oligonucleotides were selected using the PrimerBank database and are listed in Table S1. Taqman assays for Prm1 (Applied Biosystems) were performed according to manufacturer's instructions.
Basic Nuclear Protein Determination.
Basic nuclear proteins were extracted from testis as described (30) and resolved using acid-urea gel electrophoresis. Gels were stained with a Coomassie Brilliant Blue solution to visualize proteins.
Northern Blots.
Northern blot analysis has been described (31). The complete cDNA sequence of Prm1 was amplified by PCR from mouse National Institutes of Health (NIH) IMAGE clone 6774637, gel-purified, and labeled by random priming using Klenow enzyme (New England Biolabs).
Statistical Analysis.
Statistical analyses were performed with the GraphPad Prism 5.0 software package (*, P < 0.05; **, P<0.01; ***, P<0.001).
Supplementary Material
Acknowledgments.
We thank David L. Brackett and Kim Smith for providing excellent technical support during the project; Elise Rosenbaum and G. Lynn Law for assistance with early pilot studies on expression of tagged RPS17, RPS25, RPL22, and RPL36 in HeLa cells; Garth B. Brandal and Ayu W. Rahardjo for assistance with genotyping RiboTag crosses to various Cre recombinase-expressing mice; and R.D.P. for providing mouse lines and for his many helpful comments on the project and manuscript. This work was supported by National Institutes of Health Grants 1R21HD057798–01 (to P.S.A.), 1R21RR021704–02 (to D.R.M.), and GM32875 (to G.S.M.); the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)/NIH through cooperative agreement U54HD12629 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research; and the postdoctoral mobility program of the Spanish Ministry of Science and Innovation (−2008–0892) (to E.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907143106/DCSupplemental.
References
- 1.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 2.Buchstaller J, et al. Efficient isolation and gene expression profiling of small numbers of neural crest stem cells and developing Schwann cells. J Neurosci. 2004;24:2357–2365. doi: 10.1523/JNEUROSCI.4083-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hempel CM, Sugino K, Nelson SB. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nat Protoc. 2007;2:2924–2929. doi: 10.1038/nprot.2007.416. [DOI] [PubMed] [Google Scholar]
- 4.Lobo MK, et al. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 5.Rossner MJ, et al. Global transcriptome analysis of genetically identified neurons in the adult cortex. J Neurosci. 2006;26:9956–9966. doi: 10.1523/JNEUROSCI.0468-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugino K, et al. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 7.Yao F, et al. Microarray analysis of fluoro-gold labeled rat dopamine neurons harvested by laser capture microdissection. J Neurosci Methods. 2005;143:95–106. doi: 10.1016/j.jneumeth.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 8.Inada T, et al. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. Rna. 2002;8:948–958. doi: 10.1017/s1355838202026018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 10.Jarvik JW, et al. In vivo functional proteomics: Mammalian genome annotation using CD-tagging. Biotechniques. 2002;33:852–860. doi: 10.2144/02334rr02. [DOI] [PubMed] [Google Scholar]
- 11.Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroeder A, et al. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, et al. Process outgrowth in oligodendrocytes is mediated by CNP, a novel microtubule assembly myelin protein. J Cell Biol. 2005;170:661–673. doi: 10.1083/jcb.200411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun RE, et al. Protamine 3′-untranslated sequences regulate temporal translational control and subcellular localization of growth hormone in spermatids of transgenic mice. Genes Dev. 1989;3:793–802. doi: 10.1101/gad.3.6.793. [DOI] [PubMed] [Google Scholar]
- 15.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keene JD. RNA regulons: Coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 17.Kleene KC. Poly(A) shortening accompanies the activation of translation of five mRNAs during spermiogenesis in the mouse. Development. 1989;106:367–373. doi: 10.1242/dev.106.2.367. [DOI] [PubMed] [Google Scholar]
- 18.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson SJ, et al. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–772. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 22.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: A tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Zhuang X, et al. Targeted gene expression in dopamine and serotonin neurons of the mouse brain. J Neurosci Methods. 2005;143:27–32. doi: 10.1016/j.jneumeth.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 25.Bogush AI, et al. DARPP-32 genomic fragments drive Cre expression in postnatal striatum. Genesis. 2005;42:37–46. doi: 10.1002/gene.20118. [DOI] [PubMed] [Google Scholar]
- 26.Holdcraft RW, Braun RE. Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development. 2004;131:459–467. doi: 10.1242/dev.00957. [DOI] [PubMed] [Google Scholar]
- 27.Amieux PS, et al. Compensatory regulation of RIalpha protein levels in protein kinase A mutant mice. J Biol Chem. 1997;272:3993–3998. doi: 10.1074/jbc.272.7.3993. [DOI] [PubMed] [Google Scholar]
- 28.Zomzely CE, Roberts S, Brown DM, Provost C. Cerebral protein synthesis. I. Physical properties of cerebral ribosomes and polyribosomes. J Mol Biol. 1966;19:455–468. doi: 10.1016/s0022-2836(66)80016-5. [DOI] [PubMed] [Google Scholar]
- 29.Huang Y, Roelink H, McKnight GS. Protein kinase A deficiency causes axially localized neural tube defects in mice. J Biol Chem. 2002;277:19889–19896. doi: 10.1074/jbc.M111412200. [DOI] [PubMed] [Google Scholar]
- 30.Lee K, Haugen HS, Clegg CH, Braun RE. Premature translation of protamine 1 mRNA causes precocious nuclear condensation and arrests spermatid differentiation in mice. Proc Natl Acad Sci USA. 1995;92:12451–12455. doi: 10.1073/pnas.92.26.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burton KA, et al. Haploinsufficiency at the protein kinase A RI alpha gene locus leads to fertility defects in male mice and men. Mol Endocrinol. 2006;20:2504–2513. doi: 10.1210/me.2006-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.