Abstract
In addition to its cellular homeostasis function, autophagy is emerging as a central component of antimicrobial host defense against diverse infections. To counteract this mechanism, many pathogens have evolved to evade, subvert, or exploit autophagy. Here, we report that autophagy proteins (i.e., Beclin-1, Atg4B, Atg5, and Atg12) are proviral factors required for translation of incoming hepatitis C virus (HCV) RNA and, thereby, for initiation of HCV replication, but they are not required once infection is established. These results illustrate a previously unappreciated role for autophagy in the establishment of a viral infection and they suggest that different host factors regulate the translation of incoming viral genome and translation of progeny HCV RNA once replication is established.
Keywords: flaviviridae, autophagosome, LC3, ATG, translation
HCV infection is a leading cause of chronic liver disease world-wide. With 180 million persistently infected people, chronic hepatitis C infection, which induces end stage liver diseases, such as liver cirrhosis and hepatocellular carcinoma (HCC), represents a major public health problem of high socioeconomic impact (1). However, treatment options for chronic hepatitis C are limited and a vaccine against HCV is not available. Thus, efforts to elucidate the details of the host-virus relationship during HCV infection are needed to develop efficient therapeutic strategies against this major human pathogen.
HCV is an enveloped, positive strand RNA virus in the Flaviviridae family. The HCV genome is approximately 9.6 kb in length and consists of a single ORF flanked by untranslated regions (UTR) (2). Translation of the single ORF is driven by an internal ribosomal entry site (IRES) sequence present within the 5′UTR. The resulting polyprotein is processed by cellular and viral proteases into the structural proteins (core, E1, and E2) and the nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (2). HCV NS3 to NS5B proteins are necessary and sufficient to establish membrane-bound replication complexes that catalyze RNA replication (2).
Autophagy is an intracellular degradation process in which cytoplasmic components, including organelles, are directed to the lysosome by a membrane-mediated process (3). More than 31 autophagy-related genes (ATG) whose products regulate autophagy have been identified, primarily through yeast genetics, and several have mammalian counterparts (3). They regulate distinct aspects of autophagic vesicle formation and maturation, including the assembly of an isolation membrane around a portion of cytoplasm, completion of a double-membrane-surrounded autophagosome, and lysosomal fusion with degradation of its contents (3). Importantly, many studies demonstrate that autophagy is activated upon viral or bacterial infection (4). In Sindbis virus, tobacco mosaic virus, herpes simplex virus type 1 (HSV-1) and several bacterial infections, autophagy may have a protective function by restricting intracellular pathogen replication or by ensuring the survival of infected and/or uninfected cells (4). In this regard, autophagy serves as an innate host defense mechanism, and some viruses and bacteria produce virulence factors that counteract these antimicrobial processes (4–7). Conversely, certain viruses, for example, mouse hepatitis virus, poliovirus, coxsackievirus and dengue virus, actually exploit the elements of the autophagy system for their replication (8–12).
In this study, by manipulating the expression and function of key autophagy regulators, we show that these proteins are required for productive HCV infection. Moreover, by dissecting the individual steps of HCV infection, we demonstrate that autophagy is required for translation and/or delivery of incoming viral RNA to the translation apparatus.
Results
Enhanced Autophagic Vesicle Content in HCV-Infected Cells.
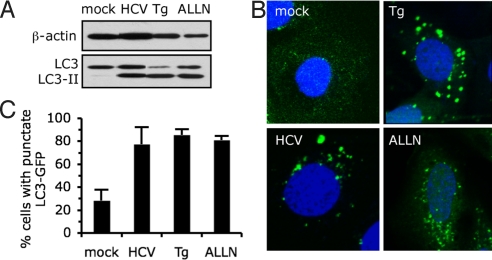
A hallmark of autophagy induction is lipidation of the microtubule-associate protein 1 light chain 3 (LC3) (13). A series of covalent transfers leads to phosphatidylethanolamine (PE) conjugation to LC3, termed LC3-II, which becomes membrane-associated and participates in autophagosome formation (3, 14, 15). To determine whether JFH-1-HCV infection regulates autophagy, we examined the conversion of endogenous LC3 to LC3 II. The intensity of LC3-II band was increased in HCV infected cells relative to uninfected (mock) cells (Fig. 1A). In addition, co-migration of LC3-II in infected cells and in cells treated with autophagy inducers, for example, the proteasome inhibitor acetyl-l-leucyl-l-leucyl-l-norleucinal (ALLN) and the ER stress inducer thapsigargin (Tg) (16, 17), confirmed that the band detected was authentic LC3-II. Another hallmark of autophagy is the redistribution of LC3 from a diffuse cytoplasmic localization to a characteristic punctate cytoplasmic pattern reflecting the recruitment of LC3 to autophagic vesicles (13, 14). Indeed, in cells transfected with a GFP-tagged LC3 expression vector (LC3-GFP), LC3-GFP redistribution into discrete dots was markedly increased by HCV infection (Fig. 1B and C), further confirming the accumulation of autophagic vesicles upon HCV infection.
Fig. 1.
Enhanced autophagic vesicle content in HCV-infected cells. (A) Analysis by immnunoblotting of endogenous LC3 lipidation in cell extracts of Huh-7 cells infected for 24 h with HCV or treated with thapsigargin (Tg) (500 nM) or with ALLN (10 μM) for 16 h, or untreated (mock), as indicated. β-actin expression was examined as a protein loading control. The gels are representative of three independent experiments. (B) Representative confocal images. LC3-GFP transfected Huh-7 cells were grown in normal medium (mock), infected for 24 h with HCV, or treated with ALLN or with Tg, as described in A. The nuclei were stained by Hoechst solution (blue). (C) Quantification of the frequency of Huh-7 cells displaying a punctate distribution of LC3-GFP. Results represent the means of two independent experiments.
The Autophagy Machinery Is Required for the HCV Life Cycle.
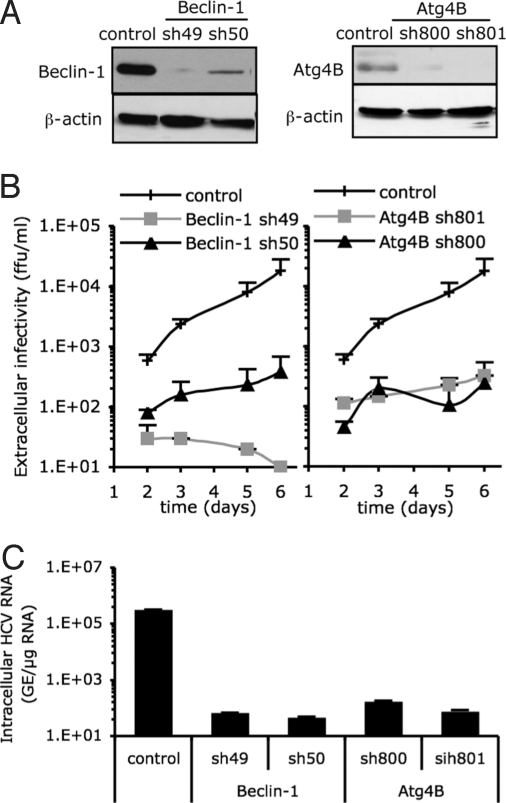
To determine whether the autophagy machinery performs an antiviral or proviral function during HCV infection, we monitored the effect of shRNA-mediated knock-down of key autophagy proteins on the establishment of HCV infection. We targeted Beclin-1 and Atg4B, because Beclin-1, in complex with Vps34, the class III phosphatidylinositol-3-kinase [PI(3)K] promotes formation of autophagic vesicles (18), and Atg4B is a cysteine-protease essential for LC3 lipidation during autophagosome formation and de-lipidation for recycling after autophagosome maturation (14, 15, 19–22). As shown in Fig. 2A, Beclin-1 and Atg4B expression was strongly inhibited in Huh-7 cells by lentivirus-based vector-mediated shRNA technology. As expected, the induction of autophagy by thapsigargin (Tg) was markedly decreased in these cells, as measured by the frequency of cells containing punctate LC3-GFP (Fig. S1), confirming the reduction of autophagic vesicle formation that has been previously described for other cell types (23, 24).
Fig. 2.
Autophagy machinery is required for HCV spread. Huh-7 cells were transduced with lentiviral vectors expressing shRNA against Beclin-1 (shRNA denoted as sh49 and sh50) or Atg4B (sh800 and sh801). The selected shRNA sequence and multiplicity of infection (MOI) used to transduce Huh-7 cells did not compromise the cell viability. At 8 days posttransduction, transduced and control cells were harvested and seeded in equal numbers. Twenty and seventy-two hours later MTT was added and cellular proliferation was monitored. The results for the Beclin-1-(sh49) transduced cells were 99.3 ± 27.3% of the nontransduced cells, set to 100. (A) The relative abundance of Beclin-1 and Atg4B proteins in the Huh-7 cells was analyzed by immunoblotting. β-actin expression was used as a protein loading control. (B) Beclin-1- and Atg4B-deficient and nontransduced (control) Huh-7 cells were infected with HCV at an MOI of 0.01. The infectivity of their virus-containing cell supernatants was determined at different times postinoculation, as indicated. Results display average infectious titers, expressed as focus-forming units (ffu)/ml (mean ± SD; n = 2). (C) The intracellular HCV RNA levels were determined by RT-qPCR 6 days postinoculation (MOI of 0.01) in Beclin-1- and Atg4B-deficient and control cells (mean ± SD; n = 2). GE, genome equivalent.
Next, Atg4B- and Beclin-1-deficient cells were infected with HCV at low multiplicity of infection (MOI) and HCV expansion was measured over the next 6 days by determining extracellular infectious viral titers. This analysis revealed that, HCV expansion was suppressed up to 100-fold in Atg4B-deficient cells and almost completely abolished in Beclin-1-deficient cells as compared to control cells (Fig. 2B). The intracellular HCV RNA content paralleled extracellular HCV infectivity with reductions of up to 10,000-fold in Atg4B- and Beclin-1-down-regulated cells (Fig. 2C). To confirm and extend these observations, we also tested the effect of shRNA-mediated inhibition of Atg12, a critical component of a ubiquitin-like conjugation system that enables elongation of the autophagosome (3). As expected, because LC3 lipidation depends on Atg5 and its conjugation with Atg12 (25, 26), both LC3-II species and the induced punctate GFP-LC3 distribution were reduced in Atg12 shRNA-treated Huh-7 cells as compared to control cells (Fig. S2A). Importantly, HCV expansion was greatly reduced in Atg12-deficient cells (Fig. S2B), reinforcing the requirement of autophagy machinery proteins for efficient HCV infection. Consistent with these observations, ectopic expression of a dominant negative Atg5 mutant (Atg5K130R) that is defective for conjugation with Atg12 and suppresses autophagic vesicle formation (25, 27) also inhibited HCV infection (Fig. S3).
Collectively, these results demonstrate that key proteins responsible for different steps of autophagic vesicle formation are required for initiation of HCV infection in Huh-7 cells and thus represent proviral factors for HCV.
The Autophagy Machinery Is Not Required for HCV Particle Entry or Secretion.
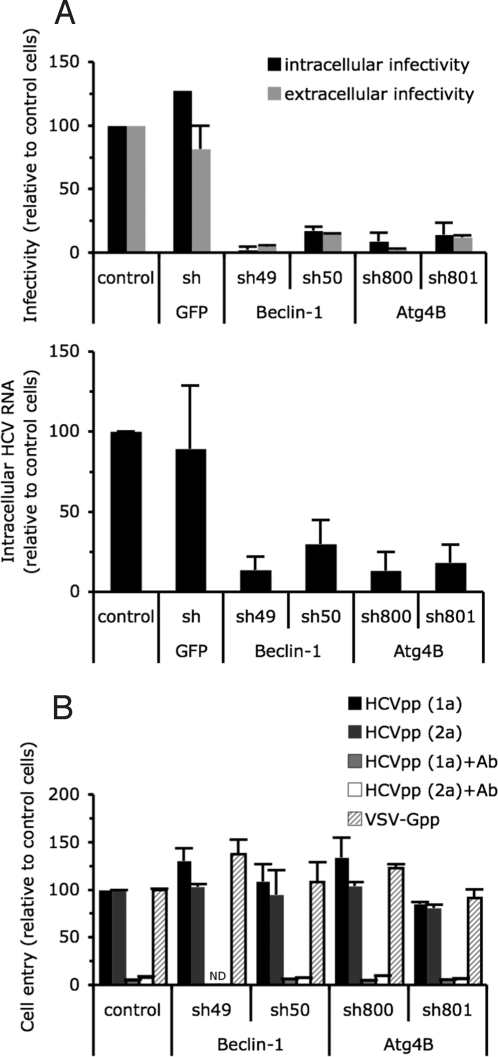
To identify the autophagy-dependent step(s) in the HCV life cycle, we performed single cycle infections in Beclin-1-, Atg4B-, and Atg12-deficient and control cells. Intracellular and extracellular (i.e., secreted) infectious particle production were reduced to a similar degree in Beclin-1-, Atg4B-, and Atg12-deficient cells, (Fig. 3A and Fig. S2C), indicating that, in contrast to the role of autophagy proteins in poliovirus particle release (8), the autophagy machinery is not required for HCV particle secretion. Moreover, accumulation of intracellular HCV RNA was also significantly reduced in autophagy protein-down-regulated cells (Fig. 3A and Fig. S2C), suggesting that the autophagy machinery is required for either virus entry, RNA translation or replication.
Fig. 3.
The autophagy machinery regulates a postentry step, upstream or at the level of HCV replication. Huh-7 cells were transduced or not (control) with lentiviral vectors expressing shRNA against Beclin-1, Atg4B, and GFP. (A) Extracellular and intracellular infectious virus and intracellular RNA production during 24 h in a single step infection at MOI of 10 (mean ± SD; n = 3). (Upper) Accumulation of extracellular and intracellular HCV infectious particles was determined 24 h after inoculation, and expressed as a percentage of that in control cells. (Lower) Intracellular HCV RNA content was determined at 24 h post inoculation and expressed as a percentage of that in control cells. (B) Cell entry of HCVpp harboring E1E2 glycoproteins derived from HCV strains H77 (1a) or JHF-1 (2a) and control particles harboring the VSV-G glycoprotein (VSV-Gpp) expressed as a percentage of that in control cells (mean ± SD; n = 4). To assess the specificity of E1E2 glycoprotein-mediated cell entry, anti-E2 antibody (Ab) was added at 20 μg/mL. ND, not determined.
To examine the dependence of HCV entry on autophagy, we studied the impact of shRNA-mediated inhibition of autophagy protein expression on HCV pseudoparticle (HCVpp) entry into Huh7 cells (28). Autophagy protein-down-regulated and control cells were infected with HCVpp that contain envelope glycoproteins from the JFH-1 (genotype 2a) and H77 (genotype 1a) strains of HCV. As shown in Fig. 3B and Fig. S2D, HCVpp entry was comparable in autophagy protein-deficient and control cells (Fig. 3B and Fig. S2D). Collectively, these results suggest that the autophagy machinery is required for HCV infection at a post-entry step, most likely HCV RNA translation and/or replication.
The Autophagy Machinery Is Required for HCV Replicon Replication.
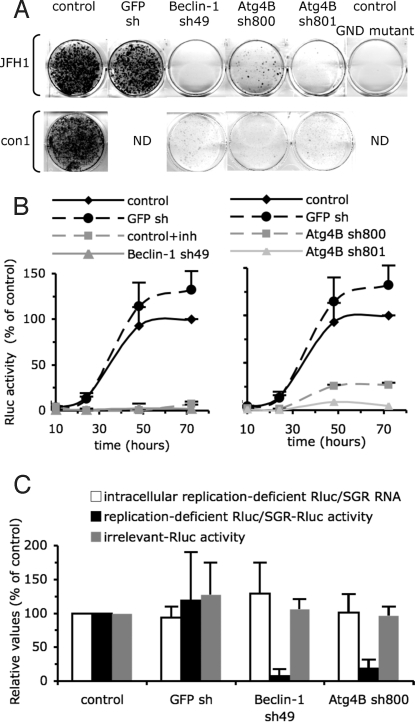
To determine whether the autophagy machinery modulates HCV RNA translation and/or replication, we examined the ability of autophagy protein-down-regulated cells to support replication of an HCV subgenomic replicon. Identical amounts of JFH-1 subgenomic replicon RNA containing the neomycin marker gene (neo/SGR) were introduced into autophagy protein-down-regulated and control cells. Replicon-replicating cells were selected for neomycin resistance as described previously (29). As shown in Fig. 4A and Fig. S4A, down-regulation of Beclin-1, Atg4B, and Atg12 strongly suppressed JFH-1 replicon replication despite comparable RNA transfection efficiency (Fig. S5A) and cell viability as determined by trypan blue staining. Similar results were obtained using a Con1 (genotype 1b) subgenomic replicon (Fig. 4A, Fig. S4A, and Fig. S5B). To rule out the possibility of nonspecific effects on neomycin sensitivity by suppression of the autophagy machinery, we confirmed these findings by using a subgenomic reporter replicon containing the renilla luciferase gene (Rluc/SGR). While the transfection efficiency was comparable in all cell populations (Fig. S5C), the Rluc activity was reduced by up to 50-fold at all time points posttransfection in Beclin-1, Atg4B, and Atg12-deficient cells (Fig. 4B and Fig. S4B). Collectively, these results indicated that autophagy machinery proteins are required for RNA translation and/or replication.
Fig. 4.
Autophagy machinery regulates translation and/or delivery of incoming viral RNA to the translation apparatus. Huh-7 cells were transduced with lentivirus expressing shRNA against Beclin-1, Atg4B, and GFP. (A) Analysis of JFH1 and Con1 neo/SGR replication. Neomycin selected cells were fixed and stained with Crystal Violet. A replication defective GND mutant (GDD-to-GND mutation in the NS5B protein) neo/SGR was used as a positive control for the neomycin selection. Results are representative of two independent experiments. (B) Analysis of Rluc/SGR translation/replication by monitoring Rluc activity at different times posttransfection. To assess the specificity of Rluc activity, control cells were treated with 5 μM of 2′-C-methyladenosine, a HCV polymerase inhibitor (denoted inh). For each independent experiment, Rluc activity was normalized to cell density and expressed as a percentage of that determined in control cells at 72 h posttransfection (mean ± SD; n = 4). (C) Translation of replication-deficient Rluc/SGR RNAs. Intracellular RNA levels and Rluc activity of replication deficient Rluc/SGR were determined at 6 h posttransfection. Rluc activities are statistically reduced in autophagy protein deficient cells compared to shRNA GFP expressing cells (P values <0.05 from paired Student's t test). In parallel transfections, Rluc activity expressed from pRL-TK plasmid (irrelevant-Rluc activity) was determined. For each independent experiment, Rluc activity was normalized to cell density and expressed as a percentage of that determined in control cells (mean ± SD; n = 3).
Next, we asked whether autophagosomes might facilitate the tethering of HCV replication complex proteins to cellular membranes, a process that is known to be required for HCV replication (2). Consistent with a previous report using the H77 strain of HCV (30), we did not observe any colocalization of HCV JFH1 proteins involved in viral replication (NS4A/4B and NS5A) with LC3-GFP in infected cells (Fig. S6), in the absence or presence of the vesicle acidification inhibitor Bafilomycin A1 (Fig. S7). These results suggest that the autophagy machinery is most likely not required to maintain an active HCV replication complex.
The Autophagy Machinery Is Required for Translation and/or Delivery of Incoming Viral RNA to the Translation Apparatus.
Interestingly, Rluc activity was compromised in autophagy protein-deficient cells >3-fold already at 5 h posttransfection, suggesting that autophagy proteins might play a role in HCV RNA translation initiation. To specifically monitor viral translation, we used a replication-defective Rluc/SGR construct that contains an inactivation mutation (GDD-to-GND) in the active site of the viral polymerase (NS5B), and we analyzed Rluc activity expressed from the HCV IRES, as previously described (31). While the level of intracellular Rluc/SGR RNA was comparable in autophagy protein-deficient and control cells (Fig. 4C and Fig. S4C), HCV IRES-dependent translation measured by Rluc activity was strongly reduced in Beclin-1, Atg4B, and Atg12-down-regulated cells as compared to control and GFP shRNA-expressing cells (Fig. 4C and Fig. S4C). Reduced Rluc expression was not due to a general effect on ectopic gene translation efficiency or protein stability, since Rluc activity derived from cap-dependent translation of a RNA polymerase II transcribed RNA was not significantly different in autophagy protein-deficient and control cells (Fig. 4C and Fig. S4C). Collectively, these results suggested that the autophagy machinery is required for HCV IRES-dependent translation of the incoming viral RNA, or for delivery of the incoming RNA to the translation apparatus, perhaps by targeting it to the appropriate cellular factor(s) or location.
The Autophagy Machinery Is Required for the Initiation of HCV Replication But Not to Maintain It.
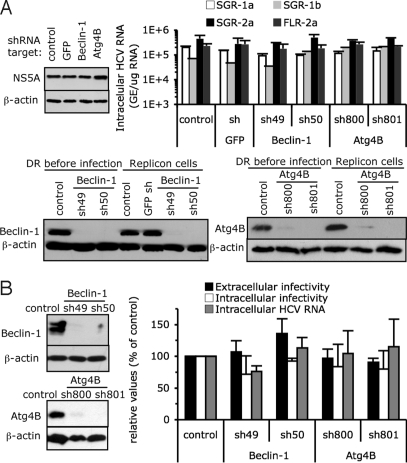
Our results point to a crucial role of the autophagy machinery for translation of incoming viral RNA and the establishment of replication either in HCV RNA transfected cells (Fig. 4 and Fig. S4) or in de novo infected cells (Figs. 2 and 3 and Figs. S2 and S3). To determine whether autophagy proteins are required only for initiation of translation and the establishment of HCV replication or if they are also required to maintain ongoing HCV translation and replication, we analyzed the impact of autophagy protein down-regulation on HCV RNA and protein content in stable replicon cells. Whereas Beclin-1 and Atg4B protein content was greatly reduced in shRNA-transduced replicon cells (Fig. 5A), HCV RNA and protein (illustrated by NS5A expression) levels were unchanged (Fig. 5A). Similar results were obtained in Huh-7 cells harboring H77 (genotype 1a) and Con1 (genotype 1b) subgenomic replicons or a JFH-1 full-length replicon (Fig. 5A), suggesting that the autophagy machinery is not required to maintain ongoing HCV replication.
Fig. 5.
Autophagy machinery does not regulate established HCV replication. (A) Replicon bearing-Huh-7 cells were transduced with lentiviral vectors expressing shRNAs against Beclin-1, Atg4B, and GFP. Protein and RNA levels were determined 10 days posttransduction. (Upper Left) NS5A protein levels in replicon cells as analyzed by immunoblotting. (Upper Right) Intracellular HCV RNA levels in H77 (1a), con-1 (1b), JFH-1 (2a) SGR, or JFH-1 full-length (FLR) replicon-bearing Huh-7 cells monitored by RT-qPCR (mean ± SD; n = 2). GE, genome equivalent. (Lower) Efficiency of down-regulation of Beclin-1, Atg4B, as analyzed by immunoblotting. Beclin-1 and Atg4B content were analyzed in established JFH-1 SGR replicon cells (denoted Replicon cells) and were compared to shRNA-treated cells before HCV infection (denoted DR before infection). β-actin expression was used as a protein loading control. (B) Huh-7 cells that were virtually all infected by HCV were transduced with lentiviral vectors expressing shRNA against Beclin-1, Atg4B. (Left) Relative levels of Beclin-1 and Atg4B in the infected cells, as analyzed by immunoblotting. β-actin expression was used as a protein loading control. (Right) Extracellular and intracellular infectivity and intracellular HCV RNA levels were determined 8 days posttransduction and are expressed as a percentage of those in control cells (mean ± SD; n = 2).
To rule out the possibility that stable replicon cells might be artefactually autophagy-independent because of their prior selection, Beclin-1, Atg4B, and Atg12 were down-regulated in nonselected infected cells virtually all of which were HCV E2 positive (Fig. S4D). While Beclin-1 and Atg4B expression (Fig. 5B) and LC3 lipidation (Fig. S4E) were strongly reduced in these cells, intracellular levels of infectious particles and HCV RNA were unchanged (Fig. 5B and Fig. S4E), confirming that the autophagy machinery is not required to maintain replication of established infection. Furthermore, production of infectious virus particles (Fig. 5B and Fig. S4E) were not significantly reduced in down-regulated cells, confirming that autophagy proteins aren't required for HCV particle secretion (Fig. 3A).
Collectively, these results indicate that the autophagy machinery is required for translation of incoming RNA and, therefore, HCV replication in de novo infected cells, but not for translation of progeny HCV RNA once replication is established.
Discussion
In this study, we analyzed the impact on HCV infection of several proteins that regulate distinct molecular events leading to autophagy vesicle formation, including its initiation (e.g., Beclin-1), and maturation by the Atg12 (e.g., Atg12 and Atg5) and LC3 (e.g., Atg4B) conjugation systems. We showed that dominant negative inhibition and down-regulation of different regulators of the autophagy pathway strongly suppress productive HCV infection. Our results are in agreement with a recent report suggesting that the down-regulation of LC3 and Atg7, a protein that mediates LC3 lipidation, decreases the HCV RNA content in cells electroporated with JFH-1 HCV RNA (32). In addition, our results demonstrate that autophagy machinery proteins modulate a post entry-step of HCV infection and, through the analysis of single step infection assays, reveal a functional interaction upstream or at of the level of RNA replication. Consistent with those results, we showed that the autophagy machinery is required for the translation and/or delivery of incoming viral RNA to the translation apparatus and, therefore, for the establishment of productive infection. Importantly, our results demonstrate that autophagy proteins are required for the initiation of HCV RNA translation/replication, but not once these processes are established, indicating that different host factors are required for translation of the incoming viral RNA versus the progeny HCV RNA once replication is established.
Our results indicate that the content of autophagy vesicles is increased in HCV-infected cells, extending recent results obtained in cells infected with the H77 strain of HCV and in cells transfected with JFH1 strain HCV RNA (30, 32). Sir et al. (32) proposed that the unfolded protein response (UPR) contributes to the induction of autophagy in HCV RNA-transfected cells. However, our results point to a role of autophagy proteins at a very early stage of the HCV life cycle, when HCV proteins are produced from the incoming RNA, which should be independent of the UPR. Interestingly, for other pathogens, pattern recognition receptors (PRR) and the innate signaling pathway have been reported to trigger autophagy in infected cells (4). Notably, cytosolic RNA-sensing protein kinase PKR and eIF2α phosphorylation regulate virus- and starvation-induced autophagy (7, 33). It is tempting to speculate that recognition of the incoming HCV RNA by RNA-sensing molecules induces autophagy and hence, favors its initial translation. Alternatively, constitutive basal autophagic vesicle formation might be required for initial HCV RNA translation. In this regard, we observed a reduction of LC3 lipidation in Atg12-deficient Huh-7 cells even in the absence of an autophagy inducer (Fig. S2A), that was associated with a reduction in susceptibility of these cells to HCV infection. Therefore, it is likely that the establishment of HCV infection depends on the basal level of autophagy rather than its induction.
Formation of a membrane-associated replication complex, composed of viral proteins, replicating RNA and altered cellular membranes, is a hallmark of all positive-strand RNA viruses investigated so far (34). HCV proteins form a multiprotein complex in an intracellular membrane complex, thought to be derived from ER membranes, which is the site of RNA replication (2, 35). Interestingly, the viral components of certain positive-strand RNA viruses (e.g., poliovirus, mouse hepatitis virus and dengue virus) that subvert the autophagy machinery for their replication, co-localize with autophagy markers, suggesting that the translation/replication complexes of those viruses might assemble on autophagic vesicles (8, 10–12). However, HCV proteins do not colocalize with autophagy proteins in infected cells suggesting that the HCV replication complex does not assemble on autophagic vesicles but rather, that HCV exploits the autophagy pathway by a currently undefined mechanism that appears to be different from other autophagy-dependent viruses. It is conceivable that the autophagy pathway supports HCV replication by sequestering or destroying HCV replication restriction factor(s). Alternatively, it is tempting to speculate that the autophagy pathway might provide an initial membranous support for translation of incoming RNA, before accumulation of viral proteins and eventual virus-induced cellular modifications. The lack of an effect of autophagy protein down-regulation on previously established HCV replication supports this hypothesis.
How the incoming HCV RNA is targeted to the appropriate cellular factors or location for initial translation remains unknown. It is believed that IRES-mediated HCV genome translation, polyprotein processing and RNA replication occur within ER-derived membranes. Notably, an internal signal sequence located at the C terminus of the core protein targets the nascent polypeptide to the ER membrane for translocation of E1 glycoprotein into the ER lumen (2). Interestingly, a model of autophagosome maturation from specialized domains of the ER has been proposed (36–38). Therefore, the possible continuum between ER and autophagic vesicles and/or autophagy-induced local ER rearrangements, through a dynamic connection between ER and autophagy vesicles (37), might be of importance, by providing a specialized platform for translation of the incoming HCV RNA.
Materials and Methods
Plasmid and Reagents.
The antibodies used for immunoblotting and immunostaining were specific for E2 (C1); NS5A (MS5); Beclin-1 (Santa Cruz Biotechnology); Atg4B (Abcam); LC3 (Novus Biologicals); actin (Sigma Aldrich). Acetyl-l-leucyl-l-leucyl-l-norleucinal (ALLN) and thapsigargin (Tg) were purchased from Sigma Aldrich. 2′-C-methyladenosine was kindly provided by W. Zhong (Gilead Sciences). The SG and Full-length JFH-1-replicon have been previously described (39). The subgenomic (SG)-Con1 replicon (S1179I) was kindly provided by C. Rice (Rockefeller University, New York, NY) (40). The SG-H77 replicon (Htat2ANeo/QR/VI/KR//KR5A/SI) bearing Huh-7 cells were kindly provided by S. Lemon (University of Texas Medical Branch, Galveston, TX) (41). The JFH-1 Rluc/SGR or GND JFH-1 Rluc/SGR plasmids carry bicistronic RNA containing the luciferase reporter gene in the first cistron and wild-type (wt) or replication-deficient (encoding a GDD-to-GND mutation in NS5B) JFH-1 subgenomic replicon in the second cistron, respectively.
Lentiviral Particle Production and Huh-7 Cell Transduction.
Vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped lentiviral particles were produced in 293T cells by co-transfection of lentiviral vector encoding short hairpin RNAs (shRNAs) (Mission, Sigma Aldrich) with vectors encoding compatible packaging proteins and VSV-G, as described previously (42). Forty-eight hours posttransfection, cell supernatants were collected, filtrated, and used to transduce Huh-7 cells. Specific shRNA clones were selected according to the target protein down-regulation efficiency and the absence of cytotoxic effects. For all of the experiments displayed, we used freshly shRNA transduced Huh-7 to avoid any compensatory cellular mechanism in autophagy deficient cells that might misleading the interpretation of the results. The sequence of the shRNA are described in SI Text.
Preparation of Viral Stock and Infection.
JFH-1 virus was generated by transfection, and viral stocks were produced by infection of Huh-7 cells at a multiplicity of infection (MOI) of 0.01, as described previously (43). High-titer stocks of cell-culture adapted virus (D183) (29) were prepared by infection of a Huh-7.5.1 cell subclone (clone 2) and used for single step infection experiments (MOI of 10). Cell extracts containing intracellular infectious HCV particles were prepared by 6 freeze-thaw cycles of infected cells as described (39). Infectivity titers in cell culture supernatants and cell extracts were determined by end-point dilution and E2-specific immunofluorescence as described (43). Intracellular HCV RNA levels were determined by HCV-specific reverse transcription-real-time quantitative PCR (RT-qPCR) and were normalized for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels as described (43).
Production of HCVpp and Cell Entry Assays.
Viral pseudoparticles named HCVpp and VSV-Gpp harbored the glycoproteins of HCV and VSV-G, respectively, and were produced by transfection in 293T cells of vectors encoding viral glycoproteins, packaging proteins, and GFP-transfer vector. For infection assays, target cells were seeded 24 h before inoculation at an MOI of 0.1. Then medium was removed and dilutions of viral supernatants were added to the cells and incubated for 4 h. Supernatants were then removed and the infected cells kept in regular medium (DMEM, 10% FCS) for 72 h before analysis of the percentage of GFP-positive cells by FACS analysis.
Reporter Replicon Replication Assays.
In vitro transcribed RNAs were introduced into Huh-7 cells by electroporation as described in SI Text. RNA transfection efficiency and cell viability were assessed by measuring the intracellular neo/SGR RNA content 6 h posttransfection and by determining cell densities posttransfection in the absence of selection, respectively. For renilla luciferase assay (wt and GND mutant Rluc/SGR), cells were lysed with cell culture lysis buffer (Promega). Lysates were harvested and mixed with luciferase assay substrate (Promega), as suggested by the manufacturer. RLU values were normalized to the number of viable cells for each sample determined by a trypan blue staining. Cell viability was confirmed by 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) cytotoxicity assays.
Supplementary Material
Acknowledgments.
We thank Takaji Wakita (National Institute of Infectious Diseases, Tokyo) for kindly providing the infectious JFH-1 molecular clone and replicon constructs, Dennis Burton (The Scripps Research Institute, La Jolla, CA) for providing the C1 anti-E2 antibody, Michael Houghton (Chiron, Emeryville, CA) for providing the MS5 anti-NS5A antibody, Michinori Kohara (Tokyo Metropolitan Institute of Medical Science, Tokyo) for providing the anti-NS4A/NS4B antibody, Inder Verma (Salk Institute, La Jolla, CA) for providing lentiviral plasmids, François-Loïc Cosset (Ecole Normale Superieure, Lyon, France) for providing the vectors necessary for HCVpp production, Charles M. Rice (Rockefeller University, New York) for providing Con1-SGR plasmid, and Stanley Lemon (University of Texas Medical Branch, Galveston, TX) for providing SGR-H77 bearing Huh-7 cells; Bryan Boyd, Josan Chung, and Christina Whitten for excellent technical assistance; Yvon Jaillais for his confocal expertise; Greg Meiffren and Urtzi Garaigorta for critical reading of the manuscript; and our colleagues for encouragement and advice. This work was supported by National Institutes of Health Grant AI079043. This is paper no. 20097IMS from the Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907344106/DCSupplemental.
References
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 3.Reggiori F. 1. Membrane origin for autophagy. Curr Top Dev Biol. 2006;74:1–30. doi: 10.1016/S0070-2153(06)74001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orvedahl A, Levine B. Eating the enemy within: Autophagy in infectious diseases. Cell Death Differ. 2009;16:57–69. doi: 10.1038/cdd.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orvedahl A, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Ogawa M, et al. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 7.Talloczy Z, Virgin HWt, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 8.Jackson WT, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong J, et al. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol. 2008;82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khakpoor A, Panyasrivanit M, Wikan N, Smith DR. A role for autophagolysosomes in DEN-3 production in HepG2 cells. J Gen Virol. 2009 doi: 10.1099/vir.0.007914-0. [DOI] [PubMed] [Google Scholar]
- 11.Lee YR, et al. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374:240–248. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabeya Y, et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 16.Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell. 2006;17:2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogata M, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: A unique autophagy-related protein. Cell Res. 2007;17:839–849. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- 19.Fujita N, et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19:4651–4659. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanida I, Ueno T, Kominami E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J Biol Chem. 2004;279:47704–47710. doi: 10.1074/jbc.M407016200. [DOI] [PubMed] [Google Scholar]
- 21.Tanida I, et al. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 22.Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem. 2003;278:51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- 23.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 24.Jagannath C, et al. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009;15:267–276. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- 25.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujita N, et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 28.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J Exp Med. 2003a;197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong J, et al. Persistent hepatitis C virus infection in vitro: Coevolution of virus and host. J Virol. 2006;80:11082–11093. doi: 10.1128/JVI.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ait-Goughoulte M, et al. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaller T, et al. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J Virol. 2007:814591–4603. doi: 10.1128/JVI.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sir D, et al. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talloczy Z, et al. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci USA. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gosert R, et al. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simonsen A, Stenmark H. Self-eating from an ER-associated cup. J Cell Biol. 2008;182:621–622. doi: 10.1083/jcb.200807061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gastaminza P, et al. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82:2120–2129. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 41.Yi M, Lemon SM. Adaptive mutations producing efficient replication of genotype 1a hepatitis C virus RNA in normal Huh7 cells. J Virol. 2004;78:7904–7915. doi: 10.1128/JVI.78.15.7904-7915.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dreux M, et al. The exchangeable apolipoprotein ApoC-I promotes membrane fusion of hepatitis C virus. J Biol Chem. 2007;282:32357–32369. doi: 10.1074/jbc.M705358200. [DOI] [PubMed] [Google Scholar]
- 43.Zhong J, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.