Abstract
The developing endosperm of rice (Oryza sativa, Os) synthesizes a large amount of storage proteins on the rough (r)ER. The major storage proteins, glutelins and prolamins, contain either intra or intermolecular disulfide bonds, and oxidative protein folding is necessary for the sorting of the proteins to the protein bodies. Here, we investigated an electron transfer pathway for the formation of protein disulfide bonds in the rER of the rice endosperm, focusing on the roles of the thiol-disulfide oxidoreductase, OsEro1. Confocal microscopic analysis revealed that N-glycosylated OsEro1 is localized to the rER membrane in the subaleurone cells, and that targeting of OsEro1 to the rER membrane depends on the N-terminal region from Met-1 to Ser-55. The RNAi knockdown of OsERO1 inhibited the formation of native disulfide bonds in the glutelin precursors (proglutelins) and promoted aggregation of the proglutelins through nonnative intermolecular disulfide bonds in the rER. Inhibition of the formation of native disulfide bonds was also observed in the seeds of the esp2 mutant, which lacks protein disulfide isomerase-like (PDIL)1;1, but shows enhanced OsEro1 expression. We detected the generation of H2O2 in the rER of the WT subaleurone cells, whereas the rER-derived H2O2 levels decreased markedly in EM49 homozygous mutant seeds, which have fewer sulfhydryl groups than the WT seeds. Together, we propose that the formation of native disulfide bonds in proglutelins depends on an electron transfer pathway involving OsEro1 and OsPDIL.
Keywords: protein disulfide isomerase, seed storage protein, esp2, hydrogen peroxide, protein body
The developing endosperm of rice (Oryza sativa, Os) synthesizes a large amount of disulfide-bond-rich proteins, including 2 major groups of storage proteins, the acid- or alkaline-soluble glutelins and the alcohol-soluble prolamins. During seed development, these storage proteins are synthesized on the rough (r)ER and translocated into the rER lumen, where oxidative protein folding proceeds. Disulfide bond formation is necessary for the sorting of the proteins to the protein bodies (PBs) (1). The assembly of prolamins into the ER-derived PBs (PB-I) is stabilized by intermolecular disulfide bonds, whereas the glutelin precursors (proglutelins) form intramolecular disulfide bonds before being transported via the Golgi from the ER to the protein storage vacuole (PSV, also designated PB-II) (2–4).
Protein disulfide isomerase (PDI) is the principal catalyst for disulfide-linked protein folding in the ER lumen, functioning as the direct donor of disulfide bonds to nascent polypeptides by a thiol-disulfide exchange reaction (5). The activity of PDI depends on 2 pairs of Cys residues, each of which is found in the CGHC motif within a thioredoxin-like redox-active domain (5). The rice genome encodes 7 OsPDI-like (L) proteins containing 2 thioredoxin-like active domains with the CxxC motif, OsPDIL1;1-1;4 and OsPDIL2;1-2;3 (6). OsPDIL1;1 is an orthologue of Zea mays (Zm)PDIL1;1, which is by far the most highly expressed in the maize endosperm (6). Studies on the OsPDIL1;1-knockout rice mutant, esp2, showed that OsPDIL1;1 has an important role for the segregation of prolamins and proglutelins in the ER (7). Glutathione constitutes the major small-molecule redox buffer in the ER. The redox state of the ER is more oxidative than that of the cytosol in eukaryotic cells. The ratio of reduced glutathione to oxidized form ([glutathione (GSH)]/[oxidized glutathione (GSSG)]) ranges from 1:1 to 3:1 in the ER, whereas it ranges from 30:1 to 100:1 in the cytosol (8). In the endosperm cells, which are devoted to the synthesis of disulfide-rich storage proteins, large amounts of reducing equivalents flux from the cytosol into the ER in the form of Cys residues during the synthesis of storage proteins, and storage proteins are sequestered from the ER lumen to the PBs. A key question is how the endosperm cells constantly establish the ER redox environment favorable for regeneration of oxidized PDIL; thus, facilitating oxidative folding of nascent polypeptides.
The flavoenzyme Ero1p was first identified in yeast by mutation studies of the temperature-sensitive conditional mutant ero1-1, which fails to support protein disulfide bond formation in the ER and consequently accumulates misfolded proteins (9, 10). Ero1p supplies oxidizing equivalents for disulfide bond formation in the ER, by relaying the oxidizing power from molecular oxygen to the reduced yeast Pdi1p (11–13). Ero1p directly oxidizes the active site of Pdi1p by thiol-disulfide exchange reactions with the oxidized shuttle Cys pair, Cys-100-Cys-105, the reduced form of which is reoxidized by the active-site Cys pair, Cys-352-Cys-355 (14–16). Ero1 proteins of different species characterized to date are all localized to the ER. However, the mechanisms for their ER retention appear to differ; yeast Ero1p is tightly associated with the ER membrane (9, 10), whereas human Ero1α and Ero1β are retained in the ER by covalent interactions with PDI and ERp44 (17). The cDNA clones encoding plant homologues of yeast Ero1p, AERO1 and AERO2, have been isolated from Arabidopsis (Arabidopsis thaliana) (18). In vitro translation analysis showed that these Arabidopsis Ero1 homologues are glycoproteins (18), but their localization in the cell has not been characterized.
In this study, we investigated the roles of OsEro1 protein as a source of protein disulfide bonds in the endosperm. The 3n endosperm cells are unique; the endosperm exhibits a high level of enzymatic activity to synthesize a vast amount of storage disulfide-rich proteins during early seed development; and the endosperm, but not the adjacent 2n embryo, is destined for programmed cell death during the subsequent seed desiccation and maturation phase. We studied the OsEro1 localization in the subaleurone cells by confocal microscopic and biochemical analyses, and demonstrated that the N-terminal region of OsEro1 functions as a rER membrane-targeting signal. We produced OsERO1-knockdown rice plants by inducing RNAi under the control of an endosperm-specific promoter, and examined the effects on the formation of disulfide bonds and OsPDIL expression. We detected the generation of H2O2 in the rER in correlation with the oxidation of sulfhydryl groups, and will discuss its physiological role in seed maturation.
Results
Subcellular Localization of OsEro1 in the Endosperm Subaleurone Cells.
A rice homologue (OsERO1) of yeast ERO1 was identified by BLAST search of the rice genomic sequence and the EST database. OsERO1 is predicted to encode a polypeptide of 474-aa residues containing 2 pairs of Cys residues, Cys-134-Cys-139 and Cys-391-Cys-394, which are suggested to be catalytically active sites in yeast Ero1p (14–16). OsEro1 shows 71 and 56% identity to Arabidopsis AERO1 and Physcomitrella patens Ero1, respectively, and 36 and 29% identity to human Ero1α and yeast Ero1p, respectively.
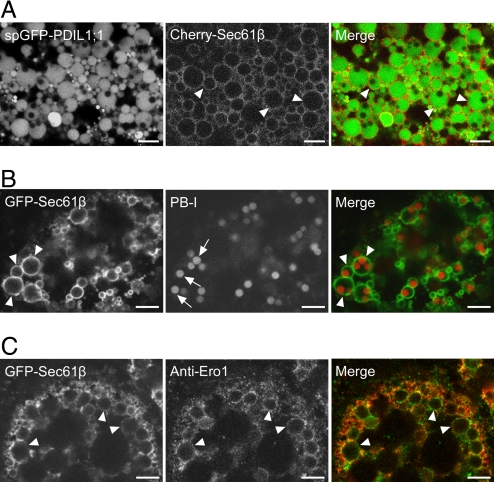
OsEro1 does not contain an obvious ER retention signal such as the C-terminal tetrapeptide KDEL. We examined subcellular localization of OsEro1 in the subaleurone cells of the rice endosperm during early seed development (for the subaleurone layers examined, see Fig. S1). First, we identified the rER structure in the subaleurone cells by using GFP-fused OsPDIL1;1 and Cherry-fused Sec61β (the single membrane-spanning β subunit of rice Sec61) as a rER lumen marker and a rER membrane marker, respectively. PDI is a highly abundant resident protein of the ER lumen (5). The Sec61αβγ complex is the essential core of the protein translocation machinery in the ER membrane and is tightly associated with membrane-bound ribosome (19). Confocal microscopic analysis revealed that spGFP-OsPDIL1;1 was evenly distributed within the dilated rER, and Cherry-Sec61β was localized to the boundary of the dilated rER (Fig. 1A). Note that PB-I was formed within the dilated rER (Fig. 1B; Fig. S2). By immunofluorescence microscopy with the anti-OsEro1 antibody, we observed that OsEro1 was mostly colocalized with GFP-Sec61β (Fig. 1C), which suggests that OsEro1 is associated with the rER membrane.
Fig. 1.
OsEro1 localizes to the rER membrane in the subaleurone cells. (A) Confocal fluorescence images of the subaleurone cells (10 DAF) expressing spGFP-OsPDIL1;1 and Cherry-Sec61β. (B) Confocal fluorescence images of the subaleurone cells (10 DAF) expressing GFP-Sec61β. PB-I was labeled with Rhodamine. (C) GFP-Sec61β-expressing subaleurone cells (14 DAF) were labeled with the anti-OsEro1 antibody and then with the Rhodamine-conjugated anti-rabbit antibody. Arrowheads and arrows indicate the rER and PB-I, respectively. (Scale bar, 5 μm.)
Characterization of the N-Terminal Region of OsEro1 on rER Membrane Targeting.
The algorithm TMpred (20) predicts that OsEro1 contains a single transmembrane domain (TMD; Ala-37 to Ser-55) with a score of 2436 (Fig. S3A). By comparison, the yeast Ero1p and human Ero1α TMpred scores are lower (786 and 1806, respectively). To examine the role of the putative TMD of OsEro1 in subcellular localization, we expressed each of 3 chimeric genes encoding modified GFPs in rice endosperm cells: the full length of OsEro1 (Met-1 to Ile-474) followed by GFP (OsEro1-GFP), the N-terminal truncated form of OsEro1 (Ser-56 to Ile-474) fused to spGFP (spGFP-OsEro1ΔN), and the N-terminal 75-aa residues from OsEro1 followed by GFP (OsEro1ΔC-GFP) (Fig. 2 A and B). Confocal microscopic analysis of the subaleurone cells revealed that OsEro1-GFP was localized to the rER membrane (Fig. 2C). In contrast, deletion of the N-terminal 55-aa residues of OsEro1 (spGFP-OsEro1ΔN) led to dispersion of the GFP fusion protein evenly throughout the rER lumen (Fig. 2D). OsEro1ΔC-GFP was targeted to the rER membrane (Fig. 2E).
Fig. 2.
The N-terminal region of OsEro1 is necessary and sufficient for rER membrane localization. (A) Schematic representation of constructs. Blue boxes, OsEro1; red boxes, predicted TMD of OsEro1 (Ala-37 to Ser-55); yellow lines, catalytic active sites of OsEro1 (Cys-134-Cys-139 and Cys-391-Cys-394); green boxes, GFP. (B) Extracts from seeds (10 DAF) expressing each of 3 chimeric genes encoding OsEro1-GFP (lane 1), spGFP-OsEro1ΔN (lane 2), and OsEro1ΔC-GFP (lane 3) were subjected to SDS/PAGE, followed by Western blot analysis with anti-GFP antibody. The predicted molecular mass of OsEro1-GFP, spGFP-OsEro1ΔN, and OsEro1ΔC-GFP are 80, 78, and 34 kDa, respectively. (C–E) Confocal fluorescence images of the subaleurone cells (10 DAF) expressing OsEro1-GFP (C), spGFP-OsEro1ΔN (D), and OsEro1ΔC-GFP (E). PB-I was labeled with Rhodamine. Arrowheads indicate the rER. The fluorescence signals of DsRed-Sec61β are visible in C, but those of DsRed-Sec61β (D) and Cherry-Sec61β (E) are not detectable. (Scale bar, 5 μm.)
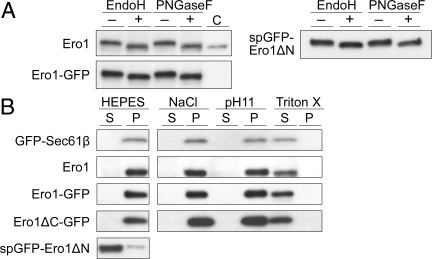
OsEro1 contains 2 potential N-glycosylation sites at Asn-381 and Asn-422. The treatment of seed extracts with either Endo H or PNGase F caused a mobility shift of a polypeptide that reacted with the anti-OsEro1 antibody (see panel Ero1 in Fig. 3A), confirming that OsEro1 is glycosylated. We analyzed the properties of membrane binding of OsEro1 by fractionation of cell extracts under various conditions. Both OsEro1 and GFP-Sec61β were recovered primarily in the membrane pellet fraction (Fig. 3B), and were solubilized from the membrane pellet by detergent (1% TritionX-100), but not by high salt (1 M NaCl) or by an alkaline pH (pH 11) (Fig. 3B). OsEro1-GFP, which was also glycosylated (Fig. 3A), and OsEro1ΔC-GFP showed membrane-binding properties similar to those of the endogenous OsEro1 (Fig. 3B). In contrast, spGFP-OsEro1ΔN, which was also glycosylated (Fig. 3A), was recovered primarily in the soluble fraction (Fig. 3B). Together, these results indicate that the N-terminal region (Met-1 to Ser-55) of OsEro1 is necessary and sufficient for targeting the protein to the rER membrane.
Fig. 3.
OsEro1 is a glycoprotein with the N-terminal region targeting to the rER membrane. (A) Extracts from seeds (10 DAF) expressing OsEro1-GFP or spGFP-OsEro1ΔN were incubated in the absence (−) or presence (+) of Endo H or PNGase F and then subjected to SDS/PAGE. Crude extracts of Escherichia coli cells transformed with the OsEro1 expression vector, expressing OsEro1 Met-1 to Ile-474 with a predicted MW of 53 kDa, were also separated by SDS/PAGE (Left, lane C). (B) Extracts from seeds (10 DAF) expressing each of 3 chimeric genes encoding OsEro1-GFP, OsEro1ΔC-GFP, and spGFP-OsEro1ΔN were fractionated by centrifugation under the following conditions: control buffer, high-salt buffer, alkaline buffer, and Triton X-100 buffer. The supernatant (S) and pellet (P) fractions were separated by SDS/PAGE. The endogenous and GFP-fused OsEro1 proteins were analyzed by Western blot analysis with antibodies against OsEro1 (Ero1, Ero1-GFP, and spGFP-Ero1ΔN) and GFP (Ero1ΔC-GFP). As a positive control, extracts from GFP-Sec61β-expressing seeds were fractionated as above, followed by Western blot analysis with anti-GFP antibody.
Generation of H2O2 in the rER of the Subaleurone Cells.
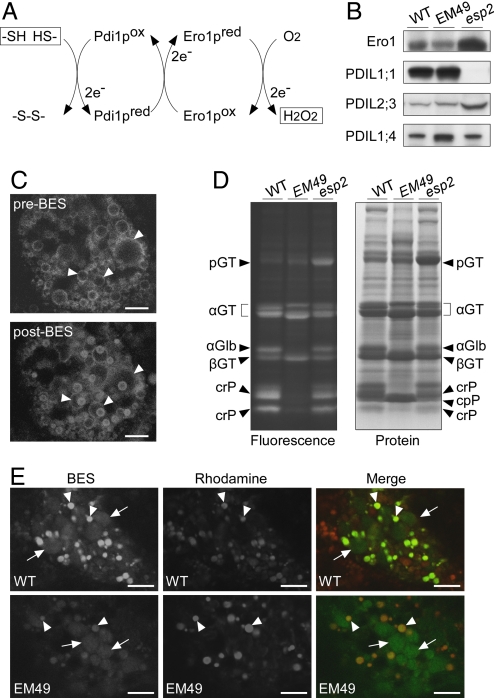
The reduced form of yeast Pdi1p is oxidized by Ero1p, which, in turn, directly reduces molecular oxygen; thus, regenerating oxidized Ero1p and yielding stoichiometric levels of H2O2 (Fig. 4A) (11–13). Western blot analysis of seed extracts detected OsPDIL proteins, OsPDIL1;1, OsPDIL2;3, and OsPDIL1;4, corresponding to predicted molecular masses of 54, 46, and 60 kDa, respectively (Fig. 4B), whose maize counterparts (ZmPDIL1;1, ZmPDIL2;3, and ZmPDIL1;3, respectively) are expressed at much higher levels than other ZmPDILs in the endosperm (6). Note that esp2 mutant seeds, in which OsPDIL1;1 was completely absent, accumulated OsEro1 and OsPDIL2;3 to markedly enhanced levels compared with the WT seeds, but accumulated OsPDIL1;4 to only the WT level (Fig. 4B).
Fig. 4.
Detection of hydrogen peroxide in the rER of the subaleurone cells. (A) An electron transfer pathway for disulfide bond formation in yeast. (B) Western blot analysis of OsEro1, OsPDIL1;1, OsPDIL2;3, and OsPDIL1;4 in the WT, EM49 and esp2 mature seeds. (C) Confocal fluorescence images of GFP-Sec61β-expressing subaleurone cells (10 DAF; pre-BES) treated with the BES-H2O2 probe (post-BES). Arrowheads indicate the rER. (Scale bar, 5 μm.) (D) Fluorescent labeling of sulfhydryl groups in total extracts from WT, EM49, and esp2 mature seeds. Fluorescence bands were visualized under UV light and then stained with Coomassie Brilliant Blue; pGT, proglutelin; αGT, glutelin α subunit; βGT, glutelin β subunit; αGlb, α-globulin; crP, Cys-rich prolamin; cpP, Cys-poor prolamin. (E) Confocal fluorescence images of BES-H2O2-treated subaleurone cells. The WT and homozygous mutant EM49 seeds (20 DAF) were selected from the same panicle of an EM49 heterozygous plant. Fluorescence images were acquired under the same conditions for direct comparison of the signal intensities. Arrowheads and arrows indicate the ER-derived PBs (PB-I) and amyloplasts, respectively. (Scale bar, 10 μm.)
A fluorescent probe, BES-H2O2, which is converted to the membrane-impermeant derivative by intracellular esterase, exhibits high selectivity for intracellular H2O2 by H2O2-mediated hydrolytic deprotection of acyl derivative of fluorescein (21). BES-H2O2 was used to determine whether H2O2 is generated in the rER of the subaleurone cells. As shown in Fig. 4C, BES-H2O2 treatment generated fluorescence signals within the rER. BES-H2O2-derived signals appeared as rings or speckles, rather than an even diffusion, and merged with Rhodamine-stained PB-I (Fig. 4 C and E). It is conceivable that both Rhodamine and BES-H2O2 dyes preferentially bind to hydrophobic PB-I in the rER lumen.
Prolamins, encoded by a multigene family, are divided into the Cys-rich and Cys-poor prolamins. The 10-kDa (λRP10) and 16-kDa (λRP16) prolamins contain 10 and 12 Cys residues, respectively, whereas the 13-kDa prolamin (λRM4) has no Cys residues (3, 4). EM49 possesses a seedling-lethal recessive mutation and is sustained only as a heterozygous mutant line (22). The homozygous mutant seeds of EM49 have fewer sulfhydryl groups than the WT seeds, accumulating Cys-rich prolamins and α-globulin (the saline-soluble storage protein) at markedly reduced levels and Cys-poor prolamins at increased levels (Fig. 4D). In the heterozygous or homozygous WT seeds of EM49, BES-H2O2 fluorescence signals appeared in the rER more strongly (see panel WT, arrowheads, in Fig. 4E) than in the amyloplasts (see panel WT, arrows, in Fig. 4E; for amyloplast identification, see Fig. S4). However, in the EM49 homozygous mutant seeds, the BES-H2O2 fluorescence intensity in the rER was weak at levels similar to those observed in the amyloplasts (see panel EM49, arrowheads and arrows, in Fig. 4E). These results indicate that the generation of H2O2 in the rER correlates with the oxidation of sulfhydryl groups.
Effects of RNAi Knockdown of OsERO1 on the Formation of Disulfide Bonds in Proglutelins.
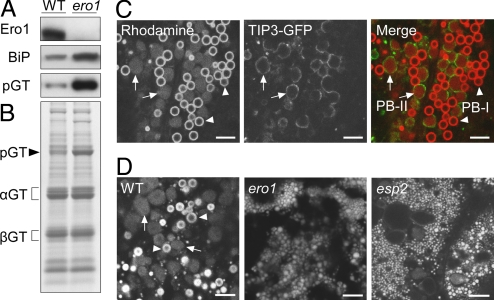
To investigate the function of OsEro1 in the subaleurone cells, we obtained OsERO1-knockdown lines (ero1) with severely decreased levels of OsEro1 in seeds by inducing RNAi in the endosperm (Fig. 5A). Note that BiP, the major ER chaperone, was strongly induced in the ero1 seeds (Fig. 5A). Compared with the WT seeds, the ero1 seeds accumulated proglutelins at a higher level (Fig. 5 A and B; Fig. S5). The WT subaleurone cells formed 2 distinct types of PBs, PB-I and PB-II, which were differentially stained with Rhodamine (Fig. 5C). Rhodamine labels PB-I (1–2 μm) as intense signals due to the preference of Rhodamine for binding hydrophobic prolamin polypeptides (see arrowheads in Fig. 5C; Fig. S2) (23). The rice tonoplast intrinsic protein OsTIP3 is specifically localized to the membrane of PB-II (3–4 μm) (Fig. S6A) (24). Rhodamine weakly and evenly stained the matrix of PB-II (see arrows in Fig. 5C; Fig. S6B), which was distinguished from the dilated rER (Fig. S6 C–F). The ero1 seeds failed to form the typical PB-I and PB-II, and instead, formed abnormal aggregates of Rhodamine-stained small particles (see panel ero1 in Fig. 5D), as observed in esp2 mutant seeds (see panel esp2 in Fig. 5D).
Fig. 5.
The RNAi knockdown of OsERO1 in seeds. Total extracts of the WT and ero1 seeds (17 DAF) were subjected to SDS/PAGE, followed by Western blot analysis with antibodies against OsEro1, BiP, and glutelin (A) or Coomassie Brilliant Blue staining (B); pGT, proglutelin; αGT, glutelin α subunit; βGT, glutelin β subunit. (C) Confocal fluorescence images of the subaleurone cells (17 DAF) expressing OsTIP3-GFP (PB-II membrane marker), stained with Rhodamine. (D) Confocal fluorescence images of Rhodamine-stained subaleurone cells (17 DAF) from the WT, ero1, and esp2 seeds. Arrowheads and arrows indicate PB-I and PB-II, respectively. (Scale bar, 5 μm.)
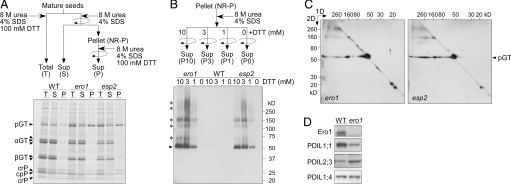
Proglutelins (57 kDa) acquire intramolecular disulfide bonds in the ER before being transported to PB-II and proteolytically processed into the acidic (α; 37–39 kDa) and basic (β; 22–23 kDa) subunits in PB-II (2). Proglutelins accumulated at a markedly lower level in the WT seeds than did the α and β subunits (Fig. 6A, lane WT, T). By comparison, proglutelins accumulated to a higher level in the ero1 seeds than did the α and β subunits (Fig. 6A, lane ero1, T), which was also the case for the esp2 seeds (Fig. 6A, lane esp2, T). The proglutelins were efficiently extracted from the WT seeds into the supernatant without a reducing agent (Fig. 6A, lane WT, S). Note that extraction of the proglutelins from the ero1 and esp2 seeds required a reducing agent (Fig. 6A, lanes ero1 and esp2, P). Although proglutelins also accumulated to a higher level in seeds with enhanced expression of SEC12 (PGT1-SEC12; Fig. S7 A and B), the PGT1-SEC12 seeds showed proglutelin extraction into the supernatant without a reducing agent (Fig. S7C). The 57-kDa proglutelins (see arrowhead in Fig. 6B) and larger apparent molecular mass (Fig. 6B, asterisks) were extracted from the nonreduced pellet fractions (NR-P; Fig. 6 A and B Upper) of the ero1 and esp2 seeds in a DTT concentration-dependent manner (Fig. 6B, lanes ero1 and esp2), whereas those were hardly detectable in the WT fractions (Fig. 6B, lanes WT). To further examine whether the NR-P fractions from the ero1 and esp2 seeds contain proglutelins with nonnative intermolecular disulfide bonds, we used a modified 2D SDS/PAGE separation (25), in which the NR-P proteins were partially reduced at 3 mM DTT in the first dimension, and then reduced at 200 mM DTT in the second dimension. The separation of the ero1 seed extracts recovered the proglutelins in a broad molecular mass range below the diagonal line, which was also the case for esp2 (Fig. 6C), indicating that the proglutelins form aggregates through nonnative intermolecular disulfide bonds in the ero1 and esp2 seeds.
Fig. 6.
Proglutelins form aggregates through nonnative intermolecular disulfide bonds in the ero1 seeds. (A) Proteins were extracted from the WT, ero1, and esp2 mature seeds, as in schematic representation (Upper). The T, S, and P fractions were subjected to SDS/PAGE, followed by Coomassie Brilliant Blue staining; pGT, proglutelin; αGT, glutelin α subunit; βGT, glutelin β subunit; crP, Cys-rich prolamin; cpP, Cys-poor prolamin. (B) The NR-P fractions were homogenized in the presence of varying concentrations of DTT (0, 1, 3, and 10 mM) for 30 min at 25 °C. After centrifugation, the resulting supernatants (P0, P1, P3, and P10; Upper) were subjected to SDS/PAGE, followed by Western blot analysis with anti-glutelin antibody. (C) The P3 fractions from the ero1 and esp2 seed extracts were subjected to 2D SDS/PAGE, followed by Western blot analysis with anti-glutelin antibody. (D) Western blot analysis of OsEro1, OsPDIL1;1, OsPDIL2;3, and OsPDIL1;4 in the WT and ero1 mature seeds (fractions T in A).
Discussion
Pathway for Disulfide Bond Formation Involving OsEro1.
This report provides key insights into how plant cells establish the redox environment in the ER for the formation of protein disulfide bonds. The RNAi knockdown of OsERO1 and knockout of OsPDIL1;1 led to aggregation of the proglutelins through nonnative intermolecular disulfide bonds (Figs. 5D and 6C), which demonstrates that OsEro1 and OsPDIL1;1 are essential for the formation of the correct pattern of disulfide bonds in the subaleurone cells. Yeast Ero1p activity is rapidly modulated by a redox-dependent feedback system through 2 noncatalytic Cys pairs (Cys-90-Cys-349 and Cys-150-Cys-295); reduction of the regulatory disulfide bonds increases Ero1p activity for the promotion of disulfide bond formation by direct oxidation of Pdi1p (26). On the basis of sequence similarity, the Cys pair Cys-90-Cys-349 of Ero1p is conserved, but the Cys-150-Cys-295 equivalent is missing in OsEro1 (Fig. S3B). Recent studies revealed that human Ero1α possesses a different mode of regulation, in which Cys-94 functions as a molecular switch by forming an active disulfide bond with Cys-99 (shuttle Cys pair) or a regulatory disulfide bond with Cys-131 (27). Such a feedback system may exist in OsEro1 regulation, because Cys-134, Cys-139, and Cys-167 of OsEro1 appear to correspond to Cys-94, Cys-99, and Cys-131 of Ero1α, respectively (Fig. S3B).
The changes in the ER redox environment induce a set of proteins involved in protein folding in the ER, including Ero1p (9, 10), Pdi1p (28), and BiP (7). In the esp2 seeds, OsEro1 was up-regulated (Fig. 4B), presumably in response to the accumulation of misfolded proglutelins. Interestingly, in the ero1 seeds, which exhibited the unfolded protein response-related induction of BiP (Fig. 5A), the protein levels of OsPDIL1;1 markedly decreased, whereas OsPDIL2;3 and OsPDIL1;4 accumulated to enhanced and WT levels, respectively (Fig. 6D), as observed in the esp2 seeds (Fig. 4B). OsPDIL1;1 and OsPDIL1;4 show a similar organization of the 2 redox-active (a and a′) and 2 redox-inactive (b and b′) domains in the order a-b-b′-a′, which is similar to the order in yeast Pdi1p and human PDI, whereas the domains of OsPDIL2;3 are in the order a-a′-b, which is similar to the order in human P5. The mechanism for the different responses of these OsPDILs to the changes in the rER redox environment is not clear, but OsPDIL1;1, OsPDIL2;3, and OsPDIL1;4 may have distinct functions in the subaleurone cells.
Although OsEro1 activity is essential for the formation of native disulfide bonds in the subaleurone cells, the proteins nevertheless formed native and nonnative disulfide bond pairings in the ero1 seeds (Fig. 6), indicating that the alternative electron transfer pathways for disulfide bond formation operate in the subaleurone cells. A possible pathway may involve quiescin-sulfhydryl oxidase, which has been identified in higher eukaryotes, and can catalyze the direct oxidation of a wide range of unfolded proteins without additional partners in vitro (29).
N-Terminal Region-Dependent Localization of OsEro1 to the rER Membrane.
The typical ER of higher plants is composed of a network of the flat lamellar and tubular cisternae throughout the cytoplasm, corresponding to the rough and smooth ER, respectively (30). Unlike the typical rER structures found in higher plants, the rER lumen (spGFP-OsPDIL1;1) and membrane marker (Cherry-Sec61β) proteins revealed the dilated rER structures in the subaleurone cells during early stages of seed development (Fig. 1A). Such a unique structure of the dilated rER allowed us to distinguish the membrane-bound OsEro1 from luminal proteins such as OsPDIL1;1 by confocal microscopic analysis (Figs. 1 and 2). In vivo mutational analyses demonstrated that the N-terminal region (Met-1 to Ser-55) of OsEro1 functions as a rER membrane-targeting signal (Figs. 2 and 3). The C-terminal region (Ser-56 to Ile-474) of OsEro1 contains the highly conserved amino acid residues essential for catalytic activity (Cys-134, Cys-139, Cys-391, and Cys-394), for FAD binding (Trp-230, His-261, and Arg-298), and for N-glycosylation (Asn-381 and Asn-422), which are all expected to occur within the rER lumen. We predict that OsEro1 has a type II topology with the N-terminal TMD (Ala-37 to Ser-55) and the C-terminal region (Ser-56 to Ile-474) exposed to the rER lumen.
Biochemical analyses revealed that yeast Ero1p is tightly associated with the luminal face of the ER membrane (9), requiring the C-terminal tail of 127-aa residues (31). By comparison, human Ero1α, which lacks the C-terminal domain tail found in Ero1p, contains a signal peptide of 23-aa residues (31, 32), and is retained in the ER by covalent interactions with PDI and ERp44 (17). OsEro1ΔC-GFP, which contains a sequence rich in Ala and Pro residues (Met-1 to Trp-36) and the putative TMD (Ala-37 to Ser-55), was localized to the rER membrane (Figs. 2E and 3B), suggesting that the N-terminal region of OsEro1 (Met-1 to Ser-55) may also act as a rER-retention signal.
Possible Roles of Hydrogen Peroxide in Seed Maturation.
The endosperm cells undergo induction of desiccation and programmed cell death during the late stages of seed maturation, the progression of which is triggered by ethylene signaling (33). Overexpression of Ero1p in yeast cells causes a depletion of GSH and increased generation of reactive oxygen species, which contributes to cell death (34, 35). The major intracellular sources of H2O2 in higher plants during normal metabolism include the Mehler reaction in chloroplasts, the mitochondrial electron transport chain, and the peroxisomal respiratory pathway. The present study suggests that disulfide bond formation in the rER lumen may lead to the production of substantial amounts of H2O2 (Fig. 4 C–E). Interestingly, wheat PER1, which has thiol-specific antioxidant activity to scavenge H2O2, is expressed in the aleurone and embryo made up of viable cells in the mature seeds, but not in the endosperm destined for cell death (36). High concentrations of H2O2 in the rER, if not scavenged promptly, can cause peroxidation of the membrane lipid, which may gradually deteriorate the membrane integrity and lead to the leakage of small molecules, including water (37). It should be noted that the amount of H2O2 in the rER significantly decreased in the homozygous mutant seeds of EM49 (Fig. 4E). The homozygous mutant seeds of EM49 contained a higher ratio of water (65 ± 3%, n = 6) than the WT seeds (33 ± 3%, n = 6) at the same stage [20 days after flowering (DAF)] of seed development, whereas the mature seeds of the homozygous EM49 mutant and WT contained a similar water content (8.5 ± 0.2 and 8.7 ± 0.4, respectively; n = 6). It is possible that H2O2 in the rER can function as a signal for inducing programmed endosperm cell death or can directly damage the rER membrane. If this hypothesis is correct, the synthesis of a large amount of storage proteins, which supply amino acids for germination and initial seedling growth, may also contribute to programmed endosperm cell death and seed desiccation by the concomitant generation of H2O2.
Materials and Methods
Plasmid Construction and Rice Transformation.
The binary vectors (listed in Table S1) were generated by the Gateway system (Invitrogen) and used to transform rice, as described in SI Materials and Methods. The plasmid for OsERO1 RNAi contains inverted repeat of a sequence encoding OsEro1 Val-119 to Lys-410. A peptide derived from rice α-globulin, Met-1 to Ser-27, was added to sGFP (S65T) (38), designated spGFP. PCR primers are listed in Table S2.
Suborganellar Fractionation and Deglycosylation Analysis.
Rice seeds (10 DAF) were homogenized in 10 volumes (vol/wt) of buffer A (50 mM Hepes-KOH, pH 7.5, 5 mM EDTA, and protease inhibitors; Roche Applied Science). After centrifugation (10,000 × g, 20 min, 4 °C), the resulting supernatant was treated with each solution of buffer A, high-salt buffer (1 M NaCl in buffer A), alkaline buffer (0.1 M Na2CO3, pH 11, in buffer A), and Triton X-100 buffer (1% [vol/vol] Triton X-100 in buffer A). After incubation on ice for 1 h, the samples were fractionated by ultracentrifugation at 100,000 × g for 1 h at 4 °C. Supernatants were collected and mixed with SDS sample buffer, and membrane pellets were solubilized in the SDS sample buffer at the same final volume as the supernatants. Proteins in the supernatant (for spGFP-OsEro1ΔN) and NaCl-treated membrane pellet (for the endogenous OsEro1 and OsEro1-GFP) fractions were incubated in the presence of Endo H or PNGase F (NEB) according to the manufacturer's instructions. For details, see SI Materials and Methods.
Protein Extraction from Rice Seeds and Fluorescent Labeling of Sulfhydryl Groups.
Total proteins were extracted from rice seeds in 20% (vol/vol) glycerol, 4% (wt/vol) SDS, 6 M urea, 100 mM DTT, and 50 mM Tris·HCl, pH 6.8 (200 μL per 10 mg of mature seed or 700 μL per 1 developing seed). Fluorescent labeling of sulfhydryl groups was performed as described in SI Materials and Methods.
For protein fractionation (Fig. 6A), proteins were extracted from rice mature seeds in nonreducing buffer B [10% (vol/vol) glycerol, 4% (wt/vol) SDS, 8 M urea, and 50 mM Tris·HCl, pH 6.8; 700 μL per seed] by vigorous shaking for 2.5 h at 25 °C. The homogenate was centrifuged at 20,400 × g for 10 min at 25 °C, and supernatants were collected (fraction S). The resulting pellets (fraction NR-P) were homogenized in buffer B containing 0.1 M DTT for 30 min at 25 °C, and the soluble fractions were collected by centrifugation as described above (fraction P). Total proteins (fraction T) were extracted from mature seeds in buffer B containing 0.1 M DTT. The S fractions were reduced with 0.1 M DTT before SDS/PAGE analysis.
SDS/PAGE and Western Blot Analysis.
The NR-P fractions were homogenized in buffer B (350 μL per 10 mg of seed; for details, see SI Materials and Methods), and aliquots were incubated in the presence of varying concentrations of DTT (0, 1, 3, and 10 mM) for 30 min at 25 °C. After centrifugation (10,000 × g, 10 min, 25 °C), the resulting supernatants (fractions P0, P1, P3, and P10) were subjected to SDS/PAGE (5–20% acrylamide gradient; Fig. 6B). The P3 fractions were subjected to 2D SDS/PAGE (5–20% acrylamide gradient; Fig. 6C) as described previously (25). The gel lane was immersed in SDS sample buffer containing 200 mM DTT for 30 min at 25 °C before SDS/PAGE in the second dimension. To visualize the diagonal line, a reducing agent-free protein molecular marker (Invitrogen) was used.
Proteins were separated by SDS/PAGE (10–20% acrylamide, unless otherwise indicated), electroblotting to a polyvinylidene difluoride membrane (Atto) was followed by immunodetection using specific antibodies. Antibodies specific for OsEro1, OsPDIL2;3, and OsPDIL1;4 were raised against fragments derived from OsEro1, Arg-57 to Ile-474 (prepared as described in SI Materials and Methods), OsPDIL2;3, Ser-150 to Ala-166, and OsPDIL1;4, Asp-132 to Gly-147. The antibodies against OsPDIL1;1, BiP, and glutelin β subunit were prepared previously (1, 7). Anti-GFP antibody was purchased from MBL. The antigen-antibody complex was visualized with horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (GE Healthcare).
Confocal Laser Scanning Microscopy.
Rhodamine labeling of PBs, immunofluorescent staining, and detection of intracellular H2O2 generation were performed as described in SI Materials and Methods. The fluorescence images of the subaleurone cells were analyzed with a confocal laser scanning microscope with laser beams of wavelengths 488 and 543 nm (TCS SP2 AOBS; Leica). The data were processed using Adobe Photoshop CS3.
Supplementary Material
Acknowledgments.
This work was supported by the Research and Development Program for New Bio-Industry Initiatives from the Bio-Oriented Technology Research Advanced Institution.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904429106/DCSupplemental.
References
- 1.Kawagoe Y, et al. The critical role of disulfide bond formation in protein sorting in the endosperm of rice. Plant Cell. 2005;17:1141–1153. doi: 10.1105/tpc.105.030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamagata H, Sugimoto T, Tanaka K, Kasai Z. Biosynthesis of storage proteins in developing rice seeds. Plant Physiol. 1982;70:1094–1100. doi: 10.1104/pp.70.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogawa M, et al. Purification of protein body-I of rice seed and its polypeptide composition. Plant Cell Physiol. 1987;28:1517–1527. [Google Scholar]
- 4.Mitsukawa N, et al. Amino acid sequencing and cDNA cloning of rice seed storage proteins, the 13kDa prolamins, extracted from type I protein bodies. Plant Biotechnol. 1999;16:103–113. [Google Scholar]
- 5.Wilkinson B, Gilbert HF. Protein disulfide isomerase. BBA-Proteins Proteom. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 6.Houston NL, et al. Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 2005;137:762–778. doi: 10.1104/pp.104.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takemoto Y, et al. The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 2002;128:1212–1222. doi: 10.1104/pp.010624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 9.Frand AR, Kaiser CA. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- 10.Pollard MG, Travers KJ, Weissman JS. Ero1p: A novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell. 1998;1:171–182. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- 11.Frand AR, Kaiser CA. Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol Cell. 1999;4:469–477. doi: 10.1016/s1097-2765(00)80198-7. [DOI] [PubMed] [Google Scholar]
- 12.Tu BP, Weissman JS. The FAD- and O2-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol Cell. 2002;10:983–994. doi: 10.1016/s1097-2765(02)00696-2. [DOI] [PubMed] [Google Scholar]
- 13.Gross E, et al. Generating disulfides enzymatically: Reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc Natl Acad Sci USA. 2006;103:299–304. doi: 10.1073/pnas.0506448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frand AR, Kaiser CA. Two pairs of conserved cysteines are required for the oxidative activity of Ero1p in protein disulfide bond formation in the endoplasmic reticulum. Mol Biol Cell. 2000;11:2833–2843. doi: 10.1091/mbc.11.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross E, Kastner DB, Kaiser CA, Fass D. Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell. 2004;117:601–610. doi: 10.1016/s0092-8674(04)00418-0. [DOI] [PubMed] [Google Scholar]
- 16.Sevier CS, Kaiser CA. Disulfide transfer between two conserved cysteine pairs imparts selectivity to protein oxidation by Ero1. Mol Biol Cell. 2006;17:2256–2266. doi: 10.1091/mbc.E05-05-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otsu M, et al. Dynamic retention of Ero1α and Ero1β in the endoplasmic reticulum by interactions with PDI and ERp44. Antioxid Redox Signal. 2006;8:274–282. doi: 10.1089/ars.2006.8.274. [DOI] [PubMed] [Google Scholar]
- 18.Dixon DP, Van Lith M, Edwards R, Benham A. Cloning and initial characterization of the Arabidopsis thaliana endoplasmic reticulum oxidoreductins. Antioxid Redox Signal. 2003;5:389–396. doi: 10.1089/152308603768295122. [DOI] [PubMed] [Google Scholar]
- 19.Gorlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann K, Stoffel W. TMbase-a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 21.Maeda H, et al. Fluorescent probes for hydrogen peroxide based on a non-oxidative mechanism. Angew Chem Int Ed. 2004;43:2389–2391. doi: 10.1002/anie.200452381. [DOI] [PubMed] [Google Scholar]
- 22.Matsusaka H, Kumamaru T, Ogawa M, Satoh H. In: Advances in Rice Genetics. Khush GS, Brar DS, Hardy B, editors. Manila, Philippines: International Rice Research Institute; 2003. pp. 441–444. [Google Scholar]
- 23.Hamada S, et al. Dual regulated RNA transport pathways to the cortical region in developing rice endosperm. Plant Cell. 2003;15:2265–2272. doi: 10.1105/tpc.013821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi H, et al. Differential localization of tonoplast intrinsic proteins on the membrane of protein body type II and aleurone grain in rice seeds. Biosci Biotechnol Biochem. 2004;68:1728–1736. doi: 10.1271/bbb.68.1728. [DOI] [PubMed] [Google Scholar]
- 25.Yano H, Wong JH, Lee YM, Cho MJ, Buchanan BB. A strategy for the identification of proteins targeted by thioredoxin. Proc Natl Acad Sci USA. 2001;98:4794–4799. doi: 10.1073/pnas.071041998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sevier CS, et al. Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell. 2007;129:333–344. doi: 10.1016/j.cell.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 27.Appenzeller-Herzog C, Riemer J, Christensen B, Sorensen ES, Ellgaard L. A novel disulphide switch mechanism in Ero1α balances ER oxidation in human cells. EMBO J. 2008;27:2977–2987. doi: 10.1038/emboj.2008.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 29.Rancy PC, Thorpe C. Oxidative protein folding in vitro: A study of the cooperation between quiescin-sulfhydryl oxidase and protein disulfide isomerase. Biochemistry. 2008;47:12047–12056. doi: 10.1021/bi801604x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staehelin LA. The plant ER: A dynamic organelle composed of a large number of discrete functional domains. Plant J. 1997;11:1151–1165. doi: 10.1046/j.1365-313x.1997.11061151.x. [DOI] [PubMed] [Google Scholar]
- 31.Pagani M, Pilati S, Bertoli G, Valsasina B, Sitia R. The C-terminal domain of yeast Ero1p mediates membrane localization and is essential for function. FEBS Lett. 2001;508:117–120. doi: 10.1016/s0014-5793(01)03034-4. [DOI] [PubMed] [Google Scholar]
- 32.Pagani M, et al. Endoplasmic reticulum oxidoreductin 1-Lβ (ERO1-Lβ), a human gene induced in the course of the unfolded protein response. J Biol Chem. 2000;275:23685–23692. doi: 10.1074/jbc.M003061200. [DOI] [PubMed] [Google Scholar]
- 33.Young TE, Gallie DR, DeMason DA. Ethylene-mediated programmed cell death during maize endosperm development of wild-type and shrunken2 genotypes. Plant Physiol. 1997;115:737–751. doi: 10.1104/pp.115.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuozzo JW, Kaiser CA. Competition between glutathione and protein thiols for disulphide-bond formation. Nat Cell Biol. 1999;1:130–135. doi: 10.1038/11047. [DOI] [PubMed] [Google Scholar]
- 35.Haynes CM, Titus EA, Cooper AA. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol Cell. 2004;15:767–776. doi: 10.1016/j.molcel.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 36.Stacy RA, Nordeng TW, Culianez-Macia FA, Aalen RB. The dormancy-related peroxiredoxin anti-oxidant, PER1, is localized to the nucleus of barley embryo and aleurone cells. Plant J. 1999;19:1–8. doi: 10.1046/j.1365-313x.1999.00488.x. [DOI] [PubMed] [Google Scholar]
- 37.Sattler SE, et al. Nonenzymatic lipid peroxidation reprograms gene expression and activates defense markers in Arabidopsis tocopherol-deficient mutants. Plant Cell. 2006;18:3706–3720. doi: 10.1105/tpc.106.044065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H. Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 1999;18:455–463. doi: 10.1046/j.1365-313x.1999.00464.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.