Abstract
In the unicellular cyanobacterium Synechococcus elongatus PCC 7942, essentially all promoter activities are under the control of the circadian clock under continuous light (LL) conditions. Here, we used high-density oligonucleotide arrays to explore comprehensive profiles of genome-wide Synechococcus gene expression in wild-type, kaiABC-null, and kaiC-overexpressor strains under LL and continuous dark (DD) conditions. In the wild-type strains, >30% of transcripts oscillated significantly in a circadian fashion, peaking at subjective dawn and dusk. Such circadian control was severely attenuated in kaiABC-null strains. Although it has been proposed that KaiC globally represses gene expression, our analysis revealed that dawn-expressed genes were up-regulated by kaiC-overexpression so that the clock was arrested at subjective dawn. Transfer of cells to DD conditions from LL immediately suppressed expression of most of the genes, while the clock kept even time in the absence of transcriptional feedback. Thus, the Synechococcus genome seems to be primarily regulated by light/dark cycles and is dramatically modified by the protein-based circadian oscillator.
Keywords: circadian clock, cyanobacteria, genome-wide expression, KaiC, light:dark
Most organisms exhibit daily cycles that are driven by endogenous circadian clocks. Until recently, transcription/translation feedback on central clock genes has been proposed as the core mechanism of circadian rhythm generation in any organism. A similar model was proposed for the functions of 3 essential clock genes, kaiA, kaiB and kaiC, in the cyanobacterium Synechococcus elongatus PCC 7942 (hereinafter, Synechococcus) under continuous light (LL) conditions (1). The model proposed the importance of feedback regulation in which KaiC inhibits its own (kaiBC) transcription, while being enhanced by KaiA (1). However, the circadian rhythm of KaiC phosphorylation persisted for at least 3 cycles after the rapid disappearance of kaiABC mRNA under continuous dark (DD) conditions even in the presence of excess transcription and translation inhibitors (2). More strikingly, incubating the 3 recombinant Kai proteins with ATP was sufficient to generate a temperature-compensated circadian KaiC phosphorylation cycle in vitro (3, 4). Thus, the core process in generating basic oscillation is not a transcription/translation feedback but is likely to be an enzymatic oscillation of KaiC phosphorylation/ATPase activities.
In Synechococcus, a random “promoter-trap” analysis method using a bacterial luciferase reporter has been adopted to monitor the promoter activities of ≈800 randomly chosen genes covering less than one-third of chromosomal genes (5). Strikingly, all promoter activities examined have shown circadian bioluminescent rhythms under LL conditions (5, 6). However, this method has technical limitations for discussing the genome-wide expression profile. First, because this method is based on initial selection of bright bioluminescent clones, it underestimates promoter activities driving dim bioluminescence. Second, it is a laborious task to identify promoter sequences because cloning and sequencing of the integrated promoter region are necessary. Third, because bacterial luciferase reactions require intracellular ATP and flavin mononucleotide (FMN) as substrates and for posttranscriptional processes, it cannot be easily applied to compare cells with different intracellular environments that affect ATP and/or FMN contents or rates of protein synthesis/degradation. An alternative is the DNA microarray method, which has successfully revealed clock-controlled genes in eukaryotic organisms (7). For cyanobacteria, Kucho et al. (8) used PCR fragment-based DNA microarrays to examine circadian transcription profiles under LL conditions in another unicellular species, Synechocystis sp. PCC 6803. However, the amplitude and precision of circadian rhythms are known to be much lower in Synechocystis (9, 10) than in Synechococcus, so most genetic and physiological studies of circadian rhythms have been performed exclusively in Synechococcus (11). More recently, Toepel et al. (12) reported a microarray analysis in another unicellular, diazotrophic cyanobacterium, Cyanothece sp. ATCC 51142 during light/dark (LD) and LL conditions. However, they did not describe any expression profiles of clock genes, and there are no reported genetic studies on clock regulation in Cyanothece, although circadian rhythms in carbohydrate granule formation and nitrogen-fixing activity have been found (13). Therefore, for a thorough integrative study of the cyanobacterial circadian system, it is essential to obtain a detailed description of the genome-wide expression profile of Synechococcus.
Recently, sequencing genomic DNAs of highly homologous Synechococcus elongatus strains PCC 6301 (14) and PCC 7942 (http://genome.ornl.gov/microbial/syn_PCC7942/ [NC_007604]) have been completed. Thus, we prepared high-density oligonucleotide microarrays (Affymetrix GeneChip) for chromosomal genes found in the genome of the PCC 6301 strain to investigate the temporal expression profiles of RNA from PCC 7942 cells grown under various conditions. Our analysis identified at least 800 clock-controlled genes among 2515 possible ORFs. Most of the genes were classified into two groups, one peaking at subjective dawn and the other peaking at subjective dusk, with most of the high-amplitude genes peaking at subjective dusk. In kaiABC-null mutant strains, most of these transcriptional rhythms disappeared as expected, while we found 17 genes possibly cycling with lower amplitude even in the absence of kai genes. Although it has been previously proposed that the kaiC gene product essentially represses all promoters (6), our microarray analysis revealed that kaiC overexpression arrested the clock at subjective dawn. Under DD conditions, accumulation levels of most gene products were dramatically lowered, regardless of whether they are circadian or continuously regulated under LL conditions. A small number of genes were up-regulated under dark conditions, but this activation was eliminated in the presence of excess transcription inhibitor without disturbing the KaiC phosphorylation cycle. We suggest that Synechococcus cells become nearly mRNA-deficient during the night phase, whereas transcription restarts from dawn so that they exhibit a clock-modified expression profile during the daytime.
Results and Discussion
Clock-Controlled Genes in Synechococcus elongatus.
Initially, we designed Affymetrix high-density oligonucleotide microarrays representing the 2515 predicted protein-coding genes on the genome of Synechococcus (14). To analyze clock-controlled mRNA accumulation profiles, we analyzed changes in the transcripts differentially expressed in the wild-type and kaiABC-null Synechococcus cells under LL conditions. On the basis of the 3-filtration method, taking into account oscillatory index parameters (amplitude, correlation P value; for the standard filtering conditions, P value of <0.1, and amplitude of >10−1) and reproducibility over 2 independent experiments (see SI Methods and Dataset S1), we extracted 800 genes as “significantly cycling” genes observed exclusively in the wild-type strains under LL conditions (Fig. 1 A and B). This number may be larger or smaller depending on the filtration cutoff values that are applied. Note that it is not surprising that a smaller number of rhythmic genes are revealed by DNA microarray analysis than by the promoter-trap assay. Time resolution is much lower by DNA microarray analysis (4 h within 48 h of sampling) compared with promoter-trap analysis (≈30 min over 4–7 days). Moreover, promoter-trap analysis can be used in viable individual bacterial colonies without being disrupted by cell collection, RNA extraction, or hybridization (5). Therefore, even very low-amplitude rhythms can be reliably analyzed in these assays. On the other hand, because promoter-trap analysis is based on translation and assembly of luciferase mRNA and proteins that undergo rapid degradation, it does not reflect the different half-lives of the target transcripts. Our microarray data confirmed high-amplitude kaiB and kaiC expression rhythms peaking at around circadian time (CT) 12 (1, 15) (Fig. 3A), and purF (5) (Fig. 4A) and psbAII (16) genes peaking during subjective dawn (Dataset S1). Moreover, we performed Northern blot analysis for 8 genes that exhibited different amplitude and peak times with a pattern that is similar to microarray profiles (Fig. S1), validating our analysis.
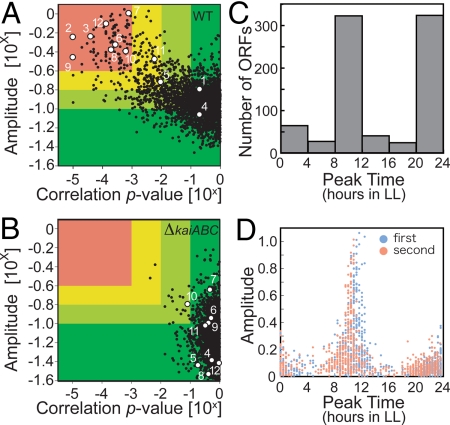
Fig. 1.
Analysis of genome-wide circadian transcription profiles under LL conditions. (A and B) Variations in the oscillatory indexes; amplitude (ordinate) and a cosine-fitting correlation score (correlation P value; abscissa) from each transcript under LL conditions in wild-type (WT; A) and in kaiABC-null (ΔkaiABC; B) strains. Definition of each score is described in SI Methods. Briefly, a higher amplitude indicates the standard deviation (SD) normalized to the mean value representing larger fluctuations, and a lower P value means better correlation to a periodic (sinusoidal) waveform with a 24 h period. Eight hundred “cycling genes” were identified with an amplitude of >0.1 and a P value of <0.1. For more stringent filtration, the lower amplitude and higher P value from 2 independent experiments were used to ensure reproducibility. The numbers in the panel indicate scores of 12 rhythmic or arrhythmic genes: point 1, kaiA; point 2, kaiB; point 3, kaiC; point 4, sasA; point 5, rpaA; point 6, cikA; point 7, rpoD5/sigC; point 8, rpoD6; point 9, purF; point 10, pilH/rre7; point 11, ctaC; and point 12, opcA. (C and D) Phase distributions of the peak expression times of 800 circadianly expressed genes. The numbers of ORFs (C) or the amplitudes from 2 independent experiments (D) are plotted on the ordinate.
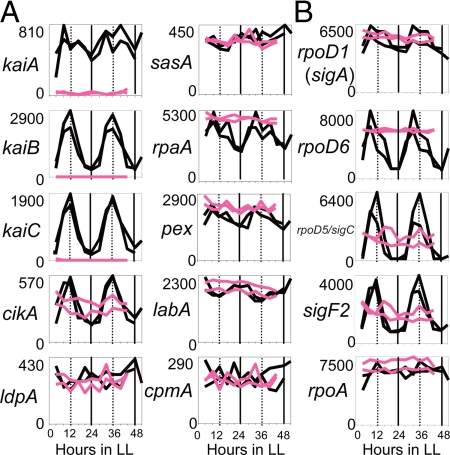
Fig. 3.
Temporal expression profiles of representative clock-related genes and sigma factor genes. Temporal profiles of clock-related genes and sigma factor genes in LL. Expression profiles of genes in wild-type (black) and kaiABC-null (red) strains from 2 independent experiments are shown. The number on the ordinate indicates relative expression level. (A) mRNA profiles of clock genes, kaiA, kaiB, kaiC, and clock-controlled genes, cikA, ldpA, sasA, rpaA, pex, labA and cpmA examined by microarray analysis. (B) Temporal profiles of the principal (Group 1) sigma factor gene, rpoD1/sigA, and high-amplitude Group 2 sigma factor genes, rpoD6, rpoD5/sigC and sigF2. For nomenclature of sigma factors, see ref. 32. rpoA encodes the alpha subunit of RNA polymerase.
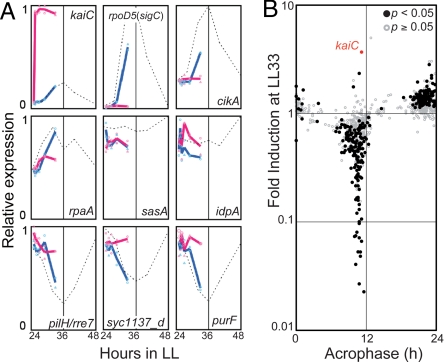
Fig. 4.
Overinduction of KaiC arrests the clock at subjective dawn. (A) Ptrc::kaiC cells were cultured in the presence (red) or absence (blue) of 100 μM IPTG from hour 25 (CT 1) for 8 h in LL after 12 h darkness and subjected to microarray analysis. Solid lines show the average mRNA level of the 2 independent experiments shown in dots. Dashed lines show the circadian expression profiles of corresponding genes in wild-type strains. Results for representative subjective dusk genes (kaiC, sigC, cikA), low-amplitude/arrhythmic genes (rpaA, sasA, ldpA) and subjective dawn genes (pilH/rre7, syc1137_d/Synpcc7942_0377, purF) are shown. (B) The expression level of each clock-controlled gene in Ptrc::kaiC cells in the presence of IPTG for 8 h was compared with that in the absence of IPTG at hour 33 in LL. The ordinate indicates the peak time of each of the 800 cycling genes as shown in Fig. 1D. Two hundred sixty-nine cycling genes that show significantly different expression between WT and Ptrc::kaiC cells are plotted on a logarithmic scale as black filled circles (Student's t test; P < 0.05) and the other genes are plotted as gray open circles.
Fig. 2A and Fig. S2A show the 800 cycling genes we extracted from the wild-type strains under LL conditions, in order of peak time. Clearly, there are 2 peaks at around CT 8–12 and CT 20–24 (Figs. 1C and 2A). The previous promoter-trap assays have revealed that ≈85% and 15% of examined promoters peaked at the subjective dusk and dawn, respectively (5). Similarly, when we used more stringent filtering conditions (P value of <0.001, and amplitude of >10−0.6) for the microarray data, 87 of 97 higher amplitude genes peaked exclusively at around CT 8–12, whereas only 10 genes peaked at subjective dusk (Figs. 1 C and D and 2B). Note that our statistical analysis does not support the previous model in which cycling genes are classified into 2 distinct categories, one as high-amplitude genes and the other as low-amplitude genes (6). Instead, transition of genes from low to high amplitude is continuous, and there is no distinct cutoff point to distinguish between the 2 groups (Figs. 1 A and D and S2D).
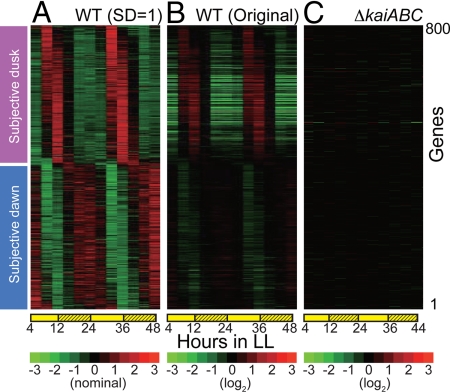
Fig. 2.
Clock-controlled expression profiles in wild-type and kaiABC-null strains under LL conditions. (A and B) Expression profiles of 800 cycling genes sorted by peak time in wild-type strains. The colors in descending order from red to black to green represent normalized data. For diagram A, the average and SD over 2 cycles are 0.0 and 1.0, respectively, whereas for diagram B the data were normalized to the mean value. (C) Expression profiles of the clock-controlled genes in kaiABC-null mutant strains. The sorting order of the genes is the same as for A and B. The data were normalized to the mean value of expression levels in wild-type strains. One of 2 independent experiments in each strain is shown (see Fig. S2 for results from the other series of experiments).
Furthermore, we estimated the numbers and the organization of operons on the genome based on the correlation coefficient of expression profiles under LL, magnitude of expression levels, and intergenic base pair length between neighboring genes (see SI Methods) to address whether temporal gene expression is locally coregulated on the genome. The threshold for each parameter was determined to cover each of the known operons that had been experimentally validated (SI Methods). Based on these criteria, we estimated that 1322 of the 2515 tested coding genes are organized into 488 operons to be cotranscribed and 1194 genes are singly transcribed (Table S1). Although we mapped phase, oscillatory index parameters (amplitude, P value) and expression level onto the chromosome location considering the estimated operonic structure (Fig. S3 A–D), we did not find convincing evidence in the genome of local circadian coregulation except for operonic control. We also calculated the correlation of temporal expression patterns in all possible combinations of the total 2515 ORFs and of the 1681 possible transcriptional units (Fig. S3 E and F). Using total ORFs, we observed multiple neighboring genes that were similarly regulated, whereas much less correlation was observed using transcriptional units, further supporting our evaluation.
Clock-Controlled Genes with Related Functions.
As summarized in Fig. S4, the ratio of rhythmically expressed genes within those belonging to the central intermediary metabolism, such as glycoprotein and polysaccharide synthesis, transcription and energy metabolism, was higher than that of rhythmic genes on the total genome (P < 0.01; Fisher's exact test). Among clock-related genes other than the kaiBC genes, we found that the circadian input histidine kinase gene cikA (17) and the circadian response regulator gene rpaA (18) showed high-amplitude expression rhythms peaking at CT 12 (Figs. 3A and S1). Although the kaiA (1, 15), pex (19), labA (20), and ldpA (21) genes did not fulfill our 2-filtration criteria for cycling genes, they each showed low-amplitude rhythms peaking at subjective dusk and dawn as illustrated in Fig. 3A. Thus, the 800 cycling genes identified through the filtering criteria were still not inclusive, and it is plausible that the Synechococcus genome contains additional cycling genes. On the other hand, transcripts of a KaiC/RpaA-interacting histidine kinase gene, sasA (22), and of light-input-involved cpmA (23) accumulated at an essentially constant level (Fig. 3A). Involvement of sigma factors in circadian output control has been suggested (5, 6, 24, 25). Genetic disruption of rpoD2 lowered the amplitude of the circadian expression of a subset of genes and slightly affected the phase of transcriptional rhythms (24, 25). We found that transcripts of 3 sigma factor genes, rpoD5/sigC, rpoD6 (syc0015_c/Synpcc7942_1557) and sigF2 (syc2309_c/Synpcc7942_1784), exhibited high-amplitude rhythms (Figs. 3B and S1). Although single mutations in each gene did not dramatically affect the genome-wide circadian profile, triple mutation of all 3 genes was not viable, suggesting that some of their functions may overlap in contributing to circadian regulation.
In addition to previously known purF and psbAII genes, we identified another ≈400 rhythmic genes peaking during subjective dawn (Figs. 1D, 2A, and S1). Among these genes, we found 4 dawn-expressed gene clusters, syc1184-1175_c/Synpcc7942_0329-0338 (atp cluster encoding ATPase synthase subunits), syc0685-0680_c/Synpcc7942_0855-0860 (pixGJ-pilH/rre7 cluster including 3 response regulator genes; see also Fig. S1A for Northern blot experiments), syc1986-1989_d/ Synpcc7942_2107-2104 (cynABDS cluster encoding putative cyanate ABC transporter subunits), and syc0135-0129_c/Synpcc7942_1421-1427 (ccmKLMNO-rbcLS cluster encoding RubisCO and its related carboxysomal proteins) (Figs. 4A and S1B). In addition to the RubisCO/carboxisome gene cluster, a light-dependent chlorophyll biosynthesis gene por was also found to be a photosynthesis-related subjective dawn gene (Fig. S1B).
Previous studies have reported that 2–9% and ≈10% of the Synechocystis (8) and the diazotrophic unicellular Cyanothece (12) genome, respectively, are regulated by the clock under LL conditions. These ratios are much lower than that of Synechococcus (> 30%). Because the Cyanothece data were limited to one day, we reevaluated the Synechocystis profiles over 2 days under LL conditions with our 3-filtration method and confirmed that the number and amplitude of cycling genes were much lower in Synechocystis than in Synechococcus (Fig. S5). In both species, most of the clock-regulated transcripts peaked at subjective dawn or dusk. Nevertheless, most of the high-amplitude genes identified in Synechococcus, such as kaiB, kaiC, rpoD5/sigC, rpoD6, purF, pilH/rre7 (syc0684_c/Synpcc7942_0856) and cikA, did not significantly cycle in Synechocystis (Fig. S4 and ref. 8), whereas a subset of genes, such as hox genes encoding bidirectional hydrogenase subunits, were rhythmic in both species.
However, it should be noted that circadian rhythm in a transcript does not always give rise to cyclic protein induction or accumulation. For example, the promoter activity and transcript level of kaiA are both rhythmic, whereas the KaiA protein is constitutively expressed under LL conditions (1, 26). Conversely, a nonrhythmic gene expression does not necessarily mean that the encoded protein is arrhythmic. For example, RpoD4 accumulates rhythmically, peaking at subjective dawn under LL conditions (25), whereas rpoD4 mRNA was not rhythmic or had very low amplitude in our microarray analysis (Dataset S1). Moreover, the kaiBC operon and the KaiB and KaiC proteins exhibit circadian cycles with a 6 h time lag (1, 27). This is also the case for the cikA gene, whose mRNA and protein levels peak at CT 12 (Fig. 3A) and at CT 20 (28), respectively. More strikingly, although kai gene transcripts are rapidly eliminated under dark conditions, the Kai proteins are stabilized and accumulate constitutively (2). All these results indicate marked and interesting discrepancies between transcriptomic and proteomic profiles. Thus, to validate the physiological relevance of the individual clock and/or diurnally controlled genes, more detailed analysis is necessary.
Global Transcription Profiles in the Absence of kai Genes.
We next examined the kai gene dependency of the genome-wide expression profile under LL conditions. As expected, in the kaiABC-null mutant strain, the number of cycling genes and their amplitude were severely reduced (Figs. 1B and 2C, and 3), although the average expression level of each individual gene in the genome was not dramatically altered by the presence or absence of the kai genes (Fig. S6A). Thus, in the kaiABC-null mutant, the clock was not arrested at subjective dawn or dusk but remained at an average level. This property is also supported by the lower correlation between expression profiles in the kaiABC-null and wild-type strains at subjective dawn or subjective dusk than at other circadian times (Fig. S6B). Interestingly, 17 and 2 genes were found to be rhythmic in the kaiABC-null mutant with a period of 24 h by using the standard (P value of <0.1, and amplitude of >10−1) and more stringent (P value of <0.01, and amplitude of >10−0.8) filtering condition (Fig. 1B; Table S2), respectively, although their amplitude was relatively low (Fig. S6; Table S2). Note that 800 and 258 genes were found to be cyclic under the same (standard and more stringent) filtering conditions in the wild-type strain. These results suggest the presence of kai-independent oscillator(s) in Synechococcus, and are reminiscent of the frequency (frq)-null mutant in Neurospora in which at least 3 oscillatory genes were found (29). Among the 17 kai-independent cycling transcripts, 6 genes (syc2457-2462_c/Synpcc7942_1470-1476) and 3 genes (syc2000-2002_c/Synpcc7942_2093-2091) were organized into 2 gene clusters. Note that the other 8 genes show much lower amplitude cycles (Fig. S7). Interestingly, some genes in the 2 clusters (sbtA, ndhD5, ndhD3, chpY/cupA, and ndhF3) and an additional transcript (syc0207_c/Synpcc7942_1346, ndhE) are involved in the CO2-concentrating mechanism (30). Other genes include those encoding the RecQ DNA helicase and tRNA methyltransferase. Further analyses are needed to reveal the physiological relevance of their cycling properties under the kai-less condition and the mechanism of the oscillation.
Overexpression of kaiC Gene Arrests the Clock at Subjective Dawn.
Previously, Nakahira et al. (6) performed promoter-trap analysis using a luciferase reporter gene set in an IPTG (isopropyl-β-D-thiogalactopyranoside)-inducible kaiC overexpressor strain under the control of an E. coli trc promoter (Ptrc::kaiC). For all tested gene promoters, bioluminescence levels were reduced to the base level of the circadian bioluminescence profile in wild-type strains to become arrhythmic under kaiC-overexpressing conditions. The results suggested that KaiC is a global gene repressor (6), but in retrospect, most of the gene promoters analyzed peaked at subjective dusk, and thus the effect of kaiC-overexpression on the subjective dawn genes is unknown. We therefore applied the microarray analysis to the Ptrc::kaiC strain from CT 1 (hour 25 in LL) to CT 9 (hour 33 in LL) in the presence or absence of IPTG (Fig. 4). In the absence of inducer, transcripts of subjective dusk genes such as kaiC, sigC, cikA and rpaA increased, whereas those of subjective dawn genes such as purF, pilH/rre7 and syc1137_d/Synpcc7942_0377 decreased during the 8 hours of subjective day (Fig. 4A, blue lines). On the other hand, after IPTG was added at hour 25 in LL (CT 1) to overexpress kaiC, expression of subjective dawn and dusk genes decreased and failed to increase, respectively (Fig. 4A, red lines; Fig. 4B). In other words, overexpression of kaiC elevates the subjective dawn genes to the peak level of the original circadian oscillation, whereas it represses the subjective dusk genes to the trough level and arrests the clock at subjective dawn. Correlation analysis of data from Ptrc::kaiC and wild-type strains also supports this idea (Fig. S6C). Note that kaiC overexpression did not increase or decrease specific nonrhythmic genes. Thus, its effect seems primarily limited to circadian control.
Dramatic Suppression of Genome-Wide Transcription in the Dark.
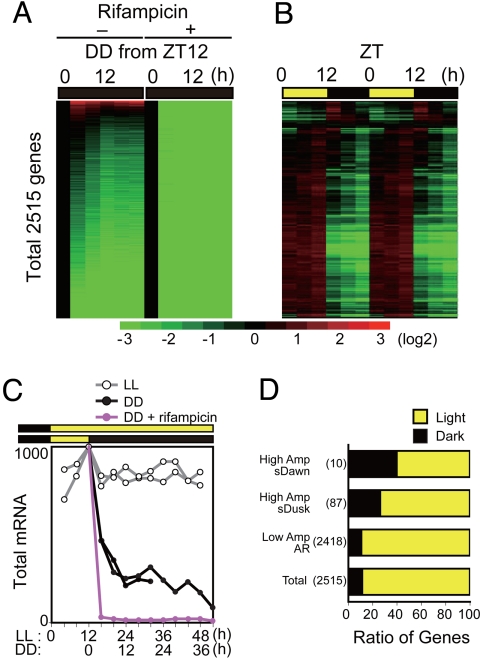
After cells were transferred to dark conditions, kai genes were dramatically suppressed, whereas the circadian KaiC phosphorylation cycle persisted (2). To address the effect of dark conditions on global expression profiles, cells were initially entrained to 12 h/12 h LD cycles, transferred to DD from hour 12 in the light (ZT 12), and subjected to microarray analysis (Fig. 5). Note that zeitgeber time (ZT) is used to indicate time in an LD cycle where ZT 0 indicates dawn. We confirmed that in the dark, kaiA and kaiBC genes were down-regulated to zero within 4 h. Strikingly, expression of most genes was also dramatically suppressed in the dark, whereas a minor subset of genes was up-regulated (Fig. 5A). The total transcript level, which was estimated from the sum of raw hybridization signals, was rapidly down-regulated in the dark (black lines) to reach ≈20% of initial levels within 12 h, whereas it remained almost constant under LL conditions (Fig. 5C, gray lines). Such a dramatic decrease in total mRNA levels caused by dark treatment has not been reported for any other tested cyanobacterial strain (31). Note that we used total RNA including rRNAs and tRNAs as a source of cDNA synthesis, and the amount of mRNAs is known to be much less than that of rRNAs. Because there were no significant differences in total RNA content and the resulting cDNA content between light-grown and dark-grown cells, rRNA amounts seem less affected by LD cycles than mRNA. Therefore, it is plausible that in our analysis the coding transcript levels were normalized to rRNA (and tRNA) levels. When a transcription inhibitor, rifampicin, was administered from ZT 12, all detectable hybridization signals were rapidly reduced to background levels within 4 h (Fig. 5 A and C). Thus, up-regulation of dark-induced genes and maintenance of residual transcript levels in the dark requires de novo transcription. Note that the KaiC phosphorylation cycle persisted under DD conditions in the presence of rifampicin (2). Nevertheless, we could not find any significantly rhythmic genes under DD conditions.
Fig. 5.
Suppression of genome-wide transcription under DD conditions. (A) Organization of Synechococcus expression profiles in DD from ZT 12 sorted by induction levels in the dark in the absence or presence of rifampicin. The data were normalized to the value at ZT 12. (B) Double plot of one LD cycle reconstructed by connecting data from hours 4–12 in LL and hours 0–12 in DD. The data were normalized to the average level over one LD cycle. (C) Total mRNA accumulation levels estimated from the sum of mRNA hybridization signals normalized to genomic DNA signals (see SI Methods) under LL (white circles) and DD in the absence (black) or presence (red) of rifampicin. Maximal total signals under LL were normalized to 1,000. (D) Population of nocturnally accumulating or diurnally accumulating transcripts in either subjective dawn (sDawn) or subjective dusk (sDusk) genes or low-amplitude/arrhythmic genes. Transcripts were defined as light-accumulating (yellow) or dark-accumulating (black) if the sum of the expression levels at ZT 4, 8 and 12 was larger or smaller than that at ZT 16, 20 and 24. Because the effect of LD transition is generally higher than the moderate changes in low-amplitude genes under LL, data from 97 higher-amplitude genes were used to identify 87 genes peaking at subjective dusk and 10 peaking at subjective dawn. Data for the 2418 remaining low-amplitude/arrhythmic (LA/AR) genes out of 2515 total genes are also shown.
Fig. 5B shows the double-plotted genome-wide expression profiles of all 2515 genes under LD cycles from data obtained from hours 4–12 under LL conditions (ZT 4–12) and hours 12–24 under DD conditions (ZT 12–24). The results clearly show that almost all genes of the genome (> 95%) rhythmically oscillated in LD cycles with much higher amplitude than under LL conditions. Next, we examined populations of nocturnally, variably accumulated transcripts in subjective dawn genes, subjective dusk genes and arrhythmic genes. As the effect of LD cycles is generally higher than that of the circadian control on accumulation levels, we used the 97 genes with the highest amplitude identified using highly stringent filtering conditions (Fig. 5D). Interestingly, 26% (23 of 87) and 40% (4 of 10) of the transcripts of subjective dusk genes and dawn genes, respectively, showed greater accumulation during the nocturnal phase. Their scores were significantly higher than those of control (low-amplitude and arrhythmic) genes or total genes (≈12%; Fig. 5D). When the 800 rhythmic genes identified by using relaxed filtering conditions were analyzed, the proportion of nocturnally accumulated transcripts in subjective dusk genes was almost the same (23%, 90 of 387 genes), whereas that of subjective dawn genes was much lower (5%, 20 of 413). Although the mechanism for this observation remains to be resolved, such a combination of different phasic outputs in daytime and during the night could give rise to a variety of expression patterns during LD cycles. As discussed above, the Kai proteins are constitutively stable and keep time during the nocturnal phase, whereas the kai genes are rapidly repressed (2). This property is not necessarily kai gene specific, considering the total protein content was not dramatically altered during DD conditions, whereas total mRNA decreased to ≈20% within 12 h (Fig. 5C). Although bacterial transcription has been considered to correlate well with proteomic profiles (e.g., ref. 31), we suggest that there is a much greater discrepancy between the 2 profiles than previously thought. Again, as for the circadian transcriptomic profiles, the physiological relevance of the dynamic diurnal/nocturnal control of Synechococcus needs to be clarified by further proteomic analysis.
Materials and Methods
Strains and Culture Conditions.
We used wild-type Synechococcus (PCC 7942), kaiABC-null and IPTG-inducible kaiC-overexpressor (Ptrc::kaiC) strains, each harboring a bacterial luciferase reporter gene set luxAB fused to a kaiBC promoter with a chloramphenicol resistance gene (6, 15). For microarray and Northern blotting analyses, Synechococcus cells were cultivated in BG-11 media in a continuous culture system (optical density of ≈0.2 at 730 nm) at 30 °C and ≈40 μmol·m−2·s−1). To synchronize the circadian clock, the culture was acclimated with 2 LD cycles and then transferred to LL or kept under DD.
Synechococcus Microarray Design and Analysis.
We designed the photolithography-based Affymetrix high-density oligonucleotide microarray (GeneChip CustomExpress Arrays) representing 2538 protein-coding genes, known RNA genes including tRNA genes, their antisense sequences, and intergenic sequences of larger than 100 base pairs on the genome of Synechococcus elongatus PCC 6301 (14; database accession no. AP008231). Because nucleotide sequences of PCC 6301 and PCC 7942 genomes show >99.9% identity (14), it is possible to evaluate transcription profiles of PCC 7942 strains by using this microarray. We adopted a 15–25-mer oligonucleotide probe set (15 each of perfectly matched oligonucleotide probes and single base pair mismatches) for each target gene. For the present article, we did not consider RNA genes, episomal genes on 3 Synechococcus plasmids or antisense sequences, but exclusively used data obtained for the predicted ORFs. For more detailed information, see SI Methods and Fig. S8.
Northern Blotting Analysis.
Northern blotting analysis was performed as described in ref. 2.
Supplementary Material
Acknowledgments.
We thank K. Kucho (Kagoshima University) for information on Synechocystis microarray experiments, M. Furumichi (Kyoto University) and Y. Nakamura (Kazusa Research Institute) for information on functional categories of Synechococcus and Synechocystis genes, S. Okamoto (Kazusa Research Institute) for advice on estimation of operonic structures, R. Yamada (RIKEN CDB) for advice on analyzing microarray data, and members of the Iwasaki laboratory (Waseda University) and the Kondo laboratory (Nagoya University) for valuable comments and advice. This work was supported in part by Japanese Society for the Promotion of Science Grants-in-Aid 17687017, 18016025, 17657002, 19657019, and 20370072 (to H. Iwasaki), 21010517 (to H. Ito), 17007534 (to M. M.), and 18006440 (to Y. M.); Ministry of Education, Culture, Sports, Science and Technology of Japan Grants-in-Aid 15GS0308 (to T. K.) and 13206027 (to M.S. and H. Iwasaki); the Uehara Memorial Foundation; the Asahi Glass Foundation; the Nakajima Peace Foundation; and the Exploratory Research for Advanced Technology (ERATO) Aihara Complexity Modeling Project (JST) (H. Iwasaki).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data and DNA sequence information have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) Database, www.ncbi.nlm.nih.gov/geo (accession no. GSE14225).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902587106/DCSupplemental.
References
- 1.Ishiura M, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 2.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription–translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 3.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 4.Terauchi K, et al. The ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2007;104:16377–16381. doi: 10.1073/pnas.0706292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, et al. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 6.Nakahira Y, et al. Global gene repression by KaiC as a master process of prokaryotic circadian system. Proc Natl Acad Sci USA. 2004;101:881–885. doi: 10.1073/pnas.0307411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duffield. DNA microarray analyses of circadian timing: The genomic basis of biological time. J Neuroendocrinol. 2003;15:991–1002. doi: 10.1046/j.1365-2826.2003.01082.x. [DOI] [PubMed] [Google Scholar]
- 8.Kucho K, et al. Global analysis of circadian expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2005;187:2190–2199. doi: 10.1128/JB.187.6.2190-2199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoki S, Kondo T, Ishiura M. Circadian expression of the dnaK gene in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 1995;177:5606–5611. doi: 10.1128/jb.177.19.5606-5611.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aoki S, Kondo T, Wada H, Ishiura M. Circadian rhythm of the cyanobacterium Synechocystis sp. strain PCC 6803 in the dark. J Bacteriol. 1997;179:5751–5755. doi: 10.1128/jb.179.18.5751-5755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki H, Kondo T. Circadian timing mechanism in the prokaryotic clock system of cyanobacteria. J Biol Rhythms. 2004;19:436–444. doi: 10.1177/0748730404269060. [DOI] [PubMed] [Google Scholar]
- 12.Toepel J, et al. Differential transcriptional analysis of the cyanobacterium Cyanothece sp. strain ATCC 51142 during light–dark and continuous light growth. J Bacteriol. 2008;190:3904–3913. doi: 10.1128/JB.00206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneegurt MA, Sherman DM, Nayar S, Sherman LA. Oscillating behavior of carbohydrate granule formation and dinitrogen fixation in the cyanobacterium Cyanothece sp. strain ATCC 51142. J Bacteriol. 1994;176:1586–1597. doi: 10.1128/jb.176.6.1586-1597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugita C, et al. Complete nucleotide sequence of the freshwater unicellular cyanobacterium Synechococcus elongatus PCC 6301 chromosome: gene content and organization. Photosynth Res. 2007;93:55–67. doi: 10.1007/s11120-006-9122-4. [DOI] [PubMed] [Google Scholar]
- 15.Iwasaki H, et al. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo T, et al. Circadian rhythms in rapidly dividing cyanobacteria. Science. 1997;275:224–227. doi: 10.1126/science.275.5297.224. [DOI] [PubMed] [Google Scholar]
- 17.Schmitz O, et al. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289:765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- 18.Takai N, et al. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci USA. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kutsuna S, Kondo T, Aoki S, Ishiura M. A period-extender gene, pex, that extends the period of the circadian clock in the cyanobacterium Synechococcus sp strain PCC 7942. J Bacteriol. 1998;180:2167–2174. doi: 10.1128/jb.180.8.2167-2174.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniguchi Y, et al. labA: A novel gene required for negative feedback regulation of the cyanobacterial circadian clock protein KaiC. Genes Dev. 2007;21:60–70. doi: 10.1101/gad.1488107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. LdpA: A component of the circadian clock senses redox state of the cell. EMBO J. 2005;24:1202–1210. doi: 10.1038/sj.emboj.7600606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwasaki H, et al. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 23.Katayama M, Tsinoremas NF, Kondo T, Golden SS. cpmA, a gene involved in an output pathway of the cyanobacterial circadian system. J Bacteriol. 1999;181:3516–3524. doi: 10.1128/jb.181.11.3516-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsinoremas NF, et al. A sigma factor that modifies the circadian expression of a subset of genes in cyanobacteria. EMBO J. 1996;15:2488–2495. [PMC free article] [PubMed] [Google Scholar]
- 25.Nair U, Ditty JL, Min H, Golden SS. Roles for sigma factors in global circadian regulation of the cyanobacterial genome. J Bacteriol. 2002;184:3530–3538. doi: 10.1128/JB.184.13.3530-3538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y, Mori T, Johnson CH. Circadian clock-protein expression in cyanobacteria: Rhythms and phase setting. EMBO J. 2000;19:3349–3357. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivleva NB, Gao T, LiWang AC, Golden SS. Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc Natl Acad Sci USA. 2006;103:17468–17473. doi: 10.1073/pnas.0606639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Correa A, et al. Multiple oscillators regulate circadian gene expression in Neurospora. Proc Natl Acad Sci USA. 2003;100:13597–13602. doi: 10.1073/pnas.2233734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price GD, Badger MR, Woodger FJ, Long BM. Advances in understanding the cyanobacterial CO2-concentrating mechanism (CCM) J Exp Bot. 2008;59:1441–1461. doi: 10.1093/jxb/erm112. [DOI] [PubMed] [Google Scholar]
- 31.Stoeckel J, et al. Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proc Natl Acad Sci USA. 2008;105:6156–6161. doi: 10.1073/pnas.0711068105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osanai T, Ikeuchi M, Tanaka K. Group 2 sigma factors in cyanobacteria. Physiol Plant. 2008;133:490–506. doi: 10.1111/j.1399-3054.2008.01078.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.