Abstract
Bacteriocins are a large and functionally diverse family of toxins found in all major lineages of Bacteria. Colicins, those bacteriocins produced by Escherichia coli, serve as a model system for investigations of bacteriocin structure–function relationships, genetic organization, and their ecological role and evolutionary history. Colicin expression is often dependent on host regulatory pathways (such as the SOS system), is usually confined to times of stress, and results in death of the producing cells. This study investigates the role of the SOS system in mediating this unique form of toxin expression. A comparison of all the sequenced enteric bacteriocin promoters reveals that over 75% are regulated by dual, overlapping SOS boxes, which serve to bind two LexA repressor proteins. Furthermore, a highly conserved poly-A motif is present in both of the binding sites examined, indicating enhanced affinity of the LexA protein for the binding site. The use of gene expression analysis and deletion mutations further demonstrates that these unique LexA cooperative binding regions result in a fine tuning of bacteriocin production, limiting it to times of stress. These results suggest that the evolution of dual SOS boxes elegantly accomplishes the task of increasing the amount of toxin produced by a cell while decreasing the rate of uninduced production, effectively reducing the cost of colicin production. This hypothesis may explain why such a promoter motif is present at such high frequencies in natural populations of bacteriocin-producing enteric bacteria.
Introduction
Bacteriocins are naturally occurring antimicrobials found in all major lineages of Bacteria and Archaea (Riley & Wertz, 2002). Colicins, those bacteriocins produced by Escherichia coli, were the first named family of bacteriocins (Gratia, 1925), and now serve as a model system for investigations of bacteriocin structure–function relationships, genetic organization, and their ecological role and evolutionary history (Braun et al., 1994; Lazdunski et al., 1998; Riley & Wertz, 2002; Smarda & Smajs, 1998). Colicins, and most other bacteriocins produced by enteric bacteria, are encoded in tightly linked gene clusters consisting of a toxin gene (cxa) and a constitutively expressed immunity gene (cxi), which encodes a protein that provides specific protection against the toxin. Many also include a lysis gene (cxl), which encodes a protein involved in toxin release (Braun et al., 1994; Cascales et al., 2007; Smarda & Smajs, 1998). Colicin gene clusters are regulated by the SOS response regulon, which plays a primary role in the response of many prokaryotes to DNA damage (Little & Mount, 1982; Walker, 1987, 1995).
The SOS response regulon has been studied extensively in E. coli (Kelley, 2006; Little & Mount, 1982; Walker, 1995). The genes encoding SOS-induced proteins are regulated by the LexA and RecA proteins, which serve as repressor and activator, respectively. In gammaproteobacteria, LexA represses transcription by binding to a 16-mer consensus sequence (CTG-N10-CAG) called the LexA binding site or SOS box (Erill et al., 2003). Individual SOS boxes vary in their DNA sequences and in their ability to bind the LexA protein (Walker, 1995).
In vitro studies of the promoter region of colicins E1 (Ebina et al., 1983), A (Lloubes et al., 1988, 1993) and K (Mrak et al., 2007) reveal that LexA binds cooperatively to the SOS regulatory region. Site-directed mutagenesis of LexA binding sites reveals that the first operator (Table 1, solid underline) is more pronounced in LexA binding affinity than the second (Table 1, dotted underline) (Lloubes et al., 1993). In vivo studies of the promoter region of colicins E1 (Salles & Weinstock, 1989) and E7 (Lu & Chak, 1996) show that mutations in either LexA binding motif result in an increased level of colicin production, suggesting that the colicin operon contains two distinct SOS boxes. In contrast, mutations within the SOS boxes of colicin K result in reduced expression (Mrak et al., 2007).
Table 1.
LexA binding sites and consensus motifs (bold type) found in the promoter regions of the genes encoding colicins E1 (ce1a) and its derivatives, colicin Ib (ciba) and recA
| Gene | Mutations | LexA binding site | |
|---|---|---|---|
| ce1a* | (+/+) | 5′ AACTGTATATAAAACCAGTGATTATATATACAGTA | 3′ |
| ce1a | (+/−) | 5′ AACTGTATATAAAACCAGTG– – – – – – – – – –†TA | 3′ |
| ce1a | (−/+) | 3′ TACTGTATATATAATCACTG – – – – – – – – – –†TT | 5′ |
| ce1a | (−/−) | 5′ AA – – – – – – – – – – – – – – – – – – – – – – – –†GTA | 3′ |
| ciba | 5′ TACTGTATATGTATCCATATACGTAAGCAGTT | 3′ | |
| recA | 5′ TACTGTATGAGCATACAGTA | 3′ | |
| Consensus‡ | 5′ TACTGTATATATATACAGTA | 3′ |
The underlined sequences highlight each of the SOS boxes.
The dashed lines mark the deleted bases for each of the mutations.
Consensus LexA-box sequence for E. coli (adapted from Erill et al., 2003).
To better understand the unique regulatory systems employed by enteric bacteriocin gene clusters, a comparative analysis of the promoter regions of all known enteric bacteriocins was conducted. The patterns of regulatory regions based on sequence similarity suggest that over 75 % of these bacteriocins employ similar toxin expression control mechanisms. To test the role of this highly conserved motif, an analysis of bacteriocin toxin expression was conducted using deletion mutations introduced into the SOS boxes, and the effects on expression were assessed. The results suggest that enteric bacteriocins have evolved a unique, finely tuned form of regulation that enables the cell to confine toxin production, and thus cell lysis, to times of severe stress, and to inhibit background levels of uninduced transcription.
Methods
Computer analysis – bacteriocin promoter phylogenetic analysis
Twenty-six bacteriocin promoter regions were examined. The promoter region was defined according to standard methods as the region extending from the bacteriocin gene start codon to the first upstream ORF (Ronen et al., 2002), and an alignment was produced using the clustal w algorithm (Thompson et al., 1994) in VectorNTI (Invitrogen). This alignment was used to infer phylogenetic trees employing three algorithms: neighbour-joining (NJ) (Gascuel & Steel, 2006), maximum-parsimony (MP) (Kolaczkowski & Thornton, 2004) and Bayesian methods (Ronquist & Huelsenbeck, 2003). The NJ and MP trees were created in MEGA 3.1 (Kumar et al., 2004). Default parameter settings were employed with the following exceptions. NJ: Jukes and Cantor model, gamma model with α=1.000, bootstrap=10 000, pairwise deletion and all nucleotides included. MP: complete deletion, CNI level=1, bootstrap=1000. For the Bayesian analysis, default parameter settings were employed with the following exceptions. MrBayes: characters used=457, Jukes and Cantor model, gamma rates, α=1.000, number of generations=1 000 000, sample frequency=100, burnin=2500. The returned split frequency standard deviation after 1 000 000 generations was 0.006. The returned potential scale reduction factor (PSRF) was 1.000. The returned posterior probability (PP) values, a measurement for clade credibility, are presented in Fig. 1.
Fig. 1.
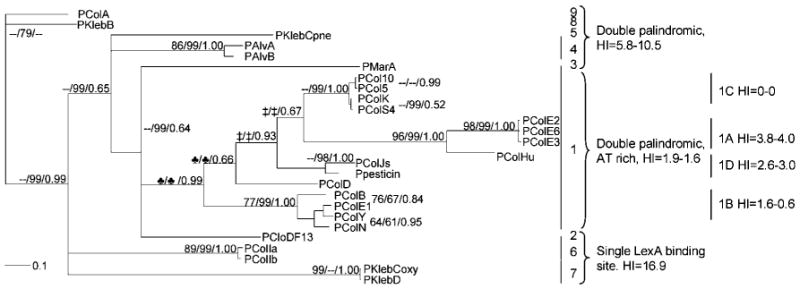
Bayesian tree indicating the structural relationship among the 300 bp promoters located just upstream of the colicin genes. Indicated are the NJ/MP/PP values obtained using the different tree-building algorithms (NJ, 10 000 bootstrap; MP, 1000 bootstrap). One major clade from pColN to pCol10 is recognized and separated from all other promoter structures. Nodes indicated with a clover-leaf symbol form one node using the NJ and MP algorithms. Similarly, nodes indicated with ‡ are one node using the NJ and MP algorithms. See Table 3.
SOS box analysis
All dual SOS boxes were aligned according to the consensus LexA binding motif N2CTGTN9CAGN2 (Fernandez De Henestrosa et al., 2000) and employing clustal w (Thompson et al., 1994). A consensus sequence was generated using WebLogo (Crooks et al., 2004), and the heterology index (HI) was calculated for each binding site according to Lewis et al. (1994).
Bacterial strains and plasmids
Table 2 lists the strains and plasmids used in this study. Construction of pDEW201 (containing the promoterless luxCDABE) and pDEW238 (containing the E. coli recA promoter) has been reported elsewhere (Van Dyk & Rosson, 1998; Van Dyk et al., 2001a). The promoters of colicins E1 and Ib were PCR-amplified and cloned into the multiple cloning site of pDEW201 (Van Dyk & Rosson, 1998). Plasmids were transformed into E. coli strains BZB1011 (Pugsley & Schwartz, 1983), DM1180 (Mount, 1977) and DM1187 (Kim & Little, 1992). Colicin inserts were confirmed by DNA sequence determination.
Table 2.
Bacterial strains and plasmids used in this study
P, promoter region.
| Strain or plasmid | Identification | Description | Reference |
|---|---|---|---|
| Strains | |||
| BZB1011 | W3110; gyrA | Pugsley & Schwartz (1983) | |
| DM1180 | lexA(Ind-); non-cleavable LexA protein | thr-1, araC14, leuB6(Am), DE(gpt-proA)62, tsx-33?, glnV44(AS), galK2(Oc), LAM-, sulA211, Rac-0, hisG4(Oc), rfbD1, mgl-51, recA441(ts), rpsL31(strR), kdgK51, xylA5, mtl-1, lexA3(Ind-) | Mount (1977) |
| DM1187 | lexA51(Def); defective LexA protein | thr-1, araC14, leuB6(Am), DE(gpt-proA)62, tsx-33?, glnV44(AS), galK2(Oc), LAM-, sulA211, hisG4(Oc), recA441(ts), rpsL31(strR), xylA5, mtl-1, thi-1?, lexA51, lexA3 | Kim & Little (1992) |
| Plasmids | |||
| pBS-E1_7 | pBS Φ(ce1a-lacZ), ampr | This study | |
| pDEW201 | (Promoterless plx), luxCDABE(-) Ampr | Van Dyk & Rosson (1998) | |
| pDEW238 | PrecA | cemtlB, ygaD, Φ(PrecA′-luxCDABE) Ampr | Van Dyk et al. (2001a) |
| pDEW-E1/7 | Pce1a (+/+) | pDEW201 Φ(Pce1a-luxCDABE) Ampr | This study |
| pDEW-E1/3738185 | Pce1a (+/−) | pDEW201 Φ(Pce1aΔ(5078–5093) Pce1aΔ(5063–5078)*-luxCDABE) Ampr | This study |
| pDEW-E1/3940222 | Pce1a (−/+) | pDEW201 Φ(Pce1aΔ(5063–5078) Δ(5078–5093)†-luxCDABE) Ampr | This study |
| pDEW-E1/3740345 | Pce1a (−/−) | pDEW201 Φ(Pce1aΔ(5063–5093) Δ(5063–5093)‡-luxCDABE) Ampr | This study |
| pDEW-Ib/18 | Pciba | pDEW201 Φ(Pciba-luxCDABE) Ampr | This study |
Δ(5078–5093) is a deletion of 13 nucleotides of Pce1a (accession no. J01566).
Δ(5063–5078) is a deletion of 13 nucleotides of Pce1a.
Δ(5063–5093) is a deletion of 26 nucleotides of Pce1a.
DNA sequencing
PCR-amplified fragments were cycle-sequenced using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer).
Growth media and chemicals
Luria–Bertani (LB) medium was used for all experiments and was supplemented at 100 mg l−1 ampicillin (Sigma). Seven concentrations (4, 2, 1, 0.5, 0.25, 0.125 and 0.0625 mg l−1) of mitomycin C (MitC; Sigma) were added to the media to induce the SOS response from a 1 g l−1 aqueous stock solution.
DNA manipulation
The 259 bp ce1a promoter was cloned into pBlueScriptII SK(+) (Stratagene) and four primers containing the desired deletions were generated:
5063R 5′ GCATAAAAGCTACGCCGCTGCATTTTC 3′;
5078F 5′ CAGTGGTTATATGTACAGTATTTATTT 3′;
5078R 5′ CTGGTTTTATATACAGCATAA 3′;
5093F 5′ GTATTTATTTTTAACTTATTG 3′ (accession no. J01566).
The plasmid containing the intact ce1a promoter region was PCR-amplified using primers 1 and 2, 3 and 4, and 1 and 4, together with HotStarTaq DNA polymerase (Qiagen). The PCR products were treated with DpnI and T4 polynucleotide kinase (New England Biolabs), and were then religated using T4 DNA ligase (New England Biolabs). The resulting vectors were transformed into E. coli strain DH10B (Stratagene) and mutations were confirmed by sequencing. Mutated promoters, i.e. Pce1a (+/−), (−/+) and (−/−) (see Table 1), were PCR-amplified, subcloned into pDEW201, and electroporated into the E. coli strains.
Reporter assay
Bacteria were grown overnight in LB supplemented with ampicillin. The cultures were diluted in LB (1 : 100) and grown to a density of ∼2 × 108 cells ml−1. A twofold dilution series of MitC was added to 96-well microtitre plates (Nunc) and equal volumes of cells were added. The plates were incubated in a temperature-controlled luminometer (Dynatech ML300). All experiments were run in duplicate and were repeated at least three times. Luminescence values are presented as arbitrary relative light units (RLU), or as the ratio of the luminescence of the induced sample to that of the uninduced control (response ratio), as described elsewhere (Van Dyk et al., 2001a).
Results
Conserved SOS box identified within enteric bacteriocin promoters
The consensus sequence of enteric bacteriocin promoters contains a σ70 promoter region (identified as −35 and −10 boxes), one or two LexA binding sites, a thymine (T)-rich region and a Shine–Dalgarno (SD) box (Riley et al., 2001). Bacteriocin promoters of 26 enteric bacteriocins were aligned using clustal w (Thompson et al., 1994). Based upon the resulting alignment, nine promoter motifs were revealed (Fig. 1).
The first motif was found in 15 bacteriocin gene clusters and is unique in possessing an A-rich region between the −35 and −10 boxes (∼10 bp), double LexA binding sites, a relatively long (∼18 bp) T-rich region, and an additional short (∼7 bp) highly conserved T-rich region (not shown). Bacteriocins with this motif can be divided into five subgroups: 1a=colicins E2, E3, E6 and Hu; 1b=colicins E1, N, B and Y; 1c=colicins 5, 10, S4 and K; 1d=colicin Js and pesticin (produced by Yersinia pestis); and 1e=colicin D. Two additional bacteriocins, cloacin DF13 and marcesin A, although possessing a similar motif, contain a single LexA binding site.
Alveocins A and B (produced by Hafnia alvei) and klebicin C (produced by Klebsiella pneumoniae) share the presence of double overlapping LexA binding sites and a variable T-rich region, but lack the short consensus T-rich region. Colicins Ia and Ib possess a single LexA binding site, which differs from the E. coli consensus sequence (Table 3), and have relatively short conserved T-rich regions. Klebicins C and D (both produced by Klebsiella oxytoca) have a single SOS box, similar in sequence to that found in E. coli, but lacking the T-rich regions. Colicin A and klebicin B (produced by K. pneumoniae) share an insertion of about 60 bp located between dual LexA binding sites and the SD box (Riley et al., 2001), and lack the T-rich region.
Table 3.
Sequences and HIs of the LexA binding sites in the promoter regions of bacteriocins produced by enteric bacteria and those of SOS-regulated genes found in the E. coli genome (Fernandez De Henestrosa et al., 2000)
| Genea | SOS box/es sequence | HIb | Accession number |
||
|---|---|---|---|---|---|
| 1c | 2d | 3e | |||
| cta (Col5) | TGCTGTACATAAAACCAGTGGTTATATGTACAGTA | 2.0 | 0.7 | X87835 | |
| cfa (Col10) | TGCTGTACATAAAACCAGTGGTTATATGTACAGTA | 2.0 | 0.7 | X82682 | |
| cka (ColK) | TACTGTACATAAAACCAGTGGTTATATGTACAGTA | 0.0 | 0.7 | Y18549 | |
| cs4a (ColS4) | CACTGTACATAAAACCAGTGGTTATATGTACAGTA | 1.6 | 0.7 | Y18684 | |
| cba (ColB) | ATCTGTACATAAAACCAGTGGTTATATGTACAGTA | 2.8 | 0.7 | M16816 | |
| cda (ColD) | CGCTGTATATAAAACCAGTGGTTTTCTATACAGTT | 3.6 | 4.8 | X14941 | |
| ce1a (ColE1) | TGCTGTATATAAAACCAGTGGTTATATGTACAGTA | 2.5 | 0.7 | AF453410 | |
| cna (ColN) | AACTGTATATAAAACCAGTGGTTATGTGTACAGTA | 1.1 | 3.3 | Y00533 | |
| cya (ColY) | TGCTGTACATAAAACCAGTGGTTATGTATACAGTA | 2.0 | 0.7 | AF197335 | |
| cua (ColU) | AACTGTATATAAAACCAGTGGTTATATATACAGTA | 1.2 | 0.7 | Y11823 | |
| ce2a (ColE2) | ATCTGTACATAAAACCAGTGGTTTTATGTACAGTA | 2.8 | 1.2 | M29885 | |
| ce3a (ColE3) | ATCTGTACATAAACCCAGTGGTTTTATGTACAGTA | 4.1 | 0.7 | J01574 | |
| ce6a (ColE6) | ATCTGTACATAAAACCAGTGGTTTTATGTACAGTA | 2.8 | 0.7 | M31808 | |
| ce7a (ColE7) | ATCTGTACATAAAACCAGTGGTTTTATGTACAGTA | 2.8 | 0.7 | X63620 | |
| chua (ColHu) | ATCTGTACATAAAACCAGTGGTTTTATGTACAGTA | 2.8 | 0.7 | AF540491 | |
| cjsa (ColJs) | TACTGTATATAAAACTAGTATTTATATATACAGTA | 7.9 | 9.3 | DQ995353 | |
| pst (pesticin) | TACTGTACATAAAAACAGTGGTTTTATGTACAGTA | 0.9 | 0.2 | NC_004837 | |
| kca (KlebC pnef) | CACTGTATAAATACACAGTGTGTATTTATACAGTG | 10.1 | 10.1 | AY578793 | |
| caa (ColA) | TACTGTATATAAACACATGTGAATATATACAGTT | 6.0 | 11.0 | X01008 | |
| kba (KlebB) | TACTGTATATAAACACATGTGTTTATATACAGTA | 6.0 | 6.0 | NC_009649 | |
| alva (AlvA) | TACTGTATATATAACAATGTATTTATATACAGTA | 7.2 | 5.4 | AY271828 | |
| Alvb (AlvB) | TACTGTATATATAGTAATTTGTTTATATACAGTA | 9.4 | 6.9 | AY271829 | |
| ciaa (ColIa) | TACTGTATATGTATCCATATGCGTAAGCAGTT | 9.3 | M13819 | ||
| ciba (ColIb) | TACTGTATATGTATCCATATACGTAAGCAGTT | 7.2 | NC_002122 | ||
| kda (KlebD) | TGCTGTGCACTCCTGAAACTGCAGTT | 22.1 | A458792 | ||
| kca (KlebC oxyg) | TGCTGTGCACTCCTGAAACTGCAGTT | 29.0 | AY578792 | ||
| ccl (CloDF13) | TACTGTGTATATATACAGTA | 7.6 | NC_002119 | ||
| kccla (KlebCcl) | CACTGTATGTATATACAGTA | 9.2 | AF190857 | ||
| c683a (Clo683) | AACTGTTAATTAACTACAGTTA | 23.0 | UPh | ||
| c647a (Clo647) | CCCTGTAGAAATGAACATCC | 7.3 | UPh | ||
| marA (MarA) | TACTGTATATAAATACAGTT | 9.2 | UPh | ||
| ydjM | CACTGTATATAAAAATCCTATACTGTACGTATCGACAGTT | 10.5 | 11.6 | AC_000091 | |
| recN | TACTGTATATAAAACCAGTTTATACTGTACACAATAACAGTAATGGTTTTTCATACAGGAAAACGACT | 1.1 | 8.7 | 29.0 | AC_000091 |
| lexA/dinF | ATCTCTGGTTTATTGTGCAGTTTATGGTTCCAAAATCGCCTTTTGCTGTATATACTCACAGCATAACTGTATATACACCCAGGG | 46.7 | 14.2 | 6.9 | AC_000091 |
| umuDC | ATCTGCTGGCAAGAACAGACTACTGTATATAAAAACAGTA | 33.1 | 2.6 | AC_000091 | |
| recA | TACTGTATGCTCATACAGTA | 15.3 | AC_000091 | ||
| ConsEi | TACTGTATATATATACAGTA | ||||
| ConsBj | TACTGTACATAAAACCAGTG | ||||
The gene names are designated cxa: colicin x activity-encoding gene, according to Cascales et al. (2007).
Calculated according to Lewis et al. (1994).
1, Distal LexA binding site.
2, Proximal LexA binding site.
3, Third LexA binding site.
Klebicin C isolated from K. pneumoniae.
Klebicin C isolated from K. oxytoca.
UP, M. A. Riley and others, unpublished data.
E. coli LexA binding site consensus sequence.
Enteric bacteriocins LexA binding site consensus sequence.
The dual, overlapping LexA binding sites, found in 77 % of the promoter regions, were aligned (Table 3). The resulting bacteriocin-based consensus sequence varied from the traditional E. coli LexA site consensus (Fig. 2). The consensus sequence forms an almost perfect palindrome, the distal LexA site is far more conserved than the proximal site, a highly conserved A motif rather than AT motif is present, and the proximal SOS box has a CAC motif in place of the CAG usually encountered in E. coli (Fernandez De Henestrosa et al., 2000).
Fig. 2.

Inter-bacteriocin consensus for 22 bacteriocin LexA binding sites. Sequences were produced using the WebLogo service at http://weblogo.berkeley.edu. Overlapping LexA binding sites are shown by the solid and dotted lines below the figure.
This unique, highly conserved region was further characterized by determination of the degree of divergence from the consensus sequence and by a predicted higher affinity for the LexA protein (Table 3). Sequences with a low HI value are more similar to the consensus, and thus are presumed to have a greater affinity for the LexA protein than sites with higher HI values (Fernandez De Henestrosa et al., 2000; Lewis et al., 1994). The majority of colicins have low HI values compared to other enteric bacteriocins (Table 3), suggesting a higher affinity for the LexA protein (Fig. 1). Furthermore, out of 31 SOS-regulated genes identified in the E. coli genome, only four have more then one SOS box in their promoters: recN, umuDC, lexA/dinF and yjdM (Fernandez De Henestrosa et al., 2000). The LexA binding sites of the proximal recN and distal umuDC sites are similar in sequence to the colicin SOS box motif (HI values of 1.1 and 2.6, respectively).
Induction patterns of colicin E1 and Ib promoters
The colicin E1 (Pce1a) and Ib (Pciba) promoters were chosen as representatives of the dual and single LexA motifs, respectively. These promoter regions were inserted upstream of the Photorhabdus luminescens luxCDABE reporter genes (Table 2) and electroporated into E. coli strain BZB1011 (Pugsley, 1985). The impact on transcription levels of seven MitC concentrations was tested (Fig. 3a). The greatest effect was observed with addition of 2 and 4 mg MitC l−1 for both Pce1a and Pciba; however, the Pce1a response ratios were always higher (Fig. 3b). When assayed without inducing agent (Fig. 3b), Pce1a had a ∼45-fold increase in light emission within 3 h (ranging from 0.1±0.06 to 4.5±0.12 RLU), while the Pciba construct showed almost no increase over the same time period (ranging from 0.2±0.03 to 0.4±0.07 RLU). The Pciba induction peak was ∼20-fold lower than that of Pce1a (335.8±30.6 compared to 16.2±4.3 RLU, respectively).
Fig. 3.
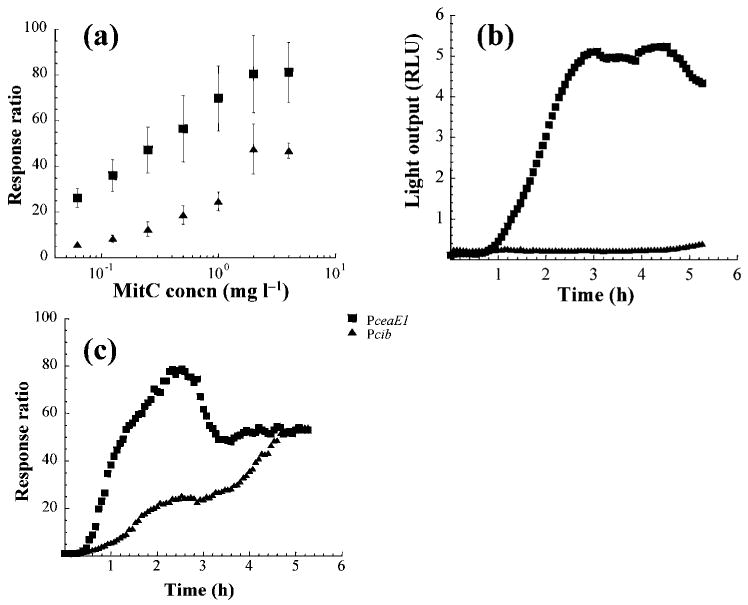
Light emission of ce1a (strain pDEW-E1/7) and ciba (strain pDEW-Ib/18) promoter fusions in response to (a) various concentrations of MitC measured at highest response ratio 2.5 h following induction, (b) no induction, and (c) 2 mg MitC l−1 over 5.5 h. Each point is the mean of at least three replicates.
Fig. 3(c) provides the response ratios resulting from induction of these constructs with 2 mg MitC l−1. The induction pattern of Pce1a includes a 30 min lag phase followed by a rapid increase in expression for 2 h, with a maximum response ratio of ∼80, followed by a decline in expression until a plateau is reached in 3 h. Pcib has a similar initial lag phase, followed by a much slower increase until a maximum response ratio of ∼50 is reached after 5 h. These observations indicate differences in regulation and promoter strength under inducing (Fig. 3c) and non-inducing conditions (Fig. 3b).
The dual LexA binding sites of the colicin E1 promoter
To assess the relative contributions of the dual SOS boxes in the ce1a promoter, three constructs with alternative LexA sequences were created (Table 2) and expressed in two E. coli strains containing mutant LexA genes (Table 2). The mutants had their first (−/+), second (+/−) or both (−/−) SOS boxes deleted (Table 2). Plasmids carrying these mutants were introduced into strain DM1180 (Mount, 1977), which encodes a non-cleavable LexA protein, resulting in complete repression of the SOS-regulated genes. Strain DM1180 served as a positive control, as light emission should be negatively proportional to the LexA repression. The second mutant host, strain DM1187 (Kim & Little, 1992), served as a negative control, as it encodes a LexA protein unable to bind to the operator, resulting in unregulated expression of the SOS-regulated genes. There should be no difference in light emission between the wild-type and mutated Pce1a constructs carried by this strain.
Levels of uninduced expression of the wild-type Pce1a in strains DM1180 and DM1187 (Table 4) differed by 99.8 %. In contrast, levels of expression of the two single (+/−, −/+) and double (−/−) mutants differed by 90.2, 36.3 and 8.6 %, respectively. These results suggest that LexA proteins bind most tightly to a promoter which contains the dual SOS box (+/+), less tightly to a promoter with a single LexA binding site (+/− and −/+ mutants), and not at all to a promoter that lacks an SOS box (−/−). Furthermore, the expression levels of the single box (−/+) promoter carried by strain DM1187 were approximately half of those measured for the wild-type promoter (+/+), the alternative single (+/−), and the double (−/−) mutants.
Table 4.
Levels of luminescence expressed under ce1a wild-type and mutated promoters carried by E. coli strains DM1180 and DM1187
| Promoter | Level of luminescence (RLU)* | Discrepancy (%)† | |
|---|---|---|---|
| DM1180 | DM1187 | ||
| ce1a (+/+) | 0.25 ± 0.04 | 112.05 ± 16.72 | 99.8 |
| ce1a (+/−) | 13.22 ± 1.81 | 134.91 ±22.13 | 90.2 |
| ce1a (−/+) | 41.72 ± 4.38 | 65.49 ± 7.83 | 36.3 |
| ce1a (−/−) | 106.49 ± 39.05 | 116.96 ± 37.35 | 8.6 |
Mean and SD of bioluminescence measured in E. coli strains DM1180 and DM1187 reported as RLU.
Discrepancy in light expression levels of strain DM1180 versus DM1187.
Response ratios of the wild-type and truncated ce1a promoters, as well as a recA promoter assayed in E. coli strain BZB1011 (Table 2), were monitored over time following the addition of 0.5 mg MitC l−1 (Fig. 4a). The recA promoter served as a positive control, as it is SOS-regulated and induced by numerous DNA-damaging agents, including MitC (Davidov et al., 2000; Rosen et al., 2000; Van Dyk et al., 2001a). Following a short lag phase, induction of the wild-type Pce1a was rapid and reached a response ratio of 80 within 2.5 h. The single mutant retaining the non-consensus LexA binding site (−/+) had a pattern of induction similar to that observed for the wild-type promoter, but with a 17-fold lower response ratio. An intermediate response was observed for the (+/−) construct, similar to that of the recA promoter (Fig. 4a). However, when no inducing agent was present, expression levels of wild-type Pce1a were an order of magnitude lower than those observed in the two single mutants, and two orders of magnitude lower than that observed in the double mutant (Fig. 4b).
Fig. 4.
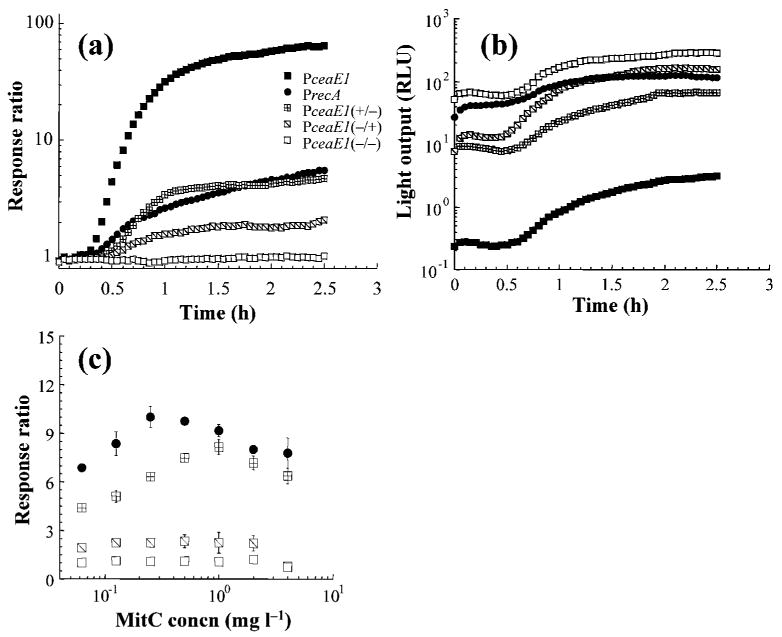
Light emission of ce1a (strain pDEW-E1/7) and recA (strain pDEW238) wild-type and mutants (strains pDEW-E1/3738185, pDEW-E1/3940222 and pDEW-E1/3740345) constructs in response to (a) 0.5 mg MitC l−1 over 2.5 h, (b) no induction, and (c) various concentrations of MitC measured at the highest response ratio 2.5 h following induction. Each point is the mean of at least three replicates.
The response ratios of the wild-type and mutated Pce1a were assayed at a range of MitC concentrations (Figs 3c and 4c), and the maximum response ratio for the Pce1a (+/+) construct was obtained with addition of 2 and 4 mg MitC l−1. In contrast, the maximum response ratios observed for the single mutant Pce1a (+/−) and PrecA were obtained with 50 and 75 % lower levels of MitC (1 and 0.25 mg l−1), respectively. The response ratios of the alternative single (−/+) and double (−/−) Pce1a mutants were unaffected by the MitC concentration.
Discussion
A comparison of 26 enteric bacteriocin promoters reveals a diversity of promoter motifs. However, the majority of these promoters (77 %) share highly conserved overlapping LexA binding sites (Table 3) abundant among colicins, yet more variable in other enteric bacteriocins. Although the dual SOS boxes are often found in bacteriocin promoters, they are rarely observed in other SOS-regulated genes in E. coli. Indeed, only four SOS-regulated genes were found to have more then one LexA binding site out of the 31 identified in the E. coli genome: umuDC, recN, ydjM and lexA/dinF. These genes have double, and even triple, LexA binding sites (Fernandez De Henestrosa et al., 2000), yet they were predicted to have quite variable binding affinity, based upon their HI values (Table 3). Experimental data further suggest that the proximal SOS box of recN does not effectively bind the LexA protein (van der Lelie et al., 1997). Nevertheless, when the promoters of these four genes are fused to a reporter gene (gfp or lux), low basal levels of expression and high response ratios have been reported upon induction (Norman et al., 2005; Van Dyk et al., 2001a, b; van der Lelie et al., 1997). Similarly, induced enteric bacteriocin promoters express low basal levels of expression and high response ratios in the current (Fig. 4) and comparable studies (Mrak et al., 2007; Norman et al., 2005; Prieto et al., 2004; Vankemmelbeke et al., 2005). Moreover, when the contributions of each of the LexA binding sites were compared, expression under a single LexA repressor resulted in higher basal levels and lower response ratios (Fig. 4), presumably indicating enhanced basal bacteriocin production.
The majority of enteric bacteriocins possess dual LexA binding sites; however, these sites deviate considerably from the E. coli SOS box consensus (Fernandez De Henestrosa et al., 2000; Lewis et al., 1994). The bacteriocin dual SOS boxes consist of a highly conserved A tract (Fig. 2) found at the distal and proximal LexA binding sites (Table 3). DNA molecules containing four to six consecutive A or T base pairs are predicted to have an intrinsic structural curvature that can impact their role in transcriptional activation by affecting promoter geometry (Barbic et al., 2003; Perez-Martin & de Lorenzo, 1997). Although increases in promoter strength due to DNA bending are usually associated with sequences upstream of the promoter region (Perez-Martin & de Lorenzo, 1997), it has been shown that bent DNA at or near the transcriptional start site is an important component of T7 RNA polymerase promoter enhancement, suggesting that in order to affect promoter activation, bent DNA should be situated in close proximity to the transcription initiation site (Ujvari & Martin, 2000).
In the case of bacteriocin promoters, this bend is predicted to occur 12 bp downstream of the −10 box. It is thus possible that changes observed in promoter strength (Table 4) are due to intrinsically bent or bendable DNA within the LexA binding sites (Perez-Martin & de Lorenzo, 1997). The highest bioluminescence levels (Table 4), in a background of DM1187 (producing a mutated LexA protein that is unable to bind to its site), were measured in Pce1a wild-type (+/+) and the single-site mutant (+/−), where the A tract is located in a 3′ position to the Pribnow box (Table 1). However, in the alternative single mutant (−/+), the A tract is not present and the TA sequence forms a rigid structure. This observation may explain the decreased levels of luminescence measured and is supported by Mrak and colleagues (Mrak et al., 2007), who showed that T substitutions in the A tract of the distal SOS box (the AAAA motif was altered to ATAT) result in a decrease in gene expression, signifying the importance of the A motif. Interestingly, the single SOS box found in the colicin Ib promoter lacks A or T tracts (Table 3), and is thus less bendable, which might explain the lower response ratio observed (Fig. 3).
Expression levels induced by the distal site are more pronounced than those of the proximal site in the colicin K promoter (Mrak et al., 2007). Additionally, the dual promoter sites of colicin E7, which both have an A tract (Table 3), show similar expression levels (Lu & Chak, 1996). Colicins K and E1 have the A tract located at the proximal SOS box only (Table 3). Moreover, SOS-regulated genes, such as dinB, yebF and yigN, containing an A or T tract fused upstream to the luxCDABE reporter operon, have a higher response ratio compared to promoters carrying the consensus AT tract, such as recA, uvrA and uvrD (Fernandez De Henestrosa et al., 2000; Van Dyk et al., 2001b). Our analysis indicates that the structure of the LexA binding sites enhances the expression of the downstream genes, and in theory occurs in conjunction with a σ70 box forming a strong promoter.
A comparison of response rates between Pce1a and Pciba reveals that the ce1a promoter induces a more rapid and higher level of gene expression compared to the ciba promoter (Fig. 3). Promoter–reporter gene fusions using gfp downstream of colicin D (Norman et al., 2005) and K (Mrak et al., 2007) promoters have been shown to respond in a similar manner, with high expression/leakage ratios and rapid responses to environmental stimuli. One hypothesis to explain these observations is that bacteriocin toxins are lethal to the producing cells and have intrinsically strong promoters. Therefore, it is vital for the cell to keep bacteriocin production under tight control; once expression is initiated it leads to cell death (Mulec et al., 2003). The highly conserved region downstream of the σ70 box compared to the upstream sequence (data not shown) in most of the colicin promoters suggests that the LexA binding sites play a major role in enhancing promoter strength and control. These sites, frequently found in the promoters of enteric bacteriocins, result in both an increase in repressor binding strength and the ability to upregulate expression. The evolution of this dual binding site system elegantly accomplishes the task of increasing toxin production when induced, without a proportional increase in leakage when repressed. This effectively reduces the cost of colicin production, and may explain how such a seemingly costly defence system is maintained at such high frequencies in enteric bacteria (Gordon & O'Brien, 2006).
Acknowledgments
The authors thank Dr Tina Van Dyk, DuPont Company, Biochemical Sciences and Engineering Division, for providing plasmids pDEW201 and pDEW238. This work was supported by NIH awards GM58433-02 and A1064588-01A2 to M.A.R.
Abbreviations
- HI
heterology index
- MitC
mitomycin C
- MP
maximum-parsimony
- NJ
neighbour-joining
- PP
posterior probability
- RLU
relative light units
- SD
Shine–Dalgarno
References
- Barbic A, Zimmer DP, Crothers DM. Structural origins of adenine-tract bending. Proc Natl Acad Sci U S A. 2003;100:2369–2373. doi: 10.1073/pnas.0437877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Pilsl H, Gross P. Colicins: structures, modes of action, transfer through membranes, and evolution. Arch Microbiol. 1994;161:199–206. doi: 10.1007/BF00248693. [DOI] [PubMed] [Google Scholar]
- Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidov Y, Rozen R, Smulski DR, Van Dyk TK, Vollmer AC, Elsemore DA, LaRossa RA, Belkin S. Improved bacterial SOS promoter ∷ lux fusions for genotoxicity detection. Mutat Res. 2000;466:97–107. doi: 10.1016/s1383-5718(99)00233-8. [DOI] [PubMed] [Google Scholar]
- Ebina Y, Takahara Y, Kishi F, Nakazawa A, Brent R. LexA protein is a repressor of the colicin E1 gene. J Biol Chem. 1983;258:13258–13261. [PubMed] [Google Scholar]
- Erill I, Escribano M, Campoy S, Barbe J. In silico analysis reveals substantial variability in the gene contents of the gamma proteobacteria LexA-regulon. Bioinformatics. 2003;19:2225–2236. doi: 10.1093/bioinformatics/btg303. [DOI] [PubMed] [Google Scholar]
- Fernandez De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- Gascuel O, Steel M. Neighbor-joining revealed. Mol Biol Evol. 2006;23:1997–2000. doi: 10.1093/molbev/msl072. [DOI] [PubMed] [Google Scholar]
- Gordon DM, O'Brien CL. Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology. 2006;152:3239–3244. doi: 10.1099/mic.0.28690-0. [DOI] [PubMed] [Google Scholar]
- Gratia A. Sur un remarquable exemple d'antagonisme entre deux souches de colilbacille. Comp Rend Soc Biol. 1925;93:1040–1041. in French. [Google Scholar]
- Kelley WL. Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon. Mol Microbiol. 2006;62:1228–1238. doi: 10.1111/j.1365-2958.2006.05444.x. [DOI] [PubMed] [Google Scholar]
- Kim B, Little JW. Dimerization of a specific DNA-binding protein on the DNA. Science. 1992;255:203–206. doi: 10.1126/science.1553548. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B, Thornton JW. Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature. 2004;431:980–984. doi: 10.1038/nature02917. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. mega3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lazdunski CJ, Bouveret E, Rigal A, Journet L, Lloubes R, Benedetti H. Colicin import into Escherichia coli cells. J Bacteriol. 1998;180:4993–5002. doi: 10.1128/jb.180.19.4993-5002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Harlow GR, Gregg-Jolly LA, Mount DW. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J Mol Biol. 1994;241:507–523. doi: 10.1006/jmbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- Little JW, Mount DW. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Lloubes R, Granger-Schnarr M, Lazdunski C, Schnarr M. LexA repressor induces operator-dependent DNA bending. J Mol Biol. 1988;204:1049–1054. doi: 10.1016/0022-2836(88)90062-9. [DOI] [PubMed] [Google Scholar]
- Lloubes R, Lazdunski C, Granger-Schnarr M, Schnarr M. DNA sequence determinants of LexA-induced DNA bending. Nucleic Acids Res. 1993;21:2363–2367. doi: 10.1093/nar/21.10.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu FM, Chak KF. Two overlapping SOS-boxes in ColE operons are responsible for the viability of cells harboring the Col plasmid. Mol Gen Genet. 1996;251:407–411. doi: 10.1007/BF02172368. [DOI] [PubMed] [Google Scholar]
- Mount DW. A mutant of Escherichia coli showing constitutive expression of the lysogenic induction and error-prone DNA repair pathways. Proc Natl Acad Sci U S A. 1977;74:300–304. doi: 10.1073/pnas.74.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrak P, Podlesek Z, van Putten JPM, Zgur-Bertok D. Heterogeneity in expression of the Escherichia coli colicin K activity gene cka is controlled by the SOS system and stochastic factors. Mol Genet Genomics. 2007;277:391–401. doi: 10.1007/s00438-006-0185-x. [DOI] [PubMed] [Google Scholar]
- Mulec J, Podlesek Z, Mrak P, Kopitar A, Ihan A, Zgur-Bertok D. A cka–gfp transcriptional fusion reveals that the colicin K activity gene is induced in only 3 percent of the population. J Bacteriol. 2003;185:654–659. doi: 10.1128/JB.185.2.654-659.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman A, Hansen LH, Sorensen SJ. Construction of a ColD cda promoter-based SOS-green fluorescent protein whole-cell biosensor with higher sensitivity toward genotoxic compounds than constructs based on recA, umuDC, or sul4 promoters. Appl Environ Microbiol. 2005;71:2338–2346. doi: 10.1128/AEM.71.5.2338-2346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martin J, de Lorenzo V. Clues and consequences of DNA bending in transcription. Annu Rev Microbiol. 1997;51:593–628. doi: 10.1146/annurev.micro.51.1.593. [DOI] [PubMed] [Google Scholar]
- Prieto AI, Ramos-Morales F, Casadesus J. Bile-induced DNA damage in Salmonella enterica. Genetics. 2004;168:1787–1794. doi: 10.1534/genetics.104.031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley AP. Escherichia coli K12 strains for use in the identification and characterization of colicins. J Gen Microbiol. 1985;131:369–376. doi: 10.1099/00221287-131-2-369. [DOI] [PubMed] [Google Scholar]
- Pugsley AP, Schwartz M. A genetic approach to the study of mitomycin-induced lysis of Escherichia coli K-12 strains which produce colicin E2. Mol Gen Genet. 1983;190:366–372. doi: 10.1007/BF00331060. [DOI] [PubMed] [Google Scholar]
- Riley MA, Wertz JE. Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol. 2002;56:117–137. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- Riley MA, Pinou T, Wertz JE, Tan Y, Valletta CM. Molecular characterization of the klebicin B plasmid of Klebsiella pneumoniae. Plasmid. 2001;45:209–221. doi: 10.1006/plas.2001.1519. [DOI] [PubMed] [Google Scholar]
- Ronen M, Rosenberg R, Shraiman BI, Alon U. Assigning numbers to the arrows: parameterizing a gene regulation network by using accurate expression kinetics. Proc Natl Acad Sci U S A. 2002;99:10555–10560. doi: 10.1073/pnas.152046799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rosen R, Davidov Y, LaRossa RA, Belkin S. Microbial sensors of ultraviolet radiation based on recA′ ∷ lux fusions. Appl Biochem Biotechnol. 2000;89:151–160. doi: 10.1385/abab:89:2-3:151. [DOI] [PubMed] [Google Scholar]
- Salles B, Weinstock GM. Mutation of the promoter and LexA binding sites of cea, the gene encoding colicin E1. Mol Gen Genet. 1989;215:483–489. doi: 10.1007/BF00427047. [DOI] [PubMed] [Google Scholar]
- Smarda J, Smajs D. Colicins – exocellular lethal proteins of Escherichia coli. Folia Microbiol (Praha) 1998;43:563–582. doi: 10.1007/BF02816372. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. clustal w: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujvari A, Martin CT. Evidence for DNA bending at the T7 RNA polymerase promoter. J Mol Biol. 2000;295:1173–1184. doi: 10.1006/jmbi.1999.3418. [DOI] [PubMed] [Google Scholar]
- van der Lelie D, Regniers L, Borremans B, Provoost A, Verschaeve L. The VITOTOX test, an SOS bioluminescence Salmonella typhimurium test to measure genotoxicity kinetics. Mutat Res. 1997;389:279–290. doi: 10.1016/s1383-5718(96)00158-1. [DOI] [PubMed] [Google Scholar]
- Van Dyk TK, Rosson RA. Photorhabdus luminescens luxCDABE promoter probe vectors. Methods Mol Biol. 1998;102:85–95. doi: 10.1385/0-89603-520-4:85. [DOI] [PubMed] [Google Scholar]
- Van Dyk TK, DeRose EJ, Gonye GE. LuxArray, a high-density, genomewide transcription analysis of Escherichia coli using bioluminescent reporter strains. J Bacteriol. 2001a;183:5496–5505. doi: 10.1128/JB.183.19.5496-5505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyk TK, Wei Y, Hanafey MK, Dolan M, Reeve MJ, Rafalski JA, Rothman-Denes LB, LaRossa RA. A genomic approach to gene fusion technology. Proc Natl Acad Sci U S A. 2001b;98:2555–2560. doi: 10.1073/pnas.041620498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankemmelbeke M, Healy B, Moore GR, Kleanthous C, Penfold CN, James R. Rapid detection of colicin E9-induced DNA damage using Escherichia coli cells carrying SOS promoter–lux fusions. J Bacteriol. 2005;187:4900–4907. doi: 10.1128/JB.187.14.4900-4907.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GC. The SOS response of Escherichia coli. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1987. pp. 1346–1357. [Google Scholar]
- Walker GC. SOS-regulated proteins in translesion DNA synthesis and mutagenesis. Trends Biochem Sci. 1995;20:416–420. doi: 10.1016/s0968-0004(00)89091-x. [DOI] [PubMed] [Google Scholar]


