Abstract
Genetic exchange as a mechanism underlying the extensive diversity of Leishmania parasites has not been shown. We report here evidence that the invertebrate stages of Leishmania are capable of a sexual cycle consistent with a meiotic process as described for African trypanosomes. Hybrid progeny were generated that bore full genomic complements from both parents, but kDNA maxicircles from one parent. Mating occurred only in the sand fly vector, and hybrids were transmitted to the mammalian host by sand fly bite. Genetic exchange likely contributes to phenotypic diversity in natural populations, and analysis of hybrid progeny will be useful for positional cloning of the genes controlling traits such as virulence, tissue tropism, and drug resistance.
Parasitic protozoa of the genus Leishmania cause a spectrum of human diseases that pose serious public health challenges for prevention, diagnosis, and treatment. The diversity of Leishmania species, with over 20 currently recognized, is thought to have arisen by gradual accumulation of divergent mutations rather than by sexual recombination. Tibayrenc et al. (1), has reported strong linkage disequilibrium in several Leishmania species, and proposed that these parasites are essentially clonal. This notion must be reconciled, however, with the accumulating examples of naturally occurring strains that share genotypic markers from two recognized species, providing circumstantial evidence for sexual recombination (2-4). Genetic exchange has been documented for the other trypanosomatids that cause human disease. Hybrid genotypes were observed in tsetse flies during co-transmission of two strains of Trypanosoma brucei (5), and in mammalian cells following co-infection with two clones of Trypanosoma cruzi differing in drug resistance markers (6). Using drug resistance markers, we provide evidence for genetic exchange in Leishmania major, and discuss the implications of these findings to Leishmania biology and experimental analysis.
One parental clone, LV39c5(HYG), was derived from strain LV39 clone 5 (RHO/SU/59/P) and was heterozygous for an allelic replacement of the LPG5A on chromosome 24 by a hygromycin B resistance cassette (LPG5A/LPG5A::†HYG) (7). The second parental clone, FV1(SAT), was derived from NIH Friedlin clone V1 (WHOM/IL/80/FN) and bore a heterozygous nourseothricin resistance (SAT) marker, integrated along with a linked firefly luciferase (LUC) reporter gene into one allele of the ~24 ribosomal RNA cistrons located on chromosome 27(8) (+/SSU::SAT-LUC). These strains were chosen as they are phenotypically identical to their respective parental WT virulent L. major, while the markers were chosen because they are functionally independent (9). The target gene modifications were chosen as they caused no effect on normal growth in vitro or in mouse infections (10), nor were epistatic interactions anticipated between these alleles.
Multiple attempts to generate hybrid parasites resistant to both antibiotics during in vitro co-culture of the parental lines were unsuccessful (see Supplementary methods). The parental clones were tested for their ability to generate parasites resistant to both drugs during co-infection in the sand fly. The growth of each parental line in Phlebotomus duboscqi, a natural vector of L. major, is shown in Supplementary Fig. 1. Promastigotes of each parent survived the initial period of bloodmeal digestion and excretion (days 1-6), and underwent metacyclogenesis at a comparable frequency (20-60%), though the FV1(SAT) parent established and maintained a 3-4 fold higher intensity of infection. The parental clones were tested for their ability to generate double drug resistant parasites during co-infection in the sand fly. Flies were fed through a membrane on mouse blood containing 3 and 1 × 106 / ml of the LV39c5(HYG) and FV1(SAT) lines, respectively, each obtained from log-phase cultures and extensively washed to remove antibiotics. A total of 102 flies from 4 independent co-infection experiments were dissected at 13-16 days post-infection, when they harbored mature infections with an average of 39,400 ± 14,700 promastigotes per midgut. Flies cannot be maintained under aseptic conditions, and more than half of the cultures established from the midgut parasites were lost to fungal contamination during the subsequent 1-2 wks of culture. In the remaining cultures, 12 (26%) grew out promastigotes that were resistant to both drugs. Clonal lines were generated from 9 of the doubly-drug resistant populations and the genotypes and phenotypes of 1-2 clones from each culture were determined (summarized in Table 1).
Table 1.
Summary of phenotype and genotype data for parental and progeny clones.
| Cell linea | Virulence profileb | Maxicircle inheritancec | 3F12 antibody | Clumping | Ploidy | Chromosomal analysis d |
|||
|---|---|---|---|---|---|---|---|---|---|
| Six chromosomese |
Chr 31f |
||||||||
| SEQ | CAPS | SEQ | CAPS | ||||||
| FV1(SAT) | f | F | + | - | 2n | ||||
| LV39.5c5(HYG) | s | L | - | + | 2n | ||||
| 1.10.B12 | ND | L | + | + | 2n | 1:1 | H | 1:1 | H |
| 1.10.D9 | ND | L | + | + | 2n | 1:1 | H | 1:1 | H |
| 1.16.A1 | f | F | + | + | 2n | 1:1 | H | L>F | H |
| 1.16.C4 | ND | F | + | + | 2n | 1:1 | H | L>F | H |
| 4.3.A12 | f | F | + | + | 2n | 1:1 | H | 1:1 | H |
| 4.3.G12 | ND | L | + | + | 2n | 1:1 | H | 1:1 | H |
| 4.7.A3 | f | F | + | + | 2n | 1:1 | H | 1:1 | H |
| 5.12.D9 | ND | F | + | + | 2n | 1:1 | H | L>F | H |
| 5.12.F11 | s | F | + | + | 2n | 1:1 | H | L>F | H |
| 5.22.A10 | f | F | + | + | 2n | 1:1 | H | 1:1 | H |
| 6.14.F9 | f | F | + | + | 2n | 1:1 | H | 1:1 | H |
| 1.14.B11 | s | L | + | + | 3n | L>F | H | L>F | H |
| 1.14.E10 | ND | L | + | + | 3n | L>F | H | L>F | H |
| 4.9.C8 | s | L | + | + | 3n | L>F | H | L>F | H |
| 4.9.E6 | s | F | + | + | 3n | L>F | H | L>F | H |
| 4.17.A3 | s | F | + | + | 3n | L>F | H | L>F | H |
| 4.17.F4 | ND | F | + | + | 3n | L>F | H | L>F | H |
| 6.16.E8 | s | F | + | + | 3n | L>F | H | L>F | H |
hybrid lines nomenclature: experiment number. fly number. clone name; for 6.14.F9 and 6.16.E8, the nomenclature refers to experiment number.ear lesion number.clone name
fast (f); slow (s); intermediate (i)
F, FV1(SAT) SNP pattern; L, LV39c5(HYG) SNP pattern
SEQ = ratio of parental SNP peaks by sequence analysis; CAPS=cleaved amplified polymorphic site analysis; H = hybrid
Chromosomes 2, 7, 21 25, 35, 36
Chromosome 31 is tetrasomic
PCR tests with primers specific for the parental markers showed that all doubly-drug resistant clones tested contained both the HYG and SAT drug resistance genes (Fig. 1A; Table 1; Supplementary Table 3). Controls showed that the marker loci had not rearranged during hybrid formation, maintaining their location within the LPG5A or SSU rRNA loci, respectively (Supplementary Fig. S2). These data indicated that the doubly-drug resistant clonal lines were genetic hybrids. To confirm that hybrid formation was compatible with transmission to the mammalian host, co-infected sand flies were allowed to bite and induce lesions in BALB/c mice. Two doubly-drug resistant clonal lines, designated 6.16.E8 and 6.14.F9, were recovered from the 8 dermal lesions examined, and were found to be similar to those directly recovered from insects, as discussed below (Table 1). Co-injection of both parental lines into mouse ears by needle never led to the recovery of doubly-drug resistant parasites from the 20 dermal lesions examined, each containing > 2.4 × 107 amastigotes, suggesting that the hybrids selected following transmission by the co-infected sand flies were generated in the fly and not in the mammalian host.
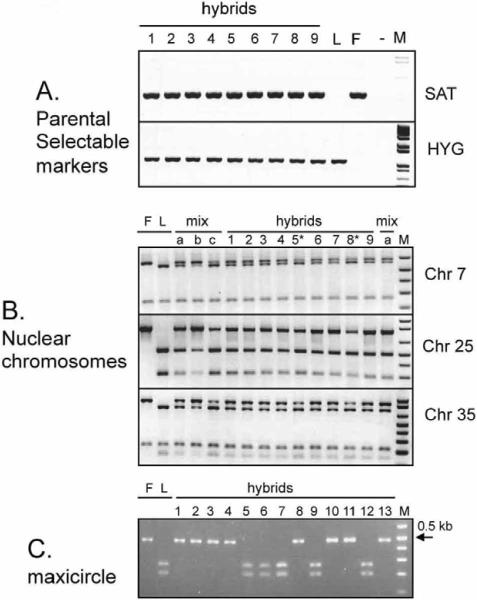
Fig. 1. Genotyping of hybrid Leishmania.
A, PCR for parental selectable drug markers SAT and HYG. Samples are F, FV1(SAT); L, LV39c5(HYG); M, 1 kb plus marker (Invitrogen, CA); -, no template control, 1, 1.10.B12; 2, 1.10.D9; 3, 4.3.G12; 4, 4.3.A12; 5, 1.14.E10; 6, 5.12.D9; 7, 5.12.F11; 8, 1.14.B11; 9, 5.22.10 B, SNP-CAPS analysis for loci on chromosomes 7, 25, and 35. F, L and M are as in panel A; mix a, b and c, parental templates mixed in different ratios (1:1, 3:1, or 1:3 F:L respectively) DNA content analysis showed that hybrids 5 and 8 are 3n (marked by *) and the remainder shown are 2n progeny. C, SNP-CAPS analysis of Maxicircle. Digestion with BfaI of the ND5 - divergent region PCR product is shown. Hybrids tested are 1, 4.7.A3; 2, 4.17.A3; 3, 4.9.E6; 4, 4.17.F4; 5, 1.10.B12; 6, 1.10.D9; 7, 4.3.G12; 8, 4.3.A12; 9, 1.14.E10; 10, 5.12.D9; 11, 5.12.F11; 12, 1.14.B11; and 13, 5.22.10.
We examined the segregation of loci not linked to chromosomes 24 and 27, the location of the HYG and SAT markers, respectively. We used SNPs developed from comparisons of the terminal ~30 kb chromosomal regions encompassing the SCG genes located on L. major chromosomes 2, 7, 21, 25, 31, 35, and 36 in the WT parent of LV39c5(HYG). SCGs comprise a family of polymorphic telomeric galactosyltransferases (11), but genes internal to these showed SNPs occurring at an overall frequency of ~0.15%, consistent with other estimates of strain variation (12, 13). For each chromosome, SNPs from 1-2 loci, located from 8.5 - 23 kb inwards of the telomeric SCGs (11) (Supplementary Tables 1, 2), were analyzed by a combination of SNP-CAPS(14) and/or direct sequencing. Each parent was homozygous for every marker tested, while all 18 progeny tested clearly inherited both parental alleles (Fig. 1B, Table 1). This provides evidence that each progeny clone inherited a full set of chromosomes from each parent and were thus full genome hybrids.
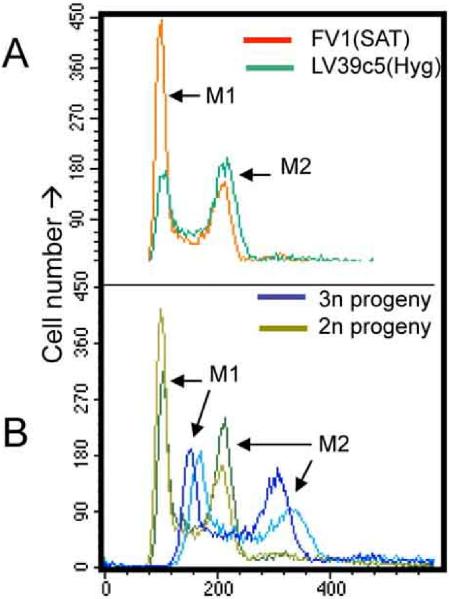
In 7/18 hybrid progeny clones, the relative ratio of the parental alleles seen in SNP-CAPS digestions differed from the expected 1:1. Instead, in each case the most intense bands or sequencing trace peaks were those associated with the LV39c5 parent, a finding seen at all loci and in all lines tested (Fig. 1B; Supplementary Fig. S3; Table 1). One possibility is the occurrence of triploid offspring, bearing two chromosomal complements from one parent (LV39) and one from the other (FV1). The DNA contents of the progeny clones were measured relative to those of the parents by flow cytometry, and the profiles compared to 2n and 4n lines studied previously (15). While the parents and most hybrids showed 2n DNA contents, the seven hybrids detected above showed 3n DNA contents (Fig. 2, Table 1); intermediate DNA contents were not observed. Thus, while all doubly drug resistant lines were full genome hybrids, a significant fraction appeared to be triploid rather than diploid, inheriting two genomic complements from the LV39 and one from FV1 parents. Similarly, for four of the 2n lines (1.16.A1, 1.16.C4, 5.12.D9, 5.12.F11) the segregation of chromosome 31 markers appeared to differ from the expected 1:1 ratio, with the LV39c5 parent again predominating in sequencing and/or SNP-CAPs analysis of the hybrids (Table 1). This may arise from the finding that Leishmania chromosomes are occasionally aneuploid (16), including tetrasomy of Leishmania chromosome 31 in the parental lines studied here. Potentially, tetrasomy may affect mitotic inheritance and segregation, as seen in autotetraploid or allotetraploid species (17). This was not pursued further here.
Fig. 2. DNA contents of parental and 2n and 3n progeny clones.
DNA contents were measured in log phase cells by flow cytometry after staining RNAse treated permeabilized cells with propidium iodide. A, parental lines (FV1(SAT), red; LV39c5(HYG), green. B, representative 2n (green; 4.3.G12, 1.10.D9) and 3n (blue; 1.14.E10; 4.9.C8) progeny. M1 refers to cells in the G1/G0 phase of the cell cycle and M2 refers to cells in G2/M phase. Small differences in the M1/M2 ratio reflect minor differences in cellular growth rate and growth phase and are not significant nor found in other studies of these lines.
SNP markers were identified to determine the inheritance of mitochondrial maxicircle kDNA (Fig. 1C; Table 1; Supplementary Tables 1-3). In contrast to the chromosomal DNA, maxicircle markers demonstrated clear and consistent uniparental inheritance, with six of the progeny clones possessing maxicircle kDNA exclusively from the LV39c5(HYG), and 12 inheriting maxicircle kDNA exclusively from FV1(SAT) (Table 1; Fig. 1C). These markers allowed us to establish that two clonal lines arising from the same fly that had identical nuclear markers in fact differed (4.9.C8/C6; 4.3.A12/G12;Table 1). kDNA minicircles were not examined.
Several studies were undertaken to compare the progeny clones for parental phenotypic traits. Metacyclic promastigotes of the FV1 line react with monoclonal antibody (mAb) 3F12, which recognizes β(1,2)Ara-terminating-mono-βGal-modified phosophoglycans, including the abundant surface LPG. The same antibody fails to bind to LV39c5 metacyclics owing to differences in the βGal chain length (18). Metacyclic promastigotes purified from all progeny clones displayed strong reactivity with mAb3F12, as determined by surface agglutination of live parasites (Supplementary Fig. S4A). Thus the mono-galactosylated 3F12 trait appeared to be inherited as a dominant trait. Another dominant trait is the `clumpy' appearance of LV39c5(HYG) when grown in standard culture media, in contrast to FV1(SAT) which grows as individual cells. All hybrid offspring appeared `clumpy' (Supplementary Fig. S4B).
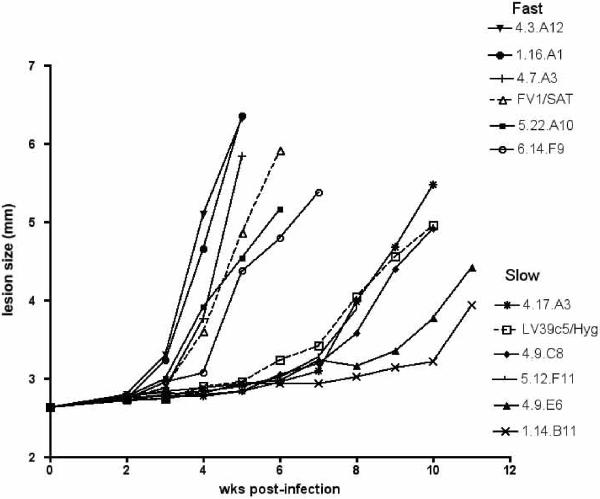
Interestingly, the parental clones differ in a virulence trait defined as the time required for the emergence of lesions in susceptible BALB/c mice (Fig. 3). Following s.c. footpad inoculation of 104 metacyclic promastigotes, purified in each case from stationary cells freshly transformed from lesion amastigotes, five of the progeny clones displayed lesion progression as fast or faster than the FV1(SAT) parent, whereas five of the progeny showed lesion development as slow or slower than the LV39c5(HYG) parent. These data indicate that despite the low nucleotide heterozygosity typically found in L. major (13), there is sufficient variation in one or both parental clones for distinct virulence phenotypes to emerge. The most parsimonious explanation is that either `slow' or `fast' growth is inherited as a dominant trait, and that one or both parental clones are heterozygous for the gene(s) controlling this trait.
Fig. 3. Lesion formation in BALB/c mice by parental and hybrid progeny clones. Mice were infected in the hind footpad by s.c. inoculation of 104 metacyclic promastigtoes.
Results are representative of three independent experiments.
All five triploid lines tested exhibited the `slow virulence' trait, which occurred in only 1/8 of the diploid lines. There may be a reduction in virulence associated with polyploidy, which accounts for the failure to detect polyploid lines in field isolates. In contrast, no significant association was seen with the maxicircle genotype and virulence or ploidy (Table 1).
Leishmania can undergo genetic exchange during growth and development in the sand fly vector, and transmit infectious stage hybrid progeny to a mammalian host. Based on the analysis of 18 hybrid clones representing a minimum of 11 independent crosses, the findings argue for inheritance of at least one full set of chromosomes from each parent, accompanied by independent, uniparental inheritance of the maxicircle kDNA derived from one parent. The inheritance patterns of nuclear DNA fit a Mendelian model of meiosis of the parental strains followed by fusion of the haploid cells. However, alternative mechanisms, modeled after the tetraploid sexual cycle described in S. cerevisiae(19), could involve fusion of parental diploid cells followed by meiosis and intracellular fusion of haploid nuclei(20, 21). Triploid offspring have also been seen in trypanosome crosses, and have been attributed to incomplete meiotic division and fusion of haploid and diploid nuclei (Supplementary Fig. S5;(20, 22)). In trypanosomes as well, maxicircle kDNA was initially thought to be inherited uniparentally; however, later studies showed it to be inherited biparentally, but subsequently to segregate out during mitosis, leading to fixation (23). It is possible that mixed maxicircle genotypes might have also been present in earlier generations of the Leishmania crosses than were examined here.
The frequency of genetic exchange involving these two parental clones would appear to be rare (~ 2.5 × 10-5 or less, after correcting for recovery of only doubly drug resistant offspring). Consistent with this, most clonal lines from a single infected fly were identical, although two flies yielded offspring with different maxicircle genotypes, suggesting two independent crossing events. Whether the frequency observed in the FV1 × LV39c5 cross here is typical for other Leishmania strains or species remains to be determined. The low frequency agrees with the general sense that gene exchange must occur rarely, as deduced from observed heterozygosities and linkage disequilibrium in natural populations (1).
Despite the infrequency of gene exchange experimentally or in nature, there are many examples of hybrid genotypes observed in field isolates involving most Leishmania species (2-4, 24-26). Potentially, these hybrids arose from rare `mating' events, yielding offspring with a strong selective advantage, such as seen in Toxoplasma gondii (27), and suggested by the clonal propagation of an emergent hybrid mucosal strain in Peru (3). Given the rarity with which mixed infections in flies are likely to occur, in conjunction with the low frequency of hybridization that we have observed in co-infected flies, any successful new genotype would be expected to propagate clonally.
While rare in nature, the frequency of experimental hybrid formation is sufficient to enable use as an experimental tool. Our studies show segregation of `virulence traits' in the FV1 × LV39c5 crosses studied here, and through positional cloning the genes responsible may be identified. Future studies will explore the possibility to carry out backcrosses, as well as crosses between species, and to develop SNP tools for genetic linkage analysis.
Supplementary Material
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, NIAID, and in part by NIAID Grant Support (SMB and NSA AI029646). We thank K. Owens for inventorying and shipping parasite strains, K. Beacht for mouse infection studies, and M. Grigg, L. Miller and A. Sher for critical review of the manuscript. Maxicircle sequences have been submitted to GenBank and had been assigned accession numbers FJ349262- FJ349263 (12SrRNA) and FJ349264-FJ349265 (Divergent region).
References
- 1.Tibayrenc M, Ben Abderrazak S, Guerrini F, Banuls A. Arch Inst Pasteur Tunis. 1993 Jul-Oct;70:375. [PubMed] [Google Scholar]
- 2.Kelly JM, Law JM, Chapman CJ, Van Eys GJ, Evans DA. Mol Biochem Parasitol. 1991 Jun;46:253. doi: 10.1016/0166-6851(91)90049-c. [DOI] [PubMed] [Google Scholar]
- 3.Nolder D, Roncal N, Davies CR, Llanos-Cuentas A, Miles MA. Am J Trop Med Hyg. 2007 Mar;76:573. [PubMed] [Google Scholar]
- 4.Ravel C, et al. Int J Parasitol. 2006 Nov;36:1383. doi: 10.1016/j.ijpara.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 5.Jenni L, et al. Nature. 1986 Jul 10-16;322:173. doi: 10.1038/322173a0. [DOI] [PubMed] [Google Scholar]
- 6.Gaunt MW, et al. Nature. 2003 Feb 27;421:936. doi: 10.1038/nature01438. [DOI] [PubMed] [Google Scholar]
- 7.Capul AA, Barron T, Dobson DE, Turco SJ, Beverley SM. J Biol Chem. 2007 May 11;282:14006. doi: 10.1074/jbc.M610869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez-Calvillo S, et al. Mol Biochem Parasitol. 2001 Sep 3;116:147. doi: 10.1016/s0166-6851(01)00310-3. [DOI] [PubMed] [Google Scholar]
- 9.Joshi PB, Webb JR, Davies JE, McMaster WR. Gene. 1995 Apr 14;156:145. doi: 10.1016/0378-1119(95)00042-5. [DOI] [PubMed] [Google Scholar]
- 10.Capul AA, Hickerson S, Barron T, Turco SJ, Beverley SM. Infect Immun. 2007 Sep;75:4629. doi: 10.1128/IAI.00735-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobson DE, Scholtes LD, Myler PJ, Turco SJ, Beverley SM. Mol Biochem Parasitol. 2006 Apr;146:231. doi: 10.1016/j.molbiopara.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Lukes J, et al. Proc Natl Acad Sci U S A. 2007 May 29;104:9375. [Google Scholar]
- 13.Ivens AC, et al. Science. 2005 Jul 15;309:436. [Google Scholar]
- 14.Thiel T, Kota R, Grosse I, Stein N, Graner A. Nucleic Acids Res. 2004;32:e5. doi: 10.1093/nar/gnh006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruz AK, Titus R, Beverley SM. Proc Natl Acad Sci U S A. 1993;90:1599. doi: 10.1073/pnas.90.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravel C, Dubessay P, Bastien P, Blackwell JM, Ivens AC. Parasitol Today. 1998 Aug 1;14:301. doi: 10.1016/s0169-4758(98)01275-7. [DOI] [PubMed] [Google Scholar]
- 17.Moody ME, Mueller LD, Soltis DE. Genetics. 1993 Jun;134:649. doi: 10.1093/genetics/134.2.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobson DE, et al. J Biol Chem. 2003 Aug 1;278:28840. doi: 10.1074/jbc.M302728200. [DOI] [PubMed] [Google Scholar]
- 19.Rose MD. Annu Rev Cell Dev Biol. 1996;12:663. doi: 10.1146/annurev.cellbio.12.1.663. [DOI] [PubMed] [Google Scholar]
- 20.Gibson W, Garside L, Bailey M. Mol Biochem Parasitol. 1992 Apr;51:189. doi: 10.1016/0166-6851(92)90069-v. [DOI] [PubMed] [Google Scholar]
- 21.Heitman J. Curr Biol. 2006 Sep 5;16:R711. doi: 10.1016/j.cub.2006.07.064. [DOI] [PubMed] [Google Scholar]
- 22.Hope M, et al. Mol Biochem Parasitol. 1999 Oct 25;104:1. doi: 10.1016/s0166-6851(99)00103-6. [DOI] [PubMed] [Google Scholar]
- 23.Turner CM, Hide G, Buchanan N, Tait A. Exp Parasitol. 1995 Mar;80:234. doi: 10.1006/expr.1995.1029. [DOI] [PubMed] [Google Scholar]
- 24.Dujardin JC, et al. Acta Trop. 1995 Aug;59:293. doi: 10.1016/0001-706x(95)00094-u. [DOI] [PubMed] [Google Scholar]
- 25.Schwenkenbecher JM, et al. Int J Parasitol. 2006 Feb;36:237. doi: 10.1016/j.ijpara.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Mauricio IL, Howard MK, Stothard JR, Miles MA. Parasitology. 1999 Sep;119(Pt 3):237. doi: 10.1017/s0031182099004710. [DOI] [PubMed] [Google Scholar]
- 27.Grigg ME, Bonnefoy S, Hehl AB, Suzuki Y, Boothroyd JC. Science. 2001 Oct 5;294:161. doi: 10.1126/science.1061888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.