Summary
A glucose transporter null mutant of the parasitic protozoan Leishmania mexicana, in which three linked glucose transporter genes have been deleted by targeted gene replacement, is unable to replicate as amastigote forms within phagolysomes of mammalian host macrophages and is avirulent. Spontaneous suppressors of the null mutant have been isolated that partially restore replication of parasites within macrophages. These suppressor mutants have amplified the gene for an alternative hexose transporter, the LmGT4 permease (previously called the D2 permease), on a circular extrachromosomal element, and they overexpress LmGT4 mRNA and protein. The suppressors have also regained the ability to transport hexoses, and they have reverted other phenotypes of the null mutant exhibiting enhanced resistance to oxidative killing, heat shock and starvation for nutrients, as well as augmented levels of the storage carbohydrate β-mannan, increased cell size and increased growth as insect stage promastigotes compared with the unsuppressed mutant. Complementation of the null mutant with the LmGT4 gene on a multicopy episomal expression vector also reverted these phenotypes, confirming that suppression results from amplification of the LmGT4 gene. These results underscore the importance of hexose transporters for the infectious stage of the parasite life cycle.
Introduction
Parasitic protozoa of the genus Leishmania are pathogens of humans and other vertebrates that are responsible for widespread disease in many tropical and subtropical regions of the globe (Herwaldt, 1999). The life cycle consists of three principal stages: flagellated procyclic promastigotes that divide within the gut of the sand fly vector; non-dividing metacyclic promastigotes that live in the mouthparts of the insect vector and are resistant to mammalian complement; and aflagellate amastigotes that live within phagolysosomal vesicles of vertebrate host macrophages. Promastigotes robustly transport and metabolize glucose and fructose that are major sources of carbon and metabolic energy (Cazzulo, 1992) in the sand fly environment, since the sand fly feeds upon plant nectar that is rich in sugars (Schlein, 1986). In contrast, the macrophage phagolysosome is thought to be relatively poor in carbohydrates (Naderer and McConville, 2008), and amastigotes rely upon oxidation of fatty acids and metabolism of amino acids to provide metabolic resources including energy and precursors for synthesis of carbohydrates via gluconeogenesis (McConville et al., 2007; Rosenzweig et al., 2008; Naderer and McConville, 2008).
Despite the more pronounced availability and utilization of carbohydrates by promastigotes compared with amastigotes, our previous studies on hexose transporters in Leishmania mexicana (Burchmore et al., 2003) have revealed that these permeases play a particularly important role in the intracellular amastigotes. A glucose transporter null mutant, Δlmgt, in which the three clustered genes encoding the glucose transporter isoforms LmGT1, LmGT2 and LmGT3 were deleted by targeted gene replacement, was unable to replicate in primary murine macrophages or as culture-form axenic amastigotes. While the null mutant grew more slowly than wild-type promastigotes in culture medium containing glucose, it was viable in this life cycle stage and able to metabolize proline, an alternate energy source present in the sand fly gut (ter Kuile and Opperdoes, 1992).
In this article, we report the identification of spontaneously derived suppressor mutants of the Δlmgt null mutant that exhibit partially restored ability to replicate within murine macrophages. These suppressor mutants have also partially regained uptake of hexoses from the medium, and they have re-established other phenotypes similar to that of wild-type parasites such as increased rate of growth in glucose-replete medium, resistance to oxidative stress, ability to withstand starvation, increased accumulation of the storage carbohydrate β-mannan and partial resistance to heat shock. These suppressors have amplified an alternate lower-affinity hexose transporter gene, LmGT4, on a circular amplicon and have increased the expression of LmGT4 mRNA and protein. Furthermore, transfection of the Δlmgt null mutants with the LmGT4 open reading frame (ORF) on an episomal expression vector confers the properties of the suppressor mutants, demonstrating that amplification of this gene alone can explain the altered phenotypes. These results underscore a crucial role for hexose transporters in the amastigote stage of the Leishmania life cycle.
Results
Glucose transporter null mutants transform into amastigotes but do not survive
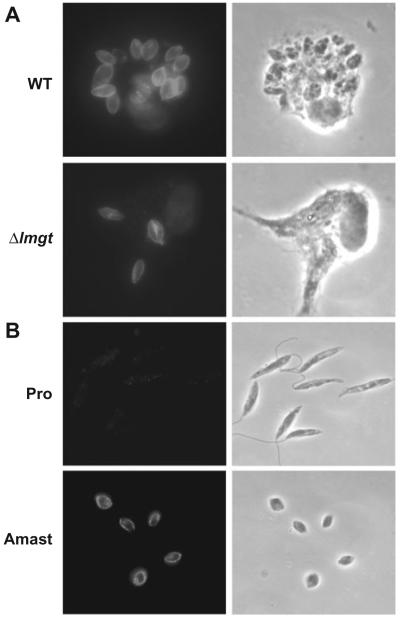
The glucose transporter null mutant Δlmgt survives poorly in murine macrophages (Burchmore et al., 2003). In initial experiments, we examined further the nature of the defect in the ability of the Δlmgt null mutants to survive within macrophages. In principle, these mutants could either (i) fail to transform from promastigotes to amastigotes and thus die within macrophages or (ii) transform efficiently into amastigotes but fail to survive as amastigotes within the environment of the phagolysosome. To discriminate between these two possibilities, we infected macrophages with wild-type and Δlmgt null mutant promastigotes and examined intracellular parasites 1, 2 and 5 days after infection. The cells were stained with a polyclonal antiserum directed against the HASPB protein, which in L. mexicana is expressed at high levels only in the amastigote stage of the life cycle (Nugent et al., 2004) (antibody kindly supplied by Dr Deborah Smith). While surviving Δlmgt parasites were much less numerous than those from the infection with wild-type parasites, those that were observed stained with the anti-HASPB antibody, as did wild-type amastigotes (e.g. Fig. 1A, 5 days post infection) and wild-type axenic culture-form amastigotes (Fig. 1B). In contrast, cultured promastigotes did not stain with this antibody (Fig. 1B), nor did metacyclic promastigotes react with this antiserum (Nugent et al., 2004). Importantly, these observations confirm that the glucose transporter null mutants do not simply fail to transform into amastigotes and die within macrophages as a result of a developmental blockade. Rather, the null mutants exhibit a defect in survival as amastigotes per se, indicating that glucose transporters are important for viability of amastigotes but not for transformation of promastigotes to amastigotes.
Fig. 1.
Immunofluorescence of intracellular amastigotes, axenic culture-form amastigotes and promastigotes with anti-HASPB antibody.
A. Immunofluorescence (left) and phase-contrast (right) images of primary murine macrophages infected with wild-type (WT) and Δlmgt null mutant parasites (5 days post infection).
B. Immunofluorescence (left) and phase-contrast (right) images of wild-type promastigotes (Pro) and axenic culture-form amastigotes (Amast).
Spontaneous suppressors of the glucose transporter null mutant
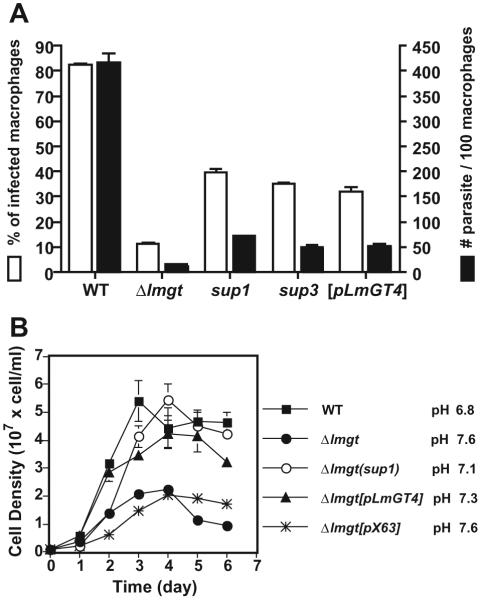
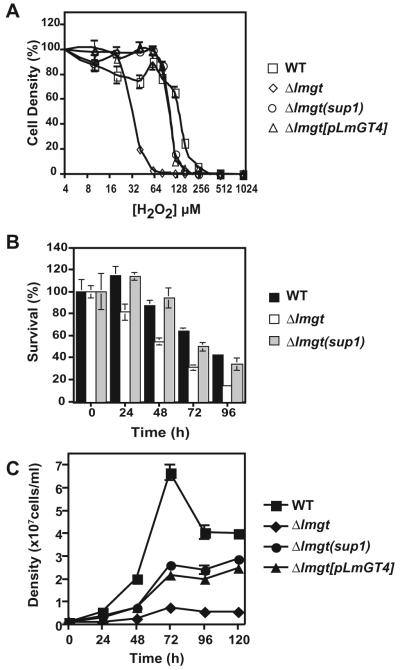
Passage of the glucose transporter null mutant Δlmgt in glucose-replete RPMI medium for about 6 months generated a spontaneously altered strain, designated Δlmgt(sup1), that exhibited partially restored survival as amastigotes within primary murine macrophages. While the original null mutant survived very poorly at 5 days post infection, the Δlmgt(sup1) mutant showed increased infectivity measured either as per cent of infected macrophages or number of parasites per macrophage (Fig. 2A). This partial restoration of infectivity was demonstrated in three replicate experiments.
Fig. 2.
Growth of amastigotes (A) and promastigotes (B) of wild type (WT), Δlmgt null mutants, Δlmgt(sup1) (sup1) and Δlmgt(sup3) (sup3) suppressor strains, Δlmgt[pLmGT4] ([pLmGT4])-complemented strain, and the Δlmgt[pX63NEO-RI] strain ([pX63]) complemented with the empty expression vector.
A. Primary peritoneal macrophages from BALB/c mice were infected with stationary-phase promastigotes at a multiplicity of 5 and incubated in Dulbecco’s modified Eagle Medium at 37°C for 5 days. Data represent the average and standard deviations of three determinations and are plotted either as per cent infected macrophages in the field (white bars) or as parasites per 100 macrophages (black bars).
B. Promastigotes were inoculated at 1 × 106 cell ml-1 and grown in RPMI medium at 26°C and samples were withdrawn and counted microscopically each day. The final pH values of each culture at day 6 are indicated next to the symbol legend at the right. The data in this and subsequent figures represent the average and standard deviations of at least three determinations.
The Δlmgt(sup1) mutant promastigotes grew as well as wild-type parasites in glucose-replete RPMI medium (Fig. 2B) suggesting that they had recovered the ability to utilize glucose as a fuel. Consistent with their ability to utilize glucose metabolically and secrete organic acids representing partially oxidized products of glucose metabolism, such as succinate, acetate, pyruvate and d-lactate (Cazzulo, 1992), wild type and the Δlmgt(sup1) mutant acidified RPMI medium (initial pH 7.25) after 6 days of growth as promastigotes (final pH of 6.8 and 7.1 respectively), whereas Δlmgt null mutants increased the pH to 7.6 (Fig. 2B).
We have observed the generation of suppressors of the Δlmgt null mutant on four independent occasions leading to the generation of Δlmgt(sup1-4) strains. The Δlmgt(sup1) and Δlmgt(sup2) suppressors were detected following about 6 months of passage as promastigotes in glucose-replete medium, employing a 1:20 dilution approximately every 3 days, and the Δlmgt(sup3) and Δlmgt(sup4) isolates were detected after about 3 months. Genomic Southern blots indicate that Δlmgt(sup1), Δlmgt(sup2) and Δlmgt(sup4) have all amplified the LmGT4 glucose transporter gene (see below), whereas the Δlmgt(sup3) mutant does not exhibit this amplification. While we did not determine the reversion frequency, these data are most consistent with the idea that suppressors occur relatively infrequently, and that the four identified represent independent events arising from the same parental Δlmgt line.
We emphasize that we did not apply any intentional selection to derive these suppressors. Rather the ability of the suppressors to grow more rapidly than the null mutant in glucose-replete medium (Fig. 2B) suggests that the associated genetic alteration provided a growth advantage to the suppressor and allowed it to overgrow the original Δlmgt population.
Δlmgt(sup1) mutants can take up hexose
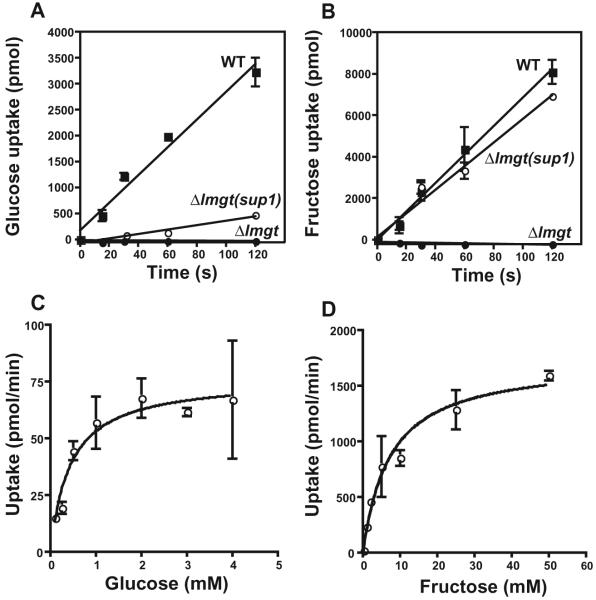
To determine whether the Δlmgt(sup1) mutants had regained the ability to take up hexoses, we measured uptake of 10 mM [14C]d-glucose and [14C]d-fructose in short-term transport assays (Fig. 3). While the original null mutant revealed no measurable uptake of these sugars over a 2 min time-course, the Δlmgt(sup1) mutant displayed pronounced uptake over the level of the null mutant (Fig. 3A and B). At this concentration of substrate, glucose uptake was partially restored, and fructose uptake was restored almost to wild-type levels. The restored uptake was saturable (Fig. 3C and D), consistent with mediation by a transporter. Multiple saturation curves for d-fructose indicated an average Km value of 8.9 ± 1.4 mM (n = 3; all errors reported as standard deviations), whereas those for d-glucose were more variable and produced an average Km value of 1.3 ± 0.78 mM (n = 3). Uptake assays (data not shown) revealed that Δlmgt(sup2) and Δlmgt(sup4) also exhibited restored fructose transport. In contrast, Δlmgt(sup3) showed no increase in fructose uptake compared with the Δlmgt null mutant.
Fig. 3.
Uptake of [14C]d-glucose (A and C) and [14C]d-fructose (B and D) by Δlmgt null mutant (●), Δlmgt(sup1) (○) and wild-type (■) parasites.
A and B. Time-courses for uptake of 10 mM sugar were performed using 3 × 107 parasites per sample. The background uptake value at t = 0 was subtracted from all data points.
C and D. Uptake assays (30 s) were performed using 2 × 107 Δlmgt(sup1) parasites over a range of substrate concentrations. Saturation curves were fitted using the Michaelis-Menten equation and Prism 4 software (GraphPad). For these curves, the Km value was 0.44 mM for glucose and 7.3 mM for fructose.
Δlmgt(sup1) mutants have amplified the LmGT4 hexose transporter gene
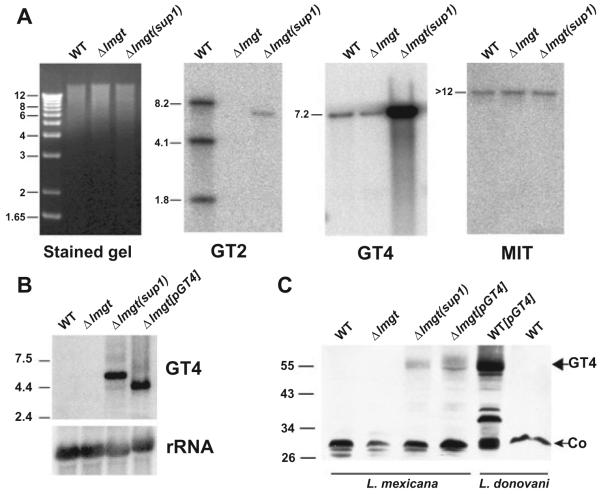
Southern blot analysis probing genomic DNA from the Δlmgt(sup1) line with a 676-nucleotide fragment representing the 3′ terminal half of the LmGT2 ORF (Fig. 4A, GT2 panel) confirmed that this line lacked the LmGT1-3 genes as expected. However, we noticed the presence of a weakly hybridizing 7 kb band in the Δlmgt(sup1) DNA, and hypothesized that this might arise through amplification of a divergent glucose transporter gene. A candidate glucose transporter family (Manolescu et al., 2007) member had been identified previously in Leishmania donovani, originally named the D2 permease (here renamed the LdGT4 transporter). The permease functions as a low-affinity hexose transporter when expressed in Xenopus oocytes and is probably expressed at considerably lower abundance compared with the LmGT1-3 family members (Langford et al., 1995). The sequence (see below) of the L. mexicana orthologue of LdGT4, LmGT4, shares 66.5% nucleotide identity with the region of the LmGT2 ORF employed as probe and thus could generate a hybridization signal upon amplification. Reprobing the genomic Southern blot with the L. donovani LdGT4 ORF (Fig. 4A, GT4 panel) revealed that the LmGT4 gene is indeed amplified in Δlmgt(sup1), and reprobing with the ORF for the MIT gene that encodes an L. donovani H+/myo-inositol co-transporter (Drew et al., 1995) (Fig. 4A, MIT panel) confirmed equal loading of DNA on each lane. Quantification indicated a 38 ± 4.4-fold (n = 5) amplification of the LmGT4 gene compared with wild-type parasites. Sequencing of two independent cDNA clones each from wild type and Δlmgt(sup1) revealed that the LmGT4 ORF was identical in both strains and that suppression was thus not due to mutation within that ORF in the Δlmgt(sup1) mutant.
Fig. 4.
Amplification of LmGT4 DNA, RNA and protein in Δlmgt(sup1) parasites.
A. Southern blot of genomic DNA from wild-type (WT), Δlmgt and Δlmgt(sup1) parasites was digested with EcoRI and BglII, and hybridized to ORFs from the LmGT2 (GT2), LmGT4 (GT4) and MIT genes. In this and subsequent blots, molecular weight markers are indicated in kilobase pairs to the left of the stained gel, and sizes of bands are indicated to the left of each blot.
B. Northern blot of total RNA (15 μg) from each parasite line was hybridized with a probe representing the L. mexicana LmGT4 ORF or rRNA (indicated to the right of the blot). Molecular weight markers are indicated at the left in kilobases.
C. Salt-extracted membranes were prepared from the L. mexicana lines (first four lanes from the left) indicated above each lane and from wild-type L. donovani (WT) and L. donovani complemented with the LdGT4 ORF (WT[pLdGT4]). A Western blot of equivalent amounts of protein (50 μg) was developed with affinity-purified antibody directed against the COOH hydrophilic terminal domain of the L. donovani LdGT4 permease. Molecular weight markers in kDa are indicated at the left. The band at ∼55 kDa (large arrow) is the LmGT4 protein and the band at ∼30 kDa (small arrow) is a cross-reacting protein that serves as a loading control (Co). Since the antibody is directed against the L. donovani LdGT4 transporter, the signal in the lane overexpressing the L. donovani LdGT4 permease (WT[pLdGT4]) is stronger than that in the lanes overexpressing the L. mexicana LmGT4 permease (Δlmgt(sup1) and Δlmgt[pLmGT4]).
Analysis of total RNA by Northern blot also revealed a 21 ± 1.7-fold (n = 2) increase in LmGT4 mRNA compared with wild-type promastigotes (Fig. 4B). Probing of salt-extracted membranes with an affinity-purified antiserum directed against the L. donovani LdGT4 permease (Langford et al., 1995) indicated a pronounced overexpression of the LmGT4 protein, although quantification of the level of overexpression was not possible due to the low level of LmGT4 protein expressed in the wild-type membranes. These observations indicate that amplification of the LmGT4 gene in Δlmgt(sup1) results in overexpression of the LmGT4 hexose transporter, concomitant with restoration of hexose transport function. Furthermore, two independently isolated suppressor mutants, Δlmgt(sup2) and Δlmgt(sup4), had a phenotype indistinguishable from Δlmgt(sup1) and had also experienced a similar amplification of the LmGT4 ORF (data not shown). In contrast, the Δlmgt(sup3) mutant showed no amplification of the LmGT4 gene. While the Δlmgt(sup2) strain emerged from glucose-replete medium, parallel passage of the Δlmgt null mutant in medium that was deficient in glucose but supplemented with either higher concentrations of amino acids or with glycerol failed to produce suppressor mutants. Hence, it appears that amplification of the LmGT4 gene is selected for by growth of the null mutant in glucose-replete medium.
Δlmgt(sup1) contains a circular episomal amplicon encompassing the LmGT4 gene
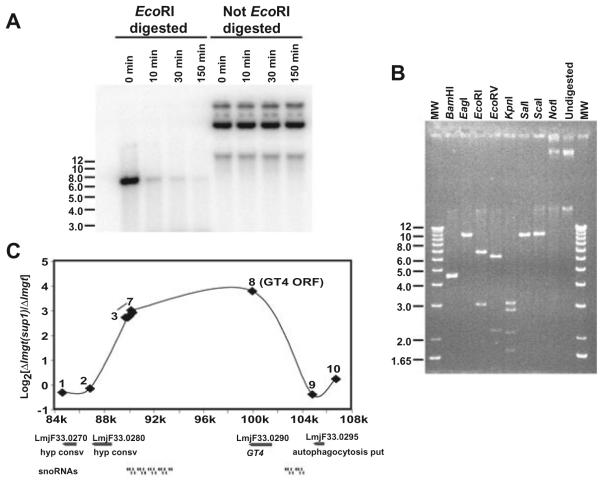
Both linear and circular amplicons have been observed within Leishmania species that are subjected to drug pressure directed against proteins encoded by genes in those amplicons (Beverley, 1991). To further examine the nature of the LmGT4 amplicon, we isolated extrachromosomal DNA from the Δlmgt(sup1) strain using an alkaline lysis protocol that enriches for circular episomes (Hanson et al., 1992). This fraction hybridized strongly to the LmGT4 ORF (Fig. 5A). This episomal DNA was not susceptible to digestion with λ exonuclease prior to digestion with a restriction enzyme, whereas EcoRI-restricted episomal DNA was degraded by this enzyme (Fig. 5A), establishing that the amplified DNA was circular. Furthermore, digestion of the episome with various restriction enzymes (Fig. 5B) produced either no alteration (NotI), a linear fragment of ∼10.5 kb (SalI, ScaI, EagI), or multiple fragments adding up to ∼10.5 kb (BamHI, EcoRI, EcoRV, KpnI; the BamHI digest exhibited a non-resolved doublet at 4.5 kb and a band of 0.8 kb that ran off the bottom of this gel, and the EcoRV digest revealed a band at ∼1.5 kb that is of low intensity in the gel). These results suggest the occurrence of an amplified circular DNA with a complexity of ∼10.5 kb. We did not carry out experiments to establish whether this DNA was organized into multimers as commonly seen in other amplified circular DNAs (Tobin et al., 1991).
Fig. 5.
Characterization of the episomal amplicon containing the LmGT4 ORF in Δlmgt(sup1). Episomal DNA from Δlmgt(sup1) was digested with (A) λ exonuclease or (B) various restriction enzymes.
A. Approximately 1 μg of purified episomal DNA was digested with 1 unit of λ exonuclease at 37°C for 0, 10, 30 and 150 min. Samples at the left were first digested with EcoRI whereas samples at the right were not pre-digested with the restriction enzyme. Digests were hybridized with the LmGT4 ORF.
B. Approximately 1 μg of episomal DNA was digested with the indicated restriction enzymes, separated on a 0.8% agarose gel and stained with ethidium bromide.
C. Characterization of the amplicon by comparative genomic hybridization. The log2 values of the ratio of fluorescence for hybridization of genomic DNAs for Δlmgt(sup1)/Δlmgt are plotted versus the position on chromosome 33 of the relevant oligonucleotides from the microarray. Numbers 1-10 refer to individual oligonucleotides representing different genes (Experimental procedures). Oligonucleotide 8 represents the L. major GT4 ORF. The numbers on the x-axis represent the position on L. major chromosome 33 in kilobases, and the diagram below the graph represents the genes present in this region as determined from the L. major genome sequence (http://www.genedb.org).
Comparison of Δlmgt(sup1) to Δlmgt DNA by comparative genomic hybridization
To further examine the nature of the genomic amplification in the Δlmgt(sup1) line, we performed comparative genomic hybridization (CGH) of genomic DNA from the Δlmgt and Δlmgt(sup1) or Δlmgt(sup3) lines to oligonucleotide microarrays representing the entire genome of the related parasite Leishmania major (Ivens et al., 2005). In these studies we compared the hybridization of DNAs of wild-type versus Δlmgt DNAs, or of Δlmgt versus Δlmgt(sup1) DNAs. These studies showed that of the 13 331 Leishmania oligonucleotides constituting this array, 74% showed sufficiently strong hybridization against L. mexicana DNA to permit their use, consistent with BLAST comparisons of the oligonucleotide data against shotgun L. mexicana genome sequence data (http://www.geneDB.org; data not shown). Quantification of the relative fluorescence intensities for hybridizations of genomic DNAs to these arrays (Fig. 5C and Table S1) revealed that a region between 10.0 and 17.9 kb in size (calculated from the positions of oligonucleotides 3 and 8 or 2 and 9 respectively) on chromosome 33 was amplified in the Δlmgt(sup1) mutant. This region encompasses the LmGT4 gene and multiple snoRNA genes and is of a size consistent with that measured from the isolated amplicon. The highest level of amplification, 13.8-fold or log2 of 3.79, is less than that determined from quantification of genomic Southern blots (Fig. 4A) and probably reflects a compression on hybridization seen in other CGH experiments (S. Beverley, unpubl. obs.).
In the course of these studies we also performed CGH comparisons of wild-type and Δlmgt DNAs, with unexpected findings. First Δlmgt exhibited aneuploidy compared with wild-type L. mexicana, including a reduction in the copy number of chromosome 31 and an increase in copy number of chromosomes 3, 16, 23 and 27 (Fig. S1). Sporadic aneuploidy in Leishmania has been reported previously, frequently in situations involving no obvious or known selective pressure, and the biological significance of these changes is unknown (Bastien et al., 1990; Cruz et al., 1993; Dumas et al., 1997; Sunkin et al., 2000). Second, there was amplification in Δlmgt parasites of a 34 kb subgenomic region corresponding to nucleotide positions 1 168 193-1 202 466 of L. major chromosome 29 (Table S1). Examination of the annotated genes within this segment of chromosome 29 (http://www.gendb.org) revealed the presence of 15 predicted genes (Fig. S1); of these 10 were hypotheticals and the annotated relationships or functions of the remaining five genes did not suggest any obvious explanation for why such amplification might have accompanied deletion of the glucose transporter locus. Importantly these two alterations (aneuploidy pattern and chromosome 29 amplification) were maintained unchanged in the Δlmgt(sup1) and Δlmgt(sup3) mutants (data not shown), suggesting they were unlikely to be responsible for the reverted growth phenotypes of the suppressor lines. Lastly, comparisons of the Δlmgt(sup1) and Δlmgt(sup3) lines showed that the latter line lacked amplification of the region containing LmGT4 or of any other sequence, consistent with Southern blot analysis (data not shown). Furthermore, uptake assays using [14C]-fructose showed no increase in uptake of this sugar over the level observed in the Δlmgt null mutant. Although we have not yet performed further characterization of the Δlmgt(sup3) mutant, this suppressor clearly differs in phenotype and genotype compared with the other three suppressor mutants described here.
Overexpression of the LmGT4 ORF in the Δlmgt null mutant replicates the Δlmgt(sup1) phenotype
The phenotype of the Δlmgt(sup1) mutant could be conferred either by amplification of the LmGT4 ORF alone or by multiple genetic alterations. To distinguish between these possibilities, we amplified the LmGT4 ORF from L. mexicana genomic DNA by PCR (DNA sequence submitted to GenBank as Accession No. EU449769), subcloned this ORF into the EcoRI site of the episomal expression vector pX63NEO-RI (Nasser and Landfear, 2004), and transfected this pX63NEO-LmGT4 plasmid into the Δlmgt null mutant. This LmGT4-complemented null mutant, designated Δlmgt[pLmGT4], restored survival in murine macrophages to levels similar to that of Δlmgt(sup1) parasites (Fig. 2A), confirming that the virulence phenotype of the suppressor mutant could be explained entirely by amplification of the LmGT4 ORF. Similarly, the LmGT4-complemented line showed restoration of growth as promastigotes in glucose-replete RPMI (Fig. 2B), re-establishment of hexose uptake (not shown), and amplification of LmGT4 DNA, RNA (Fig. 4B) and protein (Fig. 4C). Several other phenotypes of the Δlmgt(sup1) mutant (specifically uptake of fructose, accumulation of β-mannan, growth as promastigotes and amastigotes, sensitivity to H2O2, growth at elevated temperature, increase in cell volume), some of which are discussed below, were also replicated by complementation of the null mutant with the LmGT4 ORF (Figs 2, 6 and 7, and data not shown).
Fig. 6.
Suppression of various phenotypes of the Δlmgt null mutant in the Δlmgt(sup1) suppressor.
A. Killing of wild-type (□), Δlmgt (◇), Δlmgt(sup1) (○) and Δlmgt[pLmGT4] (△) parasites by H2O2. In multiple (n) experiments of the type shown here, the EC50 values for parasite killing by H2O2 were: wild type, 128 ± 11 μM (n = 7); Δlmgt, 23.8 ± 5.2 μM (n = 6); Δlmgt(sup1), 111 ± 19 μM (n = 3); Δlmgt[pLmGT4], 96.3 ± 7.2 μM (n = 3) as determined by fitting to the one-site competition model by non-linear regression using Prism 4.0a software (GraphPad Software).
B. Survival of wild-type (black bars), Δlmgt (white bars) and Δlmgt(sup1) (grey bars) parasites during starvation in PBS.
C. Growth of promastigotes of wild type (■), Δlmgt null mutants (■), and Δlmgt(sup1) suppressor (●) and Δlmgt[pLmGT4] (▲) in RPMI medium, pH 7.2 at 32°C.
In each example above, data represent the average and standard deviations of at least three determinations.
Fig. 7.
Suppression of metabolic alterations of Δlmgt null mutant in the Δlmgt(sup1) suppressor.
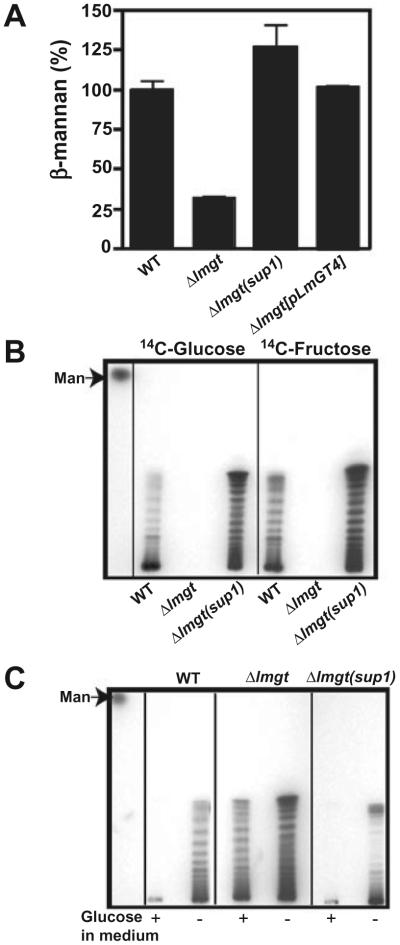
A. Quantification of β-mannan levels in 1.0 × 108 wild-type (WT), Δlmgt, Δlmgt(sup1) and Δlmgt[pLmGT4] parasites. The μg β-mannan present in wild-type sample has been normalized to 100%. Data represent the average and standard deviations of three replicate determinations.
B. Incorporation of [14C]d-glucose and [14C]d-fructose into β-mannan in each parasite strain.
C. Incorporation of [14C]l-alanine into β-mannan in the presence (+) and absence (-) of glucose in the medium in each parasite strain. The migration of mannose (Man) is indicated by the arrow.
The Δlmgt(sup1) mutant suppresses other phenotypes associated with the Δlmgt null mutant
Previous characterization of the Δlmgt null mutant revealed additional phenotypes associated with impaired survival inside macrophages. Thus the null mutants were more vulnerable than wild-type parasites to exposure to reactive oxygen species such as H2O2 (Fig. 6A), starvation for nutrients (Fig. 6B) and heat shock (Fig. 6C), all insults that parasites experience upon entry into macrophage phagolysosomes (Rodriguez-Contreras et al., 2007). The Δlmgt(sup1) mutant was relatively resistant to all these stresses, often to levels approaching that of wild-type promastigotes (Fig. 6A and B). Furthermore, complementation of the Δlmgt null mutant with the LmGT4 ORF restored resistance to oxidative stress (Fig. 6A) and elevated temperature (Fig. 6C) to the same level observed for the Δlmgt(sup1) mutant.
The Δlmgt null mutants also exhibit metabolic differences compared with wild-type parasites that are consistent with starvation for hexoses (Rodriguez-Contreras and Landfear, 2006). Thus compared with wild-type promastigotes the null mutants accumulate significantly lower levels of the storage carbohydrate and virulence factor β-1,2 mannan (Ralton et al., 2003) (Fig. 7A), they fail to incorporate radiolabelled hexoses into β-1,2 mannan even after extended incubation with the labelled sugars (Fig. 7B), and they activate incorporation of glucogenic precursors such as [14C]-alanine into β-1,2 mannan even in medium that is replete for glucose (Fig. 7C). For each of these metabolic phenotypes, the Δlmgt(sup1) mutant behaved similarly to wild-type parasites and thus suppressed the null mutant (Fig. 7A-C). Finally, measurement of protein content for each line revealed ratios of 0.66 ± 0.026 (n = 7) for Δlmgt/wild type, 0.81 ± 0.061 (n = 7) for Δlmgt(sup1)/wild type and 0.93 ± 0.036 (n = 5) for Δlmgt[pLmGT4]/wild type respectively. Hence, the Δlmgt(sup1) suppressor mutant and the LmGT4-complemented null mutant partially suppressed the reduction in cell volume and protein content previously documented in the glucose transporter null mutant (Rodriguez-Contreras et al., 2007).
Discussion
Gene amplification as a mechanism of genetic suppression in Leishmania
Gene amplification in Leishmania parasites is a well-established phenomenon that has been studied most often in the context of drug resistance but which also occurs in some non-selected isolates of various Leishmania species (Beverley, 1991). In classic studies Coderre et al. (1983) demonstrated that methotrexate-resistant mutants of L. major, derived by incrementally increasing drug pressure, amplified the dihydrofolate reductase-thymidylate synthase (DHFR-TS) gene whose product is the bifunctional enzyme that is the target of methotrexate. Most amplicons observed within Leishmania, including those encompassing the DHFR-TS gene, are circular episomes that appear to be generated by recombination involving flanking repeated DNA elements, but linear subchromosomal amplicons have also been observed occasionally (Beverley, 1991). In the current studies we present an example of a circular amplicon encoding an alternate transporter leading to suppression of a glucose transporter null mutant. Presumably this episome was generated stochastically at low frequency within the population of null mutants, and amplification of the episome followed by selection of parasites containing the amplicon was then promoted by the growth advantage these mutants exhibited in glucose-replete medium (Fig. 2B). Overgrowth of the Δlmgt null mutants by the suppressor mutants would then generate a uniform population of Δlmgt(sup1) parasites. Clonal derivatives of the Δlmgt(sup1) mutant obtained from individual colonies on agar plates showed various average levels of amplification and displayed phenotypes similar to that of the original Δlmgt(sup1) isolate (not shown). The plastic nature of the Leishmania genome suggests that it may be possible to select suppressors of various null mutants given culture conditions that would favour growth of the wild-type parasite. The prevalence of gene amplification as a mechanism may provide a relatively facile route towards genetic identification of the suppressing mutation, at least in some instances.
The CGH experiments also revealed several additional unanticipated alterations in chromosomal or subchromosomal copy number between wild-type L. mexicana and the Δlmgt null mutant, specifically a reduction in chromosome 31, an increase in chromosomes 3, 16, 23 and 27, and a pronounced increase in a subchromosomal region of chromosome 29. Alterations in ploidy of chromosomes are common among different strains of Leishmania (Bastien et al., 1990; Cruz et al., 1993; Dumas et al., 1997; Sunkin et al., 2000), and it is often unclear whether such changes in copy number are of biological significance. Similarly, we do not know whether the amplification of a region of chromosome 29 is significant, but it is possible that the increased copy number of one or more genes in this region does support the ability of the null mutant to survive as promastigotes and thus either accompanied the generation of the knockout or was selected for thereafter. Examination of the genes on this amplified segment does not suggest an obvious reason why an increase in copy number would promote viability of the null mutant, but the potential role of this amplified region may be worthy of further investigation. Nonetheless, we emphasize that the above alterations in copy number do not explain the suppressor phenotypes, since these changes are present in both the Δlmgt and Δlmgt(sup1) strains, and complementation of the Δlmgt null mutant with the LmGT4 ORF was able to replicate the suppressor phenotypes.
We have not characterized the chromosome 29 amplicon at the molecular level. However, the detection of only one circular amplicon in the Δlmgt(sup1) mutant (Fig. 5B, chromosome 33 amplicon) indicates that the chromosome 29 amplicon, which is also present in the Δlmgt(sup1) mutant, is unlikely to also be a circular episome. This raises the possibility that the chromosome 29 amplicon may be a linear subchromosomal element that would not be retained in the isolation procedure for circular episomes and hence would not be present in the fraction examined in Fig. 5B.
The LmGT4 gene as a suppressor of glucose transporter null mutants
The isolation of LmGT4 gene amplifications in multiple suppressor mutants that partially restore survival of amastigotes within macrophages underscores the role of hexose transporters in the intracellular stage of the life cycle. The ability of Δlmgt null mutants to transform into parasites with amastigote morphology that express the L. mexicana amastigote-specific marker HASPB (Fig. 1A) argues that a major defect in the null mutants is in their ability to survive within macrophages and not simply in their capacity to transform from promastigotes to amastigotes. Furthermore, wild-type axenic amastigotes fail to replicate when transferred to medium depleted of glucose, also arguing for a role for glucose transport in the amastigote stage per se. These observations are paradoxical, since amastigotes downregulate expression of glucose transporters (Burchmore and Landfear, 1998), uptake of glucose (Burchmore and Hart, 1995) and metabolism of this sugar, and upregulate β-oxidation of fatty acids (Hart and Coombs, 1982; Rosenzweig et al., 2008) as a nutrient source available within the host phagolysosome. In addition, amastigotes perform gluconeogenesis (Rodriguez-Contreras and Landfear, 2006) and can use amino acids or glycerol to synthesize carbohydrates, and they are dependent upon a functional gluconeogenic pathway for viability (Naderer et al., 2006). Nonetheless, previous studies (Rodriguez-Contreras and Landfear, 2006; Rodriguez-Contreras et al., 2007), confirmed in this article, established that the glucose transporter null mutants are relatively deficient in the virulence factor β-1,2 mannan and that they exhibit enhanced susceptibility to physiological stresses they encounter within macrophage phagolysosomes such as increased temperature and oxidative stress and decreased nutrient availability. The ability of a hexose transporter different from the LmGT1, LmGT2 and LmGT3 permeases, whose genes have been deleted in Δlmgt, to partially restore viability of the null mutant inside macrophages is also consistent with a role for hexose transporters in this life cycle stage. However, the observed amplification of the LmGT4 gene does not restore all measured phenotypes to their wild-type levels in the Δlmgt(sup1) mutant. Thus incomplete restoration of resistance of Δlmgt(sup1) and Δlmgt[pLmGT4] to elevated temperature (Fig. 6C) might explain the observation that this suppressor mutant does not replicate to wild-type levels inside macrophages (Fig. 2A).
Another curious question that remains unexplained has to do with the normal function of the LmGT4 hexose transporter. Although the LmGT4 gene is intact in the Δlmgt null mutants, these mutants are unable to transport hexoses as measured either by rapid transport assays (Fig. 3A and B) or by long-term incubations with radiolabelled hexoses (Fig. 7B). Thus the LmGT4 gene does not exhibit any measurable function in the intact parasite, at least under the conditions examined. Nonetheless, upon amplification the LmGT4 gene is able to confer hexose transport activity, increased growth as promastigotes and partially restored viability as amastigotes.
The original characterization of the LdGT4 transporter in the heterologous Xenopus oocyte system revealed the ability to take up several hexoses but with Km values much higher than those measured for LmGT1, LmGT2, or LmGT3 when these latter transporters were expressed in either oocytes or as transgenes in the Δlmgt null mutant. Thus while the Km values of LmGT1-3 for d-glucose were in the range of 0.1-1.2 mM (Burchmore et al., 2003), that of LdGT4 expressed on oocytes was estimated to be ∼150 mM (Langford et al., 1995). However the Km values for uptake of d-fructose and d-glucose in the Δlmgt(sup1) parasites, whose hexose transport is restored by amplification of LmGT4, are in the low millimolar range (Fig. 3C and D). These results suggest that Xenopus oocytes represent a suboptimal system for expression of the GT4 permease and that the true affinity of this carrier for hexoses is considerably higher than suggested by studies in the heterologous system. Nonetheless by comparison to the LmGT1-3 family members, LmGT4 is still a relatively lower-affinity transporter that is also expressed at a considerably lower level in the parasite.
Experimental procedures
Parasite culture
Promastigotes of L. mexicana were cultured in RPMI 1640 medium containing 10 mM glucose supplemented with 10% heat-inactivated fetal bovine serum (iFCS) at 26°C. For the Δlmgt[pLmGT4] promastigotes, cultures contained 100 μg ml-1 G418 to maintain the episomal expression vector; however, this drug was removed from the medium during quantification of growth under heat shock conditions (Fig. 6C). Axenic culture-form amastigotes were cultured at 32.5°C in Dulbecco’s modified Eagle’s medium modified for Leishmania (DME-L) (Iovannisci and Ullman, 1983), pH 5.5 supplemented with 30 mM MES instead of HEPES, 20% iFCS and 10 mM glucose. Infections of primary macrophages from BALB/c mice with stationary-phase promastigotes were performed as described (Rodriguez-Contreras et al., 2007) using Laboratory-Tek II chamber slides (Nalge Nunc International), except that the culture temperature was 37°C, and slides were stained with the HEMA 3 Stat Pack (Fisher Scientific) according to the manufacturer’s instruction. Stained slides were examined microscopically, and the per cent infected macrophages and intracellular parasites per 100 macrophages were determined by counting ∼250 macrophages per sample. Growth curves for promastigotes were determined by fixing parasites in 1% formaldehyde followed by counting triplicate samples on a haemacytometer grid.
Isolation and hybridization of nucleic acids
Genomic DNA was isolated using DNAzol reagent (Invitrogen) and total RNA was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, and Southern and Northern blots were performed using standard protocols (Sambrook et al., 1989). Circular amplicon DNA was isolated using an alkaline lysis protocol as described (Hanson et al., 1992). The LmGT4 ORF was PCR amplified from L. mexicana genomic DNA using primers representing the first 34 and the last 36 nucleotides of the L. major GT4 ORF. The exact 5′ and 3′ ends of the L. mexicana LmGT4 ORF were then determined by generating cDNA from total RNA (IScrip Select cDNA Synthesis Kit, Bio-Rad Laboratories) and PCR amplifying the 5′ and 3′ ends of LmGT4 cDNA as described previously for other glucose transporter cDNAs (Burchmore and Landfear, 1998). cDNAs were prepared and sequenced both from two independent clones of wild-type L. mexicana and from the Δlmgt(sup1) mutant to ensure that the LmGT4 ORF sequence was the same in both strains. Automated DNA sequencing was performed by the Core Facility of the Department of Molecular Microbiology and Immunology at the Oregon Health and Science University using an Applied Biosystems 16-capillary 3130xl automated sequence analysis system. Quantification of the LmGT4 amplification in Δlmgt(sup1) genomic DNA by Southern blot was performed using a Phosphorimager SI (Molecular Dynamics) and IP Laboratory Gel software for Apple Macintosh (Signal Analysis). Signal from the LmGT4 ORF in wild-type and Δlmgt(sup1) DNA was normalized to that of the MIT gene to obtain fold amplification. Quantification of Northern blots was performed in a similar manner, using signal from rRNA for normalization.
Phenotypic characterization
Uptake of radiolabelled sugars, measurement of parasite killing by H2O2, protein content determination and starvation in PBS were performed as described (Rodriguez-Contreras et al., 2007). For growth under heat shock conditions (32°C), mid-log-phase promastigotes were inoculated into RPMI-1640 medium, pH 7.2 at 1 × 106 cells ml-1, and cell density was monitored by microscopic examination in a haemacytometer. Quantification of β-mannan levels and incorporation of [14C]-sugars or [14C]-alanine into β-mannan were performed as detailed in Rodriguez-Contreras and Landfear (2006).
Immunofluorescence
Immunofluorescence of promastigotes and axenic culture-form amastigotes was performed as described (Ortiz et al., 2007). Rabbit polyclonal anti-HASPB antibody was diluted 1:1000 in blocking solution and incubated with each coverslip overnight in a moist chamber at 4°C. Goat anti-rabbit secondary antibody coupled to Alexa Fluor 488 (Molecular Probes) was used at a dilution of 1:2000. Slides were examined under a Nikon Microphot-FX fluorescence microscope using a 60× objective, and images were captured with MagnaFire software (Optronics).
For immunofluorescence of parasite-infected macrophages, medium was removed from the chamber slides containing infected macrophages (Rodriguez-Contreras et al., 2007) and the chamber was washed twice with 1 ml of phosphate-buffered saline (PBS). Cells were fixed for 15 min in 3.7% paraformaldehyde in PBS and then washed twice for 10 min with 10 mM glycine in PBS, followed by two more washes with PBS. Macrophages were permeabilized by incubating for 2 min in 500 μl of 2% Triton X-100 in PBS followed by two washes with PBS. Slides were incubated for 30 min with 300 μl of Blocking Solution (0.01% saponin, 0.01% sodium azide, 2% goat serum in PBS) followed by two washes with PBS and were then incubated overnight at 4°C with 300 μl of a 1:1000 dilution of anti-HASPB antibody followed by three washes of 5 min with PBS. Slides were subsequently incubated with a 1:2000 dilution of secondary antibody as described above and washed three times in PBS for 5 min. Finally slides were incubated with 4′,6-diamidino-2-phenylindole (DAPI) in PBS (2 μg ml-1) for 10 min at room temperature in the dark, washed twice with PBS and sealed with a coverslip containing Fluoromount G.
Preparation of salt-extracted membranes and immunoblotting
Salt-extracted membranes were prepared from 1 to 2 × 109 mid-log-phase promastigotes by pelleting cells, washing twice in PBS, and re-suspending the pellet in 500 μl of lysis buffer consisting of 0.1 M KH2PO4, pH 7.4, 5 mM EDTA, 10% glycerol and Complete Mini protease inhibitor cocktail (Roche). Parasites were sonicated twice for 30 s on ice using a W-225R sonicator (Ultrasonics) with a micro tip at an output setting of 5. The lysate was centrifuged for 15 min at 14 000 r. p.m. at 4°C in a Sorvall microcentrifuge to remove nuclei and unbroken cells. The supernatant was added to 750 μl of 150 mM NaHCO3 plus Complete Mini protease inhibitors, incubated on ice for 30 min and centrifuged again in a microcentrifuge as above. The supernatant was transferred to a 1.5 ml Beckman polyallomer microcentrifuge tube and centrifuged for 45 min at 264 000 g at 4°C in a Beckman TL-100 centrifuge using a TLA 100.3 rotor. The pellet was re-suspended in 150 μl of PBS, pH 7.2 containing 2% SDS, protein concentration was determined using the Bio-Rad DC protein assay kit, and samples were stored in 20 μl of aliquots at -70°C. Immediately prior to electrophoresis, samples were mixed with an equal volume of 2× Laemmli Sample Buffer (Sambrook et al., 1989) and heated to 60° for 5 min.
DNA microarrays
A DNA microarray was designed and 13 311 Leishmania oligonucleotides synthesized by Illumina (San Diego, CA) as described elsewhere (N. Akopyants et al., in preparation). The specific oligonucleotide sequences were based primarily on the L. major genome with the addition of many Leishmania infantum- and L. braziliensis-specific genes (Ivens et al., 2005; Peacock et al., 2007). Oligos were designed using the TOP algorithm (J. McPherson, unpublished), applying criteria including uniformity of GC contents and predicted Tms, minimization of homopolymer repeats and predicted secondary structures, and preference for the most 3′ location within the target. Negative controls included: (i) 10 unique non-cross-hybridizing mouse genes (designed as above), (ii) SpotReport® Oligo Array Validation System (Stratagene, CA) and (iii) buffer-only and empty positions. 70-mers were synthesized. The oligonucleotide library was printed in triplicate on Corning epoxide slides (Corning, MA) and stored desiccated at room temperature.
Array hybridization and data analysis
High-molecular-weight DNA was isolated from 2 × 109 cells as described (Brown et al., 1981) except that NaCl was used rather than NaClO4. DNA was sheared by nebulization (Invitrogen, CA) at 30 psi for 2 min, fluorescently labelled and hybridized in buffer containing 100 μg ml-1 random 20-mer oligos (60% GC content, IDT, IA) according to the 3DNA Array 900DNA™ Kit (Genisphere, Hatfield, PA). For each strain comparison two hybridizations were performed (one a dye-swap of the other) yielding a total of six data sets per comparison.
Hybridization was performed in a humid chamber overnight at 49°C. Once hybridized, slides were washed for 12 min in 2× SSC, 0.2% SDS at 40°C, 12 min in 2× SSC and finally 12 min in 0.2× SSC at room temperature. Following hybridization, arrays were scanned up to three times, with varying PMT detection settings, using an Axon GenePix 4200AL (MDS, CA). A histogram of pixel intensity distribution was generated for each image using GenePix 6.0.1.08 (MDS, CA), and analysed using BlueFuse v3.5 (BlueGnome, UK) as described (van den Ijssel et al., 2005). Samples of poor quality were excluded and the data were post-processed using block LOESS normalization (Smyth and Speed, 2003) excluding control oligos and oligos that BLAST to the genome with more then one hit. Dye swap and replicate spot were combined by taking the median fluorescence value.
Using the following format (oligonucleotide number/position on chromosome 33/gene identity), oligonucleotides 1-10, employed to monitor the amplification of the LmGT4 ORF in Fig. 5C, are: (1/84635/LmjF33.0270_ hyp_cons), (2/86860/LmjF33.0280_hyp_cons), (3/89908/LmjF33.snoRNA.0012), (4/89725/LmjF33.snoRNA.0028), (5/90041/LmjF33.snoRNA.0008), (6/90305/LmjF33.snoRNA.0015), (7/90187/LmjF33.snoRNA.0026), (8/99972/LmjF33.0290_GT4_hexose_trans porter), (9/104810/LmjF33.0295_autophagocytosis_prot_put), (10/106729/LmjF33.TRNAARG.01). The designation hyp_cons refers to a hypothetical conserved ORF.
The results of microarray studies have been deposited in the ArrayExpress database under Accession No. A-MEXP-1433.
Acknowledgements
We thank Dr Deborah Smith for a generous gift of the polyclonal antiserum directed against the L. mexicana HASPB protein and for providing information prior to publication regarding the amastigote-specific expression of this protein in L. mexicana. We thank Dr John McPherson, Natalia Akopyants and Robin Matlib for assistance in the design of the L. major oligonucleotide array, and the Washington University Microarray Facility for microarray printing and hybridizations. This work was supported by Grants AI25920 from the National Institutes of Health (to S.M.L.), AI29646 and AI21903 (to S.M.B.) and U01 AI056446 (to H.G.B.), a VA Merit Review award (to H.G.B.) and a postdoctoral fellowship from the American Heart Association (to D.R.-C.).
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Bastien P, Blaineau C, Taminh M, Rioux JA, Roizes G, Pages M. Interclonal variations in molecular karyotype in Leishmania infantum imply a ‘mosaic’ strain structure. Mol Biochem Parasitol. 1990;40:53–61. doi: 10.1016/0166-6851(90)90079-2. [DOI] [PubMed] [Google Scholar]
- Beverley SM. Gene amplification in Leishmania. Annu Rev Microbiol. 1991;45:417–444. doi: 10.1146/annurev.mi.45.100191.002221. [DOI] [PubMed] [Google Scholar]
- Brown PC, Beverley SM, Schimke RT. Relationship of amplified dihydrofolate reductase genes to double minute chromosomes in unstably resistant mouse fibroblast cell lines. Mol Cell Biol. 1981;1:1077–1083. doi: 10.1128/mcb.1.12.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchmore RJS, Hart DT. Glucose transport in promastigotes and amastigotes of Leishmania mexicana: characterization and comparison with host glucose transporters. Mol Biochem Parasitol. 1995;74:77–86. doi: 10.1016/0166-6851(95)02485-9. [DOI] [PubMed] [Google Scholar]
- Burchmore RJS, Landfear SM. Differential regulation of multiple glucose transporter genes in the parasitic protozoan Leishmania mexicana. J Biol Chem. 1998;273:29118–29126. doi: 10.1074/jbc.273.44.29118. [DOI] [PubMed] [Google Scholar]
- Burchmore RJS, Rodriguez-Contreras D, McBride K, Merkel P, Barrett MP, Modi G, et al. Genetic characterization of glucose transporter function in Leishmania mexicana. Proc Natl Acad Sci USA. 2003;100:3901–3906. doi: 10.1073/pnas.0630165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzulo JJ. Aerobic fermentation of glucose by trypanosomatids. FASEB J. 1992;6:3153–3161. doi: 10.1096/fasebj.6.13.1397837. [DOI] [PubMed] [Google Scholar]
- Coderre JA, Beverley SM, Schimke RT, Santi DV. Overproduction of a bifunctional thymidylate synthetase-dihydrofolate reductase and DNA amplification in methotrexate-resistant Leishmania tropica. Proc Natl Acad Sci USA. 1983;80:2132–2136. doi: 10.1073/pnas.80.8.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz AK, Titus R, Beverley SM. Plasticity in chromosome number and testing of essential genes in Leishmania by targeting. Proc Natl Acad Sci USA. 1993;90:1599–1603. doi: 10.1073/pnas.90.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew ME, Langford CK, Klamo EM, Russell DG, Kavanaugh MP, Landfear SM. Functional expression of a myo-inositol/H+ symporter from Leishmania donovani. Mol Cell Biol. 1995;15:5508–5515. doi: 10.1128/mcb.15.10.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas C, Ouellette M, Tovar J, Cunningham ML, Fairlamb AH, Tamar S, et al. Disruption of the trypanothione reductase gene of Leishmania decreases its ability to survive oxidative stress in macrophages. EMBO J. 1997;16:2590–2598. doi: 10.1093/emboj/16.10.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson S, Beverley SM, Wagner W, Ullman B. Unstable amplification of two extrachomosomal elements in alpha-difluoromethylornithine-resistant Leishmania donovani. Mol Cell Biol. 1992;12:5499–5507. doi: 10.1128/mcb.12.12.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart DT, Coombs GH. Leishmania mexicana: energy metabolism of amastigotes and promastigotes. Exp Parasitol. 1982;54:397–409. doi: 10.1016/0014-4894(82)90049-2. [DOI] [PubMed] [Google Scholar]
- Herwaldt BL. Leishmaniasis. Lancet. 1999;354:1191–1199. doi: 10.1016/S0140-6736(98)10178-2. [DOI] [PubMed] [Google Scholar]
- van den Ijssel P, Tijssen M, Chin SF, Eijk P, Carvalho B, Hopmans E, et al. Human and mouse oligonucleotide-based array CGH. Nucleic Acids Res. 2005;33:e192. doi: 10.1093/nar/gni191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovannisci DM, Ullman B. High efficiency plating method for Leishmania promastigotes in semidefined or completely-defined medium. J Parasitol. 1983;69:633–636. [PubMed] [Google Scholar]
- Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Kuile BH, Opperdoes FR. A chemostat study on proline uptake and metabolism of Leishmania donovani. J Protozool. 1992;39:555–558. doi: 10.1111/j.1550-7408.1992.tb04850.x. [DOI] [PubMed] [Google Scholar]
- Langford CK, Kavanaugh MP, Stenberg PE, Drew ME, Zhang W, Landfear SM. Functional expression and subcellular localization of a high-Km hexose transporter from Leishmania donovani. Biochemistry. 1995;34:11814–11821. doi: 10.1021/bi00037a020. [DOI] [PubMed] [Google Scholar]
- McConville MJ, de Souza D, Saunders E, Likic VA, Naderer T. Living in a phagolysosome; metabolism of Leishmania amastigotes. Trends Parasitol. 2007;23:368–375. doi: 10.1016/j.pt.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Manolescu AR, Witkowska K, Kinnaird A, Cessford T, Cheeseman C. Facilitated hexose transporters: new perspectives on form and function. Physiology (Bethesda) 2007;22:234–240. doi: 10.1152/physiol.00011.2007. [DOI] [PubMed] [Google Scholar]
- Naderer T, McConville MJ. The Leishmania-macrophage interaction: a metabolic perspective. Cell Microbiol. 2008;10:301–308. doi: 10.1111/j.1462-5822.2007.01096.x. [DOI] [PubMed] [Google Scholar]
- Naderer T, Ellis MA, Sernee MF, De Souza DP, Curtis J, Handman E, McConville MJ. Virulence of Leishmania major in macrophages and mice requires the gluconeogenic enzyme fructose-1,6-bisphosphatase. Proc Natl Acad Sci USA. 2006;103:5502–5507. doi: 10.1073/pnas.0509196103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser MIA, Landfear SM. Sequences required for the flagellar targeting of an integral membrane protein. Mol Biochem Parasitol. 2004;135:89–100. doi: 10.1016/j.molbiopara.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Nugent PG, Karsani SA, Wait R, Tempero J, Smith DF. Proteomic analysis of Leishmania mexicana differentiation. Mol Biochem Parasitol. 2004;136:51–62. doi: 10.1016/j.molbiopara.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Ortiz D, Sanchez MA, Pierce S, Herrmann T, Kimblin N, Bouwer HG, Landfear SM. Molecular genetic analysis of purine nucleobase transport in Leishmania major. Mol Microbiol. 2007;64:1228–1243. doi: 10.1111/j.1365-2958.2007.05730.x. [DOI] [PubMed] [Google Scholar]
- Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralton JE, Naderer T, Piraino HL, Bashtannyk TA, Callaghan JM, McConville MJ. Evidence that intracellular {beta}1-2 mannan is a virulence vactor in Leishmania parasites. J Biol Chem. 2003;278:40757–40763. doi: 10.1074/jbc.M307660200. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Contreras D, Landfear SM. Metabolic changes in glucose transporter-deficient Leishmania mexicana and parasite virulence. J Biol Chem. 2006;281:20068–20076. doi: 10.1074/jbc.M603265200. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Contreras D, Feng X, Keeney KM, Bouwer HG, Landfear SM. Phenotypic characterization of a glucose transporter null mutant in Leishmania mexicana. Mol Biochem Parasitol. 2007;153:9–18. doi: 10.1016/j.molbiopara.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig D, Smith D, Opperdoes F, Stern S, Olafson RW, Zilberstein D. Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB J. 2008;22:590–602. doi: 10.1096/fj.07-9254com. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Schlein Y. Sandfly diet and Leishmania. Parasitol Today. 1986;2:175–177. doi: 10.1016/0169-4758(86)90150-x. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Sunkin SM, Kiser P, Myler PJ, Stuart K. The size difference between Leishmania major friedlin chromosome one homologues is localized to sub-telomeric repeats at one chromosomal end. Mol Biochem Parasitol. 2000;109:1–15. doi: 10.1016/s0166-6851(00)00215-2. [DOI] [PubMed] [Google Scholar]
- Tobin JF, Laban A, Wirth DF. Homologous recombination in Leishmania enriettii. Proc Natl Acad Sci USA. 1991;88:864–868. doi: 10.1073/pnas.88.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]