Abstract
Activation of the complement system generates potent chemoattractants and opsonizes cells for immune clearance. Short-lived protease complexes cleave complement component C3 into anaphylatoxin C3a and opsonin C3b. Here we report the crystal structure of the C3 convertase formed by C3b and the protease fragment Bb, which was stabilized by the bacterial immune-evasion protein SCIN. The data suggest that the proteolytic specificity and activity depends on dimerization of C3 with C3b of the convertase. SCIN blocked the formation of a productive enzyme-substrate complex. Irreversible dissociation of C3bBb is crucial to complement regulation and was determined by slow binding kinetics of the Mg2+-adhesion site in Bb. Understanding the mechanistic basis of the central complement activation step and microbial immune evasion strategies targeting this step will aid the development of complement therapeutics.
Complement is an ancient defense mechanism, which evolved into a large protein-interaction network in mammals that initiates and performs innate immune functions and links innate with adaptive immunity1. Activation of complement is critical for protection against microbial infections; however, over-activation of complement causes host tissue damage2. The complement system is initiated either by specific recognition of target cells in the classical and lectin pathways, or spontaneously due to inherent instability of complement component C3 in the alternative pathways3. These pathways converge in the formation of C3 convertases, which cleave C3 into the small anaphylatoxin C3a and the large, reactive C3b that may covalently couple to target surfaces4, 5. In the amplification loop of the alternative pathway, pro-enzyme factor B (FB) binds to surface bound C3b and is cleaved by factor D (FD) resulting in an active convertase complex that consists of C3b and the non-covalently bound protease fragment Bb (denoted C3bBb). These convertases amplify C3b production close to the target surface resulting in rapid opsonization, of the target cell with C3b, which can then evoke B cell stimulation, phagocytosis and cell lysis1. Similarly, C3 convertases are formed in the classical and lectin pathways by C4 and C2, which are homologues of C3 and FB respectively6. Both C3 convertases (C3bBb and C4b2a) are active only towards their natural substrate C3, with limited activity towards the homologue C5 (ref. 7). In the terminal complement pathway, the substrate specificity is switched from C3 to C5 after association of one or more C3b molecules to the C3bBb or C4b2a complexes8, 9. Regulation of activity is achieved by convertase assembly and disassembly, which is mediated by complement regulators10. Essential for this regulation is that enzymatic activity towards C3 is only expressed by the assembled active convertases (C3bBb and C4b2a) and not by the pro-enzymes (FB and C2), pro-enzyme complexes (C3bB and C4bC2) or dissociated fragments (Bb and C2a). In addition, the convertases are meta-stable and dissociate irreversibly (with an inherent half-life time of 60−90 s at 37 °C)11; therefore, the protease fragments Bb or C2a do not reassociate with C3b or C4b, respectively. In this way, the protease activity of the short-lived C3 convertase complexes determines opsonization of pathogens and altered host cells, which is a pivotal step in raising complement-mediated immune responses.
In order to resist the host immune response, pathogenic bacteria and viruses have evolved a number of well-defined strategies to evade the immune system. Since the complement system is a key element in antibacterial defense, a large number of these evasion molecules turn out to be directed against complement components. Blocking the central activation step of C3 into C3b is a major bacterial complement evasion strategy: streptococci secrete molecules that degrade the C3 molecule, while many pathogens indirectly block C3 convertases by attracting host convertase regulators to their surfaces12. Previously, we described staphylococcal complement inhibitor (SCIN) from Staphylococcus aureus as the first bacterial protein that targets C3 convertases directly13. SCIN is found in 90% of S. aureus strains and specifically binds active convertases (C3bBb and C4b2a) on bacterial surfaces and prevents opsonization of bacteria and subsequent phagocytosis13. A striking characteristic of SCIN is that it stabilizes convertases on bacterial surfaces and mutational analyses indicated that this is essential for its inhibitory function14.
Structural studies of the C3 convertases are challenging, because these complexes dissociate irreversibly and exhibit short half-life times. Here we used SCIN to stabilize the C3 convertase of the alternative pathway (C3bBb) and to crystallize C3bBb in complex with SCIN. The structure gave insight into the inhibitory mode of SCIN and provided a structural basis for the enzymatic activity, substrate specificity and irreversible dissociation of the C3 convertases that are central to immune defense.
RESULTS
SCIN induces formation of stable convertase dimers
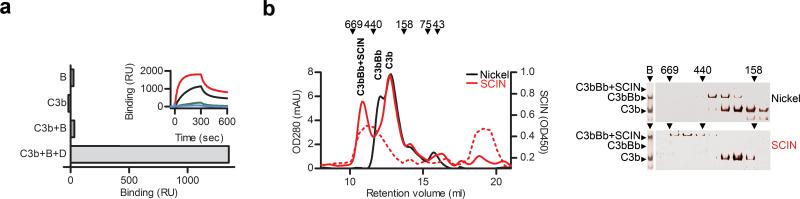
First we studied whether SCIN, an inhibitor of surface-bound convertases, also binds and stabilizes soluble convertases. Using surface plasmon resonance (SPR), we observed that surface-immobilized SCIN specifically bound C3bBb (generated in solution by mixing C3b, FB and FD) and not FB, C3b or pro-convertase C3bB (Fig. 1a). Incubating His-tagged SCIN with C3b, FB and FD in solution resulted in simultaneous C3b and Bb association to SCIN, whereas no complexes were formed in the absence of FD or FB (see Supplementary Fig. 1 online). SCIN inhibited convertases in solution as C3bBb could not cleave C3 in the presence of SCIN (Supplementary Fig. 2 online). Gel permeation chromatography of the complexes generated in the presence of SCIN demonstrated that SCIN induced formation of large complexes of ∼500 kDa, twice the size of an active convertase complex (∼240 kDa) (Fig. 1b). Immunoblotting and ELISA revealed that these complexes indeed contain C3b, Bb and SCIN (Fig. 1b and Supplementary Fig. 3 online). The SCIN-induced formation of large convertase complexes was also observed in native gel electrophoresis and we used this method to analyze the stability of complexes (Fig. 1b,c). At 20 °C SCIN-stabilized complexes had a half-life up to 4 h; whereas, at 4 °C we observed an improvement of stability with a half-life exceeding 20 h (Fig. 1c). For crystallization purposes, we optimized the stability of the complex in small-scale experiments comparing a variety of purification methods. This resulted in an effective pull-down strategy that allowed us to generate highly stable complexes in the milligram range. An N-terminal His-tagged SCIN was used to generate complexes in fluid-phase that were subsequently purified from reactants (such as FD, Ba and excess C3b and FB) using magnetic Co2+ beads. Ultrafiltration was used to remove uncomplexed SCIN and to further concentrate the sample. For crystallization, we generated 10 μM (approximately 5 mg/ml) complexes that were stable over 25 days at 4 °C (Fig. 1d). Analytical ultracentrifugation of the purified material showed a major component of 15.2 S and a minor component of ∼18 S, which likely corresponded to the SCIN-stabilized dimeric complex of ∼500 kDa and putatively a dimer of dimers, respectively (Supplementary Fig. 4 online). Thus, SCIN bound and blocked active convertases in solution and induced dimerization of C3bBb. The stabilizing capacity of SCIN allowed us to generate highly pure and stable convertase complexes.
Figure 1. SCIN induces formation of dimeric convertases.
(a) SPR analysis. Binding of soluble convertase components to surface-immobilized SCIN. Bars indicate responses at the end of injection (all components at 100 nM). Inlay, Sensorgrams of injections with C3bBb: C3b and FB at 10 nM (blue), 30 nM (green), 100 nM (black) or 300 nM (red), FD at 100 nM. (b) Gel filtration analysis and native gel electrophoresis of SCIN-inhibited convertases. (Left) Gel filtration: absorbance peaks of active convertases stabilized by Ni2+ (Nickel, black line) and SCIN-inhibited convertases (solid red line). The 178 kDa peak corresponds with free C3b. Dashed red line indicates elution positions of SCIN as determined by ELISA. Right gels, Left panel: native gel electrophoresis of active (top) and SCIN-inhibited convertase (below). Right panel: native gel electrophoresis of fractions eluted from the gel filtration column. (c) Analysis of convertase stability. C3b (500 nM), FB (500 nM), FD (250 nM) and SCIN (1 μM) were incubated at 4 °C, 20 °C or 37 °C for different time periods and subjected to native gel electrophoresis at 4 °C. The SCIN-convertase is most stable at 4 °C (> 20 h). No complexes were detected in the absence of SCIN (−). (d) Native gel electrophoresis of purified SCIN-convertase complexes after 0 to 8 days (left side, coomassie staining) and 25 days (right side, silver staining). Figures a-c are representatives of three independent experiments. Figure d is a representative of two independent experiments.
Crystal structure of the SCIN-inhibited convertase
Freshly prepared and purified SCIN-stabilized complexes were used to set up crystallization experiments at 4 °C. Crystals appeared within 1−2 days; electrophoresis showed that the crystals consisted of C3b, Bb and SCIN (Supplementary Fig. 5 online). Crystallization conditions were optimized resulting in a crystal that diffracted to 3.9-Å resolution (see Table 1 for crystallographic statistics and Supplementary Fig. 6 online for quality of the electron density). The structure was solved by molecular replacement using the available structures of the individual proteins present in the complex and refined using non-crystallographic symmetry and tight geometry restraints (see Online Methods). The final model was refined to R and Rfree factors of 25.3 and 26.8%, respectively.
Table 1.
Data collection and refinement statistics
| 3.9-Å | |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 228.6, 121.5, 280.8 |
| α, β, γ (°) | 90, 91.6, 90 |
| Resolution (Å) | 40−3.9 (4.11−3.9)* |
| Rmerge (%) | 12.8 (69.0) |
| I/σI | 10.4 (1.8) |
| Completeness (%) | 97.6 (92.0) |
| Redundancy | 3.5 (3.4) |
| Refinement | |
| Resolution (Å) | 39.8−3.9 |
| No. reflections | 137476 |
| Rwork / Rfree (%) | 25.3 / 26.8 |
| No. atoms | 67989 |
| Protein | 67266 |
| Ligand/ion | 723 |
| Water | |
| B-factors (Å2) | |
| Protein | 158 |
| Ligand/ion† | 208 |
| Water | |
| R.m.s deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.519 |
Highest resolution shell is shown in parenthesis.
Ligand/ion refers to the modelled MIDAS Mg2+ ion, waters and glycan chains.
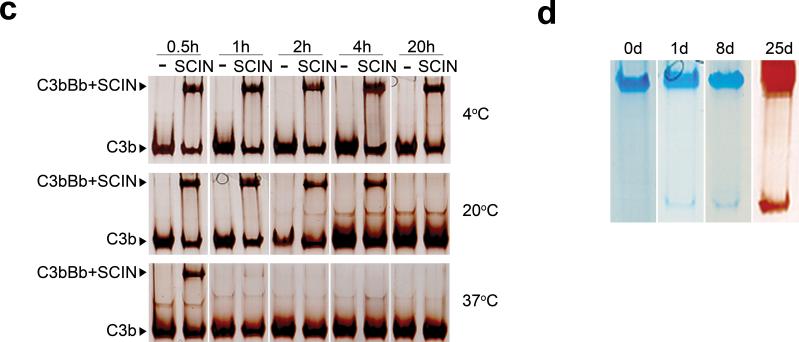
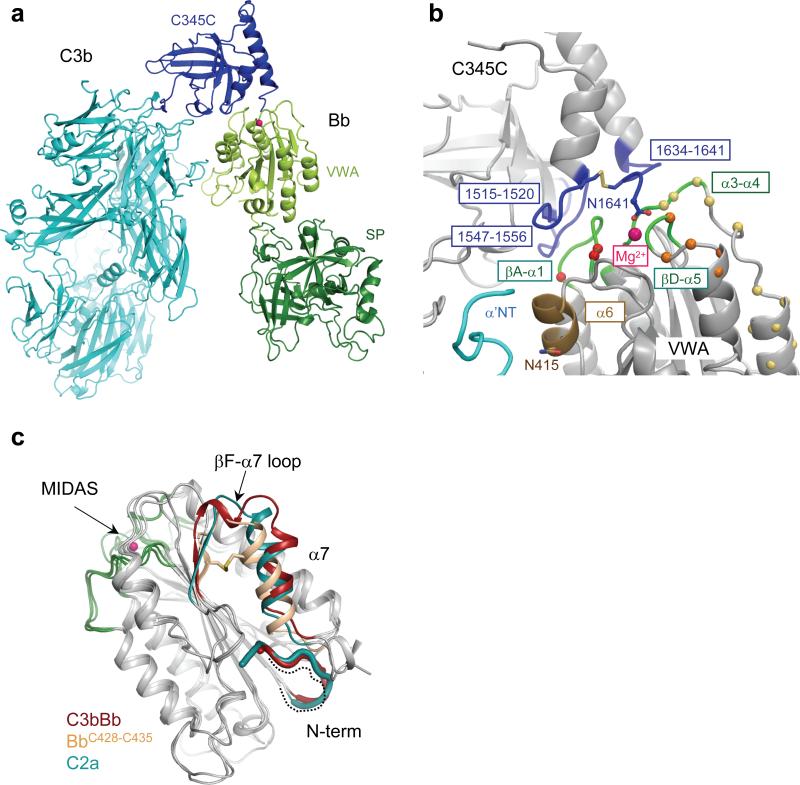
The asymmetric unit contained two complexes, each consisting of a C3bBb-SCIN dimer of 500 kDa with overall dimensions of 177×168×155 Å (Fig. 2a). The dimer was built up of a C3b-C3b homodimer that was stabilized by bridging SCIN molecules (Fig. 2 and see Supplementary Table 1 online for complex interface analysis). Each C3b molecule displayed a typical arrangement of 12 domains as observed in other structures of C3b15, 16, which consisted of a core formed by eight macroglobulin (MG) domains and a linker (LNK) domain, a ‘complement C1r/C1s, UEGF, BMP1’ (CUB) and thioester containing domain (TED) inserted between MG7 and MG8, and a C-terminal C345C domain (Fig. 2b). Two Bb molecules were positioned symmetrically on the outer edges of the C3b-C3b dimer. Bb was attached to the C-terminal C345C domain of C3b via the Von Willebrand factor type A (VWA) domain with the active site in the serine protease (SP) domain oriented outwards.
Figure 2. Crystal structure of the C3 convertase C3bBb inhibited by SCIN.
(a) Ribbon representation of the C3bBb-SCIN dimeric complex with C3b (blue and turquoise), Bb (green and golden) and SCIN (purple and orange). (b) Stereoview of the monomeric C3bBb-SCIN extracted from the dimer, coloured by protein (SCIN) or protein domain (VWA and SP of Bb; and, all 12 domains of C3b with thioester shown by red spheres); a linear representation of the domain composition is given below (with disulfide bonds, glycosylation sites and thioester marked). (c) Overlay of the four C3bBbSCIN complexes in the asymmetric unit (yellow, orange, green and magenta). The largest variation occurs in the orientation of the C345C domain (see Supplementary Fig. 8).
Inhibition of C3bBb by SCIN
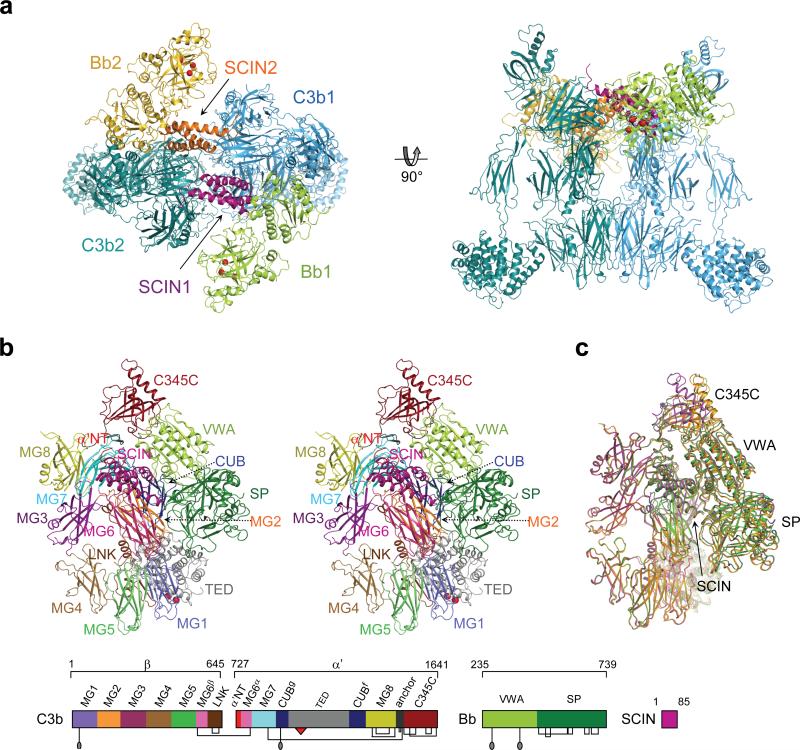
SCIN stabilized the convertase dimer by interacting with C3b and Bb of one convertase (with ∼1,400 Å2 contact areas to C3b and Bb each) and with C3b of the opposing convertase (1,800 Å2 contact area; see Supplementary Fig. 7 online, Fig. 3a and Supplementary Table 1 online). SCIN bound the first C3b at its α’ N-terminal tail (α’NT) and macroglobulin (MG) domains 6 and 7, whereas the binding site in the second C3b was formed by domains MG7 and MG8. SCIN did not block the catalytic site in Bb. Instead, SCIN bound primarily the VWA domain and the VWA-SP interface of Bb (Fig. 2b and Supplementary Fig. 7 online). Using a panel of eight different chimeras of SCIN and a non-functional homologue we previously identified two segments of SCIN that were essential for activity on bacterial surfaces14. These segments (residues 26−36 and 37−48; exchanged in chimeras Chα1C and Chα2N, respectively) coincided with the majority of contact sites for Bb and C3b of one convertase enzyme (Fig. 3b). We re-tested the same eight mutants and showed that residues 26−48 were also critical for blocking C3 conversion by convertases in solution, confirming the observed arrangement in the crystal (Fig. 3c). Next, we addressed the importance of convertase dimerization for the inhibition by SCIN. Two SCIN chimeras with altered C3b-dimerization contact sites (denoted ChN and ChC3b2) yielded stable, but monomeric C3bBb-SCIN complexes (Fig. 3d). Moreover, these two chimeras still inhibited convertase activity in solution and stabilized convertases on bacteria (Fig. 3c). These data confirmed the dimerization site as observed in the complex, but demonstrated that convertase dimerization was not essential for the inhibitory activity of SCIN. We, therefore, conclude that SCIN likely inhibits monomeric convertases either by blocking substrate binding or by preventing critical movements required for formation of active enzyme-substrate complexes.
Figure 3. Inhibition of C3bBb by SCIN.
(a) Contact sites of SCIN in the dimeric convertase. The SCIN binding pocket is shown in surface representation (left) with a ribbon representation of SCIN colour coded by molecular contact (right). (b) Amino-acid sequence alignment of SCIN and the non-functional homologue ORF-D. Convertase contact sites in SCIN are underlined. Below a schematic representation of the SCIN chimeras is given (red boxes indicate the exchanged segments). (c) Convertase inhibition by SCIN chimeras: C3 conversion by fluid-phase C3bBb (above) and Bb stabilization on bacterial surfaces (below). (d) Native gel electrophoresis of convertases in the presence of Ni2+ (lane 1), SCIN (lane 2), ChN (lane 3) or ChC3b2 (lane 4). Figures c and d are representatives of three independent experiments.
Architecture of the C3 convertase
In the SCIN-stabilized complex, we observed a loose arrangement of the C3 convertase C3bBb, in which Bb appeared to ‘dangle’ from the tip of the C3b structure. Bb contacted the C-terminal C345C domain of C3b through its VWA domain. No contacts were observed between the SP domain of Bb and C3b. This arrangement of C3bBb in the inhibited complex was consistent with recent 3D EM reconstructions of C3bB and C3bBb at 27-Å resolution17. Most prominently, the carboxy terminus residue Asn-1641 of C3b chelated the Mg2+ ion bound to the metal-ion dependent adhesion site (MIDAS) formed by three loops (βA-α1, α3-α4 and βD-α5) of the VWA domain of Bb (Fig. 4a,b)18, 19. This arrangement is in full agreement with mutagenesis data on the critical importance of the MIDAS for convertase activity20, 21. The three loops of the MIDAS contributed to the C3b-Bb interface; two of which (βA-α1 and βD-α5) were previously shown to be critical for convertase stability22 (Fig. 4b). In addition, helix α6 of the VWA domain of Bb was close to the α’NT of C3b (Fig. 4b). Previous data support the possible existence of a putative secondary interaction site that may be disrupted by SCIN, because mutation N415A in VWA-helix α6 of FB yielded convertases that were more prone to dissociation by complement regulators23. With regard to C3b, the interface was formed by loops residues1515−1520, 1547−1556 and the C-terminal tail (residues 1634−1641) in the C345C domain (Fig. 4b). Variation in the C345C orientation (see Supplementary Fig. 8 online) indicated weak interactions of C345C with the α3-α4 loop of VWA resulting in buried surface areas that differ from 600−1,200 Å2 (Supplementary Table 1 online). These variations are in agreement with previous experiments that show a limited effect of substituting the α3-α4 loop on convertase stability (Fig. 4b)22.
Figure 4. The C3b-Bb structure derived from the C3bBb-SCIN complex.
(a) Ribbon representation of the C3bBb complex with C3b (light and dark blue) and Bb (light and dark green). The Mg2+-ion is indicated by a pink sphere. (b) The C345C-VWA interface between C3b and Bb. The contact regions are coloured in blue (for C345C domain of C3b) and green (for VWA domain of Bb). The disulfide bond of C1515-C1639 in C345C domain is in stick representation. The residues mutated in FB chimeras are highlighted in spheres (red: in βA-α1 loop; beige, in α3-α4 loop and α4 helix; orange: in βD-α5 loop) based on ref. 22. (c) Overlay of VWA domains of Bb in complex with C3b, BbC428-C435 (pdb code: 1RRK)24 and C2a (pdb code: 2I6Q)26 showing the position of helix α7 and the nascent N-terminus. The MIDAS loop βA-α1, α3-α4 and βD-α5 are coloured green. The N-terminus of BbC428-C435 is missing and indicated by a dashed line.
The SCIN-stabilized C3bBb complex allowed us to study the effects of complex formation on the conformations of C3b and the proteolytic fragment Bb. The domain arrangement of C3b in the complex (Supplementary Fig. 9 online) was similar to that of other C3b structures15, 16; variations in the C345C and CUB-TED domains are common and indicate inherent flexibility of C3b. The structure of the Bb fragment in the C3 convertase was similar to that of free Bb24, which is markedly different from that of full-length pro-enzyme FB25 (Fig. 4c and Supplementary Fig. 10 online). The MIDAS arrangement in C3bBb suggested a typical high-affinity ligand-binding configuration, similar to the structures of free Bb and C2a24, 26 and activated integrin Iα domains27 (Supplementary Fig. 10 online); however, the low resolution of the diffraction data did not allow a detailed interpretation. The nascent N-terminal tail of Bb and the VWA-helix α7, which are putatively conformationally coupled to the MIDAS, adopted positions as in free C2a (Fig. 4c)26. The published structure of free Bb is in part distorted, because the construct lacked the 7 N-terminal residues and had an introduced disulfide bridge (C428-C435) that distorted helix α7 (Fig. 4c)24. Nevertheless, the VWA-SP orientations in bound versus free Bb differed by only 10°, showing limited overall effects from these distortions at the VWA-SP interface. Moreover, SCIN interacted with Bb at the VWA-SP interface; these interactions did not have an apparent effect on the VWA and SP conformations. In contrast, the VWA-SP orientations in C2a and C3bBb differed by 28°, which is possibly an inherent difference between Bb and C2a (Supplementary Fig. 10a,b online). Overall, the conformation of the components C3b and Bb in the convertase complex are similar to those of the isolated C3b and Bb proteins.
Implications for enzymatic activity and specificity
The active site in the SP domain in C3bBb showed a typical catalytic Ser-His-Asp triad and oxyanion hole. The oxyanion-hole loop (residues 672−674), however, is distorted due to a peptide flip in all structures of FB, Bb and its homologue C2a, except for one covalently inhibited Bb structure (Supplementary Fig. 10d online)24, 26, 28. In the present low-resolution structure, no conformational changes with respect to FB and Bb were apparent in the SP domain that would affect the catalytic site (Supplementary Fig. 10e online), which would indicate that C3b binding does not induce the catalytic activity of the protease fragment Bb.
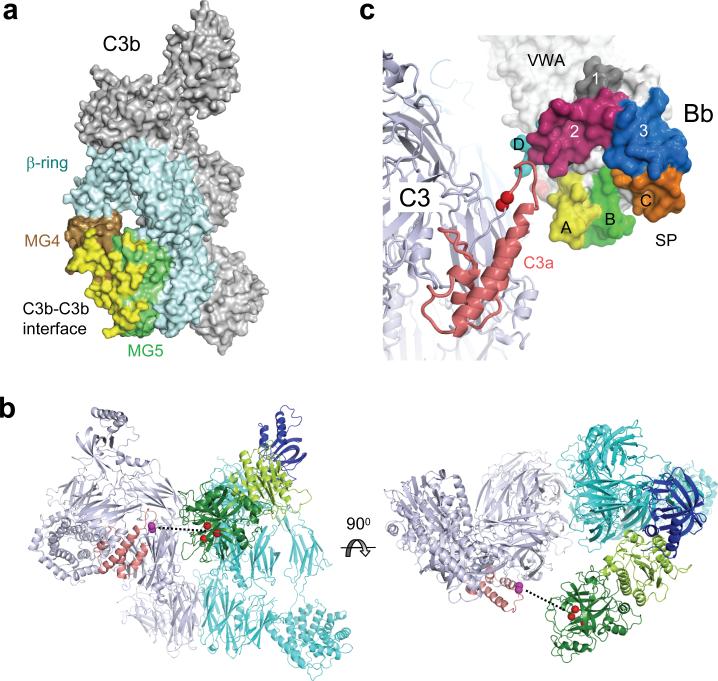
C3b likely plays a critical role in binding of the substrate C3 to the C3 convertase C3bBb; such an exosite would explain the higher activity for the natural substrate C3 over penta-peptides (KM = 6 μM27 and 210−3000 μM29 respectively; with the concentration of C3 in plasma being ∼5 μM). We observed a dimeric convertase complex that consisted of two opposing convertases formed by C3b homodimerization (Fig. 2a). The C3b-dimerization face coincided with binding sites for the inhibitors compstatin30, CRIg16 and antibody S77 (ref. 31), which block substrate binding to the C3 convertase. In the dimeric crystal structure the C3b-C3b interface is formed by the MG4−5 domains of the C3b molecules (with buried surface areas of ∼1,900−2,800 Å2) (Fig. 5a). We generated a hypothetical enzyme-substrate (C3bBb-C3) complex by superposing the substrate C3 onto C3b based on the MG4−5 domains, using the observation that the arrangements of the MG1−6 domains (which form the β-ring) are strongly conserved between C3 and C3b (Fig. 5b and Supplementary Fig. 11a online). In the resulting C3bBb-C3 model the catalytic site of C3bBb was oriented towards, but was positioned 30 Å away from the scissile loop of the substrate C3 (Fig. 5b). Furthermore, the homologous convertase C4b2a is expected to bind substrate C3 through the corresponding side of C4b, resulting in a similar C4b-C3 interface. The sequence alignment of the MG4−5 domains of C3, C4 and C5 indicated that the majority of the amino acid residues at the interaction site are conserved between C3 and C4 (Supplementary Fig. 11b online)32. C5, however, differs both in amino acid sequence and in domain-domain orientation of MG4−5; which may explain the weak interaction of C5 with C3b and poor C5 cleavage by the C3bBb complex (Supplementary Fig. 11a online)33, 34. These data indicated that the extreme specificity of the C3 convertase is possibly obtained by quasi-homodimerization of C3 to C3b of the alternative pathway convertase (C3bBb) and a similar association of C3 to the evolutionary related C4b of the classical and lectin pathways convertase (C4b2a).
Figure 5. The C3b-C3b interface and substrate-binding model.
(a) Surface representation of C3b with the C3b-C3b dimer interface shown in yellow, which is formed by domains MG4 (brown) and MG5 (green) of the β-ring of C3b (MG1−6) (cyan). (b) Model of the enzyme-substrate (C3bBb-C3) complex constructed by superimposing C3 (pdb code: 2A73)32 on the MG4−5 domains of the dimeric C3b molecule in the complex. The convertase C3bBb is colored as in Fig. 4a with the catalytic triad of Bb are indicated by red spheres. The substrate C3 is shown in grey with the anaphylatoxin domain (C3a) highlighted in coral and the scissile bond (S726-R727) indicated by magenta spheres. The dashed line indicates the distance (∼30 Å) between catalytic site and the scissile loop. (c) The relative orientation of C3a domain of C3 (substrate) and the surface loops of the SP domain of Bb forming the substrate-binding groove.
Positioning Bb of the C3bBb complex into place with respect to the substrate C3 in the C3bBb-C3 model yielded a putative docking of the scissile loop into the active site of the SP domain in a productive (N-to-C) orientation (Fig. 5c). The observed orientation of the SP domain was consistent with the predicted orientation based on the crystal structure of C2a26, with the extended surface loops (unique to the chymotrypsin-like SP domains of Bb and C2a) folding alongside the anaphylatoxin domain of C3. Two possible concurrent effects may overcome the 30-Å gap between the catalytic site and the scissile loop and result into an enzymatically active complex. First, the large positional variation of the C345C domain in various C3b structures (Supplementary Fig. 9 online), suggests that the flexibility in C345C orientation may be used to swing Bb into place for proteolysis. Second, the observed C3b-C3b interface in the inhibited complex likely represents the product release state C3bBb-C3b. Substrate binding putatively involves a larger area of C3 that includes the domains MG3, MG6−8 at the same side as MG4−5, and hence closes part of the gap between the substrate and the enzymatic complex. The differences in domain orientations of MG3, MG7 and MG8 (15°, 36° and 61°; respectively) between C332 and C3b15, 16 yield distinct shape complementarities and electrostatic surface potentials that could differentiate substrate binding from product release (Supplementary Fig. 12 online). SCIN possibly blocks both processes, it prevents swinging of Bb and it may block a tighter C3b-C3 interface. In conclusion, the data suggested that the specific proteolytic activity of the C3bBb complexes is determined by the highly specific binding of the substrate through the exosite located on the ligand C3b.
Intrinsic control of convertase activity
The C3 convertase (C3bBb) dissociates irreversibly, which is critical in controlling complement activation35. However, it is unclear what prevents reassociation of Bb and C3b. Structural comparison of C3bBb with free Bb24 and C3b15, 16 suggested that dissociation does not induce large conformational changes. Both free Bb24 (as well as the isolated VWA domain36) and likely bound Bb display a MIDAS configuration corresponding to an open, high-affinity state as observed in activated, ligand-binding integrin Iα domains27. Putative changes in the helix α7 and the N-terminal tail of the VWA domain of Bb from the bound state to the dissociated state5, 24 are not supported by the available structural data (Fig. 4c and Supplementary Fig. 10 online). SPR data showed that the pro-enzyme FB and C3b associated and dissociated in a two-step process in Mg2+-containing buffer with one fast and one slow kinetic phase that differ by orders of magnitude37. Here, we extended this analysis by measuring the binding of FB to C3b in the absence of Mg2+ (to exclude binding of the Bb fragment) and the interaction of fragment Ba with C3b (Supplementary Fig. 13 online). Our data confirmed that Ba directly binds to C3b in a Mg2+-independent manner37, 38 and showed that binding of FB in the absence of Mg2+ shifts towards a kinetic profile similar to that of Ba. We therefore conclude that the fast on/off phase in the C3b-FB interaction is Mg2+ independent and can be attributed to initial contacts via the Ba segment. This result implies that the Bb segment of FB associates and dissociates slowly. Taken together, these data indicate that the MIDAS and helix α7 configuration in Bb establishes a stable ligand binding site in the open configuration with slow association and dissociation kinetics, which is responsible for the intrinsic control of the convertase activity, the half-life time and irreversible dissociation.
DISCUSSION
We have shed important new light on the structure of the inherently instable bimolecular C3 convertase (C3bBb), which is the central protease complex of the complement system, by revealing the dimeric structure of C3bBb stabilized by SCIN from S. aureus. The structure answered crucial questions related to convertase specificity and function, relevant in understanding regulation of innate immunity. Furthermore it showed in detail how bacteria may evade the immune system, which is important to understand bacterial pathogenesis at the molecular level.
In order to protect host cells, complement activation is strictly regulated. Regulation mainly occurs at the level of C3 convertases, because the enzyme is a short-lived complex that cannot reassociate after dissociation. This first structure of C3bBb indicates that the conformation of Bb in the convertase complex is similar to that of Bb in its isolated form. Binding studies reveal that Bb cannot rebind to C3b, because it has lost its fast association binding site for C3b, which is the Ba domain. Another critical aspect of complement regulation is that the C3 convertase enzyme is highly specific for C3 and does not activate other proteins like the homologous C4 or C5. The presented structure of dimerized convertases explains that C3b of the enzyme complex (C3bBb) forms a dimer with its substrate C3 and provides an exosite that determines the enzyme specificity.
Based on our results we present a model for C3 activation by convertases. Surface-bound C3bBb binds the substrate C3 by forming a quasi-homodimer with C3b. Since Bb is bound to the flexible C345C domain in C3b it can swing towards the substrate and cleave the scissile bond in C3. The C3a domain will be released and conformational changes in C3b will induce its release from the enzyme complex. After diffusion C3b may bind covalently to hydroxyl groups on the target surface through its reactive acyl-imidazole moiety4. Possibly, covalent binding to the surface without diffusion may generate C3b2Bb complexes that exhibit C5 convertase activity; this would explain the apparent discrepancy that C5 convertase formation depends on binding one8 versus multiple C3b9 molecules to the C3 convertase complex. However, the precise arrangement of the multi-molecular enzyme complex that leads to C5 binding and cleavage is unclear.
From an immune evasion point of view, we now understand that a very large number of amino acids in SCIN are involved in the formation of the dimeric inhibited convertase. In fluid-phase, SCIN strongly binds to one convertase (C3bBb) and forms a bridge to the other convertase by binding C3b. On itself, the dimerization of convertase complexes could be an efficient convertase inhibitory strategy, by preventing substrate binding. By mutating the dimerization site in SCIN we demonstrate that a monomeric SCIN-convertase is still an inhibited and stable complex. Thus, even if dimerization can not occur on bacterial surfaces, SCIN will block the C3bBb enzyme directly. Since we observe a strong correlation between enzyme stability and inhibition by SCIN we believe that SCIN prevents swinging of Bb by fixing Bb on C3b. The dimeric nature of the convertase will contribute to complex stability and steric hindrance of complement receptor interactions with the stabilized convertase on the surface of the bacterium.
Although we can mimic these processes with purified components in solution, complement activation normally occurs on surfaces like bacteria. SCIN is a secreted bacterial molecule and we previously showed that it binds back to the surface as soon as active convertases are formed13. The secreted, versus membrane-bound, character of SCIN is critical to allow this small molecule to fit in the narrow pocket of convertase components and to bind quickly to random distributed convertases at the bacterial surface. The similar activity of SCIN mutants on surface-bound versus fluid-phase convertases suggests that the soluble complex is indeed representative for a surface-bound convertase. SCIN is an immunogenic bacterial protein, and therefore not suitable as an anti-inflammatory drug in humans. Nevertheless, the structure of the SCIN-convertase gives insight in a unique and effective convertase moulation strategy that evolved in a bacterial pathogen.
In summary, these data provide insights into the molecular mechanisms that underlie the central, amplification step of the complement system that leads to opsonization of pathogens and altered host cells. These insights are essential to understand the regulatory mechanisms of complement activation and the wide range of evasion strategies that can be used by pathogenic microorganisms. Both the structure and understanding of the convertase inhibitory strategy provide crucial insights for future development of complement inhibitors.
METHODS
Protein expression and purification
C3 was purified from freshly isolated human plasma and C3b was generated as described39. Preparation of recombinant SCIN and chimeric mutants was previously described14. The SCIN protein with an N-terminal His6 tag was prepared by overlap extension PCR. In the ChC3b2 mutant, we exchanged residue 1−13, 59−61, 64−65 and 67−68 with corresponding residues in ORF-D. Biacore, gel filtration and small-scale experiments were performed with plasma-purified FB and commercially obtained FD (Calbiochem). For large-scale experiments in crystallization trials recombinant FB and FD were used. Human FB, fused to a N-terminal His-tag containing a TEV cleavage site, was expressed in human embryonic kidney 293S GnTI− cells (HEK293ES) to enable homogeneous N-linked glycosylation40. FB was purified by metal-affinity chromatography as described25 and the His-tag was removed by TEV protease cleavage and a second column passage on Ni-NTA superflow (Qiagen) beads. Finally, FB was purified by size exclusion chromatography. Human FD was expressed in HEK293 EBNA cells without any tag. The secreted FD was purified by cation-exchange and size exclusion chromatography. Ba was recombinantly expressed in a pQE30-M15 Escherichia coli expression system (Qiagen) with a His6 tag at the N-terminus, and was purified from the inclusion bodies using a Ni-NTA superflow column. After refolding, it was subjected to anion-exchange chromatography on a MonoQ1010 column.
Binding convertase to immobilized SCIN
CM5 sensor chips were immobilized with SCIN (2526 RU) or CHIPS (3372 RU, negative control)41 using standard amine-coupling chemistry in 10 mM sodium acetate, pH 5. Purified C3b, FB, FD were passed over the surface at a flow rate of 30 μl/min in HBS-Mg (20 mM HEPES, 140 mM NaCl, 5 mM MgCl2, 0.05% Tween, pH 7.4); association and dissociation were both monitored for 300 s. Surfaces were regenerated using 0.1 M citric acid, 1 M NaCl, 1 mM EDTA, pH 5. Immobilization and binding experiments were performed at 25 °C using a Biacore 2000 instrument.
Gel Filtration and native gel electrophoresis
C3b (2 μM), FB (2 μM) and FD (1 μM, Calbiochem) were incubated in the presence of SCIN (4 μM) or NiCl2 (2.5 mM) in HBS-Mg for 1 h at 4 °C and separated on a Superdex-200 GL 10/300 equilibrated with ice-cold HBS-Mg. The column was calibrated using the HMW Calibration kit (GE Healthcare) containing Thyroglobulin (669 kDa), Ferritin (440 kDa), Aldolase (158 kDa), Conalbumin (75 kDa) and Ovalbumin (43 kDa). Alternatively, complexes were separated by native gel electrophoresis (7.5 %) at 4 °C and visualized by silver staining. C3 and FB were detected by immunoblotting13. SCIN was detected by ELISA using mouse-anti-SCIN 1C9 as a capturing antibody and biotinylated mouse-anti-SCIN 2F4 for detection13; binding was visualized by peroxidase-conjugated streptavidin and tetramethyl benzidine (TMB) substrate.
Purification of SCIN-inhibited convertases for crystallization
C3b (0.5 μM), recombinant FB (1 μM), recombinant FD (0.5 μM) and His-tagged SCIN (2 μM) were incubated in 20 ml HBS-Mg for 1 h at 4 °C. SCIN complexes were purified by incubation with 1.2 gram magnetic cobalt beads (Dynabeads Talon, Invitrogen) for 1 h at 4 °C. Beads were washed with HBS-Mg and complexes were eluted with 500 mM Imidazole in 10 mM Tris, 40 mM NaCl, 5 mM MgCl2, pH 8 (2 ml). To remove uncomplexed SCIN, complexes were washed with 200 ml buffer in a 100-kDa filter centricon device (Millipore); in a final centrifugation step complexes were concentrated to ∼5 mg/ml of purified complexes.
Analytical ultracentrifugation
A purified sample of the 500 kDa complex consisting of C3b, Bb and SCIN in 10 mM Tris pH 8.0, 40 mM NaCl and 0.05% Tween, was used for a sedimentation velocity experiment using a Beckman Coulter™ Optima™ XL-A analytical ultracentrifuge. The sample, contained in a standard 12 mm Epon charcoal-filled centerpiece with quartz windows, was diluted to obtain an optical density of ∼0.9 against a demi-water reference, through a 12 mm path length at 229 nm to optimize the signal-to-noise ratio. The sedimentation velocity run was performed at 40,000 rpm (An-50 Ti rotor; 116,400 g) 9.0 ± 0.3 °C during 2.5 h in which 140 scans were recorded at a radial step size of 30 μm (data are available from the UltraScan public Van `t Hoff Laboratory LIMS database (uslims-utrecht), ExpDataID 74, host address: bcf.uthscsa.edu). A relative low angular velocity was chosen to retain sufficient shape information based on diffusion. Sedimentation velocity data were analyzed with the enhanced van Holde-Weischet analysis42 (UltraScan43) after subtraction of the time-invariant noise which was determined in a 2-dimensional spectrum analysis44 (UltraScan). Sedimentation coefficients predicted from theory, were calculated for the subunits assuming that they are spherical, prolate ellipsoidal and oblate ellipsoidal, accounting for buffer density and viscosity as well as partial specific volumes calculated from the amino acid sequences (UltraScan43) (Supplementary Fig. 4d online). For the latter two shapes an arbitrary axial or aspect ratio of 6 was taken. Based on sedimentation velocity simulations (UltraScan), the dimeric complex (C3bBb-SCIN)2 has a predicted sedimentation coefficient that is in between 10 and 20 S (1 S ≡ 1×10−13 s).
Crystallization, data collection and structure determination
Freshly purified SCIN-stabilized complexes were set up for crystallization trials by hanging-drop vapour diffusion at 4 °C. Crystals appeared overnight and grew within 2 weeks to a typical size of 300×100×40 μm3 in multiple conditions. Over 100 crystals were screened at ESRF beamline (ID14-EH14). Most of the crystals diffracted between 4−5 Å, only one crystal diffracted better than 4 Å resolution. A complete data set was collected from this crystal, which diffracted to 3.9-Å resolution. This crystal was grown in the condition of 75 mM sodium/potassium tartrate, 8.0% PEG 3350, 50 mM Bis-Tris propane, pH 6.5. The crystal exhibited space group P21 (a = 228.6, b = 121.5, c = 280.8 Å, α = 90°, β = 91.6°, γ = 90°). The dataset was integrated and scaled with MOSFLM and SCALA45. The asymmetric unit contained four C3bBb-SCIN complexes arranged as two dimers. The structure was solved by molecular replacement by PHASER46 using the isolated structures of C3c (pdb code: 2A74)32, C3b (pdb code: 2I07)15, Bb (pdb code: 1RRK)24 and SCIN (pdb code: 2QFF)14 as the initial search models. The automatic searches for CUB, TED and C345C domains of C3b and SCIN in PHASER yielded only two copies of each domain or protein. The missing parts were manually placed by superposition and adjusted by rigid-body refinement in PHASER to obtain a complete model. One C3b molecule lacked density for its TED domain, due to disorder; occupancies were set to 0.0 for this domain. The C3b molecule in this copy (denoted as copy 4) has weaker density compared to C3b in other copies (see Supplementary Fig. 6). The model was rebuilt in COOT47 and the refinement was carried out in PHENIX48. Tight four-fold non-crystallographic symmetry (NCS) restraints were used for the following domains or molecules: MG1, MG2, MG3, MG4, MG5, MG6, LNK, α’NT and MG7, MG8, CUB of C3b, VWA and SP of Bb and SCIN. Tight three-fold NCS restraints were used for TED domains in copies of 1−3. Two-fold NCS restraints were used for C345C domains (copies of 1 and 3 as a group and copies of 2 and 4 as a group). The final Rwork and Rfree values are 25.3 and 26.8%, respectively.
Convertase inhibition assays
C3 (250 nM), C3b (50 nM), FB (500 nM), FD (500 nM) and SCIN chimeras (2 μM) were incubated in HBS-Mg for 10 min at 20 °C and C3 conversion was analyzed by SDS-PAGE under reducing conditions. Convertase stabilization on bacteria was performed as described14. HBS-MgEGTA instead of HBS-Mg2+-Ca2+ was used to solely induce the alternative pathway.
Kinetic analysis of FB and Ba interaction with C3b
Kinetic profiling was performed using SPR (Biacore 2000) at 25 °C. C3b was biotinylated at its thioester moiety and captured on a streptavidin-coated sensor chip. FB (63 nM - 2 μM) was injected for 3 min at 30 μl/min and the dissociation was monitored for 5 min. The surface was regenerated with 2 M NaCl and 0.2 M sodium carbonate pH 9.0 for 30 s each. Fragment Ba (40 nM - 40 μM) was injected for 1 min at 30 μl/min with a dissociation phase of 2 min and a short regeneration pulse (20 s) of 1 M NaCl. All binding experiments were repeated in running buffer (10 mM HEPES, pH 7.4, 0.005% Tween-20) containing either 1 mM MgCl2 or 3 mM EDTA. Signals from an untreated streptavidin surface and an ensemble of buffer blank injections were subtracted from the binding signals. Data processing was performed using Scrubber (BioLogic Software) and CLAMP49 was used for kinetic analysis.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully thank R. Romijn (U-Protein Express, Utrecht) for help with mammalian protein expression and M. Otten and M. Daha (Leiden University Medical Center) for performing haemolytic assays. We thank P. Lenting for help with Biacore analyses. We thank the European Synchrotron Radiation Facility (ESRF) for the provision of synchrotron radiation facilities; and beamline scientists of the ESRF and the European Molecular Biology Laboratory for assistance. This work was supported by the Council for Medical Sciences (MW) and Chemical Sciences (CW) of the Netherlands Organization for Scientific Research (NWO) to SHMR (NWO-MW Veni), JAGvS (NWO-MW Top), PG (NWO-CW Top) and the National Institute of Health to JDL and PG.
Footnotes
Competing interests The authors declare no financial interests.
Supplementary information includes Supplementary Figures 1−13 and Supplementary Table 1.
Accession code. Coordinates and structure factors have been deposited in the Protein Data Bank with accession code 2WIN.
Note: Supplementary information is available on the Nature Immunology website.
REFERENCES
- 1.Carroll MC. The complement system in regulation of adaptive immunity. Nat Immunol. 2004;5:981–986. doi: 10.1038/ni1113. [DOI] [PubMed] [Google Scholar]
- 2.Mollnes TE, Song WC, Lambris JD. Complement in inflammatory tissue damage and disease. Trends Immunol. 2002;23:61–64. doi: 10.1016/s1471-4906(01)02129-9. [DOI] [PubMed] [Google Scholar]
- 3.Duncan RC, Wijeyewickrema LC, Pike RN. The initiating proteases of the complement system: controlling the cleavage. Biochimie. 2008;90:387–395. doi: 10.1016/j.biochi.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 4.Law SK, Dodds AW. The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 1997;6:263–274. doi: 10.1002/pro.5560060201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gros P, Milder FJ, Janssen BJ. Complement driven by conformational changes. Nat Rev Immunol. 2008;8:48–58. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- 6.Kerr MA. The human complement system: assembly of the classical pathway C3 convertase. Biochem J. 1980;189:173–181. doi: 10.1042/bj1890173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rawal N, Pangburn MK. C5 convertase of the alternative pathway of complement. Kinetic analysis of the free and surface-bound forms of the enzyme. J Biol Chem. 1998;273:16828–16835. doi: 10.1074/jbc.273.27.16828. [DOI] [PubMed] [Google Scholar]
- 8.Kinoshita T, et al. C5 convertase of the alternative complement pathway: covalent linkage between two C3b molecules within the trimolecular complex enzyme. J Immunol. 1988;141:3895–3901. [PubMed] [Google Scholar]
- 9.Rawal N, Pangburn MK. Structure/function of C5 convertases of complement. Int Immunopharmacol. 2001;1:415–422. doi: 10.1016/s1567-5769(00)00039-4. [DOI] [PubMed] [Google Scholar]
- 10.Kirkitadze MD, Barlow PN. Structure and flexibility of the multiple domain proteins that regulate complement activation. Immunol Rev. 2001;180:146–161. doi: 10.1034/j.1600-065x.2001.1800113.x. [DOI] [PubMed] [Google Scholar]
- 11.Pangburn MK, Muller-Eberhard HJ. The C3 convertase of the alternative pathway of human complement. Enzymic properties of the bimolecular proteinase. Biochem J. 1986;235:723–730. doi: 10.1042/bj2350723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat Rev Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rooijakkers SH, et al. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat Immunol. 2005;6:920–927. doi: 10.1038/ni1235. [DOI] [PubMed] [Google Scholar]
- 14.Rooijakkers SH, et al. Staphylococcal complement inhibitor: structure and active sites. J Immunol. 2007;179:2989–2998. doi: 10.4049/jimmunol.179.5.2989. [DOI] [PubMed] [Google Scholar]
- 15.Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 16.Wiesmann C, et al. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature. 2006;444:217–220. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- 17.Torreira E, Tortajada A, Montes T, de Córdoba SR, Llorca O. 3D structure of the C3bB complex provides insights into the activation and regulation of the complement alternative pathway convertase. Proceedings of the National Academy of Sciences. 2009;106:882–887. doi: 10.1073/pnas.0810860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller-Eberhard HJ, Gotze O. C3 proactivator convertase and its mode of action. J Exp Med. 1972;135:1003–1008. doi: 10.1084/jem.135.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fishelson Z, Pangburn MK, Muller-Eberhard HJ. C3 convertase of the alternative complement pathway. Demonstration of an active, stable C3b, Bb (Ni) complex. J Biol Chem. 1983;258:7411–7415. [PubMed] [Google Scholar]
- 20.Horiuchi T, Macon KJ, Engler JA, Volanakis JE. Site-directed mutagenesis of the region around Cys-241 of complement component C2. Evidence for a C4b binding site. J Immunol. 1991;147:584–589. [PubMed] [Google Scholar]
- 21.Hourcade DE, Mitchell LM, Oglesby TJ. Mutations of the type A domain of complement factor B that promote high-affinity C3b-binding. J Immunol. 1999;162:2906–2911. [PubMed] [Google Scholar]
- 22.Tuckwell DS, Xu Y, Newham P, Humphries MJ, Volanakis JE. Surface Loops Adjacent to the Cation-Binding Site of the Complement Factor B von Willebrand Factor Type A Module Determine C3b Binding Specificity. Biochemistry. 1997;36:6605–6613. doi: 10.1021/bi963155l. [DOI] [PubMed] [Google Scholar]
- 23.Hourcade DE, Mitchell L, Kuttner-Kondo LA, Atkinson JP, Medof ME. Decay-accelerating factor (DAF), complement receptor 1 (CR1), and factor H dissociate the complement AP C3 convertase (C3bBb) via sites on the type A domain of Bb. J Biol Chem. 2002;277:1107–1112. doi: 10.1074/jbc.M109322200. [DOI] [PubMed] [Google Scholar]
- 24.Ponnuraj K, et al. Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase. Molecular cell. 2004;14:17–28. doi: 10.1016/s1097-2765(04)00160-1. [DOI] [PubMed] [Google Scholar]
- 25.Milder FJ, et al. Factor B structure provides insights into activation of the central protease of the complement system. Nat Struct Mol Biol. 2007;14:224–228. doi: 10.1038/nsmb1210. [DOI] [PubMed] [Google Scholar]
- 26.Milder FJ, et al. Structure of complement component C2A: implications for convertase formation and substrate binding. Structure. 2006;14:1587–1597. doi: 10.1016/j.str.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnan V, Xu Y, Macon K, Volanakis JE, Narayana SV. The crystal structure of C2a, the catalytic fragment of classical pathway C3 and C5 convertase of human complement. J Mol Biol. 2007;367:224–233. doi: 10.1016/j.jmb.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kam CM, et al. Human complement proteins D, C2, and B. Active site mapping with peptide thioester substrates. J Biol Chem. 1987;262:3444–3451. [PubMed] [Google Scholar]
- 30.Janssen BJ, Halff EF, Lambris JD, Gros P. Structure of compstatin in complex with complement component C3c reveals a new mechanism of complement inhibition. J Biol Chem. 2007;282:29241–29247. doi: 10.1074/jbc.M704587200. [DOI] [PubMed] [Google Scholar]
- 31.Katschke KJ, Jr., et al. Structural and Functional Analysis of a C3b-specific Antibody That Selectively Inhibits the Alternative Pathway of Complement. J Biol Chem. 2009;284:10473–10479. doi: 10.1074/jbc.M809106200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janssen BJ, et al. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 33.Fredslund F, et al. Structure of and influence of a tick complement inhibitor on human complement component 5. Nat Immunol. 2008;9:753–760. doi: 10.1038/ni.1625. [DOI] [PubMed] [Google Scholar]
- 34.Rawal N, Pangburn M. Formation of high-affinity C5 convertases of the alternative pathway of complement. J Immunol. 2001;166:2635–2642. doi: 10.4049/jimmunol.166.4.2635. [DOI] [PubMed] [Google Scholar]
- 35.Hourcade DE, Mitchell LM, Medof ME. Decay acceleration of the complement alternative pathway C3 convertase. Immunopharmacology. 1999;42:167–173. doi: 10.1016/s0162-3109(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 36.Bhattacharya AA, Lupher ML, Jr., Staunton DE, Liddington RC. Crystal structure of the A domain from complement factor B reveals an integrin-like open conformation. Structure. 2004;12:371–378. doi: 10.1016/j.str.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 37.Harris CL, Abbott RJ, Smith RA, Morgan BP, Lea SM. Molecular dissection of interactions between components of the alternative pathway of complement and decay accelerating factor (CD55). The Journal of biological chemistry. 2005;280:2569–2578. doi: 10.1074/jbc.M410179200. [DOI] [PubMed] [Google Scholar]
- 38.Pryzdial EL, Isenman DE. Alternative complement pathway activation fragment Ba binds to C3b. Evidence that formation of the factor B-C3b complex involves two discrete points of contact. J Biol Chem. 1987;262:1519–1525. [PubMed] [Google Scholar]
- 39.Lambris JD, Dobson NJ, Ross GD. Release of endogenous C3b inactivator from lymphocytes in response to triggering membrane receptors for beta 1H globulin. J. Exp. Med. 1980;152:1625–1644. doi: 10.1084/jem.152.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Haas CJ, et al. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. The Journal of experimental medicine. 2004;199:687–695. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demeler B, van Holde KE. Sedimentation Velocity Analysis of Highly Heterogeneous Systems. Analytical Biochemistry. 2004;335:279–288. doi: 10.1016/j.ab.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 43.Demeler B. UltraScan 9.7 - A Comprehensive Data Analysis Software Package for Analytical Ultracentrifugation Experiments. In: Scott DJ, Harding SE, Rowe AJ, editors. Analytical Ultracentrifugation: Techniques and Methods. Royal Society of Chemistry; Cambridge: 2005. pp. 210–230. [Google Scholar]
- 44.Brookes E, Demeler B. Parallel Computational Techniques for the Analysis of Sedimentation Velocity Experiments in UltraScan. Colloid and Polymer Science. 2008;286:139–148. [Google Scholar]
- 45.Evans P. Scaling and assessment of data quality. Acta Crystallographica Section D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 46.McCoy AJ, et al. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 49.Myszka DG, Morton TA. CLAMP: a biosensor kinetic data analysis program. Trends Biochem Sci. 1998;23:149–150. doi: 10.1016/s0968-0004(98)01183-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.