Abstract
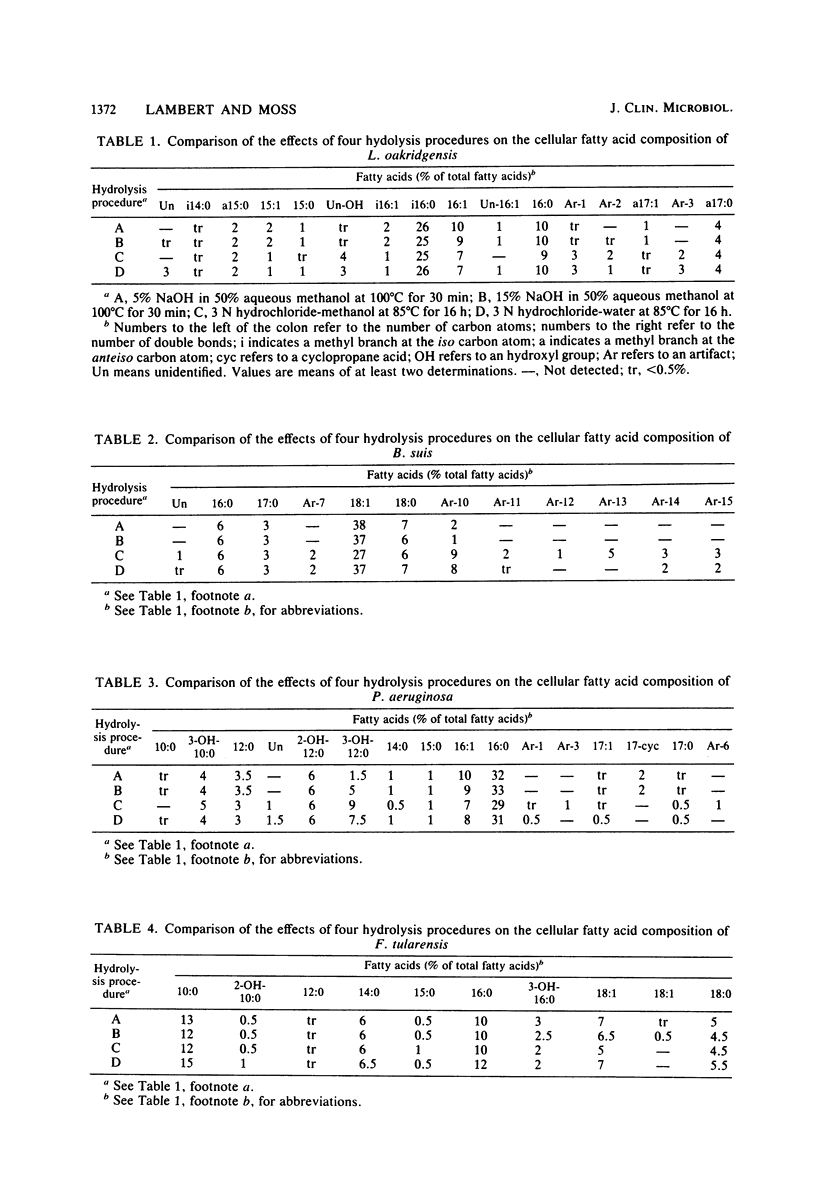
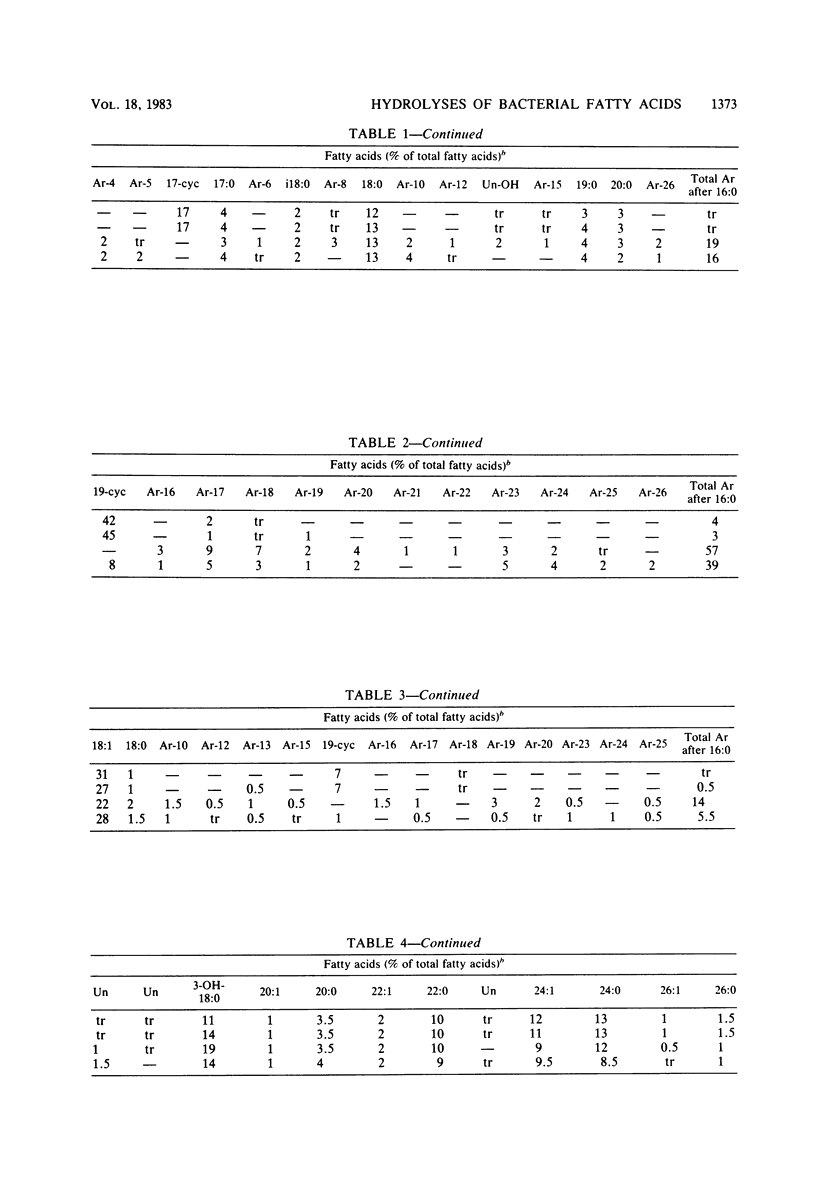
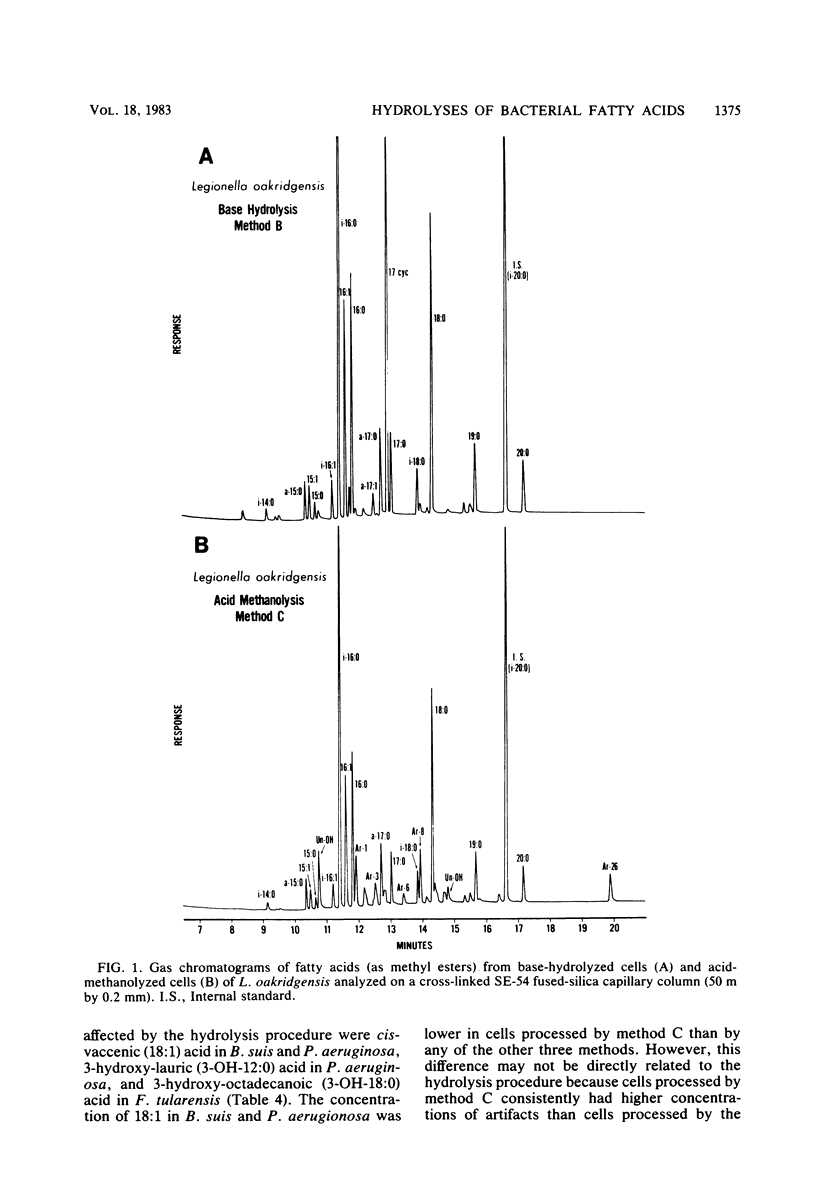
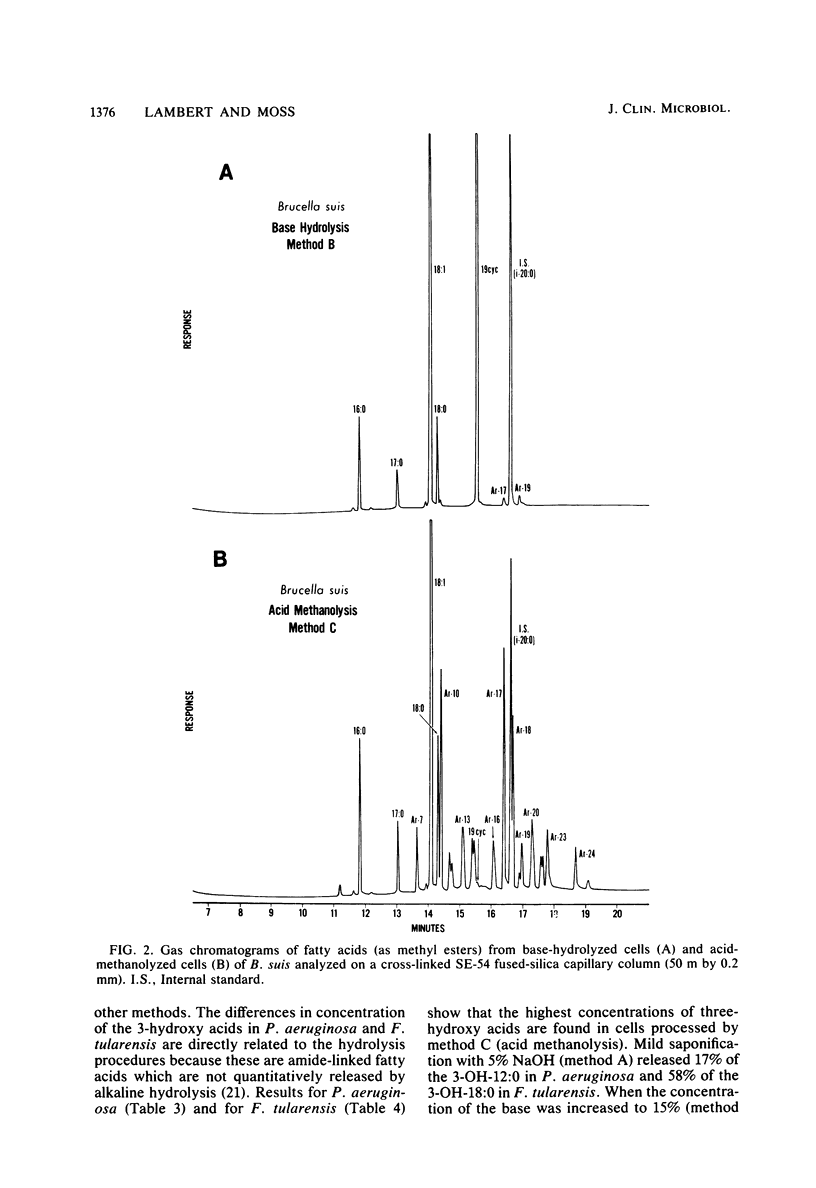
The cellular fatty acid compositions of Legionella oakridgensis, Brucella suis, Pseudomonas aeruginosa, and Francisella tularensis were compared after base hydrolysis (saponification), acid hydrolysis, and acid methanolysis procedures were used to release the fatty acids. The branched-chain, unsaturated, saturated, and ester-linked hydroxy acids were released as effectively with saponification at 100 degrees C for 30 min as with acid hydrolysis or acid methanolysis at 85 degrees C for 16 h. Although the amide-linked hydroxy acids were released more effectively by acid hydrolysis or acid methanolysis, these methods degraded the cyclopropane fatty acids, producing a number of new peaks or artifacts in the chromatograms. Cyclopropane fatty acids were not degraded by saponification, and at least 50% of the hydroxy acids were released when the cells were saponified with 15% NaOH in 50% aqueous methanol. Thus, the results show that saponification for 30 min at 100 degrees C with 15% NaOH, followed by methylation is an excellent method for routine fatty acid analysis of bacteria and for screening cultures whose identity and fatty acid composition are unknown.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dees S. B., Hollis D. G., Weaver R. E., Moss C. W. Cellular fatty acids of Brucella canis and Brucella suis. J Clin Microbiol. 1981 Jul;14(1):111–112. doi: 10.1128/jcm.14.1.111-112.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen E., Berdal B. P., Omland T. Cellular fatty acid composition of Francisella tularensis. J Clin Microbiol. 1979 Dec;10(6):928–930. doi: 10.1128/jcm.10.6.928-930.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen E., Bryn K., Bovre K. Gas chromatography of bacterial whole cell methanolysates; IV. A procedure for fractionation and identification of fatty acids and monosaccharides of cellular structures. Acta Pathol Microbiol Scand B Microbiol Immunol. 1974 Dec;82(6):753–766. [PubMed] [Google Scholar]
- Jantzen E., Knudsen E., Winsnes R. Fatty acid analysis for differentiation or Bordetella and Brucella species. Acta Pathol Microbiol Immunol Scand B. 1982 Oct;90(5):353–359. doi: 10.1111/j.1699-0463.1982.tb00131.x. [DOI] [PubMed] [Google Scholar]
- Lambert M. S., Moss C. W. Degradation of bacterial cyclopropane acids with boron trihalide reagents. Microbios. 1977;18(71):51–58. [PubMed] [Google Scholar]
- Mayberry W. R. Dihydroxy and monohydroxy fatty acids in Legionella pneumophila. J Bacteriol. 1981 Aug;147(2):373–381. doi: 10.1128/jb.147.2.373-381.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. T. Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J Clin Microbiol. 1982 Sep;16(3):584–586. doi: 10.1128/jcm.16.3.584-586.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss C. W. Gas-liquid chromatography as an analytical tool in microbiology. J Chromatogr. 1981 Jan 9;203:337–347. doi: 10.1016/s0021-9673(00)80305-2. [DOI] [PubMed] [Google Scholar]
- Moss C. W., Lambert M. A., Merwin W. H. Comparison of rapid methods for analysis of bacterial fatty acids. Appl Microbiol. 1974 Jul;28(1):80–85. doi: 10.1128/am.28.1.80-85.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrison L. H., Cherry W. B., Tyndall R. L., Fliermans C. B., Gough S. B., Lambert M. A., McDougal L. K., Bibb W. F., Brenner D. J. Legionella oakridgensis: unusual new species isolated from cooling tower water. Appl Environ Microbiol. 1983 Feb;45(2):536–545. doi: 10.1128/aem.45.2.536-545.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietschel E. T., Gottert H., Lüderitz O., Westphal O. Nature and linkages of the fatty acids present in the lipid-A component of Salmonella lipopolysaccharides. Eur J Biochem. 1972 Jul 13;28(2):166–173. doi: 10.1111/j.1432-1033.1972.tb01899.x. [DOI] [PubMed] [Google Scholar]
- Rooney S. A., Goldfine H., Sweeley C. C. The identification of trans-2-tetradecenoic acid in hydrolysates of lipid A from Escherichia coli. Biochim Biophys Acta. 1972 Jul 7;270(3):289–295. doi: 10.1016/0005-2760(72)90192-0. [DOI] [PubMed] [Google Scholar]
- Shaw N. Lipid composition as a guide to the classification of bacteria. Adv Appl Microbiol. 1974;17(0):63–108. doi: 10.1016/s0065-2164(08)70555-0. [DOI] [PubMed] [Google Scholar]
- Skarin A., Larsson L., Holst E., Mårdh P. A. Gas chromatographic study of cellular fatty acids of comma-shaped bacteria isolated from the vagina. Eur J Clin Microbiol. 1982 Oct;1(5):307–309. doi: 10.1007/BF02019977. [DOI] [PubMed] [Google Scholar]
- Vulliet P., Markey S. P., Tornabene T. G. Identification of methoxyester artifacts produced by methanolic-HCI solvolysis of the cyclopropane fatty acids of the genus Yersinia. Biochim Biophys Acta. 1974 May 29;348(2):299–301. doi: 10.1016/0005-2760(74)90241-0. [DOI] [PubMed] [Google Scholar]
- Wilkinson S. G. Artifacts produced by acidic hydrolysis of lipids containing 3-hydroxyalkanoic acids. J Lipid Res. 1974 Mar;15(2):181–182. [PubMed] [Google Scholar]


