SUMMARY
The engrailed homeoprotein is a dominantly acting or ‘active’ transcriptional repressor both in cultured cells and in vivo. When retargeted via a homeodomain swap to the endogenous fushi tarazu gene (ftz), it actively represses it, resulting in a ftz mutant phenocopy. We have mapped functional regions of engrailed using this in vivo repression assay. In addition to a region containing an active repression domain identified in cell culture assays (K. Han and J. L. Manley (1993) EMBO J. 12, 2723–2733), we find that two evolutionarily conserved regions contribute to activity. The one of these that does not flank the HD is particularly crucial to repression activity in vivo. We find that this domain is present not only in all engrailed-class homeoproteins but also in all known members of several other classes, including goosecoid, Nk1, Nk2 and msh. Thus engrailed’s active repression function in vivo is dependent on a highly conserved interaction that was established early in the evolution of the homeobox gene superfamily. We further show using rescue transgenes that the widely conserved in vivo repression domain is required for the normal function of engrailed in the embryo.
Keywords: active repression, Drosophila, embryonic development, homeodomain, transcriptional repressor, engrailed, fushi tarazu
INTRODUCTION
Transcriptional repressors that function at a distance, analogously to transcriptional activators, have been termed active repressors (Jaynes and O’Farrell, 1991; reviewed in Gray et al., 1995). Functional distinctions within this class of effectors have been proposed based on their ability to act over different distances on the DNA (Gray et al., 1994). However, mechanistic distinctions between short- and long-range repressors have yet to be made. One active repressor that has been well characterized both in cultured cells and in vivo is the product of the engrailed locus of Drosophila. The engrailed protein (en) contains a homeodomain (HD) related in DNA-binding specificity to that of the Antennapedia (Antp) class (Desplan et al., 1988), but representing a separate, conserved class with two known members in both insects and mammals. Several members of the Antp class have been shown to be transcriptional activators, including the fushi tarazu protein, ftz. Ftz is a strong, context-independent activator in cultured cells (Jaynes and O’Farrell, 1988; Winslow et al., 1989), and participates in a direct positive feedback on its own gene in Drosophila embryos (Schier and Gehring, 1992). En is an active repressor in cultured cells and dissection of this activity showed that the N-terminal half, which does not include the HD Jaynes and O’Farrell, 1991; Jaynes et al., 1990), or a small subdomain of that region (Han and Manley, 1993), can confer repression activity to heterologous DNA-binding domains.
Gene-specific transcriptional regulators generally have two activities that determine their function. First, they interact with specific sets of target genes, usually through sequence-specific DNA-binding domains. Then they affect the expression of those target genes through activation or repression domains that are often separable from domains involved in DNA binding. This distinction between targeting and effector functions can be blurred by cooperative interactions with other regulators. However, the modularity of most well-studied factors, along with the promiscuous activity of most activation and repression domains analyzed to date, suggests that the two functions may often act independently in vivo. This view is supported by recent manipulations of binding sites within the even-skipped stripe 2 enhancer (Arnosti et al., 1996). One way to test this notion and to separately address the requirements for targeting and effector function in vivo is to ‘retarget’ a regulator by exchanging targeting domains between proteins with distinct effector functions. Using such a strategy, dominant negative effects have been seen in several contexts when the active repression region from engrailed was retargeted using the DNA-binding domain of an activator. Both in mammalian cells and in transgenic mice, the DNA-binding region of the myb proto-oncogene fused with the N-terminal portion of en resulted in a dominant negative effect (Badiani et al., 1994). Similarly, by swapping HDs between ftz and en, it was shown that en domains can confer a dominant negative activity on the ftz HD. When produced ubiquitously from a heat-inducible promoter, this chimeric repressor can specifically override the activity of endogenous ftz protein, causing repression of the ftz gene and generating a ftz-mutant phenotype in Drosophila embryos (John et al., 1995). This effect was determined to be the result of active repression by three criteria. First, this action of the chimeric protein, termed EFE, depended on an N-terminal region from en implicated in active repression in culture. Second, EFE without this repression domain, while unable to significantly repress endogenous ftz gene expression, could still interact with ftz HD target sites. This was apparent from its ability to inhibit the activity of the autoregulatory ftz enhancer driving a lacZ reporter in vivo, presumably by competing for ftz binding sites, as this enhancer was previously shown to be a direct target of the ftz HD (Schier and Gehring, 1992). A third indication that the repression is active was that EFE repressed another ftz target gene (the engrailed gene) outside the domain of endogenous ftz expression. Thus, the action of EFE could not be attributed solely to preventing ftz from binding to its target genes.
Here we show, using repression of endogenous ftz as the primary assay, that the repression function of EFE is contributed by several domains, including the C-terminal region flanking the HD, but that another conserved region found in the N-terminal repression domain is particularly important. This crucial region is distinct from the region found to be most active in culture (Han and Manley, 1993) and is homologous not only to all other en class homeoproteins, but to all known members of four other homeoprotein classes: msh, Nk1, Nk2 and goosecoid. These results suggest that a highly conserved interaction, established early in homeoprotein evolution, mediates active transcriptional repression by engrailed in vivo. We further show that this domain is required for the function of wild-type engrailed protein expressed in its normal pattern in early embryos.
MATERIALS AND METHODS
Embryo preparation and staining
P-element transformations (Spradling and Rubin, 1982), cuticle preparations (Wieschaus and Nüsslein-Volhard, 1986) and in situ hybridization to fixed embryos (Edgar and O’Farrell, 1990) were performed as described previously. Antibody staining was performed essentially as described (Manoukian and Krause, 1992) using a polyclonal α-en antisera, a kind gift of Charles Girdham and Patrick O’Farrell, which had been prepared against full-length, partially purified GST-tagged en and affinity purified against a His-tagged peptide with the N-terminal 150 amino acids of en. Either alkaline phosphatase-(AP) or peroxidase-coupled secondary antibodies (Vector Laboratories) were used both for microscopic examination of fixed embryos, where either BCIP and NBT (for AP) or DAB substrates were used for staining (Boehringer Mannheim) and for quantitation of antibody signals, where the AP substrate p-nitrophenyl phosphate (Sigma) was used as described (Manoukian and Krause, 1992). Incubation times were determined to be in the linear range of the assay by incubating sets of embryos with different signal intensities for various times.
Heat shocks were administered to embryos on 35 mm collection plates by floating the plates on 37°C water inside a sealed container, in order to minimize evaporative cooling. Standard heat-shock conditions employed a 15 minute incubation followed by return to a 25°C humidified environment.
Plasmid constructions and Drosophila strains
Expression plasmids for EFE derivatives were modifications of a P-element transformation vector capable of providing inducible expression of EFE in transformed Drosophila from a heat-shock promoter (described in John et al., 1995). Modifications were made using either PCR-based methods (for Δ5 and Δ6), synthetic DNA adaptors to create deletions adjacent to unique restriction sites (for Δ234, Δ23, Δ34, Δ3 and Δ4), or a combination of the two (Δeh1, F→E, Meh1). Resulting deletion end points and amino acid substitutions are described in figure legends and the text. All regions containing synthetic or PCR-synthesized DNA were subsequently sequenced (automated) to confirm the expected structure. Appropriate restriction fragments were combined to generate the combined deletion plasmid Δ46. Details are available on request. These plasmids were introduced into flies using standard methodologies (Spradling and Rubin, 1982). Homozygous viable insertions on either the second or third chromosome were used in all analyses of repression activity.
The embryonic en expression/rescue construct p31Rg-en was constructed by combining elements from three parent plasmids: en down-stream-SV40 poly(A) region and vector sequences from pRK232 (Heemskerk et al., 1991), en intron and partial coding region from pen7.5pUC (a subclone made by J. Kassis from en genomic clones described in Kuner et al., 1985) and partial en coding, promoter and upstream regions from P[en/lac] (DiNardo et al., 1988). Additional details are either contained in figure legends or text or are available on request.
Transfections
Cell culture assays were performed using Drosophila S2 cells as described previously, with 2 µg per 60 mm culture dish of the reporter gene T3N6D-33CatB (Jaynes and O’Farrell, 1991) and 0.3 µg of ftz expression plasmid pPAc-ftz (Jaynes and O’Farrell, 1988; Winslow et al., 1989). CAT assays, as well as β-galactosidase assays for expression of the cotransfected reference gene pLac82SU (Dorsett et al., 1989), were performed as described (Jaynes and O’Farrell, 1991). Co-transfected plasmids used to express EFE and its derivatives were the same as those used for P-element transformation, wherein expression is driven by the hsp70 promoter.
Database searches and sequence comparison
Searches of GenBank and EMBL databases were performed using the BLAST (at the NCBI using the BLAST network server), FASTA and TFASTA programs of the Wisconsin Package, version 8 (Sept. 1994, Genetics Computer Group, 575 Science Drive, Madison, WI). Additional sequence analysis utilized GCG software, including PileUp, used to order the sequences of Fig. 4, and MacVector™ version 4.1.4.
Fig. 4.
The engrailed eh1 region has similarity to other classes of homeoproteins. Selected members of each of the homeoprotein classes found to contain an eh1-homologous motif are shown. These were selected to show the range of variation within each class between vertebrate and invertebrate species. The consensus shows the most highly represented amino acid at each position. Dark shading in the consensus indicates similarity in at least 30% of the individual sequences. Dark shading in the individual sequences indicates exact match to the consensus, while lighter shading indicates similarity to the consensus. The invariant Phe (F) is residue 175 of the D. melanogaster engrailed sequence. For each major class represented, all known members for which the sequence of the N-terminal region is available contain the homologous region. Species designations are followed by gene names, either as given in Duboule (1994), if included therein, or otherwise as in the GenBank and EMBL databases. S. mansoni (a flatworm) is the most divergent species found to contain a homologous sequence. C. elegans (roundworm), D. melanogaster (fruit fly), Tribolium (flour beetle), H. roretzi (an ascidian), B. rerio (zebrafish), X. laevis (frog), R. norvegicus (rat), M. musculus (mouse) and H. sapiens (human) are also represented. The msx-1s contain an msh-class homeobox (HB), while msh-2, a.k.a. Nk-4 or tinman, contains an Nk2-related HB, and XANF-1 and HESX1 are in a novel HB class. invected (inv) is the second en-class gene that is found in some insects, engs contain an en-class HB (there are three in zebrafish, Ekker et al., 1992), while S59 contains an Nk1-related HB and TTF1 contains an Nk2-related HB. See Duboule (1994) for evolutionary relationships based on HB sequence and more detailed descriptions.
RESULTS
Repression activity in vivo is determined by multiple en domains
Previous results showed (John et al., 1995) that a chimeric protein, consisting of en with its HD replaced by that of ftz, termed EFE (Fig. 1), was capable of specifically repressing ftz target genes in vivo, including its own gene, ftz. This repression required not only the ftz HD, but also a portion of en implicated in active repression in cultured cells. This was demonstrated by characterizing a deletion derivative of EFE, which removed both of the en-derived regions 3 and 4. This deletion derivative (EFEΔ34) retained the ability to localize to nuclei in embryos, as well as the ability to repress transcription when competing for binding sites in cultured cells (passive repression), but had lost most of its active repression activity in culture. Consistent with the involvement of active repression in the action of EFE in vivo, this derivative was unable to effectively repress ftz gene expression in embryos. Despite this loss of activity, it could still substantially reduce the activity of a transgenic copy of a ftz autoregulatory enhancer driving a lacZ reporter in embryos. This ftz enhancer has been implicated as a direct in vivo site of action of the endogenous ftz protein (Schier and Gehring, 1992), indicating that EFEΔ34 could still interact with a ftz HD target site in vivo. The difference in response between the endogenous ftz gene and the isolated upstream enhancer may be due to endogenous ftz expression becoming independent of direct autoregulation once it is established, while it remains subject to active repression by EFE (probably acting through the upstream enhancer). A further indication that EFE is not simply competing for ftz target sites came from the observation that another ftz target gene, the engrailed gene, was repressed not only where ftz is present (and required to activate en), but also outside the ftz domain, in the ftz-independent (odd-numbered) en stripes (again, EFE may be acting through normal ftz binding sites, but counteracting the effects of other activators). Thus the ability of EFE to repress ftz gene expression in vivo appears to require an activity in addition to targeting. The region removed in EFEΔ34 contains two interesting features. One is a previously identified homology found in all known en-class homeoproteins (Logan et al., 1992), located in region 3, and the second is a minimal active repression domain identified in cultured cell assays (Han and Manley, 1993), located in region 4 (see Fig. 1).
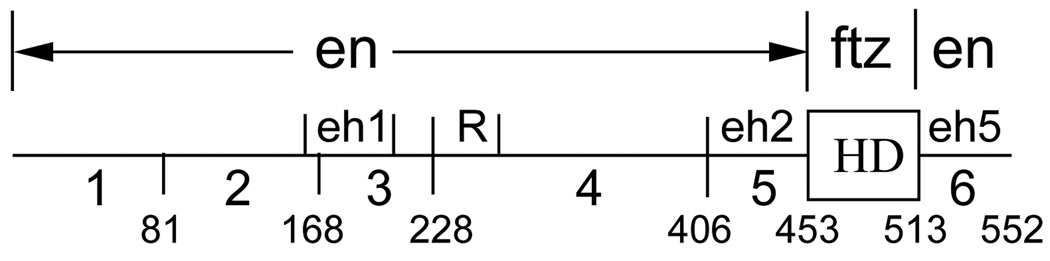
Fig. 1.
Features of the Drosophila engrailed-fushi tarazu chimera, EFE. The diagram indicates which portions of the coding sequence derive from en and which from ftz, our numerical designations of regions of en (1–6, not including the ftz HD), and the locations of known features within those regions (eh1, eh2, eh5 and R). eh1, eh2 and eh5 are peptide sequences found in all known en homologs (Logan et al., 1992) from widely divergent species, including insects and mammals, and R is an autonomous active repression domain identified in cell culture studies (Han and Manley, 1993). Homology eh1 is described further in Fig. 4, while eh2 and eh5 are part of the conserved regions flanking the en HD, which also include a sequence termed eh3 (immediately flanking the N terminus of the en HD) that has been implicated in nuclear localization (S. J. Poole, unpublished observation), and so was left intact in our analyses. Locations of region boundaries in the amino acid sequence are given below the line. Deletions and other alterations of these regions are described in detail in subsequent figures or in the text.
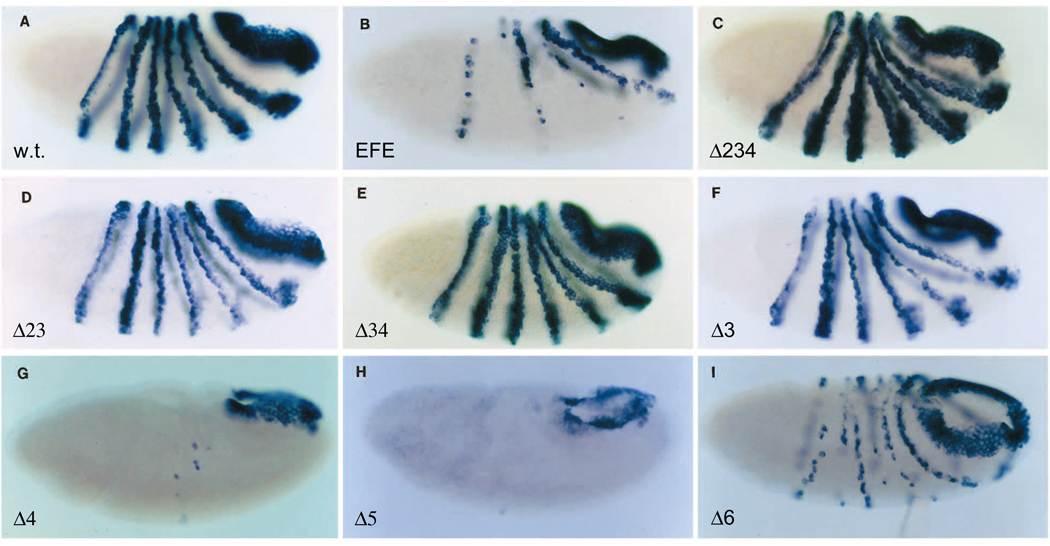
We mapped the domains responsible for the active repression activity of EFE in vivo, using the ability to repress endogenous ftz gene expression as an assay. A set of transgenic flies was constructed, each expressing a deletion derivative of EFE from a heat-inducible promoter. As described previously, a brief heat pulse induces ubiquitous expression from the transgene. Such expression of EFE causes rapid and persistent (Fig. 2B) loss of ftz expression in the trunk region (stripe 7 recovers). This results in the generation of pair-rule deletions in the cuticle pattern at the end of embryogenesis that mimic those seen in ftz mutants (John et al., 1995). Such heat treatment had no effect on endogenous ftz expression in wild-type embryos (Fig. 2A). In testing derivatives deleted for en-derived portions of EFE, we discovered that multiple regions contribute to activity. First, each of the deletions that remove region 3 caused substantial loss of activity (Fig. 2C–F). However, EFEΔ3 retains sufficient activity to cause significant repression of ftz expression (Fig. 2F), as the remaining stripes were discontinuous either laterally or dorsally. The additional deletion of region 2 caused no further loss of repression activity (Fig. 2D). In contrast, additional deletion of region 4 did cause a further reduction in activity, to the point that EFEΔ34 produced no lasting repression of the ftz gene (Fig. 2C,E; see John et al., 1995 for description of a mild, transient effect of EFEΔ34). Nonetheless, EFEΔ34, as described above, is still able to repress the autoregulatory ftz upstream enhancer, indicating that it retains targeting activity in vivo. In contrast, deletion of either region 4 or 5 alone resulted in increased repression activity (Fig. 2G,H; note that Δ5 is a partial deletion of region 5: Fig 1 and Fig 2 legends). In the case of EFEΔ4, this may be attributed to increased protein stability (for EFEΔ5, see below and Discussion). In region 6, a deletion of the most conserved 9 amino acids within the en C-terminal tail, which are the core of the C-terminal extension of the en HD, caused a partial loss of repression activity (Fig. 2I; for simplicity, we refer to this directed deletion as Δ6). The significance of this loss of activity was tested by examining both hatching rates of embryos and the cuticle defects caused by induction of EFEΔ6. Relative to lines expressing the parental EFE, the hatching rate was significantly increased and the severity of cuticle defects was clearly reduced in EFEΔ6 lines (data not shown), confirming the loss of activity due to this small deletion.
Fig. 2.
Repression of endogenous ftz gene expression by EFE and derivatives. The pattern of ftz RNA expression was monitored in embryos after induction of the indicated EFE derivative from a heat-inducible transgene. Embryos were fixed 40 minutes after induction and stained by in situ hybridization (as described in Materials and Methods) using a probe to ftz RNA sequences outside the HD. Expression was induced by giving a 15 minute heat pulse at 37°C (see Materials and Methods) beginning 2 hours 40 minutes after egg collection. Derivatives are named for the regions deleted. They remove the following amino acids of the 552 total (the ftz HD consists of amino acids 454–513): 82–399 (Δ234), 82–221 (Δ23), 169–399 (Δ34), 172–216 (Δ3), 229–399 (Δ4), 407–440 (Δ5) and 523–531 (Δ6). Thus Δ23 removes the entire eh1 homology plus additional sequences, Δ3 removes the most conserved portion of eh1 plus additional sequences, Δ5 and Δ6 are partial deletions of the most conserved portions of the homologies eh2 and eh5 (see Fig. 1 legend), Δ4 removes the R domain plus additional sequences and Δ34 removes both R and the most conserved portion of eh1.
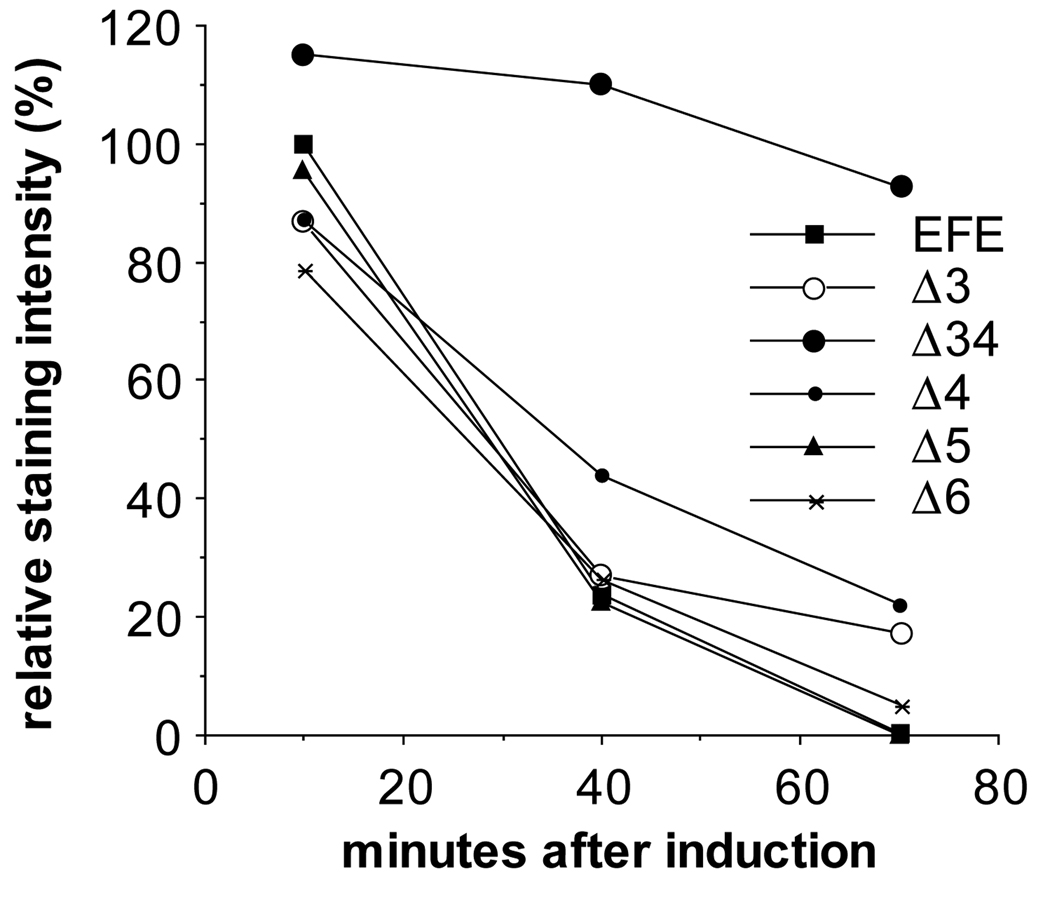
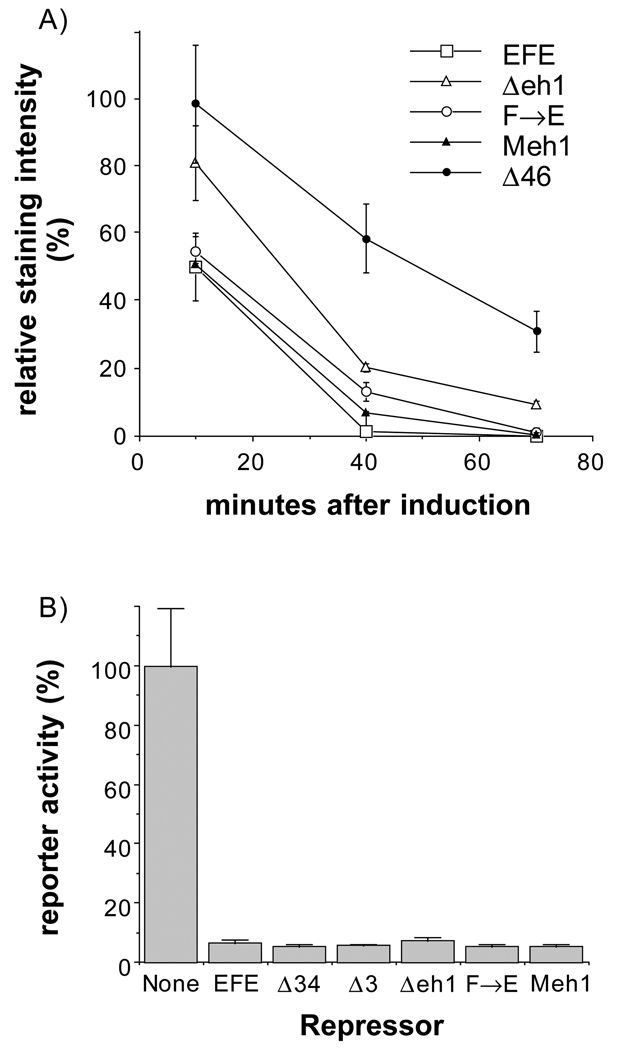
To determine whether the observed changes in activity could be attributed to differences in expression levels from the transgenes, we stained embryos following heat induction with an anti-en antiserum that reacts with the N-terminal region, including region 1 and part of region 2 (John et al., 1995). As previously shown for EFEΔ34 (John et al., 1995), each derivative retained its ability to localize to nuclei (not shown) and was expressed initially at levels comparable to EFE (Fig. 3). In addition, while most derivatives decayed with a time course very similar to that of EFE, both EFEΔ34 and EFEΔ4 were significantly more stable. experiments, in which the apparent half-life of EFE?4 was estimated to be about two-fold longer than that of EFE. These experiments also suggested that EFEΔ3 has a very slightly increased half-life (data not shown). Thus, the increased activity of EFEΔ4 may be accounted for by its increased stability, but the increased activity of EFEΔ5 is apparently not due to increased expression or stability. Likewise, the loss of activity of other derivatives cannot be explained by differences in expression or subcellular localization.
Fig. 3.
Protein levels following induction of transgene expression. Embryos were fixed at the indicated times after induction and stained with α-en antibodies (affinity purified using the N-terminal 150 amino acids of en, present in each of these derivatives) and with alkaline phosphatase (AP) coupled 2° antibodies. AP activity was quantified using the soluble-product-producing substrate PNP (Manoukian and Krause, 1992). The assay was within the linear range, since longer incubation times resulted in further linear increases in signal and both longer heat shocks with the same number of embryos and the inclusion of more embryos in parallel reactions gave approximately proportional increases in signal.
Region 3 contains similarity to other classes of homeoproteins
An obvious candidate for mediating the activity of region 3 was a previously identified region of homology among en class homeoproteins (Logan et al., 1992). In database searches using this ‘eh1’ homology region, we found that a similar motif is present in several other classes of homeoproteins (Fig. 4), which is always located N-terminal to the HD, with a variable length stretch of non-conserved amino acids intervening between the two (see Fig. 4). For the major classes, msh, goosecoid, Nk1, Nk2 and engrailed, all known members (for which complete sequence information is available) contain the motif (J. B. Jaynes, unpublished observation). In addition, two ‘novel’ class members, XANF1 and HESX1, as well as Nkx5.1, were also found to contain it (Fig. 4). In general, the apparent relatedness of these motifs parallels that of the homeodomain class to which the protein belongs, although there may be exceptions. Interestingly, there are apparently several subclasses of the motif conserved independently between vertebrates and invertebrates, including one conserved to the flatworm S. mansoni that is closely related among Nk2-class members (Fig. 4). This suggests the possibility that the different versions of the motif mediate interactions with a family of other factors. Homology within each class extends a variable distance to either side of the core motif, but similarity between the classes falls to an undetectable level within 10–20 amino acids. No reliable indications either of secondary structure or of similarity with other known domains was detected.
The eh1 homology mediates repression of ftz in vivo by EFE
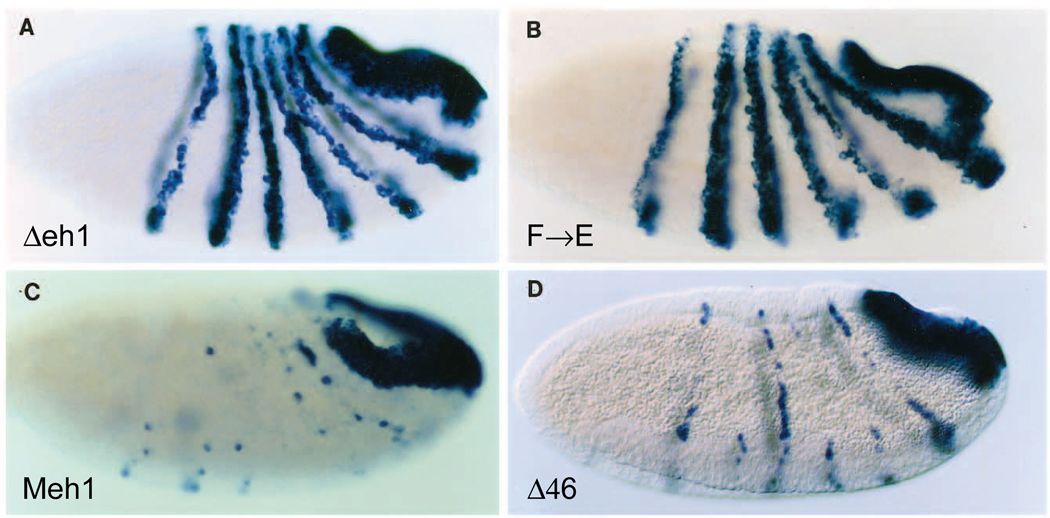
In order to test whether eh1 is required for repression by EFE, we constructed both a small deletion within eh1 and a single point mutant at the most conserved position. Both a 15 amino acid deletion removing the most conserved portion of eh1 and a change of the invariant Phe to Glu (F→E) resulted in derivatives of EFE with strongly reduced ability to repress ftz in early embryos. The levels of ftz RNA were reduced only slightly relative to wild type following induction of each of these derivatives (Fig. 5). Thus each of these changes in eh1 had an effect on EFE activity indistinguishable from that of removing region 3 entirely. To test whether the conservation of this region from flies to mammals had preserved function, we replaced the 15 amino acid region of the Drosophila protein with the corresponding region from the mouse En1 protein. This resulted in 4 non-conservative, 3 neutral and 1 conservative substitution within the region. As shown in Fig. 5, this replacement fully restored the ability of EFE to repress the endogenous ftz gene in Drosophila embryos, indicating that the function required for this activity, presumably active repression, is conserved. In contrast to the drastic effect of mutating region 3, combining two deletions that each reduce active repression in culture, Δ4 and Δ6 (Han and Manley, 1993; data not shown), resulted in a protein (EFEΔ46) still capable of repressing ftz (Fig. 5D). Examination of protein levels produced in embryos showed that, for those mutated in region 3, the less active proteins were produced at slightly higher levels than were the more active ones, while all were about equally stable (Fig. 6A). For EFEΔ46, the levels were slightly higher initially and the protein was considerably more stable than EFE, perhaps contributing significantly to its repression activity. However, EFEΔ46 is less stable than EFEΔ34 (Fig. 3), but nonetheless is a better repressor of ftz (compare Fig. 5D with Fig. 2E), indicating that it still has considerable active repression function in vivo, consistent with the strong in vivo activity of region 3.
Fig. 5.
A single point mutation in eh1 reduces, and the mouse eh1 region restores, repression activity in vivo. The ability of derivatives of EFE mutated in region 3 to repress endogenous ftz gene expression was compared to activity of a derivative that removes two other domains implicated in active repression in culture, regions 4 and 6. Endogenous ftz RNA levels were detected as in Fig. 2. (A) EFEΔeh1, which removes amino acids 172–186, the core of the eh1 homology, and (B) point mutation of the most conserved amino acid, Phe 175, to Glu each cause a drastic reduction in the ability to repress ftz, while replacement of the Δeh1 region by the corresponding mouse En1 region restores repression function (C). In contrast, combined deletion of regions 4 and 6 (D) does not reduce activity as much as the point mutation in region 3.
Fig. 6.
(A) Changes in eh1 do not reduce protein levels in vivo. Embryos from each transgenic line were stained for transgeneproduced protein following induction and the results quantified, as in Fig. 3. Note that the alterations in region 3 that reduce activity (Δeh1 and F→E) do not reduce either protein levels or stability, while EFEΔ46 is significantly more stable than EFE. (B) Competition for binding sites by EFE and derivatives in cultured cells. Drosophila S2 cells were cotransfected with a CAT (chloramphenicol acetyltransferase) reporter plasmid, which contains binding sites for the ftz HD upstream of a basal promoter and a plasmid that expresses ftz (see Materials and Methods for details). Ftz-activated reporter expression about 1000-fold above the basal level (activated level shown as 100%). The ability of EFE and the indicated derivatives to repress this activated transcription (by competing with ftz for binding sites in the reporter) was determined by cotransfection of 3 µg of the appropriate expression plasmid, which in each case is the same P-element transformation vector used to construct the corresponding transgenic fly line shown in Fig. 2. The non-repressed level was determined by cotransfection of 3 µg of ‘empty’ parental vector. CAT activity was determined and normalized to the activity of a cotransfected reference gene (see Materials and Methods for details). The graph represents the average and range of at least 2 independent transfections.
To test whether the alterations in region 3 might be affecting the ability of the ftz HD to bind to DNA, we tested several derivatives for their ability to compete for ftz binding sites in cultured cells. As shown in Fig. 6B, neither altering the eh1 domain nor removing region 3 affected the ability to repress transcription by competing for ftz binding sites. Since each of these derivatives also localize to nuclei in embryos (data not shown) and, in addition, retain the ability to partially repress endogenous ftz gene expression (seen more clearly at earlier times after induction than that shown in Fig. 2), it is likely that they retain their DNA-binding activity in vivo as well. Thus eh1 appears to mediate the effector function of EFE, that is, its ability to actively repress the endogenous ftz gene, rather than its targeting function.
eh1 mediates normal en function in the embryo
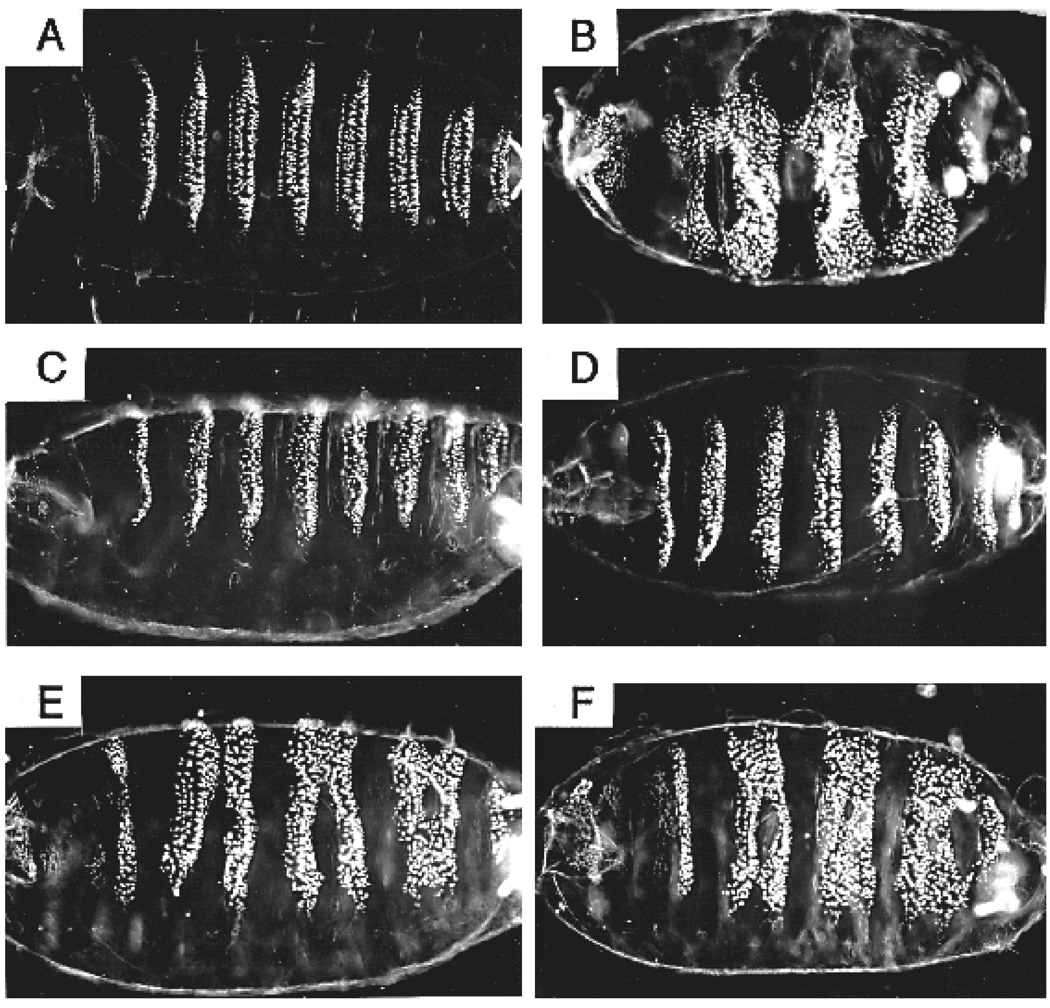
To test whether the eh1 domain is required for the function of wild-type en in vivo, we used a partial rescue assay. Although a cis-acting region capable of fully complementing an en mutant has not been identified, we were able to achieve good partial rescue of the embryonic pattern elements lost in the trunk region of en mutants by combining two previously characterized cis-acting regions. These were a portion of the en upstream region previously shown to direct part of the normal en expression pattern (DiNardo et al., 1988), and the en introns, the larger of which drives a slightly more complete en pattern in the embryo than the upstream fragment alone (Kassis, 1990). A construct containing these cis-elements driving normal en protein expression, in a strong en mutant background (enCXI, Heemskerk et al., 1991), prevents the fusion of ventral denticle bands in the thorax and abdomen that is characteristic of en mutants (Fig. 7C,D versus B; Nüsslein-Volhard and Wieschaus, 1980). This function of en is known to be mediated by the maintenance of wg expression in the cell row adjacent to each en stripe in the early embryo. The rescue construct also restores many, but not all, of the pattern elements within ventral denticle bands that are lost in en mutants. This aspect of pattern is thought to require a relatively late function of en that maintains expression, in en-expressing cells, of the hedgehog signaling molecule (for a review of these interactions, see DiNardo et al., 1994). This partial rescue of late function is consistent with the above described patterns of expression driven by each of the cis-acting elements in our construct, which fade about half way through embryogenesis, while normal en expression persists to very late stages. Thus our construct provides good rescue of early en function and partial rescue of late en function when it is used to express a wild-type en protein.
Fig. 7.
eh1 contributes to normal en function in embryos. The function of eh1 in the context of wild-type en protein was tested using a partial rescue assay. Transgenes expressing either normal en (C,D) or the point-mutated derivative enF→E (E,F) in the normal en domain (see text) were crossed into a strong en mutant (CXI) background. Cuticles were prepared from unhatched eggs 40 hours after egg laying (normal embryos hatch after about 24 hours). Mutant embryos were identified by their characteristic phenotype, which is partially but not completely rescued. (A) A normal cuticle pattern of 3 thoracic and 8 abdominal denticle bands arrayed along the ventral surface. (B) The en mutant has pair-wise fusions of these denticle bands. Several transgenic lines expressing wild-type en showed a similar range of phenotypic rescue (average rescue in two independent lines homozygous for the rescue transgene is shown in C,D), characterized by a restoration of naked cuticle between fused bands, as well as nearnormal denticle patterns within the bands (row 1 denticles are rarely rescued). In contrast, lines expressing enF→E showed only very weak rescue, with the range of phenotypes (E,F) apparently overlapping that seen in mutant-only embryos, based on the relative numbers of the various phenotypes seen in collections from heterozygous rescue lines (e.g. enCXI / CyO; [P(enF→E)] / TM3-Sb, compared with enCXI / CyO; TM3-Sb / D). Each panel is a ventral or ventrolateral view, with anterior to the left.
To test whether the eh1 region is required for this rescue activity, we used the same cis-elements to drive expression of two altered en proteins. The first is enΔeh1, which is deleted for the 15 amino acid core of the homology region, and the second is the point mutated enF→E derivative. Both of these constructs gave similar profiles of partial rescue that were much weaker than that produced by the wild-type rescue construct. Each resulted in restoration of some naked cuticle between ventral denticle bands (Fig. 7E,F and data not shown), but the degree of rescue did not approach that shown by a high percentage of embryos with the wild-type construct (Fig. 7C,D). A comparison of RNA levels expressed by the rescue constructs showed similar levels and patterns of expression for each, with the enΔeh1 line showing a slightly longer persistence of expression than the enF→E and the wild-type rescue lines, which were indistinguishable (data not shown). When we compared the range of phenotypes shown by heterozygous rescue lines (in which about 2/3 of the rescued embryos have a single copy of the rescue construct and 1/3 have two copies) we see little, if any, overlap between the phenotypes of the eh1-mutated and the wild-type constructs. In contrast, the range of phenotypes of en mutant embryos does overlap that of embryos rescued by the eh1-mutated constructs. This suggests that one copy of the wild-type construct provides more rescue activity than two copies of the eh1-mutated construct. Thus, eh1 contributes substantially to the ability of en to carry out its normal embryonic functions.
DISCUSSION
A conserved repression domain
Analysis of en repression function in vivo has shown that one domain is of critical importance. In this assay, en is retargeted in vivo to the endogenous ftz gene, by replacing the en HD with that of ftz, resulting in repression of the ftz gene by the chimera (called EFE). Previous work indicates that targeting activity is provided by the ftz HD, since normal en protein is unable to significantly affect ftz gene expression. EFE appears to repress ftz expression by an active mechanism, as a derivative containing the ftz HD but deleted for en domains 3 and 4 is unable to repress ftz, but is still capable of interacting with a ftz upstream enhancer in a reporter transgene. That EFE is utilizing an active mode of repression in vivo is also indicated by its ability to repress another ftz target gene (en) outside the region of ftz expression (John et al., 1995). The en-derived region that is most crucial to this repression activity (region 3) contains the single conserved domain not closely associated with the HD in the primary sequence. Either deleting the core of this homology region, which was previously noted in all en-class homeoproteins, or mutating the most conserved amino acid, Phe 175, strongly reduces repression activity in vivo, equivalent to deleting all of region 3 (Fig 5, Fig 3).
The repression activity apparently conferred by the eh1 homology region suggested that active repression is a conserved function of the en homeoprotein family and led us to conduct a directed search for similarity to other protein domains in the database. We found that regions homologous to eh1 are present in all known members of several other classes of HD-containing proteins. In each case, the similarity is found upstream of the HD, with a variable-length stretch of non-related amino acids between, typically 80 or more. Representative members of each class are shown in Fig. 4. In each class for which several members are known, a distinct subtype of the eh1 motif is recognizable. For example, within the classes expressed predominantly in mesodermal derivatives, msh and Nk2, a Val is found at the 2nd position after the invariant Phe and, while the Nk2 members have a basic residue at the next position (3rd following the Phe), most other classes, including msh, contain an acidic residue at this position. Such patterns of conservation suggest independently conserved functional divergence among the classes, such as might occur in conjunction with the evolution of a family of interacting proteins, with specific members of this family diverging to interact with specific members of the eh1 family. This view allows that there may be significant divergence of function associated with each distinct partner in the putative interacting family. It may be that as the interacting partner of each eh1 class diverged, the transcriptional activity was modified and perhaps reversed. Such a possibility is underscored by the recent finding that one of the proteins containing eh1 similarity, Nkx2.5, can function as an activator of transcription when its C-terminal region is deleted (Chen and Schwartz, 1995). However, it is likely that the specific transcriptional activity of eh1 is conserved among the en homologs, since replacing the Drosophila eh1 with mouse eh1 restores repression function in Drosophila (Fig. 5). Active repression function may extend at least as far as the goosecoid class, as the Drosophila gsc protein (Goriely et al., 1996) was recently found to be an active repressor in cultured cells (C. Mailhos and C. Desplan, personal communication).
We note that there are possible similarities between eh1 and other conserved motifs, some of which have been proposed previously. For example, eh1 resembles the conserved octapeptide found downstream of some paired domains (reviewed in Noll, 1993; this was noted in Allen et al., 1991, along with the similarity between Hlx, H2.0 and en eh1, which was termed the ‘Hep’ motif). It also is similar to the first helix of the HD (not part of the helix-turn-helix region directly involved in DNA binding), as well as the last helix (helix 6) of the paired domain. However, in our database searches, these other potential homologies were not reliably recognized as significant against a background of apparently random similarities.
While it remains possible that eh1 has an effect on targeting to the ftz gene, we consider this unlikely in light of the following. First, it would be doing so in the context of the ftz HD, which has been implicated in direct targeting to the ftz upstream enhancer without the aid of an eh1-homologous region (that is, in the context of normal ftz protein). The ftz HD seems to be sufficient for such targeting, since EFEΔ34, which does not contain eh1, is still able to repress the activity of that enhancer in vivo, presumably by competing for ftz binding sites (John et al., 1995). Second, en is not targeted to the ftz gene in vivo, since ectopic en expression does not repress ftz (John et al., 1995) and since en mutants do not affect ftz expression (Carroll et al., 1988). Therefore, eh1 would have to be aiding the ftz HD in targeting to the ftz gene in some nonspecific way. Third, eh1 does not affect DNA binding in cultured cells, since derivatives that remove it repress transcription in a passive repression assay to the same extent as the parental EFE, that is when competing for binding sites with the activator, ftz (Fig. 6B). In addition, each of the derivatives altered in region 3 retain the ability to partially repress endogenous ftz expression (Fig 2, Fig 5), suggesting that they still interact with the ftz gene in vivo. This repression activity is seen more clearly immediately after induction of transgene expression, before ftz expression is allowed to recover (our unpublished observations). Taken together, the results indicate that eh1 is providing an effector function in vivo (active repression) in the context of EFE, rather than a targeting function.
Multiple en domains contribute to active repression
In addition to eh1, a conserved region that normally flanks the C terminus of the en HD (and thus flanks the ftz HD in EFE) also contributes to repression activity (Fig. 2). This is interesting in light of the involvement of conserved regions flanking the HDs of HOX gene products in determining their functional specificities in vivo (Mann and Hogness, 1990; Lin and McGinnis, 1992; Zeng et al., 1993). Differences in these regions might affect transcriptional activity, leading to different activities on common target genes, rather than, or in addition to, providing selective targeting to distinct sets of target genes.
The requirements for repression by EFE in vivo may be somewhat different from those in cultured cells, where region 3 was not found to contribute significantly to repression activity (although it appeared to confer an extremely weak activity when fused to regions 5 and 6; Han and Manley, 1993). However, region 4, which contains the single strong repression domain previously localized in en (Han and Manley, 1993), also appears to contribute to repression activity in vivo. Removing this region in addition to region 3 clearly reduces repression activity more than removing region 3 alone (Fig. 2; John et al., 1995). Removing region 4 alone results in a more stable protein (Fig. 3), possibly masking a reduction in potency, as EFE?4 represses ftz more completely than the parental EFE. The increased stability of EFEΔ4 relative to EFE, while it is only about two-fold, may well account for the increased repression activity, since effective repression of ftz probably involves the destabilization of a positive feedback loop. This is suggested by the all-or-none decision that each cell makes following EFE induction with regard to ftz expression, as seen in the discrete pattern of expressing and non-expressing cells in the embryos (Fig. 2). Thus, it may be necessary to keep ftz levels below a threshold for a period of time in order to disengage the feedback loop and a two-fold increase in protein stability may therefore have a strong impact on the number of cells that lose ftz expression. Since region 3 is clearly crucial for effective repression in vivo, it is likely that in vivo repression requires a function in addition to that identified in the cell culture transfection studies. Further analyses will be required to determine whether this additional activity is mechanistically distinct from that operating in the transfection assays.
The conserved region that flanks the N terminus of the HD has an interesting effect in vivo. Without increasing the stability of the protein, deleting this region (Δ5) actually increases repression activity (Fig. 2). As this region does not appear to provide either activation or anti-repression function in cell culture assays (our unpublished observation), it may have an effect on targeting in vivo. Perhaps region 5 mediates an interaction normally involved in targeting by en and this targeting function interferes with that of the ftz HD. Then removing it might allow increased ftz repression by EFE.
The involvement of multiple domains in repression by en, the lack of apparent activation activity in any of our derivatives (our unpublished observations; however, Han and Manley, 1993, found that an en derivative containing regions 2, 5 and 6 could weakly activate in culture) and the involvement of highly conserved motifs in repression activity in vivo, taken together, suggest that repression may be the primary effector function of en. Genetically, en is a repressor of several genes in Drosophila, including even-skipped (Harding et al., 1986; John et al., 1995), cubitus interruptus (Eaton and Kornberg, 1990; Schwartz et al., 1995), wingless (Heemskerk et al., 1991), patched (Hooper et al., 1989), Ultrabithorax (Carroll et al., 1988) and decapentaplegic (Sanicola et al., 1995). However, the idea that en might be a dedicated repressor in vivo conflicts, superficially, with results from ectopic expression assays in embryos, in which en has been shown to induce expression of its own gene (Heemskerk et al., 1991), as well as with the positive regulatory effects of en on hedgehog (Tabata et al., 1992) and polyhomeotic (Serrano et al., 1995). That these latter interactions might be indirect, through repression of a repressor, is suggested by our results. However, it remains possible that protein-protein interactions allow en to have a net positive regulatory effect on some direct target genes. It is worthy of note in this context that a similar positive autoregulatory effect of even-skipped, a repressor in both cell culture assays (Jaynes and O’Farrell, 1988; Han et al., 1989) and in vitro (Biggin and Tjian, 1989), has recently been explained by indirect effects in vivo, involving repression of other repressors (Fujioka et al., 1995).
eh1 is required for normal en function in embryos
To test whether the conservation of eh1 in en is related to known functions in patterning the Drosophila embryo, we used an in vivo assay for function of the normal en protein. Since a cis-regulatory region sufficient to fully complement an en mutant has not been identified, we combined two known regulatory regions to generate early embryonic expression, sufficient to complement the major pattern defects of en mutants in the trunk region. This is the place and time in development where en function has been most extensively studied and where en is known to affect its own expression and expression of other downstream genes, including hedgehog (in en-expressing cells) and wingless (in the adjacent cell row), which encode signaling molecules important in the patterning processes that are disrupted in en mutants (reviewed in DiNardo et al., 1994). When wild-type en protein is expressed in strong en mutants using these cis-elements, it rescues the severe defects in the trunk region, as indicated by the reappearance of evenly spaced denticle bands on the ventral surface of the larval cuticle (Fig. 7). However, when either the enΔeh1 deletion mutant or the single point mutant enF→E is similarly expressed, very weak rescue is obtained, indicating the importance of eh1 in the normal function of en during embryogenesis. The correlation of eh1 activity in the context of EFE, where it is required for strong repression activity in vivo, to its function in normal patterning processes indicates that active repression is required for the normal functioning of en in the embryo.
Acknowledgments
Many thanks to Miki Fujioka for helpful advice and valuable assistance, to Tadaatsu Goto, Alexander Mazo and Claude Desplan for helpful discussions, to Alexander Mazo for help in database searches to identify eh1 homologs, to C. Desplan and C. Mailhos for sharing information prior to publication, to Charles Girdham and Pat O’Farrell for polyclonal anti-en antiserum, to Steve DiNardo, T. Goto, M. Fujioka, Robert Finkelstein and A. Mazo for comments on the manuscript, and to Aleyamma John for her excellent technical assistance. This work was supported by NIH grant R01-GM50231-01A2.
Footnotes
Note added in proof
Consistent with our interpretation of Δ5, which deletes essentially the eh2 homology, as removing a targeting function that interferes with targeting by the ftz HD in EFE, L. T. C. Peltenburg and C. Murre (1996, EMBO J. 13, 3385–3393) have recently shown that the en eh2 region mediates interaction with extradenticle, another HD protein.
REFERENCES
- Allen JD, Lints T, Jenkins NA, Copeland NG, Strasser A, Harvey RP, Adams JM. Novel murine homeo box gene on chromosome 1 expressed in specific hematopoietic lineages and during embryogenesis. Genes Dev. 1991;5:509–520. doi: 10.1101/gad.5.4.509. [DOI] [PubMed] [Google Scholar]
- Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122:205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- Biggin MD, Tjian R. A purified Drosophila homeodomain protein represses transcription in vitro. Cell. 1989;58:433–440. doi: 10.1016/0092-8674(89)90424-8. [DOI] [PubMed] [Google Scholar]
- Carroll SB, DiNardo S, O’Farrell PH, White RA, Scott MP. Temporal and spatial relationships between segmentation and homeotic gene expression in Drosophila embryos: distributions of the fushi tarazu, engrailed, Sex combs reduced, Antennapedia, and Ultrabithorax proteins. Genes Dev. 1988;2:350–360. doi: 10.1101/gad.2.3.350. [DOI] [PubMed] [Google Scholar]
- Chen CY, Schwartz RJ. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J. Biol. Chem. 1995;270:15628–15633. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- Desplan C, Theis J, O’Farrell PH. The sequence specificity of homeodomain-DNA interaction. Cell. 1988;54:1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S, Heemskerk J, Dougan S, O’Farrell PH. The making of a maggot: patterning the Drosophila embryonic epidermis. Curr. Opin. Genet. Dev. 1994;4:529–534. doi: 10.1016/0959-437x(94)90068-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S, Sher E, Heemskerk JJ, Kassis JA, O’Farrell PH. Two-tiered regulation of spatially patterned engrailed gene expression during Drosophila embryogenesis. Nature. 1988;332:604–609. doi: 10.1038/332604a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Viglianti GA, Rutledge BJ, Meselson M. Alteration of hsp82 gene expression by the gypsy transposon and suppressor genes in Drosophila melanogaster. Genes Dev. 1989;3:454–468. doi: 10.1101/gad.3.4.454. [DOI] [PubMed] [Google Scholar]
- Duboule D, editor. Guidebook to the Homeobox Genes. New York: Oxford University Press; 1994. [Google Scholar]
- Eaton S, Kornberg TB. Repression of ci-D in posterior compartments of Drosophila by engrailed. Genes Dev. 1990;4:1068–1077. doi: 10.1101/gad.4.6.1068. [DOI] [PubMed] [Google Scholar]
- Edgar B, O’Farrell PH. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell. 1990;62:469–480. doi: 10.1016/0092-8674(90)90012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker M, Wegner J, Akimenko MA, Westerfield M. Coordinate embryonic expression of three zebrafish engrailed genes. Development. 1992;116:1001–1010. doi: 10.1242/dev.116.4.1001. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Jaynes JB, Goto T. Early even-skipped stripes act as morphogenetic gradients at the single cell level to establish engrailed expression. Development. 1995;121:4371–4382. doi: 10.1242/dev.121.12.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriely A, Stella M, Coffinier C, Kessler D, Mailhos C, Dessain S, Desplan C. A functional homologue of goosecoid in Drosophila. Development. 1996;122:1641–1650. doi: 10.1242/dev.122.5.1641. [DOI] [PubMed] [Google Scholar]
- Gray S, Szymanski P, Levine M. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev. 1994;8:1829–1838. doi: 10.1101/gad.8.15.1829. [DOI] [PubMed] [Google Scholar]
- Gray S, Cai H, Barolo S, Levine M. Transcriptional repression in the Drosophila embryo. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1995;349:257–262. doi: 10.1098/rstb.1995.0111. [DOI] [PubMed] [Google Scholar]
- Han K, Levine MS, Manley JL. Synergistic activation and repression by Drosophila homeobox proteins. Cell. 1989;56:573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- Han K, Manley JL. Functional domains of the Drosophila Engrailed protein. EMBO J. 1993;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding K, Rushlow C, Doyle HJ, Hoey T, Levine M. Cross-regulatory interactions among pair-rule genes in Drosophila. Science. 1986;233:953–959. doi: 10.1126/science.3755551. [DOI] [PubMed] [Google Scholar]
- Heemskerk J, DiNardo S, Kostriken R, O’Farrell PH. Multiple modes of engrailed regulation in the progression towards cell fate determination. Nature. 1991;352:404–410. doi: 10.1038/352404a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper JE, Scott MP. The Drosophila patched gene encodes a putative membrane protein required for segmental patterning. Cell. 1989;59:751–765. doi: 10.1016/0092-8674(89)90021-4. [DOI] [PubMed] [Google Scholar]
- Jaynes JB, O’Farrell PH. Activation and repression of transcription by homoeodomain-containing proteins that bind a common site. Nature. 1988;336:744–749. doi: 10.1038/336744a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaynes JB, Vincent J, O’Farrell PH. Drosophila Homeodomain-containing Proteins Regulate Transcription. In: Mahowald AP, editor. Genetics of Pattern Formation and Growth Control. New York: Wiley-Liss Inc; 1990. pp. 47–64. [Google Scholar]
- Jaynes JB, O’Farrell PH. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 1991;10:1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John A, Smith ST, Jaynes JB. Inserting the Ftz homeodomain into Engrailed creates a dominant transcriptional repressor that specifically turns off Ftz target genes in vivo. Development. 1995;121:1801–1813. doi: 10.1242/dev.121.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis JA. Spatial and temporal control elements of the Drosophila engrailed gene. Genes Dev. 1990;4:433–443. doi: 10.1101/gad.4.3.433. [DOI] [PubMed] [Google Scholar]
- Kuner JM, Nakanishi M, Ali Z, Drees B, Gustavson E, Theis J, Kauvar L, Kornberg T, O’Farrell PH. Molecular cloning of engrailed: a gene involved in the development of pattern in Drosophila melanogaster. Cell. 1985;42:309–316. doi: 10.1016/s0092-8674(85)80126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, McGinnis W. Mapping functional specificity in the Dfd and Ubx homeo domains. Genes Dev. 1992;6:1071–1081. doi: 10.1101/gad.6.6.1071. [DOI] [PubMed] [Google Scholar]
- Logan C, Hanks MC, Noble-Topham S, Nallainathan D, Provart NJ, Joyner AL. Cloning and sequence comparison of the mouse, human, and chicken engrailed genes reveal potential functional domains and regulatory regions. Devel. Genetics. 1992;13:345–358. doi: 10.1002/dvg.1020130505. [DOI] [PubMed] [Google Scholar]
- Mann RS, Hogness DS. Functional dissection of Ultrabithorax proteins in D. melanogaster. Cell. 1990;60:597–610. doi: 10.1016/0092-8674(90)90663-y. [DOI] [PubMed] [Google Scholar]
- Manoukian AS, Krause HM. Concentration-dependent activities of the even-skipped protein in Drosophila embryos. Genes Dev. 1992;6:1740–1751. doi: 10.1101/gad.6.9.1740. [DOI] [PubMed] [Google Scholar]
- Noll M. Evolution and role of Pax genes. Curr. Opin. Genet. Dev. 1993;3:595–605. doi: 10.1016/0959-437x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Sanicola M, Sekelsky J, Elson S, Gelbart WM. Drawing a stripe in Drosophila imaginal disks: negative regulation of decapentaplegic and patched expression by engrailed. Genetics. 1995;139:745–756. doi: 10.1093/genetics/139.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF, Gehring WJ. Direct homeodomain-DNA interaction in the autoregulation of the fushi tarazu gene. Nature. 1992;356:804–807. doi: 10.1038/356804a0. [DOI] [PubMed] [Google Scholar]
- Schwartz C, Locke J, Nishida C, Kornberg TB. Analysis of cubitus interruptus regulation in Drosophila embryos and imaginal disks. Development. 1995;121:1625–1635. doi: 10.1242/dev.121.6.1625. [DOI] [PubMed] [Google Scholar]
- Serrano N, Brock HW, Demeret C, Dura JM, Randsholt NB, Kornberg TB, Maschat F. polyhomeotic appears to be a target of engrailed regulation in Drosophila. Development. 1995;121:1691–1703. doi: 10.1242/dev.121.6.1691. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:341–347. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Tabata T, Eaton S, Kornberg TB. The Drosophila hedgehog gene is expressed specifically in posterior compartment cells and is a target of engrailed regulation. Genes Dev. 1992;6:2635–2645. doi: 10.1101/gad.6.12b.2635. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C. Looking at embryos. In: Roberts DB, editor. Drosophila, A practical approach. Oxford, England: IRL Press; 1986. pp. 199–228. [Google Scholar]
- Winslow GM, Hayashi S, Krasnow M, Hogness DS, Scott MP. Transcriptional activation by the Antennapedia and fushi tarazu proteins in cultured Drosophila cells. Cell. 1989;57:1017–1030. doi: 10.1016/0092-8674(89)90340-1. [DOI] [PubMed] [Google Scholar]
- Zeng W, Andrew DJ, Mathies LD, Horner MA, Scott MP. Ectopic expression and function of the Antp and Scr homeotic genes: the N terminus of the homeodomain is critical to functional specificity. Development. 1993;118:339–352. doi: 10.1242/dev.118.2.339. [DOI] [PubMed] [Google Scholar]