Abstract
Alterations in DNA methylation (DNAm) in cancer have been known for 25 years, including hypomethylation of oncogenes and hypermethylation of tumor suppressor genes1. However, most studies of cancer methylation have assumed that functionally important DNAm will occur in promoters, and that most DNAm changes in cancer occur in CpG islands2,3. Here we show that most methylation alterations in colon cancer occur not in promoters, and also not in CpG islands but in sequences up to 2 kb distant which we term “CpG island shores.” CpG island shore methylation was strongly related to gene expression, and it was highly conserved in mouse, discriminating tissue types regardless of species of origin. There was a surprising overlap (45-65%) of the location of colon cancer-related methylation changes with those that distinguished normal tissues, with hypermethylation enriched closer to the associated CpG islands, and hypomethylation enriched further from the associated CpG island and resembling non-colon normal tissues. Thus, methylation changes in cancer are at sites that vary normally in tissue differentiation, and they are consistent with the epigenetic progenitor model of cancer4, that epigenetic alterations affecting tissue-specific differentiation are the predominant mechanism by which epigenetic changes cause cancer.
These experiments focused on three major questions. First, taking a comprehensive genome-wide approach to DNAm, where are DNA methylation changes that distinguish tissue types? For this purpose, we examined three normal tissue types representing the three embryonic lineages, liver (endodermal), spleen (mesodermal) and brain (ectodermal), obtained from 5 autopsies. A difference from previous methylation studies of tissues, besides the genome-wide design, is that here they were obtained from the same individual, thus controlling for potential inter-individual variability. Second, where are DNAm alterations in cancer, and what is the balance between hypomethylation and hypermethylation? For this purpose, we examined 13 colorectal cancers and matched normal mucosa from the patients. Third, what is the functional role of these methylation changes? For this purpose, we performed a comparative epigenomics study of tissue methylation in the mouse, as well as gene expression analyses.
To examine DNAm genome-wide, we performed comprehensive high-throughput array-based relative methylation (CHARM) analysis, which is a microarray-based method agnostic to preconceptions about DNAm, including location relative to genes and CpG content5. The resulting quantitative measurements of DNAm, denoted with M, are log ratios of intensities from total (Cy3) and McrBC-fractionated DNA (Cy5): positive and negative M values are quantitatively associated with methylated and unmethylated sites, respectively. For each sample we analyzed ∼4,6 milion CpG sites across the genome using a custom designed NimbleGen HD2 microarray, including all of the classically defined CpG islands as well as all non-repetitive lower CpG density genomic region of the genome. We included 4,500 control probes to standardize these M values so that unmethylated regions are associated, on average, with values of 0. CHARM is 100% specific at 90% sensitivity for known methylation marks identified by other methods (e.g., in promoters), while including the approximately half of the genome not identified by conventional region pre-selection5. The CHARM results were also extensively corroborated by quantitative bisulfite pyrosequencing analysis.
Results
Most tissue-specific differential methylation in normal tissues occurs not in CpG islands but in “CpG island shores” usually outside of promoters
Because CHARM is not biased for CpG island or promoter sequences, we could obtain objective data on tissue-specific methylation. We identified 16,379 tissue differential methylation regions (T-DMRs), defined as M values for one tissue consistently different than the others at a false discovery rate (FDR) of 5% (see Methods). The median size of a T-DMR was 255 bp. Previous studies of tissue- or cancer-specific DNAm have focused on promoters and/or CpG islands, which have been defined as regions with the fraction of C and G > 0.5, and the observed to expected ratio of CpG > 0.6 2,6. It has been previously reported that differences in DNAm of promoters in somatic cells is relatively low in conventionally defined CpG islands and higher at promoters with intermediate CpG density7,8. However, two recent studies identified a relatively small fraction, 4-8%, of CpG islands with tissue-specific methylation 9,10. We also found that DNAm variation is uncommon in CpG islands (Supplementary Fig. 1 online).
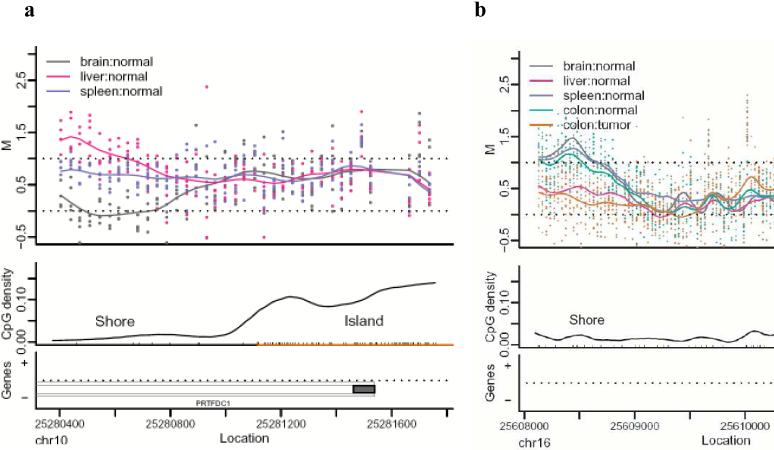
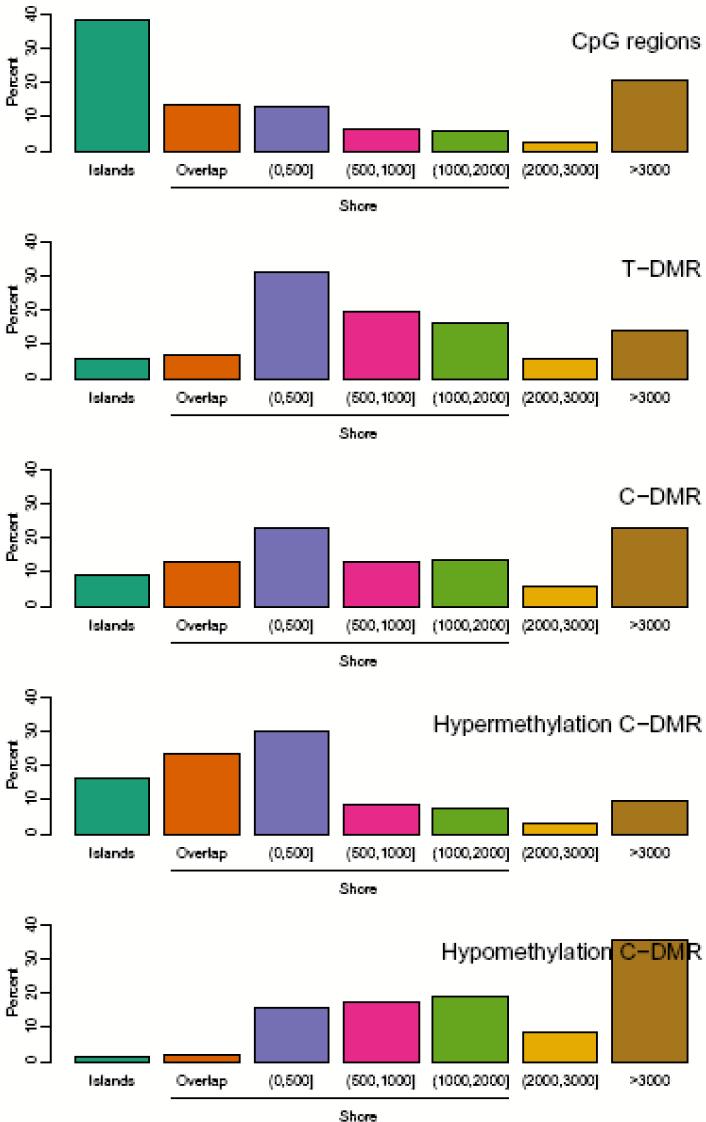
The genome-wide approach of CHARM also enabled us to discover a surprising physical relationship between CpG islands and DNAm variation, namely that 76% of T-DMRs were located within 2 kb of islands in regions we now denote as “CpG island shores.” An example is shown in Fig. 1a, representing a T-DMR in the PRTFDC1 gene, a brain-specific phosphoribosyltransferase which is relatively hypomethylated in the brain. Note that the spreading of M values among the tissues begins ∼200 bp from the CpG island and at a point where the CpG density associated with the island has fallen to 1/10 the level in the island itself (Fig. 1a). Thus the association of T-DMRs with CpG island shores is not due to an arbitrary definition of CpG islands but to a true association of these DNAm differences near but not in the regions of dense CpG content. Supplementary Data 1 describes all T-DMR regions, and Supplementary Fig. 2 provides plots like Fig. 1a for the top 50 T-DMRs ordered by statistical significance, with the complete set provided at http://rafalab.jhsph.edu/t-dmr3000.pdf. The distribution of T-DMRs by distance from the respective islands shows that DNAm variation is distributed over a ∼2 kb shore, and that while CpG islands are enriched on the arrays, because of their high CpG content (33% of CHARM probes are in islands), only 6% of T-DMRs are in islands, compared to 76% in shores; an additional 18 % of T-DMRs were located > 2 kb from the respective islands (Fig. 2). Not surprisingly, the localization of T-DMRs also occurred largely outside of promoters (96%), since CpG islands are localized largely within promoters14. Furthermore, more than half (52%) of T-DMRs were greater than 2 kb from the nearest annotated gene. The distribution of the distance to islands remained essentially unchanged using FDR cutoffs of 0.01, 0.05, 0.10 (Supplementary Fig. 3).
Figure 1.
Most tissue-specific differential DNA methylation is located at CpG island shores. (a) An example of a T-DMR located at a CpG island shore in the PRTFDC1 gene. The upper panel is a plot of M value versus genomic location for brain (grey), liver (pink), and spleen (purple). Each point represents the methylation level of an individual sample for a given probe. The curve represents averaged smoothed M values, described in detail in the Methods. Due to the scale and standardization used, M values which range from −0.5 to 0.5 represent unmethylated sites as defined by the control probes, and values from 0.5 to 1.5 represent baseline levels of methylation. The middle panel provides the location of CpG dinucleotides with black tick marks on the x-axis. CpG density was calculated across the region using a standard density estimator and is represented by the smoothed black line. The location of the CpG island is denoted on the x-axis as an orange line. The lower panel provides gene annotation for the genomic region. The thin outer grey line represents the transcript, the thin inner lines represent a coding region. Filled in grey boxes represent exons. On the y-axis, plus and minus marks denote sense and antisense gene transcription respectively. (b) An example of a C-DMR that is located in a CpG island shore and overlaps a T-DMR. Brain (grey) is hypomethylated relative to liver (pink) and spleen (purple) tissues. Hypomethylation of colon tumor (orange) is observed in comparison to matched normal colon tissue (green) and overlaps the region of brain hypomethylation.
Figure 2.
Distribution of distance of T-DMRs and C-DMRs from CpG islands. Islands (teal) are regions which cover or overlap more than 50% of a CpG island. Overlap (orange) are regions which overlap 0.1-50% of a CpG island. (0,500] (purple) are regions which do not overlap islands but are located ≤ 500 bp of islands. (500,1000] (magenta) are regions located > 500 and ≤ 1000 bp from an island. (1000,2000] (green) are regions > 1000 bp and ≤ 2000 bp from an island. (2000,3000] (yellow) are regions located > 2000 bp and ≤ 3000 bp from an island. Greater than 3000 (brown) are regions > 3000 bp from an island. The percentage of each class is provided for CpG regions (the CHARM arrays themselves, null hypothesis), tissue-specific differentially methylated regions (T-DMRs), cancer-specific differentially methylated regions (C-DMRs), and the latter subdivided into regions of cancer-specific hypermethylation and hypomethylation.
We confirmed the array-based result that the differential methylation was in CpG island shores rather than in the associated islands by performing bisulfite pyrosequencing analysis on over 100 CpG sites in the islands and shores associated with four genes, three T-DMRs and one C-DMR. At all 101 sites, the DMR was confirmed to lie within the shore rather than the island (Supplementary Table 1 online). For example, PCDH9, which encodes a brain-specific protocadherin, was relatively hypomethylated in the brain at all 6 sites examined in the CpG island shore, but unmethylated in both brain and spleen at all 18 sites examined in the associated island. (Supplementary Table 1 online). Differential methylation of an additional 4 CpG island shores was also confirmed by bisulfite pyrosequencing of 39 total CpG sites and all showed statistically significant differences in DNAm (P < 0.05) (Supplementary Table 2). These data verify the sensitivity of CHARM for detecting subtle differences in DNAm. Furthermore, they confirm that most normal differential methylation takes place at CpG island shores.
Most cancer-specific single-copy differential methylation is at CpG island shores, with similar degrees of hypomethylation and hypermethylation
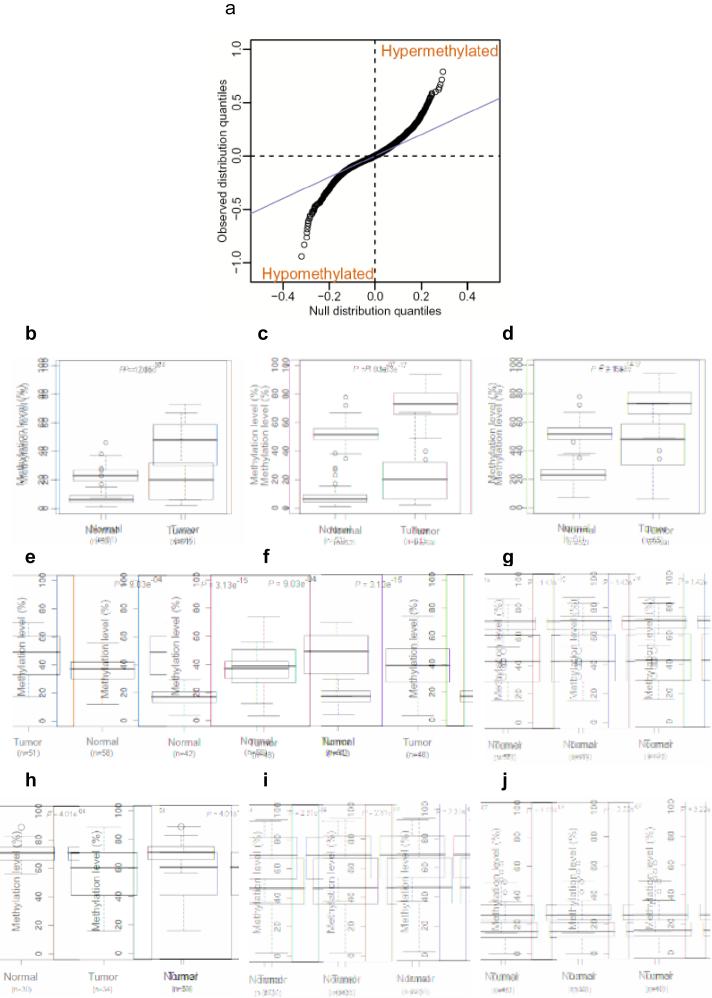
We used the same comprehensive genome-wide approach to address cancer-specific DNA methylation, focusing on colorectal cancer, a paradigm for cancer epigenetics because of the availability of matched normal mucosa from the same patients, the cell type from which the tumors arise. We analyzed DNAm on 13 colon cancers and matched normal mucosa from the same patients, identifying 2,707 regions showing differential methylation in cancers (C-DMRs) with an FDR of 5% (Supplemetary Data 2 and Supplementary Fig.4 online, and the complete set provided at http://rafalab.jhsph.edu/c-dmr-all.pdf). These C-DMRs were similarly divided between those showing hypomethylation in the cancer (compared to the normal colon) and those showing hypermethylation, 1199 (44%) and 1508 (56%), respectively. Note that the CHARM arrays, like other tiling arrays, do not contain repetitive sequences, so the abundance of hypomethylation is not due to enrichment for repetitive DNA, which has been shown to be hypomethylated in cancer11. This similarity in amount of hypomethylation and hypermethylation is also shown dramatically in a QQ plot, in which quantiles for the observed average difference between tumor and normal sample Ms are plotted against quantiles from a null distribution constructed with the control (M = 0) regions (Fig. 3a).
Figure 3.
Similar numbers of sites of hypomethylation and hypermethylation in colon cancer. (a) A quantile-quantile plot shows a similar number of sites of hypomethylation and hypermethylation in colon cancer. The quantiles of the differences in M values between tumor and normal colon tissues are plotted against the quantiles of a null distribution formed using the differences seen in the control regions. Points deviating from the diagonal are not expected by chance, and a similar proportion is seen for hypomethylation and hypermethylation in cancer. (b-j) Bisulfite pyrosequencing confirms the prevalence of 5 hypermethylated and 4 hypomethylated C-DMR shores in a large set of colon tumor and normal mucosa samples. Box-plots represent DNA methylation level measured using bisulfite pyrosequencing. (b), distal-less homeobox 5 (DLX5); (c), leucine rich repeat and fibronectin type III domain containing 5 (LRFN5); (d), homeobox A3 (HOXA3); (e), SLIT and NTRKlike family, member 1 (SLITRK1); (f), FEZ family zinc finger 2 (FEZF2), (g), transmembrane protein 14A (TMEM14A); (h), glutamate-rich 1 (ERICH1); (i), family with sequence similarity 70, member B (FAM70C); (j), prostate transmembrane protein, androgen induced 1 (TMEPAI), (n) equals the number of samples analyzed by pyrosequencing.
While both hypomethylation and hypermethylation in cancer involved CpG island shores, there were subtle differences in the precise regions that were altered. Thus, the hypermethylation often extended to include portions of the associated CpG islands in 11% of cases (Fig. 2), which could account for the island hypermethylation frequently reported in cancer, even though that is not the predominant site of modification. In contrast, the hypomethylation often extended away from the associated CpG islands in 34% of cases (Fig. 2).
To confirm differential methylation in colon tumors, we performed additional bisulfite pyrosequencing validation of 9 C-DMRs, including 5 regions exhibiting hypermethylation and 4 regions with hypomethylation, in an average of 50 primary cancer and normal mucosal samples per gene. For all of the genes, the pyrosequencing data matched the CHARM data (P values ranging from 10-4 to 10-17) (Fig. 3b-j and Supplementary Table 3). Thus, CHARM was precise in identifying both T-DMRs and C-DMRs.
Our screening process was effective at identifying known targets of altered DNAm in cancer. For example, 10 of the 25 most statistically significant C-DMRs have previously been reported to show altered DNAm in cancer, e.g. WNK2, hypermethylated in glioblastoma12 and HOXA6, hypermethylated in lymphoid malignancies13. However, we identified hundreds of genes not previously described as well. For example, for hypermethylation, GATA-2 is an important regulator of hematopoetic differentiation14, and RARRES2 expression is decreased in intestinal adenomas15. For hypomethylation, the results were also striking, e.g. DPP6 is a biomarker for melanoma16, MRPL36 is a DNA helicase that confers susceptibility to breast cancer17, and MEST is a known target of hypomethylation and loss of imprinting in breast cancer18. Note also that while previous T-DMR screens have been focused on CpG islands, which we show account for only 8% of T-DMRs, our screen did identify CpG island loci validated by others as well, e.g. PAX6, OSR1, and HOXC12. Thus, cancer, like normal tissues, involves changes in DNAm in CpG island shores, with comparable amounts of hypomethylation and hypermethylation, but with subtle differences in the precise distribution of these alterations with respect to the associated CpG island. These differences will have important functional implications for gene expression, as discussed later.
Gene expression is linked to non-CpG island methylation, even up to 2 kb from transcriptional start sites
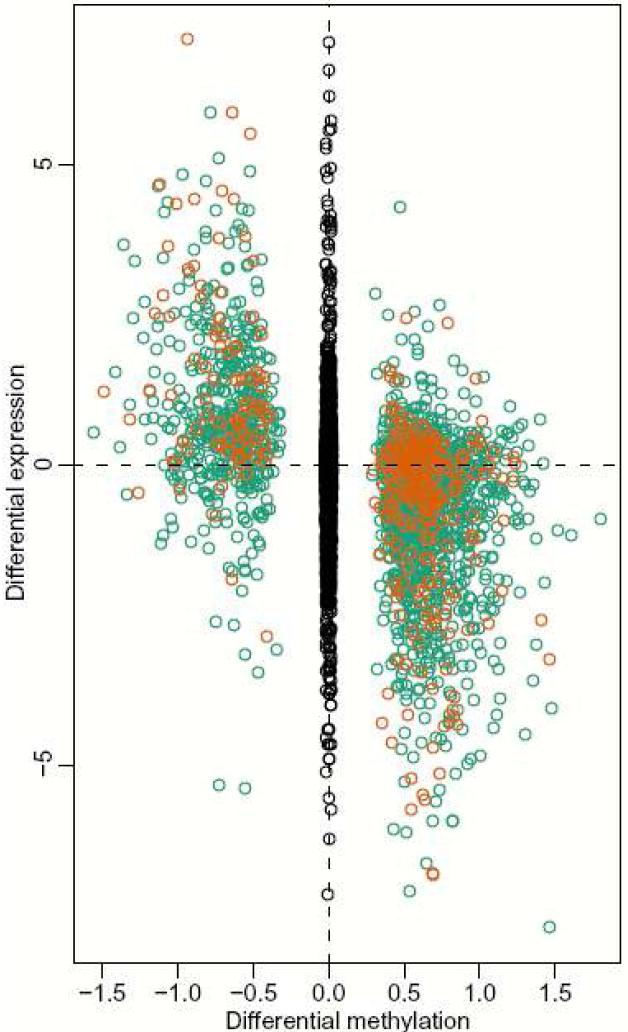
Because of the unexpected discovery of CpG island shores, it was important to explore the functional relationship between their differential methylation and expression of associated genes. To address tissue-specific and cancer-specific DNAm, we analyzed gene expression across the genome in 5 primary brains and livers from the same autopsy specimens in the very cases on which we had genome-wide methylation data, and performed a similar analysis of 4 colon cancers and matched normal mucosa from the same patients, again on those from which we had performed genome-wide methylation analysis. Methylation of T-DMRs showed a strong inverse relationship with differential gene expression, even though these DMRs are not CpG islands but are CpG island shores. The relationship between DNAm and gene expression was greater for DMRs in which one of the two measured points was ∼0 methylation (“some-to-none,” compared to “some” methylation to “more” or “less” methylation), particularly for hypomethylation (Fig. 4). The significant association of gene expression with T-DMRs was true even when the DMR was 300-2000 bp from the TSS, e.g. 0.84 and 0.35 (P < 10-37 and 10-4), for “some-none” and “some-more/less” methylation, respectively, comparing liver to brain (Fig. 4). Moreover, when we related T-DMRs to changes in gene expression from over 242 samples, representing 20 different tissue types, we found 5,352 of the 8,910 genes that were differentially expressed across the 20 tissues were within 2 kb of a T-DMR, much more than expected by chance (P < 10-15). For C-DMRs, as well, even though there were fewer of them than T-DMRs, there was a significant association of gene expression with DNAm, P < 10-6 and 10-3 for hypermethylation and hypomethylation, respectively, again much more strikingly when one of the two measured points had no methylation (Supplementary Fig. 5 online).
Figure 4.
Gene expression is strongly correlated with T-DMRs at CpG island shores. For each brain versus liver T-DMR we found the closest annotated gene on the Affymetrix HGU133A microarray, resulting in a total of 2,041 gene/T-DMR pairs. Plotted are log (base 2) ratios of liver to brain expression against delta M values for liver and brain DNAm. Orange dots represent T-DMRs located within 300 bp from the corresponding gene's transcriptional start site (TSS). Green dots represent T-DMRs that are located from 300-2000 bp from the TSS of an annotated gene. Black dots, in the middle, represent log ratios for all genes further than 2 kb from an annotated TSS.
We validated the inverse relationship between DNAm and transcription at 8 CpG island shores, 2 T-DMRs and 6 C-DMRs in tissues and colon cancers, respectively, using quantitative real-time PCR. Both of the T-DMRs were in shores, 1 located 844 bp upstream of the promoter and 1 within the gene body. Similarly, all six of the C-DMRs assayed were in shores, with 5 located in the gene promoter and 1 within the gene body (Supplementary Table 4 online). These quantitative data provide additional support for a strong relationship between differential methylation in CpG island shores and transcription of associated genes. Note that this functional relationship between gene expression and shore methylation applies to shores located within 2 kb of an annotated transcriptional start site, but leaves open the possibility of additional regulatory function for shores located in intragenic regions or gene deserts.
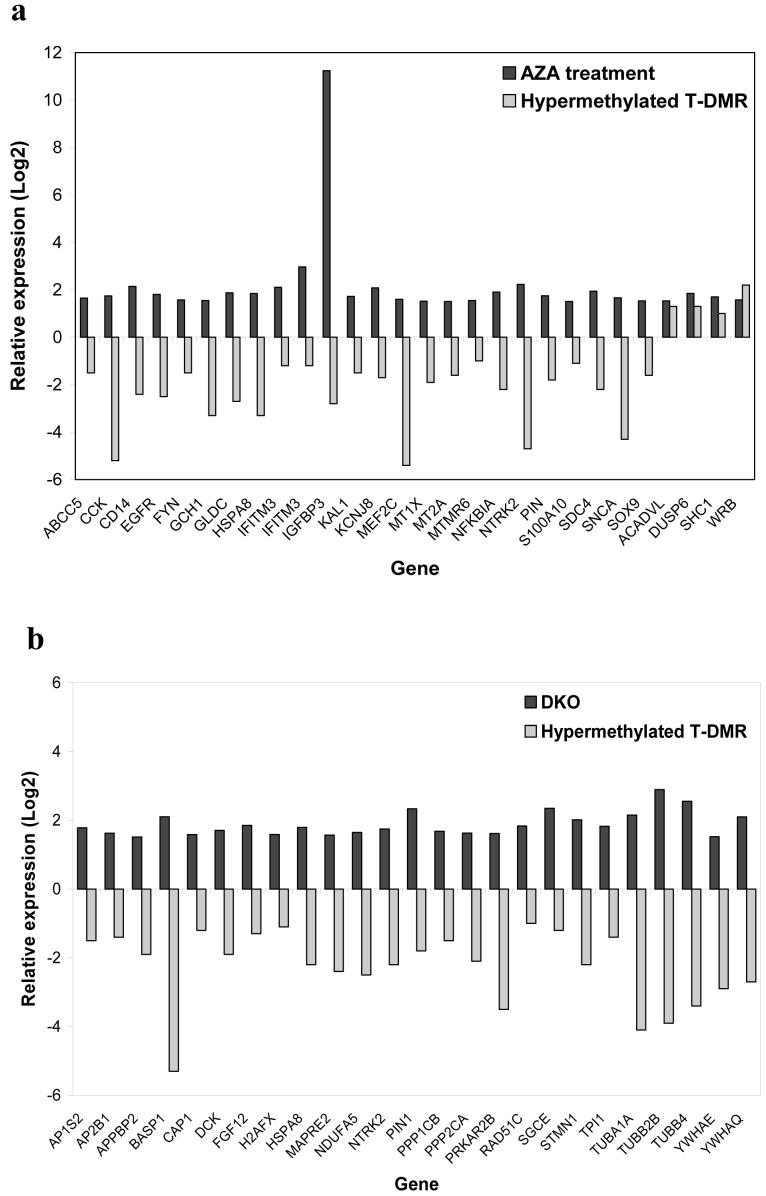
Most genes downregulated in association with shore hypermethylation are also activated by 5-aza-2′-deoxycytidine and DNA methyltransferase knockout
The previous data, while compelling, are associative in nature. For a more functional analysis, we thereforecompared DNA methylation and gene expression data from tissues studied in the current work, to a rigorous analysis using hundreds of expression microarray experiments published earlier19, which tested the effects on gene expression of 5-aza-2′-deoxycytidine (AZA) and also double DNA methyltransferase 1 and 3B somatic cell knockout (DKO). We compared genes from the present study with DMRs meeting an FDR < 0.05, and also showing differential expression in the relatively hypomethylated tissues at P < 0.05, to genes showing significant P values after AZA or DKO. Of 28 DMRs that show relative hypermethylation with gene silencing in tissues, 24 were activated by AZA (Figure 5a and Supplementary Data 3 online). Similarly, of 25 DMRs that showed relative hypermethylation with gene silencing in tissues, all 25 were activated by DKO (Figure 5b and Supplementary Data 3 online). Thus both chemical and genetic demethylation cause changes in gene expression similar to those associated with increased methylation of CpG island shores.
Figure 5.
Genes downregulated in association with T-DMR shore hypermethylation are activated by 5-aza-2′-deoxycytidine treatment of colon cancer cell line HCT116 and knockout of DNA methyltrasferase 1 and 3b in HCT116. (a) Genes significantly upregulated (p<0.05) after treatment of HCT116 cells with 5-aza-2′-deoxycytidine (AZA) (black) that are also associated with a relatively hypermethylated T-DMR showing a significant change in gene expression (p<0.05) (grey). 24/28 genes are activated by AZA. (b) Genes significantly upregulated (p<0.05) after knockout of DNA methyltransferases 1 and 3b (DKO) in HCT116 cells (black) that are also associated with a relatively hypermethylated T-DMRs showing a significang change in gene expression (p<0.05) (grey). 25/25 genes are activated by DKO. Plotted are log (base 2) ratios of expression of AZA/untreated, DKO/HCT116, and relatively hypermethylated/hypomethylated tissue.
Differential methylation in normal tissues and in cancer is associated with alternative transcription
What might be the function of differential methylation at CpG island shores? An intriguing possibility is alternative transcription. Intriguingly, both the T-DMRs and C-DMRs often involved alternative transcripts, defined by cap analysis gene expression (CAGE)20,21: 68% and 70% of the T-DMRs and C-DMRs respectively were not within 500 bp of an annotated transcriptional start site but were within 500 bp of an alternative transcriptional start site. By chance we expect only 58% to have this relationship (P < 10-15). These results suggest that DNA methylation might regulate alternative transcription in normal differentiation and cancer. We therefore performed 5-rapid amplification of cDNA end (RACE) experiments, in order to confirm the presence of alternative transcripts and their differential expression in cancer. We examined 3 colon tumor and matched normal mucosa at the PIP5K1A locus, a C-DMR that is hypomethylated in colon tumors, and confirmed that an alternative RNA transcript is produced in colon tumors compared to their matched normal counterparts (Supplementary Fig. 6 online). Thus a major function for differential methylation during differentiation may be alternative transcription, and the role of altered DNAm in cancer may in part be disruption of the regulatory control of specific promoter usage.
DNAm, even far from genes, completely discriminates tissues regardless of human or mouse origin
A compelling argument for the functional importance of differential DNAm of CpG island shores would be their conservation across species. One might expect DMRs near transcriptional start sites to be conserved because the genes are conserved. However, when we examined the relationship between gene-distant T-DMRs (2-10 kb away from an annotated gene) and sequence conservation using the phastCons28way table from the UCSC genome browser, we found 48% of differentially methylated regions were sequence-conserved. Furthermore, 91% of DMRs were located within 1 kb of a highly conserved region (p < 0.001).
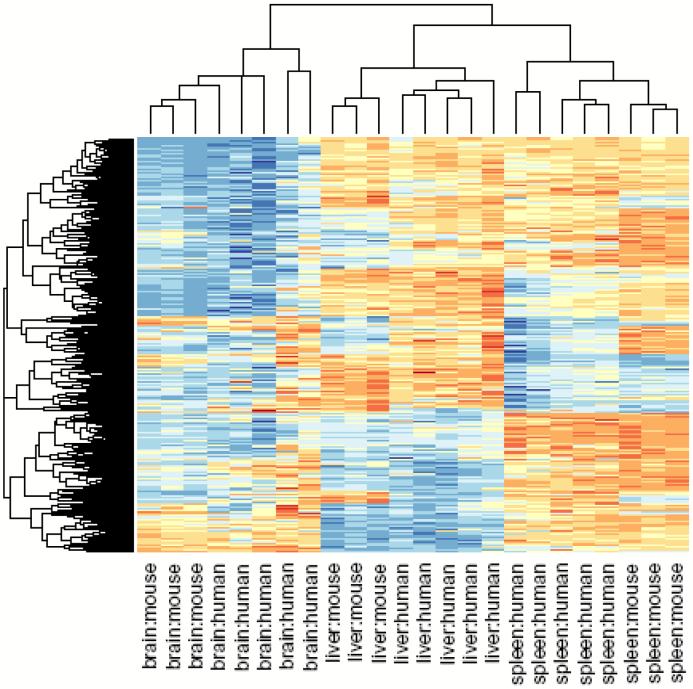
To address whether the DNA methylation itself is conserved across species, we created a mouse CHARM array with ∼2.1 million features independently of the human array. We then isolated tissue replicates from each of 3 mice, corresponding to the tissues examined in the human T-DMR experiments, and then mapped these methylation data across species using the UCSC LiftOver tool (http://genome.ucsc.edu). The inter-species correspondence of tissue-specific methylation was dramatic, and unsupervised clustering perfectly discriminated among the tissues, regardless of the species of origin (Fig. 6; P < 10-9). Interestingly, perfect discrimination among the tissues was found even when we limited the analysis to gene-distant DMRs (Supplementary Fig. 6 online). Thus, DNAm itself is highly conserved across 50 Myr of evolution (approximately 51% of mapped DNAm sites were conserved). We also noticed relatively little heterogeneity in tissue specific methylation in the mouse compared to the human (height of the cluster bars in Fig. 6), suggesting a genetic-epigenetic relationship since the mice are inbred.
Figure 6.
Clustering of human tissue samples using mouse T-DMRs results in perfect discrimination of tissues. The M values of all tissues from the 1,963 regions corresponding to mouse T-DMRs that mapped to the human genome were used for unsupervised hierarchical clustering. By definition, the mouse tissues are segregated. Surprisingly, all of the human tissues are also completely discriminated by the regions that differ in mouse tissues. The three major branches in the dendrograms correspond perfectly to tissue type regardless of species. Columns represent individual samples, and rows represent regions corresponding to mouse T-DMRs. The heatmap displays M values, with red being more methylated and blue less.
The locations of cancer-specific DNA methylation strikingly overlaps those those that distinguish normal tissues
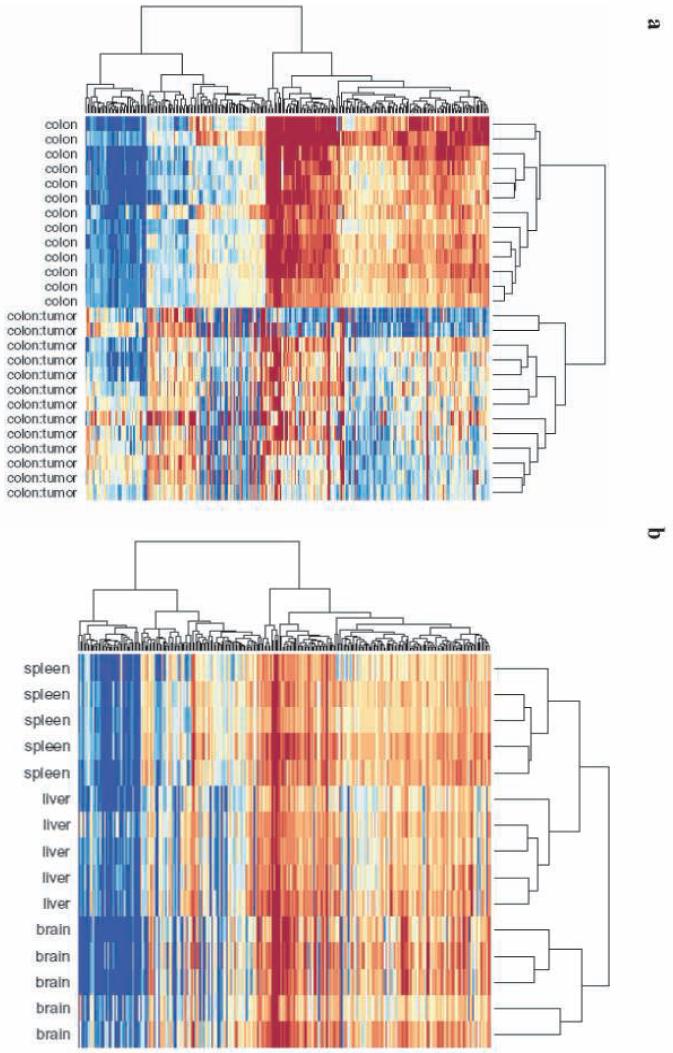
Because both C-DMRs and T-DMRs were located at CpG island shores, we then asked if their locations were similar to each other. We focused on DMRs in which the methylation difference was from no methylation to some methylation, based on the gene expression data above showing a strong relationship between “none-to-some” methylation and gene silencing. Surprisingly, 52% of the C-DMRs overlapped a T-DMR, compared to only 22% expected by chance (p < 10-14), when using a FDR of 5% for defining T-DMRs. While these data are strikingly significant, the definition of a T-DMR based on FDR of 5% is conservative. We therefore also asked directly whether C-DMRs are enriched for tissue variation in DNAm, by computing an averaged F-statistic (comparison of cross-tissue to within-tissue variation) at each C-DMR. The cross-tissue variation in normal tissues was significant at 64% of the C-DMRs, compared to 20% of randomly selected CpG regions on the array matched for size (p < 10-143). When we define DMRs using an FDR of 5%, 1229 of 2707 C-DMRs overlap a T-DMR, of which 265, 448, and 185 are brain-, liver-, and spleen-specific, and 331 show variation among all of the tissues (Supplementary Data 4 online). Thus the colon C-DMRs were highly enriched for overlap with liver T-DMRs (P < 10-15), and liver is embryologically closest to colon of the autopsy tissues studied. For example, the C-DMR located in the CpG island shore upstream of the heparan sulfate D-glucosaminyl 3-O-sulfotransferase 4 (HS3ST4) gene is hypomethylated in colon cancer compared to normal colon, and coincides with a T-DMR that distinguishes liver from other tissues (Fig. 1b). The correspondence between C-DMRs and T-DMRs was so striking that when we performed unsupervised clustering of the normal brain, liver and spleen, using the M values from the C-DMRs, there was perfect discrimination of the tissues (Fig. 7).
Figure 7.
Clustering of normal tissue samples using C-DMRs results in perfect discrimination of tissues. The M values of all tissues from the 2,707 regions corresponding to C-DMRs were used for unsupervised hierarchical clustering. (a) By definition, the colon tumors and matched normal mucosa are segregated. The two major branches in the dendrograms correspond perfectly to tissue type. (b) Surprisingly, all of the normal brains, spleens and livers are also completely discriminated by the regions that differ in colon cancer. The three major branches in the dendrograms correspond perfectly to tissue type. Columns represent individual samples, and rows represent regions corresponding to C-DMRs. The heatmap displays M values, with red being more methylated and blue less.
Interestingly, most tissue-specific methylation difference more commonly involves hypomethylation, although this varies by tissue type (50-79% of DMRs representing hypomethylation) (Supplementary Table 5 online), and cancer-specific methylation differences slightly more frequently involve hypermethylation (56%:44%), (Supplementary Table 5 online). For both T-DMRs and C-DMRs, when there is differential methylation, it is common that at least one of the tissues is completely unmethylated (68% and 37%, respectively). Furthermore, hypomethylated C-DMRs were twice as likely to resemble another tissue type, such as liver, than were hypermethylated C-DMRs (82% vs. 61%, P < 10-31), even though hypermethylated C-DMRs overlapped T-DMRs 1.5-fold more frequently than did hypomethylated C-DMRs (54% vs. 35%, P < 10-21).
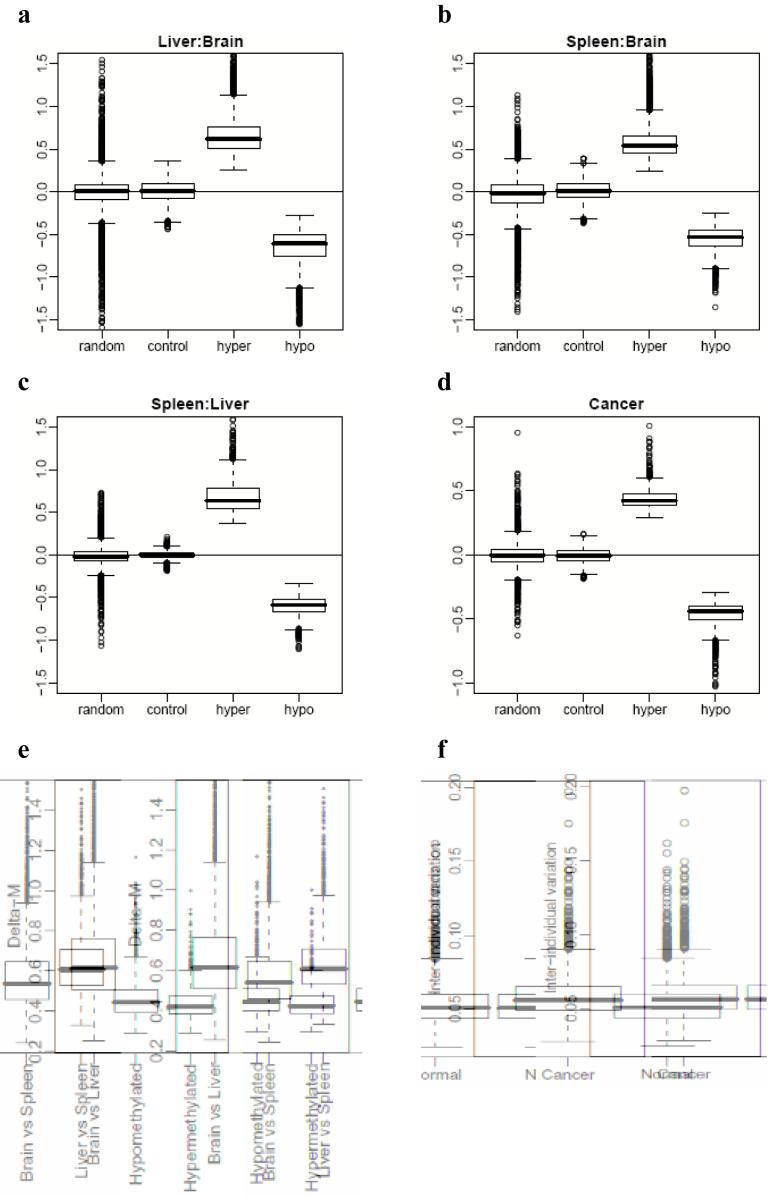
To further explore the relationship between differentiation and type of methylation change, we performed Gene Ontology (GO) analysis for both hypomethylated and hypermethylated C-DMRs in the cancers (see Methods). The GO analysis showed enrichment for pluripotency-associated genes for both hyper- and hypomethylated C-DMRs (P < 0.01) (Supplementary Table 6 online). Interestingly, hypomethylated C-DMRs were also enriched for genes associated with differentiated cellular functions for lineages other than the colon (P < 0.01) (Supplementary Table 6 online). Thus, cancer-specific DNA methylation predominantly involves the same sites that show normal DNAm variation among tissues, particularly at genes associated with development. Next, we examined the magnitude of differential methylation and variation in C-DMRs and T-DMRs. The ΔM values for tissue and cancer DMRs differ dramatically from nomethylation controls or randomly selected regions. Note that the latter have an average value comparable to controls but with significant tails (since by definition they may contain DMRs themselves) (Figure 8a-d). The ΔM values for normal tissues were comparable across the tissues, but the ΔM values between normal and cancer were on average approximately half the ΔM between normal tissue pairs (Fig. 8e), which is logical given that the cancers are compared with their tissue of origin. Another difference between cancer and normal is an increase in the inter-individual variation in M among the colon cancers, which is on average ∼50% greater than the inter-individual variation among the normal colons (Fig. 8f), a result which may help to explain tumor cell heterogeneity. Given the strong inter-individual variability we found in cancer, we identified 205/2707 C-DMRs that are consistently differentially methylated between the colon tumor and matched normal mucosa from all 13 individuals examined (Supplementary Data 4). These regions provide a smaller, more focused set of regions for biomarker discovery and carcinogenesis studies.
Figure 8.
Magnitude of differential methylation and variation in C-DMRs and T-DMRs. (a-d) Boxplots of average delta M values over all DMRs, compared to randomly chosen regions and unmethylated control regions, matched for length. (a) Liver versus brain, (b) spleen versus brain, (c), spleen versus liver, (d) colon cancer versus normal colonic mucosa. (e) Differences in DNA methylation are greater in magnitude among normal tissues than are differences between colon tumors and matched normal mucosa. For all DMRs we computed the average delta M. We then stratified these values into T-DMRs, hypermethylated C-DMRs and hypomethylated C-DMRs. T-DMRs were further stratified according to brain versus liver, brain versus spleen, and liver versus spleen pairwise comparisons. The box-plots represent absolute values of the delta Ms. (f) Inter-individual variation in M is larger among colon tumors than matched normal mucosa. For each C-DMR we computed the average inter-individual standard deviation of the Mvalues. The box-plots represent these values for normal colon mucosa and colon tumors.
Discussion
We have performed a genome-wide analysis of DNA methylation across tissue types, between cancer and normal, and between human and mouse, revealing several surprising relationships between all three areas, supported by extensive bisulfite pyrosequencing and functional analysis. The first is that most tissue-specific DNAm occurs not at CpG islands, but at CpG island shores. The identification of these regions opens the door to functional studies, such as the mechanism of targeting DNAm to these regions and the role of differential methylation of shores. Supporting a functional role for shores, gene expression was closely linked to T-DMR and C-DMR methylation, particularly for switches from “none” to “some” methylation. The relationship between shore methylation and gene expression was confirmed by 5-aza-2′-deoxycytidine and DNA methyltransferase knockout experiments altering expression of the same genes. Another intriguing mechanism for shores supported by this study is alternative transcription, supported by mapping and RACE experiments.
While 76% of T-DMRs were in CpG island shores, at least for the three tissues examined here, 24% were not adjacent to conventionally defined CpG islands. However, these regions were nevertheless shores of CpG-enriched sequences (For an example, see Supplementary Fig. 8 online). We are currently developing a novel algorithm for CpG island definition based on Hidden Markov modeling that will likely increase the fraction of T-DMRs in CpG island shores. The “CpG clusters” recently identified by Glass et al.22 are not CpG island shores (only 4% of shore DMRs map to them), although the shores of these clusters, like the shores of CpG islands, are enriched for DMRs. Note, though, that the variation in DNAm is still not within the dense CpG regions as defined by any of these definitions, but in CpG shores.
The second major finding of the study is that T-DMRs are highly conserved between human and mouse, and the methylation itself is sufficiently conserved to completely discriminate tissue types regardless of species of origin. This was true even for T-DMRs located >2 kb from transcriptional start sites. The incorporation of epigenetic data, such as DNAm, in evolutionary studies as done here, should greatly enhance the identification of conserved elements that regulate differentiation. We also found greater DNAm heterogeneity in human than in mouse (at least in an inbred strain), even for DMRs located >2 kb from a gene promoter. This result suggests that the conservation of DNAm between human and mouse may have a strong genetic basis, consistent with a greater degree of tissue DNAm homogeneity in the inbred mouse strain.
The third major finding of the study is that most cancer-related changes in DNAm, i.e. C-DMRs, at least for colon cancer, correspond to T-DMRs, and that these changes are similarly divided between hypomethylation and hypermethylation, and also involve CpG island shores. Thus epigenetic changes in cancer largely involve the same DMRs as epigenetic changes in normal differentiation. These results have important implications for studies such as the Cancer Genome Atlas, in that most altered DNA methylation in cancer does not involve CpG islands, even though that is the main focus of previous and current study. Similarly, high throughput sequencing efforts based on reduced representation analysis of CpG islands are unlikely to identify most DNAm variation in normal tissues or in cancer.
Finally, GO annotation analysis suggests that DNAm changes in cancer reflect both pluripotency-associated genes and differentiated cellular functions for lineages other than the colon. These data are consistent with the epigenetic progenitor model of cancer4, that epigenetic alterations affecting tissue-specific differentiation are the predominant mechanism by which epigenetic changes cause cancer. The genes identified in this analysis will themselves be of considerable interest for further study, as well as the potential regulatory regions that did not lie in close proximity to annotated genes.
Methods
Samples
We obtained snap frozen colon tumors and dissected normal mucosa, from the same patients, courtesy of Bert Vogelstein. Human post-mortem brain, liver, and spleen tissues, from the same individual, were donated by The Stanley Medical Research Institute brain collection.
CHARM DNA methylation analysis
We performed McrBC fractionation followed by CHARM array hybridization for all human tissue samples as previously described5. For each probe, we computed average M values across the 5 samples in each tissue type. Differential methylation was quantified for each pairwise tissue comparison by the difference of averaged M values (ΔM). Replicates were used to estimate probe-specific standard deviations which provided standard errors (SE) for Δ M. We formed z-scores: Δ M/SE(:ΔM) and contiguous statistically significant values were grouped into regions. Because millions of z-scores are examined, statistical confidence calculation needed to account for multiple comparisons. We therefore computed false discovery rates (FDR) and reported a list with an FDR of 5% 23,24. Assessment of statistical significance of the regions was assessed as described in the Supplementary Methods. C-DMRs were determined using the same procedure described above with the following exception: since we observed greater heterogeneity in the cancer samples (Fig. 8f), we did not divide Δ M by the standard errors as this would penalize regions of highly variable M values. For all microarray analysis, we used RMA for processing25 then averaged the samples in each tissue, and computed the difference (equivalent to average log ratio). Mouse T-DMRs were determined using the same statistical procedures as described above for the T-DMRs and were then mapped to the human genome using the UCSC liftOver tool (http://genome.ucsc.edu/). To correct for possible “array” effects, each T-DMR was standardized within species by dividing the mean M across all samples in species and divided by SD across all samples in species. A list of all mouse T-DMRs is provided in Supplementary Data 5. Overlap of C-DMRs with T-DMRs was determined by adding the number of regions.
Bisulfite pyrosequencing
Bisulfite pyrosequencing was performed as previously described26. Supplementary Table 7 online provides genomic location of CpG sites measured. Primer sequences are provided in Supplementary Table 8 online.
Quantitative real-time PCR
RNA for quantitative real time PCR was prepared using Trizol (Invitrogen). Complementary DNA was prepared using the QuantiTect RT kit (QIAGEN). TaqMan assays (Applied Biosystems) were used to determine relative gene expression and experiments analyzed on a 7900HT detection system. Taqman assay identification numbers are provided in Supplementary Table 9. Human ACTB was used as an endogenous control. Relative expression differences were calculated using the ΔΔCt method27.
Acknowledgements
We thank Dr. Bert Volgelstein for providing colon tumors and matched normal mucosa samples. Postmortem brain, liver and spleen tissue was donated by The Stanley Medical Research Institute collection courtesy of Drs. Michael B. Knable, E. Fuller Torrey, and Robert H. Yolken. This work was supported by NIH grants P50HG003233 (A.F.), R37CA54358 (A.F.), and R01GM083084 (R.I.).
Footnotes
Full methods and any associated references are available in the online Supplementary information.
Accession codes NCBI GEO: Gene expression microarray data has been submitted under accession number [xxx]
Supplementary Information is linked to the online version of the paper at www.nature.com/ng.
Supplementary Material
References
- 1.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 2.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–5. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–16. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 4.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 5.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:771–9. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 7.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 9.Illingworth R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen L, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–36. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–13. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 12.Hong C, et al. Epigenome scans and cancer genome sequencing converge on WNK2, a kinase-independent suppressor of cell growth. Proc Natl Acad Sci U S A. 2007;104:10974–9. doi: 10.1073/pnas.0700683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strathdee G, et al. Inactivation of HOXA genes by hypermethylation in myeloid and lymphoid malignancy is frequent and associated with poor prognosis. Clin Cancer Res. 2007;13:5048–55. doi: 10.1158/1078-0432.CCR-07-0919. [DOI] [PubMed] [Google Scholar]
- 14.Cantor AB, et al. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J Exp Med. 2008;205:611–24. doi: 10.1084/jem.20070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segditsas S, et al. Putative direct and indirect Wnt targets identified through consistent gene expression changes in APC-mutant intestinal adenomas from humans and mice. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaeger J, et al. Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin Cancer Res. 2007;13:806–15. doi: 10.1158/1078-0432.CCR-06-1820. [DOI] [PubMed] [Google Scholar]
- 17.Seal S, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are lowpenetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–41. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 18.Pedersen IS, et al. Frequent loss of imprinting of PEG1/MEST in invasive breast cancer. Cancer Res. 1999;59:5449–51. [PubMed] [Google Scholar]
- 19.Gius D, et al. Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell. 2004;6:361–71. doi: 10.1016/j.ccr.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 20.Shiraki T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A. 2003;100:15776–81. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wakaguri H, Yamashita R, Suzuki Y, Sugano S, Nakai K. DBTSS: database of transcription start sites, progress report 2008. Nucleic Acids Res. 2008;36:D97–101. doi: 10.1093/nar/gkm901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glass JL, et al. CG dinucleotide clustering is a species-specific property of the genome. Nucleic Acids Res. 2007;35:6798–807. doi: 10.1093/nar/gkm489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statist Soc B (Methodol.) 1995;57:289–300. [Google Scholar]
- 24.Storey John D. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann Statist. 2003;31:2013–2035. [Google Scholar]
- 25.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 26.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–75. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.