Abstract
Background
Recent trial data have challenged the hypothesis that cholesteryl ester transfer protein (CETP) and high-density lipoprotein cholesterol (HDLC) have causal roles in atherothrombosis. One method to evaluate this issue is to examine whether polymorphisms in the CETP gene that impact on HDLC levels also impact on the future development of myocardial infarction.
Methods and Results
In a prospective cohort of 18,245 initially healthy American women, we examined over 350,000 singe nucleotide polymorphisms (SNPs) first to identify loci associated with HDLC and then to evaluate whether significant SNPs within these loci also impact upon rates of incident myocardial infarction during an average 10-year follow-up period. Nine loci on 9 chromosomes had one or more SNPs associated with HDLC at genomewide statistical significance (P<5×10−8). However, only SNPs near or in the CETP gene at 16q13 were associated with both HDLC and risk of incident myocardial infarction (198 events). For example, SNP rs708272 in the CETP gene was associated with a per-allele increase in HDLC levels of 3.1 mg/dL and a concordant 24 percent lower risk of future myocardial infarction (age-adjusted HR 0.76, 95%CI 0.62–0.94), consistent with recent meta-analysis. Independent and again concordant effects on HDLC and incident myocardial infarction were also observed at the CETP locus for rs4329913 and rs7202364. Adjustment for HDLC attenuated did not eliminate these effects.
Conclusion
In this prospective cohort of initially healthy women, SNPs at the CETP locus impact upon future risk of myocardial infarction, supporting a causal role for CETP in atherothrombosis, possibly through an HDLC mediated pathway.
Keywords: HDL cholesterol, myocardial infarction, atherosclerosis, genetic association
Cholesteryl ester transfer protein (CETP) promotes the transfer of cholesteryl esters from high-density lipoprotein cholesterol (HDLC) to other lipoprotein particles, and individuals genetically deficient for CETP often have extremely high HDLC levels1, 2. In part on this basis, and because of consistent epidemiologic evidence that elevated levels of HDLC are protective against cardiovascular disease, inhibitors of CETP were developed with the hope that raising HDLC through this mechanism would reduce vascular event rates. However, in the recently reported ILLUMINATE trial of torcetrapib conducted among more than 15,000 individuals at high risk for cardiovascular disease, mortality rates were increased in the active as compared to placebo treatment group3 leading to considerable debate as to whether CETP and/or HDLC are in fact causal for heart disease and should remain viable pharmacologic targets4, 5.
One approach to understanding causal pathways is to ascertain if genetic polymorphism known to impact on an intermediate phenotype (such as plasma lipid levels) also impacts on vascular risk6. For example, recent work has found that those with polymorphism in PCSK9 not only have reduced levels of low-density lipoprotein cholesterol (LDLC), but also reduced rates of vascular events7. Similarly, recent reports have shown that polymorphism within several other LDL related pathways determine both plasma LDLC levels and vascular risk8–11. By contrast, data demonstrating that polymorphism in genes known to affect plasma HDLC levels also affect vascular event rates have been inconsistent, particularly with regard to CETP9, 11–14. However, such evidence, if available from a prospective cohort study free of selection bias, would support continued investigation into CETP and HDLC as targets for therapy.
To address this issue, we evaluated more than 350,000 single nucleotide polymorphisms (SNPs) across the human genome to first determine genetic loci associated with HDLC and then evaluate whether any significant SNPs within these loci in turn impact upon rates of incident myocardial infarction during an average 10-year follow-up period.
Methods
We evaluated the role of polymorphisms that impact upon HDLC as potential determinants of incident myocardial infarction among participants in the prospective Women’s Genome Health Study (WGHS)15. In brief, participants in the WGHS include initially healthy American women aged 45 and older with no prior history of cardiovascular disease, cancer, or other major chronic illness who provided a baseline blood sample during the enrollment phase of the Women’s Health Study16 between 1992 and 1995, and who gave consent for blood based analyses related to risks of incident chronic diseases.
All study participants were followed up through March 2007 for incident myocardial infarction events that were adjudicated by an endpoints committee using standardized criteria and full medical record review. The endpoint of myocardial infarction was confirmed if symptoms met World Health Organization Criteria and if the event was associated with abnormal levels of cardiac enzymes or diagnostic electrocardiographic criteria. Only confirmed endpoints were included in this analysis.
All study participants had baseline blood samples assayed for total cholesterol, HDLC, direct LDLC, apolipoprotein A-1, and apolipoprotein B-100, and high-sensitivity C-reactive protein (hsCRP) in a core laboratory certified by the national Heart Lung and Blood Institute / Centers for Disease Control and Prevention Lipid Standardization Program; coefficients of variation were < 3 percent for total cholesterol, HDL and LDL cholesterol, and < 5 percent for apolipoproteins A-1 and B-100.
As described elsewhere, DNA extracted from the baseline WGHS blood samples underwent SNP genotyping using the Illumina Infinium II assay to query a genomewide set of 318,237 SNP markers (Human HAP300 panel) as well as an additional focused panel of 45,751 SNPs selected to enhance coverage of genomic regions without regard to allele frequency in which we had a strong a priori interest owing to presence of genes thought to be of relevance to metabolic, lipid, inflammatory, and other biological functions15. To reduce the potential for population stratification, we evaluated only WGHS participants who were of European ancestry. As previously reported in the WGHS, a principal component analysis using 1,443 ancestry-informative SNPs was used to confirm self-reported ancestry in 99.7 percent of the sample17, leaving 18,245 participants with both self-reported and genetically inferred European ancestry for this study who also had both HDLC and genotype data available. Further, in an additional principal component analysis performed for the exclusion of within European stratification based on 124,931 SNPs chosen to have pairwise disequilibrium r2 < 0.4, no correction for within European ancestry was required17.
For the current analysis, we first ascertained those loci that contained one or more SNPs that associated with plasma HDLC at a genomewide level of statistical significance (P < 5×10−8) and that had Hardy-Weinberg P >10−6, minor allele frequency greater than 1 percent, and genotyping call rates greater than 90 percent. To evaluate for associations between any of these SNPs and plasma HDLC, we assumed an additive model of inheritance and initially conducted univariate linear regression analysis to test the null hypothesis that HDLC levels did not differ by the number of inherited copies of the SNP minor alleles; in these initial analyses, we adjusted plasma HDLC levels on an a priori basis for age, smoking, body mass index, hormone therapy, and menopausal status, and limited the evaluation to non-diabetic women who were not taking lipid-lowering agents.
For any locus containing at least one SNP that was associated with HDLC at a genomewide level of statistical significance, we next sought evidence of association between the significant SNPs in that loci and incident myocardial infarction. Association testing of these SNPs with incident myocardial infarction was performed with age-adjusted Cox proportional hazards models as well as models that additionally adjusted for HDLC, and in fully adjusted models that further controlled for smoking status (current, not current), blood pressure (Framingham categories), diabetes (yes/no), parental history of myocardial infarction before age 60 years (yes/no), LDLC, and log transformed triglycerides (mg/dL).
Haplotype analysis was performed using the haplo.glm program from the haplo.stats analysis package in R18–20 within blocks of linkage disequilibrium as defined previously21. Briefly, this program provided a method for logistic regression of myocardial infarction dependent on inferred haplotypes in a pre-specified block of linkage disequilibrium. The key feature of the haplo.glm algorithm is its use of an expectation maximization procedure to optimize the likelihood of both the logistic model fit and the haplotype inference in an iterative fashion.
To replicate associations at the 2q24.3 locus, we used HDLC measurements from PRINCE cohort for which genotype information was available through the PARC consortium22–25. The genotype data derive from Illumina Human HAP300 genotyping among 670 PRINCE participants, of whom 168 (25.1%) were female.
The study protocol was approved by the Institutional Review board of the Brigham and Women’s Hospital, Boston, MA.
Statuement of responsibility
All authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the manuscript as it is written.
Results
Effects of polymorphism on HDLC
Nine loci on 9 chromosomes were identified that contained one or more SNPs that were associated with HDLC at a genomewide level of statistical significance (P < 5×10−8) (Table 1). Eight of these nine loci contain genes known to impact upon HDLC metabolism, while one locus near the genes COBLL1 and GRB14 at 2q24.3 appears to be novel 8, 11, 24, 26, 27. The per-allele shifts in HDLC for the two SNPs with genomewide significance at 2q24.3, rs10490694 and rs7607980, were 1.35 mg/dL (P=3.9 ×10−9) and 1.29 mg/dL (P=1.5×10−8), respectively. These SNPs were in high linkage disequlibirum (LD; r2=0.98) with minor allele frequencies 12.3% and 12.5%, respectively. The minor allele frequencies of the two SNPs were comparable and their associations with HDLC were significant with consistent direction of effects in separate genetic analysis in the PRINCE population. Specifically, the per-allele shifts were 2.9 mg/dL (P=0.0008) and 3.1 mg/dL (p=0.0004), respectively. The effects were larger among the 168 PRINCE women (both SNPs 7.3 mg/dL) than the 501 men (1.5 mg/dL and 1.7 mg/dL, respectively), and an interaction with gender was observed (P-interaction=0.002 and 0.003, respectively).
Table 1.
Genetic loci with one or more SNPs associated with HDLC at a genome-wide level of statistical significance.
| Locus smallest | |||
|---|---|---|---|
| Locus | N SNPs* | P-value | candidate genes |
| 2q24.3 | 2 | 3.9×10−9 | COBLL1, GRB14 |
| 8p21.3 | 11 | 1.4×10−17 | LPL |
| 9q31.1 | 1 | 1.6×10−8 | ABCA1 |
| 11q23.3 | 6 | 2.8×10−12 | APOA1, APOA4, APOA5, APOC3 |
| 15q22.1 | 12 | 1.4×10−23 | LIPC |
| 16q13 | 20 | 3.7×10−93 | CETP |
| 18q21.1 | 1 | 1.4×10−9 | LIPG |
| 19q13.32 | 1 | 2.6×10−11 | APOC1, APOC2, APOC4, APOE |
| 20q13.12 | 3 | 1.9×10−14 | PLTP |
Number of locus SNPs with P<5×10−8
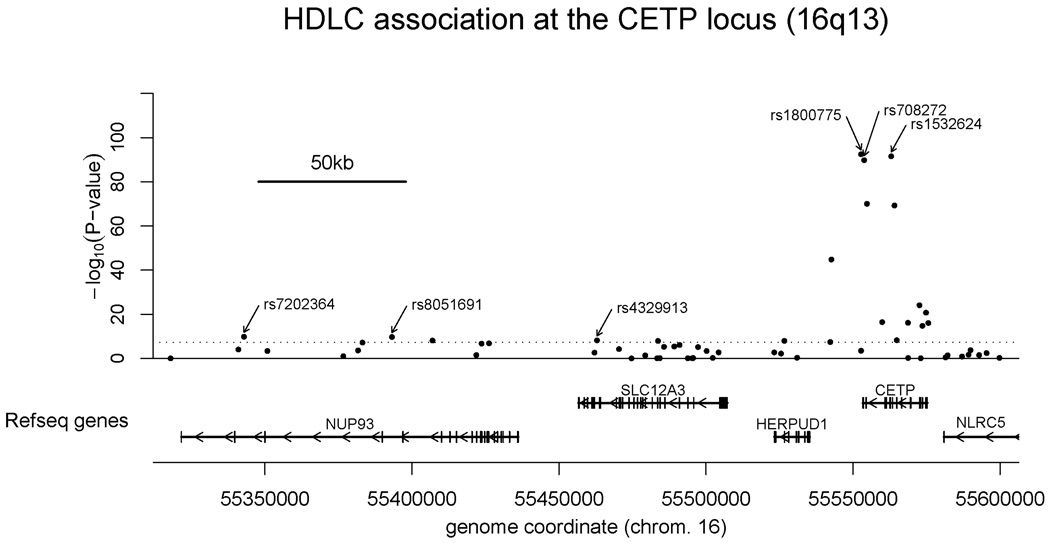
At the CETP locus, where genomewide associations with HDLC were both most significant and numerous, 20 SNPs were associated with plasma HDLC at a genomewide level of statistical significance (P-values ranging from 3.8 × 10−8 to 3.7×10−93) (Table 2). All of these were also highly associated with apolipoprotein A-1 levels (data not shown). These SNPs spanned about 242 kb of chromosome 16 with most clustered around the CETP gene but three mapping to the neighboring NUP93 gene encoding a nuclear pore protein, and another six in mapping to the neighboring SLC12A3 and HERPUD1 genes, respectively encoding a sodium transporter and a protein of uncertain function localized to the endoplasmic reticulum (Figure 1).
Table 2.
SNPs in the CETP gene with genomewide effects on plasma HDL cholesterol.
| SNP* | BP | MAF | M/m | HW-p | HDL-C (mg/dL) | per allele | P | ||
|---|---|---|---|---|---|---|---|---|---|
| MM | Mm | Mm | HDL shift (mg/dL) | ||||||
| rs1800775 | 55552736 | 0.486 | C/A | 0.44 | 49 (41–59) | 52 (43–62) | 55 (46–67) | 3.09 | 3.7×10−93 |
| rs1532624 | 55562979 | 0.430 | C/A | 0.57 | 50 (42–60) | 52 (44–63) | 56 (46–68) | 3.14 | 3.0×10−92 |
| rs708272 | 55553788 | 0.428 | G/A | 0.28 | 50 (42–60) | 52 (44–63) | 56 (46–67) | 3.08 | 1.7×10−90 |
| rs1864163 | 55554733 | 0.255 | G/A | 0.27 | 54 (45–65) | 50 (42–61) | 48 (40–58) | −3.10 | 9.2×10−71 |
| rs7499892 | 55564090 | 0.177 | G/A | 0.96 | 53 (44–64) | 50 (41–60) | 47 (40–57) | −3.50 | 5.5×10−70 |
| rs9989419 | 55542639 | 0.395 | G/A | 0.35 | 54 (45–65) | 51 (43–62) | 50 (42–60) | −2.19 | 1.6×10−45 |
| rs5880 | 55572591 | 0.053 | G/C | 0.16 | 52 (44–63) | 49 (41–59) | 45 (39–52) | −3.48 | 8.2×10−25 |
| rs1800777 | 55574819 | 0.036 | G/A | 0.28 | 52 (44–63) | 49 (40–58) | 43 (37–47) | −3.91 | 1.9×10−21 |
| rs12597002 | 55559904 | 0.300 | C/A | 0.96 | 53 (44–64) | 51 (43–62) | 50 (42–60) | −1.39 | 3.6×10−17 |
| rs4784744 | 55568685 | 0.349 | G/A | 0.90 | 53 (44–64) | 52 (43–62) | 50 (41–61) | −1.32 | 7.4×10−17 |
| rs289744 | 55575602 | 0.302 | A/C | 0.99 | 51 (43–62) | 53 (44–63) | 54 (45–64) | 1.37 | 8.8×10−17 |
| rs5882 | 55573592 | 0.318 | A/G | 0.42 | 51 (43–62) | 53 (44–63) | 53 (45–64) | 1.30 | 1.7×10−15 |
| rs7202364 | 55342890 | 0.267 | A/G | 0.40 | 52 (43–62) | 53 (44–63) | 53 (44–65) | 1.10 | 1.5×10−10 |
| rs8051691 | 55393212 | 0.267 | C/A | 0.44 | 52 (43–62) | 52 (44–63) | 53 (44–65) | 1.09 | 1.9×10−10 |
| rs5883 | 55564853 | 0.054 | G/A | 0.13 | 52 (43–62) | 54 (45–64) | 58 (49–67) | 1.94 | 5.6×10−9 |
| rs4329913 | 55462932 | 0.236 | G/A | 0.79 | 52 (43–63) | 52 (43–62) | 50 (42–60) | −1.03 | 6.6×10−9 |
| rs1529929 | 55406996 | 0.411 | G/A | 0.09 | 51 (43–62) | 52 (43–63) | 53 (44–64) | 0.89 | 8.5×10−9 |
| rs2217332 | 55526648 | 0.145 | G/A | 0.74 | 52 (44–63) | 51 (42–62) | 51 (41–62) | −1.23 | 1.1×10−8 |
| rs13306677 | 55483695 | 0.096 | G/A | 0.90 | 52 (43–62) | 53 (44–64) | 53 (45–62) | 1.47 | 1.3×10−8 |
| rs247615 | 55542263 | 0.232 | A/G | 0.76 | 52 (44–63) | 52 (43–62) | 51 (43–61) | −0.98 | 3.8×10−8 |
M= major allele, m = minor allele
MAF = minor allele frequency
HW-p = Hardy Weinberg P-value
In order of decreasing significance for association with HDLC
Figure 1.
Genomic context for CETP. Plot of genomic region surrounding SNPs with genomewide association with HDLC near the CETP gene. Upper panel: SNPs are shown according to their physical location and -log10 of their association P-values with adjusted HDLC. Lower panel: Genes from RefSeq release 30. Only one isoform is indicated when multiple splicing variants are known. Annotated SNPs are discussed in this manuscript.
As also shown in Table 2, several of the CETP SNPs were common (minor allele frequencies between 30 and 48 percent), and several had substantive effects on plasma levels of HDLC. For example, median HDLC levels for homozygous major allele carriers, heterozygotes, and homozygous minor allele carriers at rs70872 [minor allele frequency 0.428] were 50, 52, and 56 mg/dL, respectively (P = 1.7×10−90) such that each minor allele increased HDLC levels by an average 3.1 mg/dL or approximately 6 percent. By contrast, other CETP SNPs that were associated with plasma HDLC levels at a genome-wide level of significance were less common or had only marginal absolute effects on plasma HDLC.
Effects of polymorphism on incident myocardial infarction
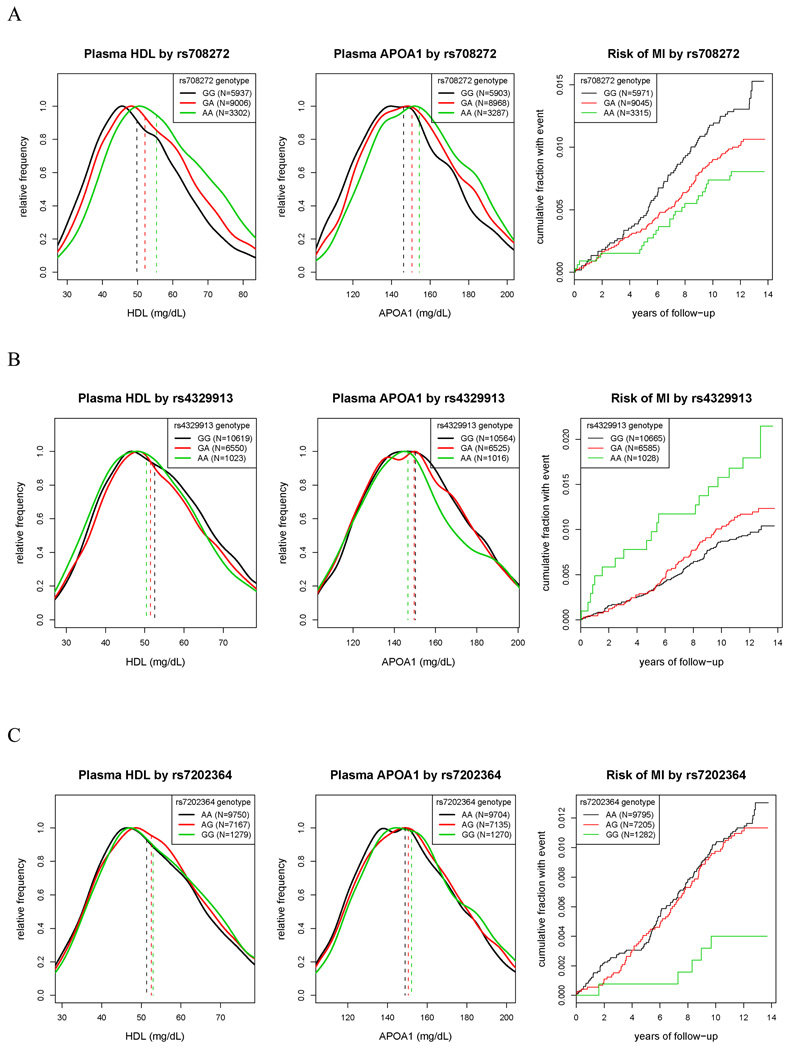
During follow-up, 198 incident myocardial infarction events were confirmed by the Endpoints Committee. Among the 9 loci in Table 1, only one at 16q3 (the site encompassing the CETP gene) contained SNPs that were both associated with HDLC and also had significant impact on rates of incident myocardial infarction. For example, as shown in Table 3, polymorphism at rs708272 in the CETP gene was associated with a per-allele increase in HDLC levels of 3.1 mg/dL and a concordant 24 percent lower risk of future myocardial infarction (age-adjusted hazard ratio 0.76, 95%CI 0.62–0.94; P=0.01) (Figure 2A). The direction and magnitude of effect for rs708272 is consistent with recent meta-analysis involving 38 prior studies 28, indicating external validation.
Table 3.
Hazard ratios (95%C) for the risk of incident myocardial infarction according to CETP genotype. Data are provided on an individual genotype basis, and on a per-allele basis, adjusted for age and adjusted for multiple risk factors.
| Genotype HR | Per allele | Per allele | Per allele | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | N | (age-adjusted) | (age-adjusted) | (HDLC-adjusted) | (fully-adjusted*) | |||||
| MM | Mm | Mm | HR | P | HR | P | HR | P | ||
| rs708272 | 18245 | GG | GA | AA | ||||||
| 1.0 | 0.75 | 0.59 | 0.76 | 0.01 | 0.84 | 0.094 | 0.83 | 0.099 | ||
| (0.56–1.02) | (0.38–0.92) | (0.62–0.94) | (0.68–1.03) | (0.66–1.04) | ||||||
| rs4329913 | 18190 | GG | GA | AA | ||||||
| 1.0 | 1.26 | 1.94 | 1.34 | 0.008 | 1.29 | 0.021 | 1.29 | 0.031 | ||
| (0.94–1.69) | (1.19–3.16) | (1.08–1.66) | (1.04–1.6) | (1.02–1.63) | ||||||
| rs1532624 | 17719 | CC | CA | AA | ||||||
| 1.0 | 0.77 | 0.61 | 0.78 | 0.018 | 0.86 | 0.15 | 0.83 | 0.12 | ||
| (0.57–1.04) | (0.40–0.95) | (0.64–0.96) | (0.7–1.06) | (0.66–1.05) | ||||||
| rs1800775 | 18211 | CC | CA | AA | ||||||
| 1.0 | 0.81 | 0.67 | 0.82 | 0.048 | 0.9 | 0.31 | 0.88 | 0.24 | ||
| (0.59–1.11) | (0.45–1.00) | (0.67–1) | (0.74–1.1) | (0.7–1.09) | ||||||
| rs7202364 | 18194 | AA | AG | GG | ||||||
| 1.0 | 0.94 | 0.32 | 0.79 | 0.045 | 0.81 | 0.077 | 0.76 | 0.042 | ||
| (0.71–1.25) | (0.13–0.79) | (0.62–0.99) | (0.64–1.02) | (0.59–0.99) | ||||||
| rs8051691 | 18237 | CC | CA | AA | ||||||
| 1.0 | 0.94 | 0.32 | 0.79 | 0.047 | 0.81 | 0.079 | 0.76 | 0.043 | ||
| (0.71–1.25) | (0.13–0.79) | (0.62–1) | (0.64–1.02) | (0.59–0.99) | ||||||
Fully adjusted model controlled for age (years), body mass index (kg/m2), smoking status (current, not current), hormone replacement therapy use (yes/no), blood pressure (Framingham categories), diabetes (yes/no), parental history of myocardial infarction before age 60 years (yes/no), LDLC, HDLC, and log transformed triglycerides (mg/dL).
Figure 2.
Distribution of HDLC (left), ApoA1 (middle), and Kaplan Maier estimates of cumulative incidence of myocardial infarction (right) according to the genotype of SNPs at the CETP locus. For comparison, in the left and middle panels, the distribution of HDLC and ApoA1 levels for each genotype is scaled to have maximum value of one. Vertical dashed lines indicate median values of lipid fractions by genotype. A) SNP rs708272; B) SNP rs4329913; C) SNP rs7202364
Adjustment for HDLC level only partially attenuated the effect of rs708272 on myocardial infarction risk (age and HDLC adjusted hazard ratio 0.84, 95%CI 0.68–1.03; P=0.09), and in subsequent analyses further associations were observed between rs708272 and LDLC and triglycerides, albeit on a much more modest basis. For example, for rs708272, LDLC levels were 123, 122, and 120 mg/dL among GG, GA, and AA participants, respectively (P = 0.0009), while corresponding triglyceride levels were 123, 120, and 114 mg/dL, respectively (P < 0.0001). CETP genotype at rs708272 was not associated with age, obesity, smoking, blood pressure, or other major clinical covariates (Table 4). Adjustment for these additional factors (as well as for body mass index, smoking, HRT use, blood pressure, diabetes, and parental history of myocardial infarction before age 60 years) again only marginally attenuated risk from rs708272 (fully adjusted hazard ratio 0.83, 95%CI 0.6–1.04; P=0.1).
Table 4.
Characteristics of study participants according to rs708272 genotype
| GG | GA | AA | P | |
|---|---|---|---|---|
| N (%) | 5937 (32.5) | 9006 (49.4) | 3302 (18.1) | |
| Age (yrs) | 52.8 (48.9–58.9) | 53.0 (49.0–59.0) | 52.8 (48.9–58.8) | 0.3 |
| BMI (kg/m^2) | 24.9 (22.5–28.3) | 24.9 (22.4–28.3) | 25.0 (22.6–28.3) | 0.2 |
| Current smokers (%) | 685 (0.12) | 1073 (0.12) | 404 (0.12) | 0.6 |
| Diabetes | 150 (0.025) | 253 (0.028) | 71 (0.022) | 0.1 |
| Hypertension | 1476 (0.25) | 2201 (0.24) | 783 (0.24) | 0.5 |
| Total Cholesterol (mg/dl) | 208 (183–235) | 209 (184–237) | 210.0 (186.0–237.0) | 0.005 |
| HDL-C (mg/dl) | 49.7 (41.5–59.6) | 52.2 (43.5–62.9) | 55.5 (46.4–67.4) | <0.0001 |
| LDL-C (mg/dl) | 122.9 (102.1–145.7) | 122.2 (100.4–145.5) | 119.9 (99.4–143.3) | 0.0009 |
| Non=HDL-C (mg/dl) | 155.9 (130.8–183.8) | 155.0 (129.5–183.1) | 151.8 (127.1–179.1) | <0.001 |
| ApoA1 (mg/dl) | 146.3 (130.0–164.9) | 150.4 (133.3–169.1) | 154.1 (137.0–173.1) | <0.001 |
| ApoB (mg/dl) | 103.3 (84.2–122.0) | 100.1 (84.1–121.6) | 98.0 (82.0–118.4) | <0.001 |
| Triglycerides (mg/dl) | 123 (85–183) | 120.0 (85.0–176.0) | 114.0 (81.0–167.8) | <0.0001 |
| Incident MI Events (N)/Event rate* | 80/1.12 | 92/0.85 | 26/0.65 | 0.008 |
incident myocardial infarction per 1,000 person years. P value (age–adjusted) from Table 2.
As also shown in Table 3, independent effects on both HDLC and on incident myocardial infarction were also observed at rs4329913 (approximately 90 kb from the CETP gene; Figure 2B) and rs7202364 (approximately 210 kb from the CETP gene; Figure 2C). Three other SNPs showed similar concordant effects on both HDLC and myocardial infarction risk, rs180775 and rs1532624 (both of which were in high LD with rs708272) and rs8051691 (which was in high LD with rs7202364). For these SNPs, adjustment for HDLC or for traditional risk factors again attenuated but did not eliminate effects on risk of incident myocardial infarction (Table 3).
Haplotype analysis was performed to evaluate whether combinations of SNPs at the CETP locus might be more strongly associated with myocardial infarction than individual SNPs. Haplotypes were inferred for neighboring SNPs in blocks with little evidence of historical recombination21, 29 and tested for association with myocardial infarction. These analyses did not reveal stronger associations than were found in the single SNP analysis.
Discussion
Evaluating variation at 9 loci with genomewide significance for HDLC among 18,245 initially healthy American women followed prospectively over an average period of 10 years, we found that common SNPs exclusively in or near the CETP gene were also associated with risk of incident myocardial infarction. Effects on HDLC and myocardial infarction were concordant such that SNP alleles associated with increased plasma HDLC (or ApoA1) levels were also associated with decreased vascular risk, and vice versa. SNP-based risk was attenuated but not eliminated by adjustment for either plasma HDLC or traditional risk factors.
At the same time, our genomewide design identified a novel candidate locus for HDLC at 2q24.3. Analysis in the PRINCE population validated the 2q24.3 associations with HDL-C level and furthermore suggested potential gender specific effects. This region contains two genes, COBLL1 and GRB14, the latter of which may be the better candidate for a functional role in determining HDLC levels. GRB14 encodes and adaptor protein known to form inhibitory interactions with the insulin receptor and other growth receptors and is expressed in the liver, skeletal muscle, and adipose tissue30, 31. Mice deficient of GRB14 have altered patterns of glucose metabolism32, a biological process that may also be linked to HDLC levels.
As prior epidemiologic work relating polymorphism in the CETP gene to vascular risk has been controversial12–14 and as two recent studies have not found significant relationships9, 11, external validation of our findings is important for interpretation. In this regard, in a recent comprehensive meta-analysis that included data from 38 prior studies employing a candidate gene approach 28, the same rs708272 SNP uncovered in the WGHS using a genome-wide approach was associated with a 4.5 percent per-allele increase in HDLC and a directionally concordant 5 percent per allele reduction in coronary risk (odds ratio 0.95, 95%CI 0.92 – 0.99). For direct comparison, in our data, rs708272 was associated with a 6 percent per allele increase in HDLC and a 17 percent per allele concordant reduction in myocardial infarction after multivariate adjustment (HR 0.83, 95%CI 0.66–1.04). Thus, for the primary observation in this large-scale GWAS linking CETP genotype not only to HDLC but also to incident vascular events, external validation is available for rs708272 when prior studies are statistically pooled28.
While the direction of effect for rs708272 in our data is consistent with that of the meta-analysis, the absolute magnitude of genetic impact on vascular events is somewhat larger. We believe there are several potential explanations for this effect. First, our data are limited to incident myocardial infarction whereas data in the recent meta-analysis includes many forms of coronary heart disease, often inclusive of angina and coronary revascularization, events that may have more to do with atherosclerotic progression than with acute plaque rupture.
Second, our data derive from a prospective cohort study of initially healthy women in which event status was determined solely by occurrence of disease rather than by any selection criteria imposed by the investigators or the patients that could result in inadvertent confounding or bias. In both theory and practice, inadvertent bias introduced in the conduct of even the best case-control studies may, in some situations, be greater in magnitude than the effect under study, particularly when that effect is modest to small in absolute magnitude.
Third, we limited our analysis to a Caucasian population, and thereby reduced the potential for ethnicity-based stratification to adversely impact upon the validity of our data. In this regard, it should be noted that several early candidate gene studies and prior meta-analyses of CETP polymorphism were unable to control for such effects and that substantial heterogeneity between studies has previously been observed12.
We believe it of interest that one of the SNPs most significantly associated with myocardial infarction in our data was rs708272, the SNP that defines the B2 allele of the CETP TaqIB polymorphism (and is the core candidate CETP SNP defined in the recent comprehensive meta-analysis28). Although using a different genotyping technology, the current data also extend to women early candidate gene work suggesting that the TaqIB polymorphism is associated with myocardial infarction in men with low HDLC levels33. In this regard, we note that some reports suggest that the Taq1B polymorphism is acting through linkage disequilibrium to a second SNP in the promoter of the CETP gene at position −629 from the transcription start site34, 35. This SNP is equivalent to rs1800775, which was the single most strongly associated SNP in our analysis with HDLC (Table 1) and thus again is consistent with previous reports.
The second SNP within 16q13, rs4329913, which was independently associated with a decrease in HDLC and an increase in the risk of myocardial infarction, is relatively far from the transcription start site for the CETP gene and within the transcribed region for a gene encoding a solute transporter, SLC12A3. Neither SLC12A3 nor a second neighboring gene HERPUD1 are obvious candidates for regulation of HDLC levels, perhaps suggesting the effect of rs4329913 is mediated by long range linkage disequilibrium to a causal variant nearer the CETP gene or long range effects on transcription at the entire locus. In our data, linkage disequilibrium between rs4329913 and at least one other SNP (rs1800777) nearer the CETP gene was as high as D’=0.85, reinforcing the potential for long range linkage effects. Finally, the third SNP in 16q13 independently associated with both HDLC and incident myocardial infarction (rs7202364), is even further from the transcription site for the CETP gene and within the NUP93 gene, another unlikely candidate for effects on HDLC and again suggesting effects mediated through linkage to the CETP gene.
In population based epidemiologic studies typically conducted among men of middle-age or older, it has been observed that the risk of coronary heart disease decreases approximately 2 percent for each 1 percent increase in HDLC36, 37. However, in our fully adjusted data, the reduction in risk associated with polymorphism in CETP was somewhat greater than this prediction. One possible explanation of this effect is that lifelong elevations of HDLC may confer greater protection from vascular disease than would be anticipated from changes in HDLC that might occur from pharmacologic or dietary interventions begun at midlife. A second possibility is that gender specific differences exist in the relationships between CETP polymorphism, HDLC, and vascular risk that were brought out by our study of women. Alternatively, it is possible that the biologic effects of CETP polymorphism on myocardial infarction risk are not captured solely through the intermediate phenotype of HDLC (or through the assessment of HDLC at a single point in time). In this regard, adjustment for HDLC only moderately attenuated the magnitude of the per allele relationships we observed between genotype and myocardial infarction risk. Further, as shown in Table 3, modest associations between rs708272genotype and both LDLC and triglycerides were also observed in our data. Nonetheless, even after additional adjustment for these lipid fractions (as well as for a large number of other major risk factors), the per allele hazard ratio for rs708272 was still 0.83. This lack of further attenuation suggests that CETP may well impact upon atherothrombosis through additional intermediate pathways and/or intermediate phenotypes that go beyond its primary effects on HDLC.
In sum, in these prospective data, we found specific polymorphisms in or near the CETP gene that impact on future risk of myocardial infarction. As such, these data support continued investigation of agents that target CETP as a potential method for vascular risk reduction.
Acknowledgements and Funding
This work was supported by grants (HL 043851, HL 080467, CA 047988) from the NIH (Bethesda, MD), the Donald W. Reynolds Foundation (Las Vegas, NV), the Fondation Leducq (Paris, Fr.), and the Fonds de la Recherche en Santé du Quebec (to G.P.). Collaborative scientific support for genotyping was provided by Amgen, Inc. We are grateful to Ronald Krauss for sharing genotype data from the PARC II study, funded by the NIH (HL 069757).
Footnotes
Conflict of Interest Disclosures
PR discloses investigator initiated grants from the NIH, Reynolds Foundation, and Amgen. DC, RZ, and NC disclose support from the NIH and the Reynolds Foundation. AP and JM are employees of Amgen, Inc.
References
- 1.Brown ML, Inazu A, Hesler CB, Agellon LB, Mann C, Whitlock ME, Marcel YL, Milne RW, Koizumi J, Mabuchi H, Takeda R, Tall AR. Molecular basis of lipid transfer protein deficiency in a family with increased high-density lipoproteins. Nature. 1989;342:448–451. doi: 10.1038/342448a0. [DOI] [PubMed] [Google Scholar]
- 2.Inazu A, Brown ML, Hesler CB, Agellon LB, Koizumi J, Takata K, Maruhama Y, Mabuchi H, Tall AR. Increased high-density lipoprotein levels caused by a common cholesteryl-ester transfer protein gene mutation. N Engl J Med. 1990;323:1234–1238. doi: 10.1056/NEJM199011013231803. [DOI] [PubMed] [Google Scholar]
- 3.Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJ, Komajda M, Lopez-Sendon J, Mosca L, Tardif JC, Waters DD, Shear CL, Revkin JH, Buhr KA, Fisher MR, Tall AR, Brewer B. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med. 2007;357:2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 4.Rader DJ. Illuminating HDL--is it still a viable therapeutic target? N Engl J Med. 2007;357:2180–2183. doi: 10.1056/NEJMe0707210. [DOI] [PubMed] [Google Scholar]
- 5.Tall AR, Yvan-Charvet L, Wang N. The failure of torcetrapib: was it the molecule or the mechanism? Arterioscler Thromb Vasc Biol. 2007;27:257–260. doi: 10.1161/01.ATV.0000256728.60226.77. [DOI] [PubMed] [Google Scholar]
- 6.Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 7.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 8.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C, Orho-Melander M. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 9.Keavney B, Palmer A, Parish S, Clark S, Youngman L, Danesh J, McKenzie C, Delepine M, Lathrop M, Peto R, Collins R. Lipid-related genes and myocardial infarction in 4685 cases and 3460 controls: discrepancies between genotype, blood lipid concentrations, and coronary disease risk. Int J Epidemiol. 2004;33:1002–1013. doi: 10.1093/ije/dyh275. [DOI] [PubMed] [Google Scholar]
- 10.Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, Zhao JH, Song K, Yuan X, Johnson T, Ashford S, Inouye M, Luben R, Sims M, Hadley D, McArdle W, Barter P, Kesaniemi YA, Mahley RW, McPherson R, Grundy SM, Bingham SA, Khaw KT, Loos RJ, Waeber G, Barroso I, Strachan DP, Deloukas P, Vollenweider P, Wareham NJ, Mooser V. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boekholdt SM, Sacks FM, Jukema JW, Shepherd J, Freeman DJ, McMahon AD, Cambien F, Nicaud V, de Grooth GJ, Talmud PJ, Humphries SE, Miller GJ, Eiriksdottir G, Gudnason V, Kauma H, Kakko S, Savolainen MJ, Arca M, Montali A, Liu S, Lanz HJ, Zwinderman AH, Kuivenhoven JA, Kastelein JJ. Cholesteryl ester transfer protein TaqIB variant, high-density lipoprotein cholesterol levels, cardiovascular risk, and efficacy of pravastatin treatment: individual patient meta-analysis of 13,677 subjects. Circulation. 2005;111:278–287. doi: 10.1161/01.CIR.0000153341.46271.40. [DOI] [PubMed] [Google Scholar]
- 13.Curb JD, Abbott RD, Rodriguez BL, Masaki K, Chen R, Sharp DS, Tall AR. A prospective study of HDL-C and cholesteryl ester transfer protein gene mutations and the risk of coronary heart disease in the elderly. J Lipid Res. 2004;45:948–953. doi: 10.1194/jlr.M300520-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Zhong S, Sharp DS, Grove JS, Bruce C, Yano K, Curb JD, Tall AR. Increased coronary heart disease in Japanese-American men with mutation in the cholesteryl ester transfer protein gene despite increased HDL levels. J Clin Invest. 1996;97:2917–2923. doi: 10.1172/JCI118751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ridker PM, Chasman DI, Zee RY, Parker A, Rose L, Cook NR, Buring JE. Rationale, design, and methodology of the Women's Genome Health Study: a genome-wide association study of more than 25,000 initially healthy american women. Clin Chem. 2008;54:249–255. doi: 10.1373/clinchem.2007.099366. [DOI] [PubMed] [Google Scholar]
- 16.Ridker PM, Cook NR, Lee IM, Gordon D, Gaziano JM, Manson JE, Hennekens CH, Buring JE. A randomized trial of low-dose aspirin in the primary prevention of cardiovascular disease in women. N Engl J Med. 2005;352:1293–1304. doi: 10.1056/NEJMoa050613. [DOI] [PubMed] [Google Scholar]
- 17.Ridker PM, Pare G, Parker A, Zee RY, Danik JS, Buring JE, Kwiatkowski D, Cook NR, Miletich JP, Chasman DI. Loci related to metabolic-syndrome pathways including LEPR,HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women's Genome Health Study. Am J Hum Genet. 2008;82:1185–1192. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lake SL, Lyon H, Tantisira K, Silverman EK, Weiss ST, Laird NM, Schaid DJ. Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Hum Hered. 2003;55:56–65. doi: 10.1159/000071811. [DOI] [PubMed] [Google Scholar]
- 19.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Version 2.6.1. Vienna, Austria: R Foundation for Statistical Computing; 2007. R: A language and environment for statistical computing [computer program] [Google Scholar]
- 21.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 22.Krauss RM. Pharmacogenomics and Risk of Cardiovascular Disease (PARC) Available at: http://www.pharmgkb.org/contributors/pgrn/parc_profile.jsp.
- 23.Albert MA, Danielson E, Rifai N, Ridker PM. Effect of statin therapy on C-reactive protein levels: the pravastatin inflammation/CRP evaluation (PRINCE): a randomized trial and cohort study. Jama. 2001;286:64–70. doi: 10.1001/jama.286.1.64. [DOI] [PubMed] [Google Scholar]
- 24.Chasman DI, Pare G, Zee RY, Parker AN, Cook NR, Buring JE, Kwiatkowski DJ, Rose LM, Smith JD, Williams PT, Reider MJ, Rotter JI, Nickerson DA, Krauss RM, Miletich JP, Ridker PM. Genetic loci associated with plasma concentration of LDL-C, HDL-C, triglycerides, ApoA1, and ApoB among 6382 Caucasian women in genome-wide analysis with replication. Circulation: Cardiovascular Genetics. 2008;1:21–38. doi: 10.1161/CIRCGENETICS.108.773168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reiner AP, Barber MJ, Guan Y, Ridker PM, Lange LA, Chasman DI, Walston JD, Cooper GM, Jenny NS, Rieder MJ, Durda JP, Smith JD, Novembre J, Tracy RP, Rotter JI, Stephens M, Nickerson DA, Krauss RM. Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am J Hum Genet. 2008;82:1193–1201. doi: 10.1016/j.ajhg.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spirin V, Schmidt S, Pertsemlidis A, Cooper RS, Cohen JC, Sunyaev SR. Common Single-Nucleotide Polymorphisms Act in Concert to Affect Plasma Levels of High-Density Lipoprotein Cholesterol. Am J Hum Genet. 2007;81 doi: 10.1086/522497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tall AR, Abreu E, Shuman J. Separation of a plasma phospholipid transfer protein from cholesterol ester/phospholipid exchange protein. J Biol Chem. 1983;258:2174–2180. [PubMed] [Google Scholar]
- 28.Thompson A, Di Angelantonio E, Sarwar N, Erqou S, Saleheen D, Dullaart RP, Keavney B, Ye Z, Danesh J. Association of cholesteryl ester transfer protein genotypes with CETP mass and activity, lipid levels, and coronary risk. Jama. 2008;299:2777–2788. doi: 10.1001/jama.299.23.2777. [DOI] [PubMed] [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 30.Cariou B, Capitaine N, Le Marcis V, Vega N, Bereziat V, Kergoat M, Laville M, Girard J, Vidal H, Burnol AF. Increased adipose tissue expression of Grb14 in several models of insulin resistance. Faseb J. 2004;18:965–967. doi: 10.1096/fj.03-0824fje. [DOI] [PubMed] [Google Scholar]
- 31.Daly RJ, Sanderson GM, Janes PW, Sutherland RL. Cloning and characterization of GRB14, a novel member of the GRB7 gene family. J Biol Chem. 1996;271:12502–12510. doi: 10.1074/jbc.271.21.12502. [DOI] [PubMed] [Google Scholar]
- 32.Cooney GJ, Lyons RJ, Crew AJ, Jensen TE, Molero JC, Mitchell CJ, Biden TJ, Ormandy CJ, James DE, Daly RJ. Improved glucose homeostasis and enhanced insulin signalling in Grb14-deficient mice. Embo J. 2004;23:582–593. doi: 10.1038/sj.emboj.7600082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu S, Schmitz C, Stampfer MJ, Sacks F, Hennekens CH, Lindpaintner K, Ridker PM. A prospective study of TaqIB polymorphism in the gene coding for cholesteryl ester transfer protein and risk of myocardial infarction in middle-aged men. Atherosclerosis. 2002;161:469–474. doi: 10.1016/s0021-9150(01)00673-6. [DOI] [PubMed] [Google Scholar]
- 34.Corbex M, Poirier O, Fumeron F, Betoulle D, Evans A, Ruidavets JB, Arveiler D, Luc G, Tiret L, Cambien F. Extensive association analysis between the CETP gene and coronary heart disease phenotypes reveals several putative functional polymorphisms and gene-environment interaction. Genet Epidemiol. 2000;19:64–80. doi: 10.1002/1098-2272(200007)19:1<64::AID-GEPI5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 35.Klerkx AH, Tanck MW, Kastelein JJ, Molhuizen HO, Jukema JW, Zwinderman AH, Kuivenhoven JA. Haplotype analysis of the CETP gene: not TaqIB, but the closely linked -629C-->A polymorphism and a novel promoter variant are independently associated with CETP concentration. Hum Mol Genet. 2003;12:111–123. doi: 10.1093/hmg/ddg013. [DOI] [PubMed] [Google Scholar]
- 36.Gotto AM, Jr, Brinton EA. Assessing low levels of high-density lipoprotein cholesterol as a risk factor in coronary heart disease: a working group report and update. J Am Coll Cardiol. 2004;43:717–724. doi: 10.1016/j.jacc.2003.08.061. [DOI] [PubMed] [Google Scholar]
- 37.Lewington S, Whitlock G, Clarke R, Sherliker P, Emberson J, Halsey J, Qizilbash N, Peto R, Collins R. Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55,000 vascular deaths. Lancet. 2007;370:1829–1839. doi: 10.1016/S0140-6736(07)61778-4. [DOI] [PubMed] [Google Scholar]