Abstract
Context: Activating mutations in the RET protooncogene have been demonstrated in multiple endocrine neoplasia 2 and sporadic medullary thyroid carcinoma (MTC). However, the complete genetic etiology underlying MTC tumorigenesis remains unclear.
Objective: Our objective was to define more precisely the chromosomal regions and uncover novel genes associated with MTC tumorigenesis.
Design and Setting: In this study, we used high resolution array-based comparative genomic hybridization to define tumor-associated copy number alterations (CNA) in 30 primary MTCs: 20 sporadic tumors (50% of which harbored RET mutation), and 10 hereditary.
Results: We identified 98 CNA, including 76 genomic allelic losses, two gains, and 20 copy number variations associated with MTC. Across sporadic and hereditary groups, there was a similar and overlapping pattern of predominant allelic loss. There were 29 regions containing at least 30% CNA in the 30 tumor samples. The most frequent allelic loss occurred in four loci, 7q36.1, 12p13.31, 13q12.11, and 19p13.3-11. No regions were found to be uniquely altered in the hereditary tumors. There were 21 CNA specific to sporadic MTC, with loss of 11q23.3 uniquely altered in RET negative tumors. Pathway analysis found cellular growth and proliferation as the most significant overall target, and cell death as the most significant pathway targeted in sporadic MTC.
Conclusions: Our findings underscore the importance of candidate tumor suppressor genes together with RET alterations in MTCs. Despite of RET status, all MTC might share similar oncogenetic mechanisms. Dysfunction of cell proliferation and cell death may both be involved in MTC tumorigenesis.
A high density array comparative genomic hybridization study defining genomic chromosomal alterations associated with medullary thyroid carcinoma reveals partial similar gene profiles in different medullary thyroid carcinomas subgroups, and supports the importance of candidate tumor suppressor genes in medullary thyroid carcinoma tumorigenesis.
Medullary thyroid carcinoma (MTC) originates from the calcitonin-producing neuroendocrine C cells of the thyroid gland. Approximately 25% of MTC occurs as a component of multiple endocrine neoplasia syndrome type 2 (MEN2). Activating mutations of RET protooncogene have been reported in nearly all hereditary MTC and in 25–40% of sporadic tumors (1). The role of RET and RET pathway in MTC pathogenesis is well documented (1,2). However, further evidence has suggested the importance of the genetic background to influence the tumor phenotype and progression (3). Thus, despite a well-established role for RET in MTC pathogenesis, additional initiating and driver mechanisms involved in MTC tumorigenesis remain to be uncovered.
Karyotyping, loss of heterozygosity, and chromosomal-based comparative genomic hybridization studies demonstrated the involvement of different chromosomal alterations in MTC, with the most frequent alterations observed at 1p, 3q, 3p, 4, 9q13-22, 11c-q12, 13q, 19, and 22q (4,5,6,7,8,9,10,11). Although analysis of recurrent DNA alterations can lead to the identification of cancer driver genes, the process is often hampered by the low resolution of many platforms. In this study, we used a high-resolution array-comparative genomic hybridization platform to define chromosomal alterations associated with MTC, providing more precise boundaries for frequently altered chromosomal regions and greatly reducing the number of potential alteration-driving genes.
Patients and Methods
Patients and tumor samples
Snap-frozen primary tumor tissues from 30 patients with MTC were collected at The University of Texas M.D. Anderson Cancer Center and used in this study under an Institutional Review Board-approved protocol. All MTCs were stained for calcitonin and additional neuroendocrine markers: carcinoembryonic antigen, chromogranin, and synaptophysin. Clinical information collected included age of onset, tumor size, lymph node metastasis, and distant organ metastasis.
DNA extraction and RET sequencing
Genomic DNA was extracted from dissected tissues containing at least 80% of neoplastic cells using the DNeasy Tissue kit (QIAGEN, Germantown, MD) according to the manufacturer’s instructions. DNA sequencing was performed for RET exons 10, 11, and 13–16.
Array-comparative genomic hybridization
Genomic DNA was run on Agilent Human Genome comparative genomic hybridization microarray 244K platform (Agilent Technologies, Palo Alto, CA), which is a 60-mer oligonucleotide-based microarray set containing 236,381 distinct biological features with average resolution of 6.4 kb. Briefly, 3 μg of genomic DNA from each tumor and control genomic DNA from 6–12 sex-matched healthy individuals (Promega, Madison, WI) was labeled with Cy3-dUTP or Cy5-dUTP, respectively. The hybridization was carried out at 65 C for 40 h. Each hybridized array was scanned with Agilent’s dual laser-based scanner (Agilent Technologies).
Microarray data analysis
Comparative genomic hybridization analytic software v3.5 (Agilent Technologies) was used for data normalization and visualization. A log2 ratio greater than 0.5 was considered as DNA amplification and less than −0.5 as DNA loss. To avoid the possibility of a single probe giving false-positive signal, only those regions with aberrations occurring in more than three continuous probes were considered valid. To circumvent the detection of random events further, we included only allelic imbalances occurring in 30% or more of the tumors. Hierarchical clustering was done with Pearson absolute distance metric and average linkage using ArrayAssist version 4.0 software (Stratagene, La Jolla, CA). Genes located in the copy number alteration (CNA) regions were uploaded to Ingenuity Pathway analysis (Ingenuity Systems, www.ingenuity.com).
Quantitative real-time PCR
Quantitative real-time PCR was performed with the StepOne Real-Time PCR System using the StepOne Software v2.0 (Applied Biosystems, Foster City, CA). Quantization of genomic copy number was based on the comparative ΔΔCt method. When available, assays on matched germline DNA and tumor DNA were also performed to confirm somatic alteration.
Results
Defining genetic alterations associated with MTC
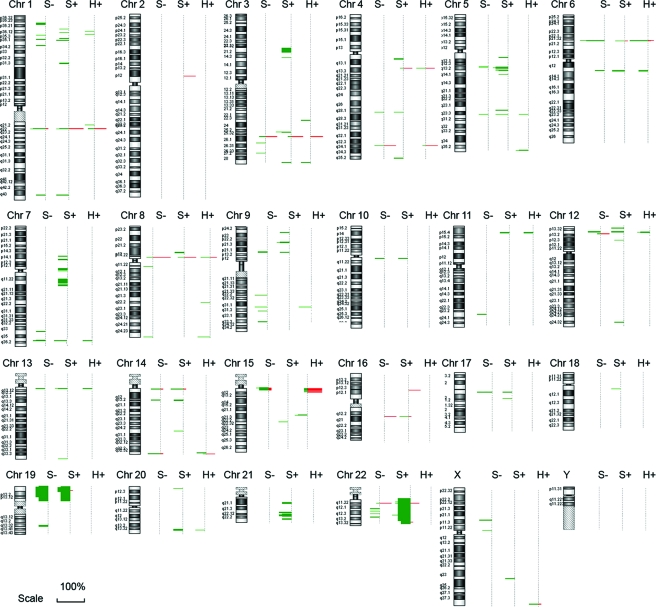
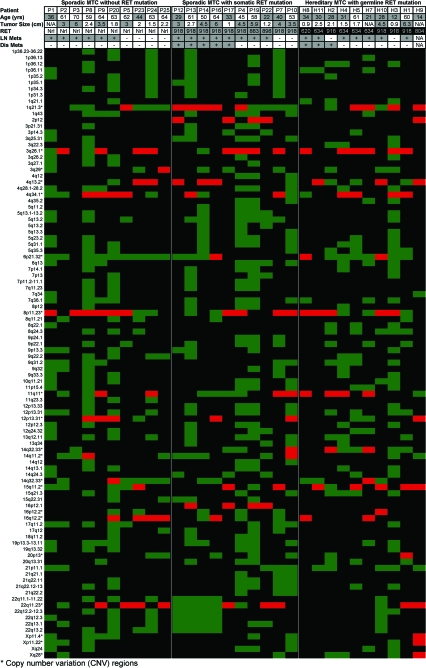
In this study, we analyzed 30 primary MTC tumors that were evenly categorized into three groups based on RET mutation status: 10 sporadic MTC tumors without a RET mutation (S−), 10 sporadic tumors with a somatic RET mutation (S+), and 10 hereditary tumors (H+). In the sporadic group, there were eight RET M918T, one A883F, and one delD898-E902 mutation. The hereditary group contained both MEN2A (four RET C634R, one C620R, one V804L) and MEN2B (four M918T) mutations. CNA were included in our analysis if they appeared in 30% or more of the tumors within a single group. We defined 49 CNA (DNA CNA, defined as gains and losses of chromosomal segments) in S− tumors, 74 in S+ tumors, and 32 in H+ tumors (Fig. 1). In total, the analysis mapped 98 loci with CNA, including 76 regions with allelic losses and two with allelic gains (Fig. 2). The remaining 20 loci were regions of copy number variation (CNV) (Fig. 2), which is considered as a DNA segment that is 1 kb or larger and is present at variable copies of a particular gene(s) in comparison with a reference genome. Quantitative real-time PCR performed for 10 loci validated array-derived DNA CNA (data not shown).
Figure 1.
Overview of genomic changes in 30 primary MTCs determined by high-density comparative genomic hybridization array demonstrates predominant allelic loss. Genetic gains (red bars) and losses (green bars) are shown at each genomic position. The length of each bar indicates the percentage of the tumors with the indicated change. The vertical thickness indicates the magnitude of the chromosomal area involved.
Figure 2.
Heat map representations of the data from 30 MTCs across all the cytobands harboring genetic alterations ordered by genome position. This figure shows age, tumor size, RET mutation status (exons 10, 11, 13–16), and the presence of lymph node (LN) or distant (Dis) metastasis (mets) in 30 tumors. The heat map shows genomic loci with a loss (green) or gain (red) in 30% of the specific tumor group (S−, S+, and H+). This data set highlights the predominant allelic loss in MTC, irrespective of RET status. In total, 98 aberrations were detected, including 76 chromosomal losses, two gains, and 20 CNVs.
Common CNA in MTC
Overall, we observed 29 regions with at least a 30% CNA frequency in the 30 tumor samples. As expected CNVs comprised the majority of these alterations (15 of the 29 CNA). The remaining CNA were all allelic losses (Table 1). In all cases, a specific CNA was present in at least one tumor from each of the three genetically defined groups. Among the three groups, the RET positive sporadic tumors (S+) had the greatest number of imbalances (57), and the hereditary group had the fewest (36). The most frequent allelic loss occurred in four loci, 7q36.1, 12p13.31, 13q12.11 and 19p13.3-11.
Table 1.
Summary of CNA with 30% or greater frequency in all MTCs
| Cytoband | Physical map
|
Mapped genes (n) | Total CNA (n)
|
Overall CNA frequency (%) | |||
|---|---|---|---|---|---|---|---|
| Start point | End point | S− | S+ | H+ | |||
| 12p13.31 | 7696275 | 8062485 | 7 | 5 | 4 | 4 | 43 |
| 7q36.1 | 151686560 | 152069929 | 2 | 4 | 4 | 3 | 37 |
| 13q12.11 | 19254966 | 19420048 | 2 | 4 | 4 | 3 | 37 |
| 19p13.3-11 | 226925 | 19692685 | 644 | 4 | 5 | 2 | 37 |
| 5q13.2 | 68868713 | 70422356 | 6 | 3 | 6 | 1 | 33 |
| 5q31.1 | 132290783 | 132440481 | 3 | 3 | 4 | 3 | 33 |
| 6q13 | 74180226 | 74456183 | 5 | 3 | 4 | 3 | 33 |
| 9q31.2 | 109184070 | 109357673 | 1 | 4 | 2 | 4 | 33 |
| 21p11.1 | 10117898 | 10144936 | 5 | 2 | 3 | 5 | 33 |
| 22q13.1 | 37696964 | 37715431 | 1 | 3 | 6 | 1 | 33 |
| 9p22.1 | 18983768 | 19243114 | 3 | 2 | 4 | 3 | 30 |
| 17q11.2 | 26108458 | 26265366 | 2 | 5 | 3 | 1 | 30 |
| 22q12.2-3 | 22731196 | 30657843 | 71 | 3 | 4 | 2 | 30 |
| 22q13.2 | 37718669 | 41848706 | 92 | 4 | 4 | 1 | 30 |
Start and end positions are based on NCBI build 36 (hg18).
CNA specific to sporadic MTC
Twenty-one CNA were specific to sporadic MTC (Table 2). Surprisingly, there were no CNVs included among these allelic imbalances and no CNA was found to be specifically associated with hereditary MTC. The majority of CNA specifically associated with sporadic MTC occurred independent of RET mutation (13 of 21; Table 2). Furthermore, the presence of a RET mutation did not affect the overall number and/or size of CNA (S−, 25 ± 19; vs. S+, 30 ± 16; P = 0.5). Seven specific CNA occurred only in RET-positive sporadic tumors (Table 2). Finally, a single allelic loss at 11q23.3 was unique to RET-negative tumors.
Table 2.
Summary of 21 CNA specific to sporadic MTC tumors
| Cytoband | Physical map
|
Mapped genes (n) | CNA no. and frequency (%)
|
|||
|---|---|---|---|---|---|---|
| Start point | End point | S− | S+ | Frequency | ||
| Imbalances independent of RET status | ||||||
| 1p36.22-23 | 8867740 | 10433514 | 24 | 3 | 2 | 25 |
| 1p35.1 | 32695245 | 32893524 | 3 | 2 | 4 | 30 |
| 1p34.3 | 39124587 | 39304462 | 3 | 3 | 2 | 25 |
| 1p31.3 | 61892291 | 62734672 | 6 | 1 | 3 | 20 |
| 3p21.31 | 47018884 | 50073930 | 30 | 1 | 3 | 20 |
| 3q27.1 | 185265380 | 185329933 | 1 | 3 | 1 | 20 |
| 7p11.1-2 | 55695924 | 57405095 | 10 | 2 | 3 | 25 |
| 7q11.23 | 71887076 | 75407752 | 74 | 2 | 3 | 25 |
| 14q12 | 30525697 | 30878740 | 4 | 1 | 3 | 20 |
| 15q22.31 | 64420772 | 64430751 | 1 | 2 | 3 | 25 |
| 17q12 | 34217158 | 34217217 | 1 | 2 | 3 | 25 |
| 18q11.2 | 22018316 | 22032151 | 1 | 1 | 3 | 20 |
| 21q22.11 | 33658674 | 33833374 | 4 | 1 | 4 | 25 |
| Imbalances, specific to S+ | ||||||
| 4q12 | 56874636 | 57555635 | 16 | 0 | 3 | 15 |
| 5q11.2 | 56437065 | 56573822 | 1 | 0 | 3 | 15 |
| 5q13.2 | 70622715 | 70698853 | 0 | 0 | 3 | 15 |
| 9p24.1 | 6610160 | 6848238 | 2 | 0 | 3 | 15 |
| 16p12.1 | 21925400 | 21925459 | 0 | 0 | 4 | 20 |
| 21q21.1 | 17667404 | 17854835 | 1 | 0 | 3 | 15 |
| 21q22.2 | 39553689 | 39729222 | 4 | 0 | 3 | 15 |
| Imbalances, specific to S− | ||||||
| 11q23.3 | 118321536 | 118342357 | 2 | 3 | 0 | 15 |
Start and end positions are based on NCBI build 36 (hg18).
Clustering and pathway analysis
We performed unbiased hierarchical clustering analysis, first using all 98 CNA shown in Fig. 2, and second using the entire genomic array dataset. Neither analysis found any clear links between any of the tumor properties and CNA profile or RET mutation status (data not shown). A K-means clustering analysis failed to segregate by the tumors based on mutation status when samples were distributed into the three most similar groups (data not shown).
We performed an Ingenuity Pathways Analysis including all CNA with at least 30% frequency in all MTC (844 genes, Table 1) showing cellular growth and proliferation as the most significant pathways (data not shown). A second analysis using the genes mapped by CNA occurring only in sporadic tumors (187 genes; Table 2) showed cell death as one of the most significant pathways (data not shown).
Discussion
In this study we used high-density array-comparative genomic hybridization to define new genomic CNA and genes potentially involved in MTC tumorigenesis.
Overall, we identified 98 discrete regions with allelic imbalances in MTC tumor samples. Among these regions, 20 were identified as CNVs. Unfortunately, the lack of matched germline DNA for most of the samples prevented defining these alterations as germline. Having matched germline DNA is preferable to overcome the genetic variation of the individual patient. To reduce the noise of genetic variations maximally, we used a pool of normal genomic DNA and focused on the most frequent aberrations (greater than 30%). However, in all cases where DNA was available, no copy number differences were observed between tumor and germline DNA. Therefore, we believe that most of these CNVs likely represent normal sequence variation. The importance of this observation lies in the fact that many of the regions previously identified as MTC-specific gains map to areas containing CNVs. We found that 76 of 78 somatic CNA had allelic loss, underscoring the significance of the overall allelic loss and the possible roles of tumor suppressor genes (TSGs) in MTC. Functional losses of TSGs have been reported in human MTC tumors (4,5,11,12,13), and previous studies on knockout mice models also showed the importance of TSG in MTC development and progression (12,14), reinforcing the significance of our findings.
A majority of the somatic CNA (57 of 78) were identified in all three MTC subgroups (S−, S+, and H+). The most common allelic losses occurred at 12p13.31, 7q36, 13q12.11, and 19p13 (Table 1). Allelic losses of 12p, 13q, and 19p have been previously identified in MTC (9,10,11), whereas loss of 7q36 has never been associated to MTC. We detected large chromosomal loss of 22q and 1p (Fig. 2), which have also been reported by previous loss of heterozygosity and comparative genomic hybridization studies (4,5,10,11). The similar distributions of the CNA across the three defined MTC subgroups and the absence of genetic abnormalities distinctive to hereditary MTC suggest an underlying common molecular signature that functions in C cell tumorigenesis. This concept is also supported by unsupervised clustering analysis of the comparative genomic hybridization data set, which failed to segregate the tumors into genetically defined groups.
Although the majority of somatic CNA were distributed among all the tumor groups, we found 21 CNA specific to sporadic MTC. This finding is most likely related to the fewer overall CNA observed in hereditary group vs. sporadic MTC. Pathway analysis found the involvement of cell death pathways in the etiology of sporadic MTC, confirming the low rate of apoptotic cell death in MTC (15,16).
What remains unclear from our study is how C cell tumorigenesis proceeds in the absence of an activating RET mutation. Allelic loss at 11q23.3 was the only region specifically associated with RET negative sporadic MTC (S−). Of note, previous studies have found 11q23.3 commonly deleted in neuroblastomas (17), despite a recent study suggesting that single nucleotide polymorphisms at chromosome band 6p22 seem to be important in the initiation of neuroblastoma (18). Specific single nucleotide polymorphisms at precise chromosomes may contribute in the initiation of MTC as well.
However, because we only sequenced the hot spots RET mutations, the possibility of novel mechanisms of activation cannot be ruled out.
In conclusion, we have precisely defined 98 CNA associated with MTC, which dramatically narrows the span of the candidate regions and genes. The high frequency of many of these alterations suggests that these regions may play an important role in C cell tumorigenesis. Furthermore, the fact that 97% of the somatic CNA are represented by allelic losses underscores the possible involvement of additional TSGs in the mechanism of MTC tumorigenesis. The fact that similar gene profiles were found in different MTC subgroups reinforces the concept that a RET mutation by itself is insufficient to drive tumorigenesis. Whether these genetic events involve genes in cell proliferation and growth as suggested by pathway analysis remains to be addressed.
Acknowledgments
We are grateful to the Kosberg Foundation and to John and Jennifer Ball, who generously supported this research, and to the Cancer Genomics Core Resource at The University of Texas, M. D. Anderson Cancer Center, which is supported by National Cancer Institute Grant CA16672.
Footnotes
Disclosure Statement: The authors have nothing to disclose.
First Published Online September 2, 2008
Abbreviations: CNA, Copy number alteration(s); CNV, copy number variation; MEN2, multiple endocrine neoplasia syndrome type 2; MTC, medullary thyroid carcinoma; TSG, tumor suppressor gene.
References
- Drosten M, Pützer BM 2006 Mechanisms of disease: cancer targeting and the impact of oncogenic RET for medullary thyroid carcinoma therapy. Nat Clin Pract Oncol 3:564–574 [DOI] [PubMed] [Google Scholar]
- Messina M, Robinson BG 2007 Technology insight: gene therapy and its potential role in the treatment of medullary thyroid carcinoma. Nat Clin Pract Endocrinol Metab 3:290–301 [DOI] [PubMed] [Google Scholar]
- Cranston AN, Ponder BA 2003 Modulation of medullary thyroid carcinoma penetrance suggests the presence of modifier genes in a RET transgenic mouse model. Cancer Res 63:4777–4780 [PubMed] [Google Scholar]
- Khosla S, Patel VM, Hay ID, Schaid DJ, Grant CS, van Heerden JA, Thibodeau SN 1991 Loss of heterozygosity suggests multiple genetic alterations in pheochromocytomas and medullary thyroid carcinomas. J Clin Invest 87:1691–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan LM, Gardner E, Smith BA, Mathew CG, Ponder BA 1993 Genetic events in tumour initiation and progression in multiple endocrine neoplasia type 2. Genes Chromosomes Cancer 6:166–177 [DOI] [PubMed] [Google Scholar]
- Huang SC, Koch CA, Vortmeyer AO, Pack SD, Lichtenauer UD, Mannan P, Lubensky IA, Chrousos GP, Gagel RF, Pacak K, Zhuang Z 2000 Duplication of the mutant RET allele in trisomy 10 or loss of the wild-type allele in multiple endocrine neoplasia type 2-associated pheochromocytomas. Cancer Res 60:6223–6226 [PubMed] [Google Scholar]
- Koch CA, Brouwers FM, Vortmeyer AO, Tannapfel A, Libutti SK, Zhuang Z, Pacak K, Neumann HP, Paschke R 2006 Somatic VHL gene alterations in MEN2-associated medullary thyroid carcinoma. BMC Cancer 6:131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CA, Huang SC, Moley JF, Azumi N, Chrousos GP, Gagel RF, Zhuang Z, Pacak K, Vortmeyer AO 2001 Allelic imbalance of the mutant and wild-type RET allele in MEN 2A-associated medullary thyroid carcinoma. Oncogene 20:7809–7811 [DOI] [PubMed] [Google Scholar]
- Hemmer S, Wasenius VM, Knuutila S, Franssila K, Joensuu H 1999 DNA copy number changes in thyroid carcinoma. Am J Pathol 154:1539–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisk T, Zedenius J, Lundberg J, Wallin G, Kytola S, Larsson C 2001 CGH alterations in medullary thyroid carcinomas in relation to the RET M918T mutation and clinical outcome. Int J Oncol 18:1219–1225 [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Theodosopoulos G, Martin-Schulte K, Richardson AL, Philips J, Röher HD, Delbridge L, Robinson BG 2003 Genome-wide copy number imbalances identified in familial and sporadic medullary thyroid carcinoma. J Clin Endocrinol Metab 88:1866–1872 [DOI] [PubMed] [Google Scholar]
- van Veelen W, van Gasteren CJ, Acton DS, Franklin DS, Berger R, Lips CJ, Höppener J 2008 Synergistic effect of oncogenic RET and loss of p18 on medullary thyroid carcinoma development. Cancer Res 68:1329–1337 [DOI] [PubMed] [Google Scholar]
- Wang DG, Liu WH, Johnston CF, Sloan JM, Buchanan KD 1998 Bcl-2 and c-Myc, but not bax and p53, are expressed during human medullary thyroid tumorigenesis. Am J Pathol 152:1407–1413 [PMC free article] [PubMed] [Google Scholar]
- Coxon AB, Ward JM, Geradts J, Otterson GA, Zajac-Kaye M, Kaye FJ 1998 RET cooperates with RB/p53 inactivation in a somatic multi-step model for murine thyroid cancer. Oncogene 17:1625–1628 [DOI] [PubMed] [Google Scholar]
- Hinze R, Gimm O, Taubert H, Bauer G, Dralle H, Holzhausen HJ, Rath FW 2000 Regulation of proliferation and apoptosis in sporadic and hereditary medullary thyroid carcinomas and their putative precursor lesions. Virchows Arch 437:256–263 [DOI] [PubMed] [Google Scholar]
- Mise N, Drosten M, Racek T, Tannapfel A, Pützer BM 2006 Evaluation of potential mechanisms underlying genotype-phenotype correlations in multiple endocrine neoplasia type 2. Oncogene 25:6637–6647 [DOI] [PubMed] [Google Scholar]
- Guo C, White PS, Weiss MJ, Hogarty MD, Thompson PM, Stram DO, Gerbing R, Matthay KK, Seeger RC, Brodeur GM, Maris JM 1999 Allelic deletion at 11q23 is common in MYCN single copy neuroblastomas. Oncogene 18:4948–4957 [DOI] [PubMed] [Google Scholar]
- Kushner BH, Cheung NK 2008 Neuroblastoma—linking a common allele to a rare disease. N Engl J Med 358:2585–2593 [DOI] [PubMed] [Google Scholar]