Abstract
Background
Large-scale protein interaction maps provide a new, global perspective with which to analyse protein function. PSIMAP, the Protein Structural Interactome Map, is a database of all the structurally observed interactions between superfamilies of protein domains with known three-dimensional structure in the PDB. PSIMAP incorporates both functional and evolutionary information into a single network.
Results
We present a global analysis of PSIMAP using several distinct network measures relating to centrality, interactivity, fault-tolerance, and taxonomic diversity. We found the following results: Centrality: we show that the center and barycenter of PSIMAP do not coincide, and that the superfamilies forming the barycenter relate to very general functions, while those constituting the center relate to enzymatic activity. Interactivity: we identify the P-loop and immunoglobulin superfamilies as the most highly interactive. We successfully use connectivity and cluster index, which characterise the connectivity of a superfamily's neighbourhood, to discover superfamilies of complex I and II. This is particularly significant as the structure of complex I is not yet solved. Taxonomic diversity: we found that highly interactive superfamilies are in general taxonomically very diverse and are thus amongst the oldest. Fault-tolerance: we found that the network is very robust as for the majority of superfamilies removal from the network will not break up the network.
Conclusions
Overall, we can single out the P-loop containing nucleotide triphosphate hydrolases superfamily as it is the most highly connected and has the highest taxonomic diversity. In addition, this superfamily has the highest interaction rank, is the barycenter of the network (it has the shortest average path to every other superfamily in the network), and is an articulation vertex, whose removal will disconnect the network. More generally, we conclude that the graph-theoretic and taxonomic analysis of PSIMAP is an important step towards the understanding of protein function and could be an important tool for tracing the evolution of life at the molecular level.
Keywords: Structural Interactome, Protein Interaction, Interactomics, Graph-theory, Interaction Rank, Taxonomic Diversity, PSIEYE, PSIMAP.
Background
Large-scale protein interaction maps [1-9] have increased our understanding of protein function, extending 'functional context' to the network of interactions which span the proteome [10-13]. Functional genomics has fuelled this new perspective and has directed research towards computational methods of reconstructing genome-scale interaction maps.
One group of computational methods uses the abundant genomic sequence data, and is based on the assumption that genomic proximity and gene fusion result from a selective pressure to genetically link proteins which physically interact [14-16]. With the exception of conserved operons and gene fusion, however, genomic proximity is more generally indicative of indirect functional associations between proteins [17] than direct interactions between the gene products.
A second group of methods, based on the assumption that protein-protein interactions are conserved across species, was originally applied to genomic comparisons [18]. Just as common function can be inferred between homologous proteins, 'homologous interaction' can be used to infer interaction between homologues of interacting proteins. This method has been validated in a comparison between PSIMAP, which contains observed protein domain interactions in the Protein Data Bank (PDB) [19] and experimentally determined domain interactions in yeast [20]. The method has also been systematically validated at the sequence level using BLAST [21], and has been improved by the use of a statistical domain level representation of the known protein interactions [22,23].
PSIMAP, the Protein Structural Interactome Map [20], is a database of all the structurally observed interactions between protein domains of known three-dimensional structure in the PDB. It can be constructed using any reliable protein domain definition, where domains are defined as evolutionarily conserved structural and functional protein units. Here we use the domain definitions provided by SCOP (Structural Classification of Proteins) [24], which uses structural and functional homology to manually define evolutionarily distinct protein domain families and superfamilies. Alternatively, other domain definitions (such as CATH [25], FSSP [26], Pfam [27], etc.) can be used.
Domains from a multi-domain PDB entry are empirically denoted as interacting with each other if at least 5 residue pairs are within 5 Angstroms (see Figure 1). Although the data in the PDB is relatively limited in comparison to the available sequence data, it is much more comprehensive when compared to the available protein interaction data [28].
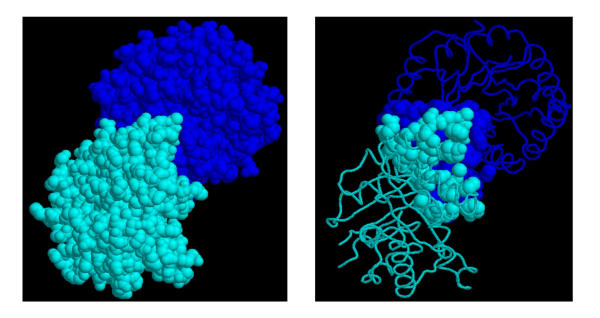
Figure 1.
Two interacting domains. Given two domains with coordinates of their residues (left), PSIMAP detects all residue pairs of the two domains within a given distance threshold (right). The two domains shown are classic TIM barrel folds from triosephosphate isomerase (7 tim).
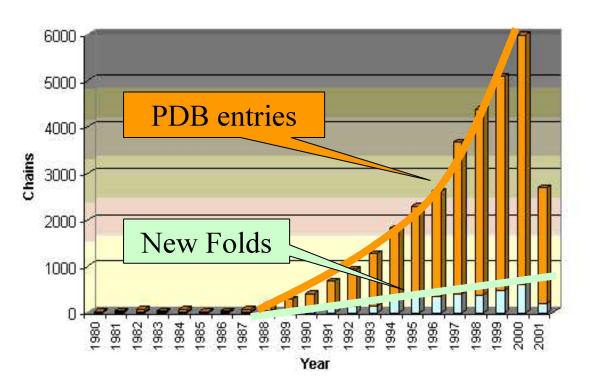
PSIMAP provides an overview of all the observed domain-domain interactions at the superfamily level. Considering interactions at this level is important with respect to the stability of the network; while the number of PDB entries is growing superlinearly, the number of new folds is only increasing linearly (see Figure 2). It is probable that there are no more than 2,000 distinct protein topologies in nature [29-33]. Because of the slow growth in the number of new superfamilies and superfamily interactions over time (data not shown) PSIMAP represents the first global overview of interactions at this level. For example the recent conservative superfamily assignment of 56 genomes covered between 40–67% of the total detected genes in eukaryotes and eubacteria (~100,000 genes) and between 31–54% of the total detected genes in archaebacteria (~10,000 genes) [34]. As a significant portion of the unassigned genes may represent trans-membrane proteins not structurally determined due to experimental difficulty, it is reasonable to suggest that the PDB, and PSIMAP, covers many of the existing globular superfamilies in nature.
Figure 2.
PSIMAP is based on the Protein Data Bank [19], which grows exponentially. PSIMAP is nonetheless relatively stable, as it considers interactions at superfamily level, which grows only linearly.
By viewing interaction between superfamilies, which encompass extremely distant evolutionary relationships [24], PSIMAP represents domain interaction within a broad evolutionary context. The analysis of PSIMAP's network topology presented here necessarily incorporates this evolutionary perspective.
Using different numbers of residue-residue contacts within different distances (contact threshold and distance threshold respectively) has a striking effect on the total number of superfamily-superfamily interactions defined. An analysis of the empirical domain interaction criterion is shown in Figure 3. Above the 4 Angstrom distance threshold, different contact thresholds yield qualitatively similar results, giving a roughly linear increase in the number of superfamily-superfamily interactions observed as the contact threshold is decreased. At the 4 Angstrom distance threshold, however, the contact threshold has the biggest effect on the number of domain–domain interactions observed, giving a roughly exponential increase in the number of superfamily-superfamily interactions observed as the contact threshold is decreased. This suggests that most domain–domain interactions occur in this approximate distance range (between 4 and 5 Angstroms). Using a contact threshold of 5 is very discriminative at the 4 Angstroms distance threshold, so the "5 by 5 rule" (defined previously [20]) is a reasonably safe choice of interaction criteria. Additionally, Tsai et al [35] show that extracting domain interaction from the PDB is a robust process.
Figure 3.
Number of superfamily-superfamily interactions observed using different residue-residue contact count and residue contact distance thresholds to analyse domain-domain contacts in the PDB. Below four Angstroms almost all superfamilies are 'isolated' from the interaction network, making very few residue-residue contacts with other superfamilies at this range. At four Angstroms, the number of interactions observed is critically dependant on the number of residue contacts threshold used, while at five Angstroms the contact count threshold has less effect.
By using a structural domain definition to extract domain-domain interactions from the PDB, it is possible to assign covalently linked domains as interacting. These 'intra-interactions' are in the minority, accounting for approximately 30% of the 20370 domain-domain interactions observed. For a breakdown of the 1232 observed superfamily-superfamily interactions (generated using SCOP version 1.61) see table 1. The validity of assigning superfamily interaction solely on the basis of observed intra-domain (covalently linked) interaction is extensive. Domain fusion has been successfully used to predict protein interaction from sequence information alone [17] and as a hypothesis for the evolution of homo [36] and hetero [14] dimers. In addition, it has been observed that intra-domain interfaces have strong similarities to inter-domain interfaces within multi-domain proteins [35,37,38]. Finally, such multi domain proteins can be identified as independent, interacting domains in ancestral genomes [14].
Table 1.
Breakdown of superfamily-superfamily interactions according to inter- and intra-interactions for homomers and heteromers. Intra-interactions are in the minority.
| Superfamily | Homomer | Heteromer | TOTAL |
| Intra-Interaction | 23 | 248 | 271 |
| Inter-Interaction | 471 | 260 | 731 |
| Both | 90 | 140 | 230 |
| TOTAL | 584 | 648 | 1232 |
To check for potential sampling error in the PDB, we checked if the absolute number of domains in a superfamily is correlated to the number of observed interactions that that superfamily makes. We did not find significant evidence for this correlation: omitting four outliers, the correlation coefficient between the number of interactions and number of domains in a superfamily is only 0.16. This suggests that a superfamily's interactivity is independent of its occurrence in the PDB.
Visualizing structurally observed protein domain interaction at the superfamily level gives a very robust network which incorporates both a broad evolutionary perspective of protein interaction with the conserved structural and functional features of protein domains. PSIMAP, therefore, represents a rich, stable overview of the protein interactome.
Results
Protein interaction databases such as BIND [39] and DIP [40] provide web interfaces which allow the examination of a small number of individual proteins and their interactions. They do not support the large-scale visualisation of protein interaction networks. This need has been addressed by several visualisation systems [41-43]. Protein interaction networks are large, however, requiring more than simple visualisation for effective data mining [12]. Consequently, there have been several global analyses of protein interaction networks (for example [44-46]). Here, we employ an integrated package (PSIEYE [47]), which complements these approaches by integrating several graph-theoretic and taxonomic measures with network visualisation and exploration. Our analysis has been motivated by several questions of particular biological interest (outlined below). While these questions are relevant to the analysis of any biological network, it is important to note the additional evolutionary perspective provided by PSIMAP when analysing this network.
1. Which superfamilies can directly or indirectly interact with each other forming subnetworks (does PSIMAP contain evolutionarily distinct interaction networks)?
2. Which superfamilies can disrupt a pathway in the network if removed (highlighting critical pathways or distinct functional contexts for superfamilies in PSIMAP)?
3. Are there multiple indirect interactions between superfamilies (making the overall superfamily interaction network topology robust, highlighting sets of superfamilies with common functional roles)?
4. How many interaction partners has a superfamily acquired over the course of evolution?
5. How well connected is the neighbourhood of a superfamily (how is a superfamily related to the rest of the network)?
6. How central is a superfamily in the network (which superfamilies make the most fundamental contribution to the overall network)?
7. Is there a core in the network (has the network grown from a core, critical set of interactions)?
8. How is the network distributed within and between taxonomic groups with respect to the above measures (how diverse is the interaction network in nature)?
The last question is particularly relevant for PSIMAP, as it is based on a reliable definition of homology applied to all the available multi-domain protein structures in the PDB. PSIMAP forms a global interaction network across many species, which can be extended using sequence-based homology searches [48]. We applied graph-theoretic and taxonomic measures to PSIMAP using the PSIEYE tool to answer the above questions.
The PSIMAP algorithm generates a large network consisting of 937 superfamilies and 538 interactions, with 512 distinct components ranging in size from the single largest component of 320, which will be the basis for further analysis, to some 400 isolated non-interacting single superfamilies, distributed according to a power-law. These and all subsequent analysis are based on PSIMAP produced from SCOP version 1.59. To analyse PSIMAP we will follow two strands: First, we looked at the network topology of the map in terms of location and interactivity. Second, we characterized the taxonomic diversity of the superfamilies. The former analysis can be broken down into two distinct aspects: location and the interactivity.
Location
Previously, it has been shown that central proteins in an interaction network are often functionally critical and their removal correlates to lethality [44]. Wuchty and Stadler define three types of centrality and apply them to metabolic and protein interaction networks [45].
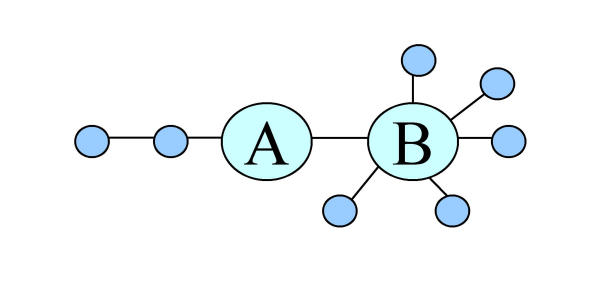
We follow this approach and use two measures of network centrality, namely eccentricity and sum of distances. The eccentricity of a vertex (used to represent a superfamily in PSIMAP) is the path distance to the farthest vertex in the network. The vertices with the minimum eccentricity form the center of the network. In contrast to eccentricity, the sum of distances averages the path distance to all other vertices in the network. The barycenter is the vertex or vertices with the minimal sum of distances. Given these definitions, the center and barycenter are not necessarily the same as shown in Figure 4, where vertex A is the center, but its neighbour B is the barycenter.
Figure 4.
Eccentricity and sum of distance. The vertex A is the center of the network, having the least distance to travel to its furthest node. Vertex B is the barycenter, having the least overall distance to every other node.
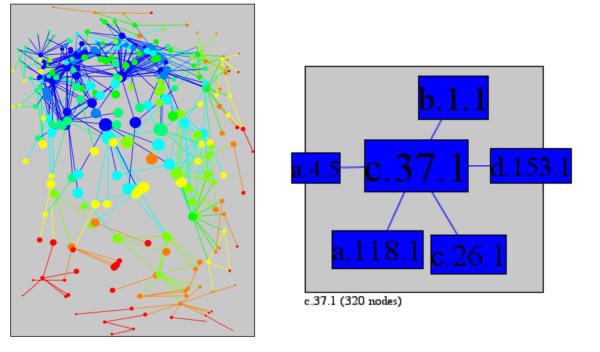
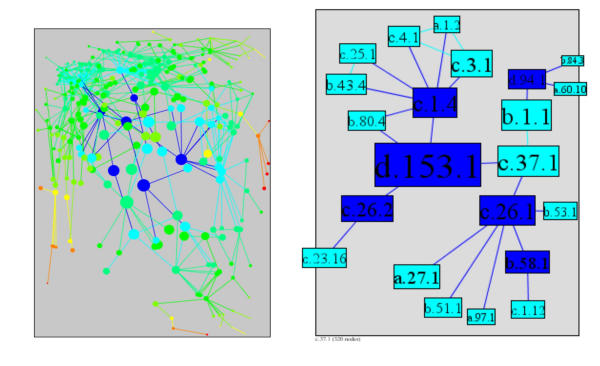
In PSIMAP, the P-loop containing nucleotide triphosphate hydrolases (c.37.1) is the barycenter (see Figure 5) with the minimum sum of distances (taking 947 steps to reach the 320 superfamilies in the largest component). It is followed in the measurement of minimum sum of distances by Immunoglobulin (b.1.1), N-terminal nucleophile aminohydrolases (NTN hydrolases) (d.153.1), ARM repeat (a.118.1), Nucleotidylyl transferase (c.26.1) and Winged helix DNA-binding domain (a.4.5). These superfamilies are involved in a broad and comprehensive range of critical cellular functions, such as regulation of gene expression, cellular transport, control of the cytoskeleton, phosphorylation, nuclear division, signalling, A/GTPase activity, immunity, and carbon and nitrogen metabolism. These nearly ubiquitous and critical functions associated with superfamilies close to the barycenter reflect their critical position in the network as these superfamilies are, on average, closely associated with every other superfamily in the network's main component. By contrast, the most peripheral superfamily, with the maximum sum of distances (taking 3248 steps to reach the 320 superfamilies in the main component) is the GroEL-like chaperone, ATPase domain superfamily (a.129.1). This superfamily has a very specific function, mediating the folding and organisation of other polypeptides in order that they form the correct oligomeric structure [49].
Figure 5.
The sum of distance, the sum of all the shortest paths from a superfamily to every other superfamily in the network. Blue indicates low sum of distance (central) and red high sum of distance (rim). The 6 superfamilies with lowest sum of distance are c.37.1, P-loop containing nucleotide triphosphate hydrolases; b.1.1, Immunoglobulin; d.153.1, N-terminal nucleophile aminohydrolases (Ntn hydrolases); a.118.1, ARM repeat; c.26.1, Nucleotidylyl transferase; a.4.5, Winged helix DNA-binding domain
The center of the network is in the same neighbourhood as the barycenter with two superfamilies (NTN hydrolases, d.153.1 and nucleotidylyl transferase, c.26.1) in common within the six highest of both centrality measures. There are six centers in the PSIMAP network with equally small eccentricity. They are PK beta-barrel domain-like (b.58.1), Nucleotidylyl transferase (c.26.1), Ntn hydrolases (d.153.1), FMN-linked oxidoreductases (c.1.4), HPr-like (d.94.1) and Adenine nucleotide alpha hydrolases (c.26.2). As with the superfamilies which rank highly in the barycenter measurement, these superfamilies are involved in highly critical cellular functions including glycolysis; galactose / fructose metabolism and nucleotide, amino acid, lipopolysaccharide, NAD and ATP synthesis. In comparison to the barycenter superfamilies, members of the center are related to more specific enzymatic activities with an emphasis on energy metabolism and macromolecular synthesis. Conversely, members of the barycenter mediate their function via structural interactions, involving molecular switching, signalling, transport, DNA binding and protein-protein interaction. Additionally, the majority of the observed enzymatic functions in the barycenter can be attributed to the ubiquitous P-loop domain.
The slight shift in topology between the center and the barycenter in PSIMAP reflects a slight shift in the functional characteristics of the overlapping subgroups of superfamilies in the topological region. Intuitively, we hypothesize that those critical superfamilies which have general functions or a predominantly structural mode of action will have a greater number of interaction partners (which is a requirement for the highest sum of distance in the barycenter). More specific but none the less critical enzymatic functions on the other hand will be associated with many different pathways, but may mediate indirect functional roles via common metabolites and thus need not make direct physical interactions with many different members of the network.
Global overviews of the center and barycenter are given in figures 5 and 6. The colour-coding of these figures indicates that the majority of superfamilies have medium eccentricity, yet small sum of distances. Intuitively, the low sum of distance means that the majority of superfamilies are member of or attached to a well-connected core and can thus reach all other superfamilies via short average paths. Eccentricity does not take this aspect of connectivity into account and most superfamilies have medium eccentricity.
Figure 6.
A superfamily's eccentricity is the maximal distance to any other superfamily in the network. Low eccentricity values (central) are coloured in blue, high values (rim) in red. The 6 most central superfamilies are b.58.1, PK beta-barrel domain-like; c.26.1, Nucleotidylyl transferase; d.153.1, N-terminal nucleophile aminohydrolases (Ntn hydrolases); c.1.4, FMN-linked oxidoreductases; d.94.1, HPr-like; c.26.2, Adenine nucleotide alpha hydrolases.
Interactivity
PSIEYE provides three measures of interactivity: connectivity, cluster index, and interaction rank. The connectivity of a vertex is simply the number of interaction partners it has. The superfamilies shown in table 2 are the 19 most interactive in PSIMAP.
Table 2.
The 19 most interactive superfamilies.
| Connectivity | SCOP ID | Superfamily |
| 46 | c.37.1 | P-loop containing nucleotide triphosphate hydrolases |
| 38 | b.1.1 | Immunoglobulin |
| 14 | c.1.8 | (Trans)glycosidases |
| 12 | d.58.1 | 4Fe-4S ferredoxins |
| 11 | a.3.1 | Cytochrome c |
| 11 | a.4.5 | Winged helix DNA-binding domain |
| 11 | c.3.1 | FAD/NAD(P)-binding domain |
| 11 | d.15.4 | 2Fe-2S ferredoxin-like |
| 10 | b.40.4 | Nucleic acid-binding proteins |
| 10 | d.142.1 | Glutathione synthetase ATP-binding domain-like |
| 9 | a.39.1 | EF-hand |
| 9 | c.1.4 | FMN-linked oxidoreductases |
| 9 | c.2.1 | NAD(P)-binding Rossmann-fold domains |
| 8 | c.55.1 | Actin-like ATPase domain |
| 8 | d.144.1 | Protein kinase-like (PK-like) |
| 8 | d.15.1 | Ubiquitin-like |
| 7 | d.145.1 | FAD-binding domain |
| 7 | d.92.1 | Metalloproteases (zincins) catalytic domain |
Figure 7 shows the most highly connected superfamilies in PSIMAP form a single connected component. Thus, the high connectivity, core superfamilies do not break down into distinct clusters, but rather form one single, central kernel at the heart of the network.
Figure 7.
The 19 most highly connected superfamilies form a connected component and are also highly diverse as the colour coding shows (red = high diversity). Clockwise from 9 o'clock: c.1.4, FMN-linked oxidoreductases; c.3.1, FAD/NAD(P)-binding domain; d.58.1, 4Fe-4S ferredoxins; d.15.4, 2Fe-2S ferredoxin-like; d.145.1, FAD-binding domain; a.3.1, Cytochrome c; b.1.1, Immunoglobulin; c.1.8, (Trans)glycosidases; c.37.1, P-loop containing nucleotide triphosphate hydrolases; a.118.1, ARM repeat; d.92.1, Metalloproteases (zincins), catalytic domain; d.144.1, Protein kinase-like (PK-like); d.15.1, Ubiquitin-like; b.40.4, Nucleic acid-binding proteins; a.39.1, EF-hand; a.4.5, Winged helix DNA-binding domain; c.55.1, Actin-like ATPase domain; c.2.1, NAD(P)-binding Rossmann-fold domains; d.142.1, Glutathione synthetase ATP-binding domain-like.
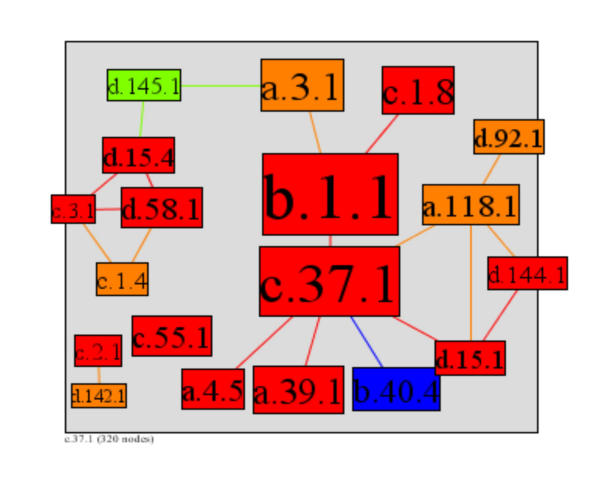
The majority of the most highly connected superfamilies contain families of functionally important enzymes, with only three main exceptions. They are: 1) domains from the Immunoglobulin superfamily (b.1.1), frequently found as domain linkers in genomic sequences and structures, having diverse structural roles and interacting with many different proteins; 2) domains from the EF hand all-alpha superfamily (a.39.1), a structural motif (with an average size of around 40 amino acids) involved in calcium binding and the diverse regulatory functions associated with calcium; 3) the winged helix DNA-binding domain (a.4.5), which has an extremely diverse set of functions related to DNA binding. For example, the winged helix domain associates with many different small molecule binding domains to form functionally diverse families of transcription factors in prokaryotes and eukaryotes [50].
The most highly interactive superfamilies take part in a wide range of critical cellular reactions, mostly relating to energy metabolism and catabolism as well as signalling and structural roles. For example, the iron-sulfur proteins (d.58.1 and d.15.4) transfer electrons in a wide variety of metabolic reactions, indicating a very early origin in protein evolution. The PK-like superfamily (d.144.1) encompasses enzymes that belong to a very extensive family of proteins involved in almost all aspects of eukaryotic signal transduction pathways, including regulation of the cell cycle, differentiation, homeostasis and the immune response. Members of this superfamily share a conserved catalytic core common with both serine/threonine and tyrosine protein kinases [51], and have related but uncharacterised counterparts in archae as well as functional homologues in viruses. Ubiquitin-like superfamily domains are found in an extremely broad range of protein families, having structural roles in proteolysis (including the unfolded protein response pathway), and linking cytoskeleton proteins to proteins in the plasma membrane, as well as having roles in signal transduction. Raf-like and Ras-binding activity, guanine nucleotide exchange activity and GTP activated Phosphatidylinositol 3-kinase activity as part of the phosphatidylinositol 3-kinase complex [52]. This superfamily is also involved in DNA repair mechanisms, chromosome segregation, viral infection, splicing, autophagy and the regulation of membrane physical properties and cell development.
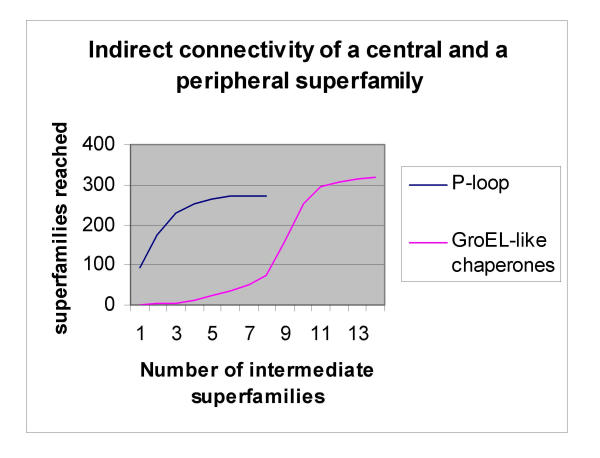
Connectivity can be extended to include the next layer of interaction partners via intermediate partners (indirect connectivity). The most highly connected and central superfamily, the P-loop (c.37.1), can reach 92 superfamilies via one intermediate and already 253 via four intermediate superfamilies. In contrast, the peripheral superfamily, GroEL-like chaperones, ATPase domain (a.129.1) can reach only one other superfamily via one intermediate and only 11 other superfamilies via four intermediate superfamilies (figure 8). Thus, the outreach of the P-loop is far greater than the GroEL-like chaperones, reflecting the centrality and connectivity of the P-loop.
Figure 8.
Central and highly connected superfamilies such as the P-loop reach a much larger number of other superfamilies via intermediate superfamilies than peripheral superfamilies such as the GroEL-like chaperones.
Indirect connectivity is useful in that it links the measures of connectivity to the location measures discussed above. Due to the particular structure of the network, the most highly interactive and central superfamilies also have higher indirect connectivity than peripheral superfamilies. Indirect connectivity shifts our perspective from a purely local view of interactivity towards a more global view. However, connectivity and indirect connectivity do not quantify interaction density, a measure to describe the extent to which a superfamily's interaction partners interact with each other.
Cluster index [53] is a measure of interaction density, and is defined as the number of interactions between a vertex's neighbours divided by the total number of possible interactions between them. A cluster index of 0 means that none of a vertex's neighbours interact, whereas a cluster index of 1 indicates that they all interact with each other.
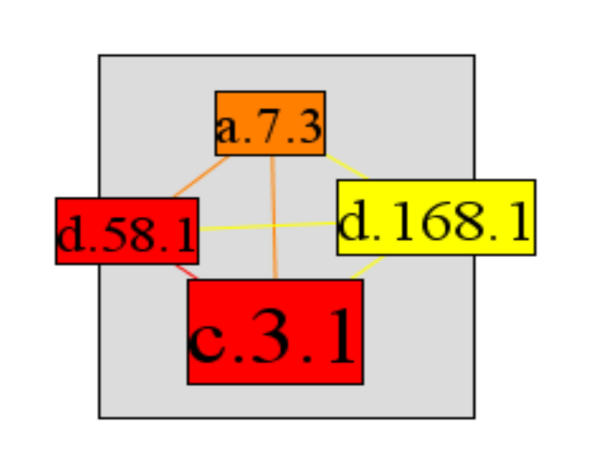
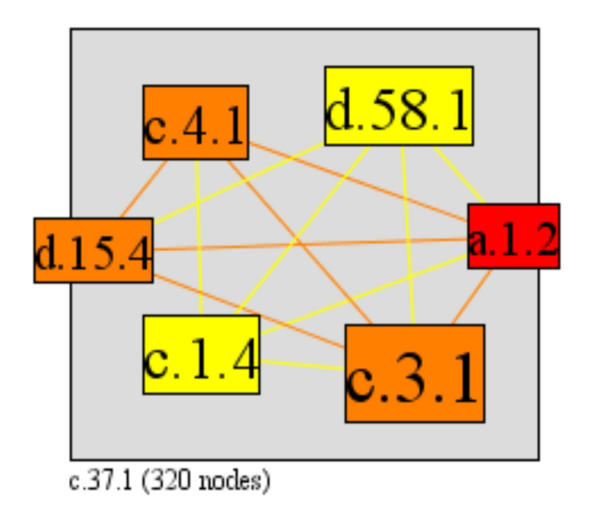
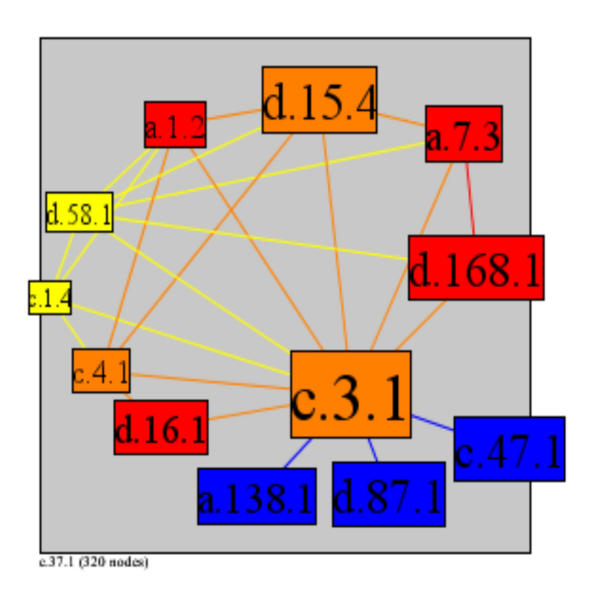
A high cluster index is more likely for low connectivity superfamilies, as the number of possible interactions between neighbours increases quadratically with an increasing number of interaction partners. This is highlighted by looking at the cluster index of the P-loop (c.37.1), which is the most highly interactive superfamily with 46 interaction partners, but which has a very low cluster index (0.011). In contrast, the succinate dehydrogenase/fumarate reductase catalytic domain (d.168.1) has the highest possible cluster index of 1, as all of its three interaction partners, the 4Fe-4S ferredoxins (d.58.1), succinate dehydrogenase/fumarate reductase C-terminal domain (a.7.3), and FAD/NAD(P)-binding domain (c.3.1) interact with each other (Figure 9). There are exceptions to this general trend, however, as some superfamilies have both a relatively high number of interaction partners and a high cluster index. The alpha-helical ferredoxin superfamily (a.1.2) has five interaction partners and a very high cluster index of 0.8. This superfamily interacts with FAD/NAD(P)-binding domain (c.3.1), FMN-linked oxidoreductases (c.1.4), 2Fe-2S ferredoxin-like (d.15.4), Nucleotide-binding domain (c.4.1), and 4Fe-4S ferredoxin (d.58.1) (Figure 10). One of this set of is another superfamily with a relatively high number of interactions and a medium cluster index; the FAD/NAD(P)-binding domain superfamily (c.3.1) with 11 partners and a cluster index of 0.236 (Figure 11).
Figure 9.
Succinate dehydrogenase/fumarate reductase catalytic domain (d.168.1) has the highest possible cluster index of 1, as all its three interaction partners 4Fe-4S ferredoxins (d.58.1), Succinate dehydrogenase/fumarate reductase C-terminal domain (a.7.3), and FAD/NAD(P)-binding domain (c.3.1) interact with each other.
Figure 10.
Superfamily a.1.2 has 5 interaction partners and very high cluster index of 0.8 (0 minimum, 1 maximum), its 5 neighbours have many interactions among each other. (a.1.2, alpha-helical ferredoxin; c.3.1, FAD/NAD(P)-binding domain; c.1.4, FMN-linked oxidoreductases; d.15.4, 2Fe-2S ferredoxin-like; c.4.1, Nucleotide-binding domain; d.58.1, 4Fe-4S ferredoxins)
Figure 11.
Superfamily c.3.1 has 11 interaction partners and medium cluster index. Clockwise from 5 pm: c.3.1, FAD/NAD(P)-binding domain; c.47.1, Thioredoxin-like; d.87.1, FAD/NAD-linked reductases dimerisation (C-terminal) domain; a.138.1, Multiheme cytochromes; d.16.1, "FAD-linked reductases C-terminal domain"; c.4.1, Nucleotide-binding domain; c.1.4, FMN-linked oxidoreductases; d.58.1, 4Fe-4S ferredoxins; a.1.2, alpha-helical ferredoxin; d.15.4, 2Fe-2S ferredoxin-like; a.7.3, Succinate dehydrogenase/fumarate reductase C-terminal domain; d.168.1, Succinate dehydrogenase/fumarate reductase catalytic domain
The interaction partners of the 3 superfamilies d.168.1, a.1.2, and c.3.1, noted above overlap considerably to form a well-connected subnetwork. Analysis of the members of this subnetwork reveals that they correlate closely to various members of the mitochondrial respiratory chain. In particular, they match subunits of complex I and complex II, indicating that perhaps this subnetwork is representative of the two complexes and the interactions between their subunits. This could be highly significant as, whilst the structure of the complex II has been solved, the structure of complex I has not yet been elucidated.
The respiratory chain involves a series of membrane-bound proteins that use a series of electron transfer steps to create a proton gradient across the mitochondrial membrane. This proton gradient is then used as the driving force for ATP synthesis. Complex I is the first protein complex in the respiratory chain. It is part of a redox reaction, catalysing the oxidation of NADH from the citric acid cycle along with the reduction of ubiquinone. The oxidation of NADH is coupled to electron transfer via a Flavin MonoNucleotide (FMN) prosthetic group, which acts as a first acceptor of electrons from NADH. Electron transfer is carried on through complex I by several iron-sulphur (FeS) clusters in the protein. Complex II (succinate ubiquinone oxidoreductase) is the second protein complex in the chain. It is involved in a different redox reaction, catalysing the oxidation of succinate (also a product of the citric acid cycle) to fumarate, along with the reduction of ubiquinone to ubiquinol. Succinate is oxidised by using the bound FAD on the 70 kDa subunit as an electron acceptor. As in complex I, several FeS clusters, which are found in the 27 kDa subunit, help in electron transfer through the protein.
To answer whether complex I and complex II relate to the above networks, we mapped known complex I and complex II protein subunits to their SCOP superfamilies via PSI-Blast [54]. Assigned complex I superfamilies account for the majority of the superfamilies in the smaller network (Figure 10), which shows alpha-helical ferredoxin (a.1.2) and its neighbours, and on the left half of the larger subnetwork (Figure 11), which shows the FAD/NAD(P)-binding domain and its neighbours. Complex II superfamilies account for at least 5 of the other superfamily nodes in the subnetwork in Figure 11. To be more precise, the proteins P15960, P34943, P42028, P15690, which are known subunits of complex I map to SCOP superfamilies d.15.4, c.4.1, d.58.1, and a.1.2, respectively, all of which are members of the smaller subnetwork. Furthermore, the other 2 superfamilies of this network, FMN linked oxidoreductase (c.1.4) and FAD/NAD (P) binding domain (c.3.1), are functionally significant to complex I. Additionally, we found that proteins Q09545 and Q09508 of complex II map to d.15.4, a.1.2, a.7.3, d.168.1, c.3.1. As with the prior example, other superfamily members of this network, multiheme cytochromes (a.138.1), FAD/NAD-linked reductases, dimerisation (C-terminal) domain (d.87.1), and thioredoxin-like (c.47.1), are all functionally related to the action of complex II.
The above findings show that 9 out of 11 neighbours of the FAD/NAD (P) binding domain belong to or are related to either complex I or complex II, or both. A subnetwork has been identified around this highly connected superfamily that has a comparatively high cluster index. This stresses both the importance of the superfamily and also the importance of connectivity and cluster index as a measure that is especially useful in uncovering complexes.
Interaction Rank
Both connectivity and cluster index have shortcomings: Connectivity does not consider interactions in a vertex's neighbourhood; cluster index favours low connectivity vertices. To get a better measure for the wider neighbourhood of a vertex, we have developed the idea of interaction rank, which treats interaction networks as Markov processes. In this analysis, each edge in the network is equated with a state transition in a Markov process. A similar approach has been used for the analysis of clusters in a network [55]. For example, a superfamily with a certain number of interaction partners, p, corresponds to a state, v, in the Markov process with p possible successor states w ∈ N(v), (where N(v) is the set of v's neighbours). A priori, each of the transitions is chosen with the same likelihood, giving a 1/|N(v)| chance for v to 'interact' with w ∈ N(v), where |N(v)| is the size of the set. If we enumerate all vertices from v1 to vn, we can capture this Markov process as a transition matrix M = (mij), where for all 1 ≤ i, j ≤ n entries, mij = 1/|N(vi)| if vi is connected to vj or 0 otherwise. If we compute the steady state transition probabilities of this process, we can rate vertices according to this notion of 'interactivity'. We call this rating 'interaction rank'. Essentially, the more interaction partners a superfamily has, the better its interaction rank. Also, the better connected a superfamily's neighbourhood, the better the interaction rank. These two trends are intuitively a consequence of the increased probability of indirectly returning back to a superfamily via the interconnections between its interaction partners. In this way, interaction rank combines aspects of connectivity and cluster index. It does so at a global scale incorporating information about the topology of the whole network. In this respect, interaction rank can point to the hubs of a network in terms of its overall structure, and can overcome some of the shortcomings of connectivity and cluster index.
To compute the steady state of the transition matrix, M, we need to find a configuration, x, such that M x = λ x for a maximal real number λ. In other words, we have to compute an eigenvector x for M for the maximal eigenvalue λ. There are standard libraries to do this, but since we require just the eigenvector for the largest eigenvalue, we used the power method, i.e. for a random initial configuration xo we iteratively compute Mn x0 for increasing n > 0 until Mnx0 converges. The elements of the resulting eigenvector represent the steady state probabilities of the Markov process M and constitute the interaction rank of the corresponding superfamilies.
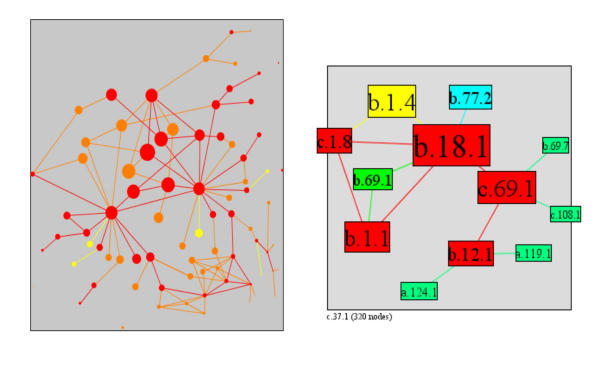
Let us consider examples of superfamilies with high interaction rank. The top 25% superfamilies in PSIMAP's main component, according to interaction rank, form a connected component, and thus define the core of the whole interaction network (figure 12). While a large number of neighbours usually implies a good interaction rank, there are examples such as alpha/beta-hydrolases (c.69.1) and the galactose oxidase, central domain (b.69.1) with few interaction partners, yet high interaction rank, as they have highly scoring neighbours. The alpha/beta-hydrolases (c.69.1) superfamily has only four interaction partners (figure 12), but has a good interaction rank as its neighbourhood consists of two high ranking nodes (Galactose-binding domain-like (b.18.1) and Lipase/lipooxygenase domain (PLAT/LH2 domain) (b.12.1)) and two medium ranking nodes (Prolyl oligopeptidase, N-terminal domain (b.69.7) and HAD-like (c.108.1)). Similarly, the Galactose oxidase, central domain, b.69.1, has a medium interaction rank despite it only having two interaction partners; however, these two partners have a very high interaction rank, which is reflected in b.69.1.
Figure 12.
The top 25% superfamilies according to interaction rank form a highly connected component (left). The superfamily alpha/beta-Hydrolases (c.69.1) has only four interaction partners, but has nonetheless a good interaction rank, as its neighbourhood consists of two good nodes (Galactose-binding domain-like (b.18.1) and Lipase/lipooxygenase domain (PLAT/LH2 domain) (b.12.1)) and two medium nodes (Prolyl oligopeptidase, N-terminal domain (b.69.7) and HAD-like (c.108.1)). From top-left to bottom-right: b.1.4, beta-Galactosidase/glucuronidase domain; b.77.2, delta-Endotoxin (insectocide), middle domain; c.1.8, (Trans)glycosidases; b.18.1, Galactose-binding domain-like; b.69.7, Prolyl oligopeptidase, N-terminal domain; b.69.1, Galactose oxidase, central domain; c.69.1, alpha/beta-Hydrolases; c.108.1, HAD-like; b.1.1, Immunoglobulin; b.12.1, Lipase/lipooxygenase domain (PLAT/LH2 domain); a.119.1, Lipoxigenase; a.124.1, Phospholipase C/P1 nuclease
To summarise, we define a transition matrix M reflecting possible interactions between superfamilies. From the transition matrix we can computer the interaction rank of each superfamily and hence complement the measures of connectivity and cluster index. In contrast to connectivity, which considers only the direct neighbourhood of a superfamily, interaction rank takes the whole network topology into account. In contrast to cluster index, which favours vertices with few interaction partners, interaction rank increases with the number of interaction partners. Furthermore, interaction rank is capable of including additional probabilistic experimental information regarding likelihood of interaction by simply updating the transition matrix accordingly. This will be a powerful basis to customize interaction rank for a researcher's specific experiments and settings.
Taxonomic Diversity
All the above measures rate vertices according to the structure of the network. Here we introduce a measure that rates superfamilies according to their taxonomic diversity. Taxonomic diversity is related to age – the more diverse a superfamily, the older it is. We have addressed the question of whether a superfamily's taxonomic diversity, and thus its age can be related to its interactivity or location in the network. This would effectively enable us to predict age from the network structure.
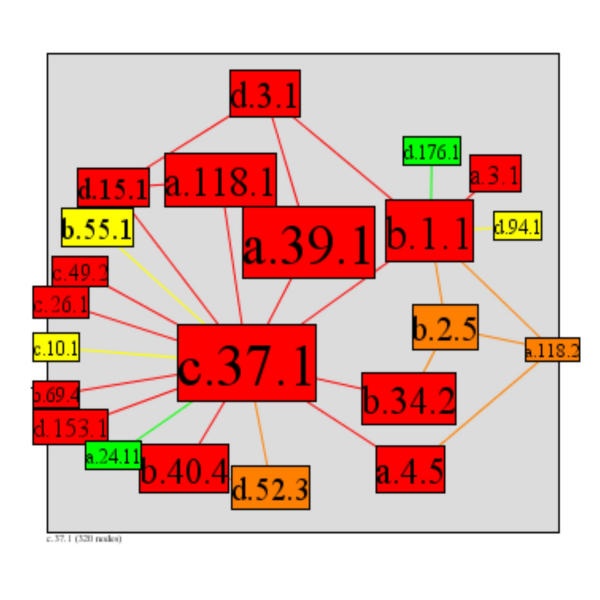
To define taxonomic diversity, we used the NCBI taxonomic database [56] to count the number of species in which a superfamily's domains occur. As this species-level measure depends highly on the structure of the taxonomy (for example there are many more eukaryotes than prokaryotes), we complemented this count species-level count by also measuring the diversity at kingdom level. Kingdom-level diversity simply indicates whether a superfamily occurs in 1, 2, 3, or 4 of the superkingdoms of archaea, bacteria, eukaryotes, and viruses. Using diversity measures, we can identify the oldest interactions and extract information about the evolution of the interaction network. The 10%, 20%, 30%, and 40% most highly diverse superfamilies in PSIMAP's main component are shown in Figure 13. Equating diversity to age, the series shows how the network developed through evolution. We further examined the core of the network: the 18 most highly diverse as shown in table 3 and their interactions as shown in Figure 14. These superfamilies can be considered the oldest, as they are the most highly diverse. It is important to note that these oldest superfamilies form one connected and (presumably ancient) component and do not break-up into different components.
Figure 13.
Evolution of the interaction network. PSIMAP's main component with the top 10%, 20%, 30% and 40% according to diversity.
Table 3.
The 18 most highly diverse and hence oldest superfamilies. The species column indicates the number of species this superfamily occurs in, the superkingdom column indicates whether the superfamily occurs in eukaryota (E), archaea (A), bacteria (B), and viruses (V), connectivity refers to the number of interaction partners.
| Species | Superkingdoms | Connectivity | SCOP ID | Superfamily |
| 270 | VEBA | 46 | c.37.1 | P-loop containing nucleotide triphosphate hydrolases |
| 193 | EBA | 9 | c.2.1 | NAD(P)-binding Rossmann-fold domains |
| 154 | EBA | 8 | c.55.1 | Actin-like ATPase domain |
| 143 | VEBA | 8 | d.144.1 | Protein kinase-like (PK-like) |
| 135 | EBA | 18 | c.3.1 | FAD/NAD(P)-binding domain |
| 128 | EBA | 38 | b.1.1 | Immunoglobulin |
| 98 | EB | 9 | a.39.1 | EF-hand |
| 90 | VEBA | 11 | a.4.5 | Winged helix DNA-binding domain |
| 66 | EBA | 11 | d.15.4 | 2Fe-2S ferredoxin-like |
| 65 | EB | 8 | d.15.1 | Ubiquitin-like |
| 63 | EBA | 12 | d.58.1 | 4Fe-4S ferredoxins |
| 62 | EBA | 10 | d.142.1 | Glutathione synthetase ATP-binding domain-like |
| 59 | EB | 11 | a.3.1 | Cytochrome c |
| 48 | EB | 6 | a.118.1 | ARM repeat |
| 37 | EBA | 9 | c.1.4 | FMN-linked oxidoreductases |
| 32 | EBA | 7 | d.145.1 | FAD-binding domain |
| 93 | EBA | 14 | c.1.8 | (Trans)glycosidases |
| 55 | EBA | 7 | d.92.1 | Metalloproteases (zincins) catalytic domain |
Figure 14.
The 22 most highly diverse superfamilies form a connected component. This subnetwork can be considered the oldest network and is the backbone of the overall network. The highly diverse superfamilies are also highly interactive as the colour coding shows (red = high connectivity). From top-left to bottom right: d.3.1, Cysteine proteinases; d.15.1, Ubiquitin-like; a.118.1, ARM repeat; d.176.1, Sulfite oxidase, middle catalytic domain; a.3.1, Cytochrome c; b.55.1, PH domain-like; a.39.1, EF-hand; b.1.1, Immunoglobulin; d.94.1, HPr-like; c.49.2, ATP syntase (F1-ATPase), gamma subunit; c.26.1, Nucleotidylyl transferase; c.10.1, RNI-like; b.69.4, Trp-Asp repeat (WD-repeat); d.153.1, N-terminal nucleophile aminohydrolases (Ntn hydrolases); a.24.11, Bacterial GAP domain; b.40.4, Nucleic acid-binding proteins; d.52.3, Prokaryotic type KH domain (pKH-domain); a.4.5, Winged helix DNA-binding domain; b.34.2, SH3-domain; b.2.5, p53-like transcription factors; a.118.2, Ankyrin repeat; c.37.1, P-loop containing nucleotide triphosphate hydrolases
Next, we want to relate the concept of taxonomic diversity to the other graph-theoretic measures. Can we predict the taxonomic diversity from structural properties in the network alone? At first glance, results appear to reject this: Eccentricity, sum of distance, and cluster index are correlated to neither of the diversity measures. Also, connectivity and interaction rank are only correlated with 0.25 to diversity at kingdom level. However, they show a reasonable correlation to diversity (both 0.56). Figure 15 shows this relationship in a scatter plot for connectivity. For diversity at kingdom level, both connectivity and interaction rank allow for the conclusion that superfamilies with high values occur in at least 3 superkingdom classes, while low values may or may not be spread across many kingdoms. Something similar holds in relation to diversity: Highly connected superfamilies and ones with a high interaction rank tend to occur in many species. However, superfamilies with low connectivity and interaction rank may or may not occur in many species. As a result, we can conclude that all highly interactive superfamilies are among the oldest.
Figure 15.
Diversity vs. Connectivity. Highly connected superfamilies occur in many species and hence can be considered to be very old. The opposite does not hold, i.e. there are old superfamilies with only few interaction partners.
Fault-tolerance, Attacks, and Convergent Evolution
It has been argued that many protein interaction networks are scale-free networks [44,57]. The scale-free property means that the vertex connectivity follows a power-law, i.e. there are few nodes that are highly connected, and many with low connectivity. This is also the case for PSIMAP. There are over 400 superfamilies that have no interaction partners and the connectivity of the most highly connected superfamilies quickly tails off as discussed above (P-loop (46), Immunoglobulin (38), (Trans)glycosidases (14), 4Fe-4S ferredoxins (12), Cytochrome c (11),...). Formally, the graph of number of interaction partners (y-axis) and superfamilies (x-axis) has a trend line of y = 58.014x-0.7152, which fits very well with the squared correlation coefficient R2 = 0.9353 (data not shown). This confirms the power law property for PSIMAP's largest component.
Scale-free networks such as PSIMAP have special properties. On the one hand, they are very fault-tolerant, in that the removal of a random vertex is not likely to disconnect a component. They are, however, prone to attacks, in that the removal of the most highly connected vertices severely affects the network. One small component of the PSIMAP interaction network consists of a methionine synthase domain (a.46.1) interacting with a methionine synthase activation domain (d.173.1) interacting with a cobalamin (vitamin B12)-binding domain (c.23.6) interacting with cobalamin (vitamin B12)-dependent enzymes (c.1.19), which in turn interacts with both a diol dehydratase gamma subunit (a.23.2) and a diol dehydratase beta subunit (c.51.3), as shown in figure 16. In this subnetwork, the cobalamin (vitamin B12)-binding domain and the cobalamin (vitamin B12)-dependent enzyme have a common role, as their removal disconnects the component, i.e. without superfamilies c.1.19 and c.23.6, methionine synthase domains a.46.1 and d.173.1 cannot interact with the diol dehydratase subunits a.23.2 and c.51.3. In graph-theory, such vertices are called 'articulation vertices', and, by definition, their removal disconnects the network.
Figure 16.
Example for component and for cut node (c.1.19 and c.23.6) a.46.1, Methionine synthase domain; d.173.1, Methionine synthase (activation domain); c.23.6, Cobalamin (vitamin B12)-binding domain; c.1.19, Cobalamin (vitamin B12)-dependent enzymes; a.23.2, Diol dehydratase, gamma subunit; c.51.3, Diol dehydratase, beta subunit
The superfamilies and interactions shown in this subnetwork incorporate information from 17 different PBD files, which give three distinct sets of domain interactions:
1) Methionine synthase: PDB entries 1k7y and 1k98 link SCOP superfamilies a.46.1, d.173.1 and c.23.6.
2) Methylmalonyl-CoA mutase: PDB entries 1cb7, 1ccw, 1e1c, 1i9c and 1-7req link SCOP superfamilies c.23.6 and c.1.19.
3) Glycerol dehydratase: PDB entries 1dio, 1eex, 1egm and 1egv link SCOP superfamilies c.1.19, a.23.2, and c.51.3.
Proteins in set one (methionine synthase) are linked to proteins in set two (methylmalonyl-CoA mutase) via the common superfamily, c.23.6 (Cobalamin binding domain) [58]. While the link between these two sets of proteins does not represent a direct physical interaction, it highlights the evolutionary connection between the two proteins (c.23.6 physically interacts with both d.173.1 and c.1.19). The link also highlights the functional coupling of the two proteins mediated by the common cofactor, showing they are involved in related metabolic pathways and diseases [59].
Methionine synthase and methylmalonyl-CoA mutase have well described functions in higher organisms, while proteins in the third set, (glycerol dehydratase) are described as bacterial. The link between the methylmalonyl-CoA mutase and glycerol dehydratase is made via the common superfamily c.1.19 (cobalamin dependent enzymes). In this case we suspect that c.1.19 facilitates a true physiological interaction pathway between the superfamilies present in both species.
As argued above, the pathway in Figure 16 is very dependent on both the cobalamin (vitamin B12)-binding domain and the cobalamin (vitamin B12)-dependent enzymes being present in the network, as these two vertices are so-called articulation vertices, whose removal disconnects the component. If for this reason certain superfamilies are particularly important, then is there any evidence for back-up mechanisms? One way how to ensure that connectivity is maintained despite the removal of vertices in the network is to have multiple and entirely different paths connecting two superfamilies. Then the interruption of one path does not interrupt the network as a whole. In graph-theory a sub-graph in which all pairs of vertices are connected by at least two entirely different paths is called a bi-connected component. Any vertex in a bi-connected component can be removed without breaking the network into separate components.
Figure 17 shows some bi-connected components in PSIMAP. Shown left is the main component with 320 superfamilies which contains one large bi-connected component of 115 superfamilies. This means that nearly all of the superfamilies in PSIMAP are not articulation vertices, i.e. e.g. removing any of these 115 superfamilies will not disconnect the largest component. Furthermore, the overview in Figure 17 highlights the comparatively few articulation vertices, which connect the main bi-connected component to the rest of the network, in pink. The P-loop is such a vertex, which links the main bi-connected component to the second largest biconnected component. Thus the P-loop is an articulation vertex and removal of the P-loop will separate these two bi-connected components.
Figure 17.
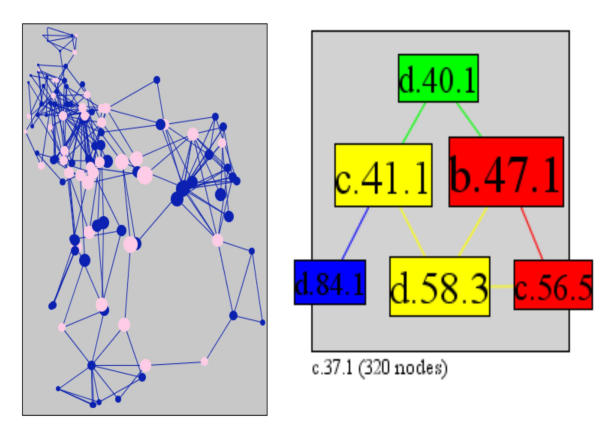
Left: The largest component in PSIMAP contains a large bi-connected component (Left). The superfamilies, which connect this bi-connected component to the rest of the network are coloured pink. On the right is bi-connected component with the four superfamilies b.47.1, Trypsin-like serine proteases; d.40.1, CI-2 family of serine protease inhibitors; d.58.3, Protease propeptides/inhibitors; c.41.1, Subtilisin-like. The colour indicates their overall connectivity.
The right figure in Figure 17 shows a smaller bi-connected component consisting of the four superfamilies trypsin-like serine proteases (b.47.1), CI-2 family of serine protease inhibitors (d.40.1), protease propeptides/inhibitors (d.58.3), and subtilisin-like (c.41.1). Similarly to the P-loop above, the removal of the subtilisin-like superfamily will disconnect the four superfamilies from the rest of the network reachable through the subtilisin-inhibitor (d.84.1). The bi-connected component shows that both the subtilisin-like and trypsin-like serine protease superfamilies can bind both the CI-2 serine protease inhibitors and the protease propeptides/inhibitors. This indicates that both protease superfamilies and both inhibitor superfamilies have a similar function, highlighting the two instances of functionally convergent evolution [60]. Thus, the graph-theoretic analysis of bi-connected components uncovers an instance of convergent evolution.
Conclusion
PSIMAP, a map of protein interactions at superfamily level, is computed using data from PDB and SCOP, and therefore provides a structural, robust, coarse-grained view of the interactome. In this paper, we have evaluated and justified PSIMAP, we have described the development of PSIEYE, a tool for large-scale interaction network analysis and visualization, and we have used PSIEYE to analyze PSIMAP and investigate several biologically significant questions.
We have evaluated and justified PSIMAP: First, we justified a threshold of 5 amino acid contacts at less than 5 Angstrom by considering interactions over the whole parameter space. Second, we justified interaction of covalently linked domains due to the use of SCOP. Third, we justified the approach of interaction at superfamily level by showing that superfamily size and number of interaction partners are not correlated.
We have developed PSIEYE, a tool for large-scale interaction network analysis and visualization: We have implemented a host of graph-theoretic measures such as connectivity, cluster index, eccentricity, sum of distance, and bi-connectivity to characterise proteins and their interactions in the maps. We complemented these measures with our novel approach of using interaction rank, which views interactions as a Markov process. This allowed us to rank proteins by their interactivity, effectively combining aspects of connectivity and cluster index at a global scale. We have discussed how to compute interaction rank by computing the stable state of the Markov process. The interaction rank approach has also the advantage that it can be customized by taking additional information on the possibility and probability of specific interactions into account thus combining the large scale structural interaction map with e.g. experimentally determined data.
We analysed PSIMAP: We applied the graph theoretic and taxonomic network measures to answer biological questions.
First, we compared the superfamilies regarding their location within the network. We found that the center and barycenter of PSIMAP do not coincide and we characterised the function of the superfamilies at the center as enzymatic activity, with an emphasis on energy metabolism and macromolecular synthesis and at the barycenter as very general. This is also due to the superfamilies at the center being not as highly connected.
Second, we analysed PSIMAP with respect to the notion of cluster index and we related a high number of interaction partners and relatively high cluster index to potential complexes. To document this, we verified that a substantial part of the highly-connected neighbourhoods of three superfamilies belong to complex I and II. The subnetwork and connections between the various superfamilies is especially interesting, as it is one of the largest yet least well characterised protein complexes in the cell. This new information regarding potential interactions and arrangements between the subunits might lead to novel insights into the structure and evolution of complex I and it complements approaches such as the method proposed by Bader and Hogue [61].
Third, we have shown how to characterise the evolution of interaction networks. We identified the most highly diverse superfamilies and showed that starting from the 10% most highly diverse superfamilies, progressing to 20%, 30% and 40%, the network does not fragment into different components, but progressively extends itself. This behaviour very closely reflects preferential attachment as observed in scale-free networks. Additionally, we investigated whether graph-theoretic measures can be used to predict the diversity of a superfamily, and showed that only two measures, connectivity and interaction rank, have such a correlation. A detailed scatter plot clearly shows that highly interactive superfamilies are also highly diverse and thus among the oldest. Finally, the concept of bi-connected components was used in the identification of a particular subnetwork. Our example shows two superfamilies, the subtilisin-like and the trypsin-like serine protease superfamilies, as instances of functionally convergent evolution [60], as they both share the same interaction partners. Overall, PSIMAP and its graph-theoretical analysis unravel important aspects of the evolution of protein interaction networks. Forth, we followed a novel approach to the fault-tolerance of interaction networks. We applied the notion of articulation vertices, whose removal disconnects the network, and of biconnectivity, where at least two completely different paths exist between vertices, to PSIMAP. We obtained the remarkable result that there are only very few articulation vertices in PSIMAP and that 1/3 of the superfamilies in PSIMAP's main component belong to a single bi-connected component. This means that the network is very fault-tolerant as removal of any of superfamily that is not an articulation vertex does not disconnect the network. This verifies that PSIMAP is a very robust network.
The analyses we carried out for PSIMAP are general in nature and can be applied to other experimental interaction data such as BIND or DIP. Combination and further analysis of the network components of these two types of protein interaction data will lead to critical understanding of the interactome.
Overall, our graph-theoretic analysis of PSIMAP allowed us answer a number of biological questions. In particular, the analysis sheds light onto the evolution of the network, it uncovers the core of the network, identifies complexes, and the most important superfamilies in terms of the network's structure.
Authors' contributions
DB has generated the PSIMAP and taxonomic data used, evaluated the PSIMAP parameters, and analysed the fault-tolerance example, PD implemented interaction rank through Eigenvector analysis, RH analysed the complex I and II data, JP developed the original PSIMAP, analysed the connectivity example and suggested fundamental questions, MS developed PSIEYE, the tool used for the analysis, conceived interaction rank and the other graph-theoretic measures, generated the examples using PSIEYE.
Acknowledgments
Acknowledgements
Jong Park was partly supported by the Ministry of Information and Communication of South Korea under grant number IMT2000-C3-4. JP acknowledges the support of MRC-DUNN in the previous period of stay.
Contributor Information
Dan Bolser, Email: dmb@mrc-dunn.cam.ac.uk.
Panos Dafas, Email: panos@soi.city.ac.uk.
Richard Harrington, Email: rah@mrc-dunn.cam.ac.uk.
Jong Park, Email: biopark@kaist.ac.kr.
Michael Schroeder, Email: msch@soi.city.ac.uk.
References
- Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith S, et al. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc Natl Acad Sci USA. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, et al. A comprehensive analysis of protein-protein interactions in: Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Walhout AJ, et al. Protein Interaction Mapping in C. elegans Using Proteins Involved in Vulval Development. Science. 1999;5450:116–121. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M, et al. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast. 2000;17:95–110. doi: 10.1002/1097-0061(20000630)17:2<95::AID-YEA16>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–82. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Ito T, et al. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci U S A. 2000;97:1143–7. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajolet M, et al. A genomic approach of the hepatitis C virus generates a protein interaction map. Gene. 2000;242:369–79. doi: 10.1016/S0378-1119(99)00511-9. [DOI] [PubMed] [Google Scholar]
- Rain JC, et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–5. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, et al. From molecular to modular cell biology. Nature. 1999;6761:C47–C54. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- Vidal M. A Biological Atlas of Functional Maps. Cell. 2001;104:333–340. doi: 10.1016/s0092-8674(01)00221-5. [DOI] [PubMed] [Google Scholar]
- Fellenberg M, et al. in Intelligent systems for molecular biology. La Jolla, CA: AAAI Press; 2000. Integrative Analysis of Protein Interaction Data. [PubMed] [Google Scholar]
- Lappe M, et al. Generating protein interaction maps from incomplete data: application to fold assignment. Bioinformatics. 2001;17:S149–56. doi: 10.1093/bioinformatics/17.suppl_1.s149. [DOI] [PubMed] [Google Scholar]
- Marcotte EM, et al. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–3. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- Dandekar T, et al. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–8. doi: 10.1016/S0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- Enright AJ, et al. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- Huynen M, et al. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10:1204–10. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, et al. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci U S A. 1999;96:4285–8. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lappe M, Teichmann SA. Mapping protein family interactions: intramolecular and intermolecular protein family interaction repertoires in the PDB and yeast. J Mol Biol. 2001;307:929–38. doi: 10.1006/jmbi.2001.4526. [DOI] [PubMed] [Google Scholar]
- Matthews LR, et al. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or "interologs". Genome Res. 2001;11:2120–6. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik J, Schachter V. Protein-protein interaction map inference using interacting domain profile pairs. Bioinformatics. 2001;17:S296–305. doi: 10.1093/bioinformatics/17.suppl_1.s296. [DOI] [PubMed] [Google Scholar]
- Deng M, et al. Inferring domain-domain interactions from protein-protein interactions. Genome Res. 2002;12:1540–8. doi: 10.1101/gr.153002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG, et al. SCOP: A Structural Classification of Proteins Database for the Investigation of Sequences and Structures. Journal of Molecular Biology. 1995;247:536. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Orengo CA, et al. CATH – a hierarchic classification of protein domain structures. Structure. 1997;5:1093–108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. in Proteins. 1997;28:405–420. doi: 10.1002/(SICI)1097-0134(199707)28:3<405::AID-PROT10>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Aloy P, Russell RB. Interrogating protein interaction networks through structural biology. Proc Natl Acad Sci USA. 2002;99:5896–5901. doi: 10.1073/pnas.092147999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992;357:543–4. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- Orengo CA, Jones DT, Thornton JM. Protein superfamilies and domain superfolds. Nature. 1994;372:631–4. doi: 10.1038/372631a0. [DOI] [PubMed] [Google Scholar]
- Alexandrov NN, Go N. Biological meaning, statistical significance, and classification of local spatial similarities in nonhomologous proteins. Protein Sci. 1994;3:866–75. doi: 10.1002/pro.5560030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZX. How many fold types of protein are there in nature? Proteins. 1996;26:186–91. doi: 10.1002/(SICI)1097-0134(199610)26:2<186::AID-PROT8>3.3.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Zhang CT. Relations of the numbers of protein sequences, families and folds. Protein Engineering. 1997;10:757–761. doi: 10.1093/protein/10.7.757. [DOI] [PubMed] [Google Scholar]
- Gough J, et al. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001;313:903–19. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, et al. Protein-protein interfaces: architectures and interactions in protein-protein interfaces and in protein cores. Their similarities and differences. Crit Rev Biochem Mol Biol. 1996;31:127–52. doi: 10.3109/10409239609106582. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Choe S, Eisenberg D. Domain swapping: entangling alliances between proteins. Proc Natl Acad Sci U S A. 1994;91:3127–31. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. The structure of interfaces between subunits of dimeric and tetrameric proteins. Protein Eng. 1989;3:77–83. doi: 10.1093/protein/3.2.77. [DOI] [PubMed] [Google Scholar]
- Jones S, Marin A, Thornton JM. Protein domain interfaces: characterization and comparison with oligomeric protein interfaces. Protein Eng. 2000;13:77–82. doi: 10.1093/protein/13.2.77. [DOI] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. BIND – a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics. 2000;16:465–77. doi: 10.1093/bioinformatics/16.5.465. [DOI] [PubMed] [Google Scholar]
- Xenarios I, et al. DIP: the Database of Interacting Proteins. Nucleic Acids Research. 2000;28:289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BH, et al. Visualization and analysis of protein interactions. Bioinformatics. 2003;19:317–318. doi: 10.1093/bioinformatics/19.2.317. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Ouzounis CA. BioLayout-an automatic graph layout algorithm for similarity visualization. Bioinformatics. 2001;17:853–854. doi: 10.1093/bioinformatics/17.9.853. [DOI] [PubMed] [Google Scholar]
- Mrowka R. A Java applet for visualizing protein-protein interaction. Bioinformatics. 2001;17:669–670. doi: 10.1093/bioinformatics/17.7.669. [DOI] [PubMed] [Google Scholar]
- Jeong H, et al. Lethality and centrality in protein networks. Nature. 2001;6833:41. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Wuchty S, Stadler PF. Centers of complex networks. J Theor Biol. 2003;223:45–53. doi: 10.1016/S0022-5193(03)00071-7. [DOI] [PubMed] [Google Scholar]
- Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nature Biotechnology. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- Schroder M, et al. PSIEYE: A tool for the graph-theoretic analysis of protein interaction networks. Bioinformatics. [submitted]
- Park J, Bolser D. Conservation of Protein Interaction Network in Evolution. Genome Informatics Series. 2001:135–140. [PubMed] [Google Scholar]
- Hemmingsen SM, et al. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988;333:330–4. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, Aravind L. Regulatory Potential, Phyletic Distribution and Evolution of Ancient, Intracellular Small-molecule-binding Domains. Journal of Molecular Biology. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. Protein kinases 6: The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. Faseb Journal. 1995;9:576. [PubMed] [Google Scholar]
- Djordjevic S, Driscoll PC. Structural insight into substrate specificity and regulatory mechanisms of phosphoinositide 3-kinases. Trends in Biochemical Sciences. 2002;27:426–432. doi: 10.1016/S0968-0004(02)02136-9. [DOI] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of 'small-world' networks. Nature. 1998;393:440–2. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongen Sv. Graph Clustering by Flow Simulation. PhD thesis University of Utrecht, Centre for Mathematics and Computer Science. 2000.
- Wheeler DL, et al. Database resources of the National Center for Biotechnology: Information. Nucleic Acids Res. 2000;28:10–4. doi: 10.1093/nar/28.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Fell DA. The small world inside large metabolic networks. Proc R Soc Lond Biol Sci. 2001;1478:1803–1810. doi: 10.1098/rspb.2001.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen B, et al. Homocysteine remethylation during nitrous oxide exposure of: cells cultured in media containing various concentrations of folates. J Pharmacol Exp Ther. 1992;261:1096–105. [PubMed] [Google Scholar]
- Allen RH, et al. Metabolic abnormalities in cobalamin (vitamin B12) and folate: deficiency. Faseb J. 1993;7:1344–53. doi: 10.1096/fasebj.7.14.7901104. [DOI] [PubMed] [Google Scholar]
- Doolittle RF. Convergent evolution: the need to be explicit. Trends Biochem Sci. 1994;19:15–8. doi: 10.1016/0968-0004(94)90167-8. [DOI] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in: large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]