Summary
Mutation of BLM helicase results in the autosomal recessive disorder Bloom syndrome (BS). Patients with BS exhibit hyper-recombination and are prone to almost all forms of cancer. BLM can exhibit its anti-recombinogenic function either by dissolution of double Holliday junctions or by disruption of RAD51 nucleofilaments. We have now found that BLM can interact with the pro-recombinogenic protein RAD54 through an internal ten-residue polypeptide stretch in the N-terminal region of the helicase. The N-terminal region of BLM prevented the formation of RAD51-RAD54 complex, both in vitro and in vivo. Using the fluorescence recovery after photobleaching (FRAP) technique, we found that RAD54 and BLM rapidly and concurrently, yet transiently, bound to the chromatinized foci. Presence of BLM enhanced the mobility of both soluble and chromatinized RAD51 but not RAD54. The BLM-RAD54 interaction could occur even in absence of functional RAD51. The N-terminal 1-212 amino acids of BLM or an ATPase-dead mutant of the full-length helicase enhanced the ATPase and chromatin-remodeling activities of RAD54. These results indicate that apart from its dominant function as an anti-recombinogenic protein, BLM also has a transient pro-recombinogenic function by enhancing the activity of RAD54.
Keywords: Homologous recombination, Fluorescence recovery after photobleaching, RAD51, RecQ helicase
Introduction
Bloom helicase (BLM) is a 1417 amino acid 3′ to 5′ helicase whose mutation leads to an autosomal recessive disorder called Bloom syndrome (BS). Patients with BS are characterized by a very high incidence of different types of cancer (both solid tumors and leukemia), chromosomal instability, skin disorders, proportional dwarfism and immunodeficiency (Ouyang et al., 2008; Sharma et al., 2006). At the molecular level, loss of BLM expression leads to abnormal replication events and a high rate of homologous recombination (HR), manifested by increased incidence of sister chromatid exchange (SCE). BLM might regulate HR via its intrinsic ability to suppress inappropriate recombination and/or by modulating the functions of other proteins involved in the process. In support of the first model, BLM can catalyze branch migration of Holliday junctions in vitro, unwind D-loops and promote regression of model replication fork (reviewed by Ouyang et al., 2008; Sharma et al., 2006). The second model is supported by the fact that BLM along with its interacting partners topoisomerase IIIα, BLAP75 and RMI2 can resolve double Holliday junctions (Raynard et al., 2006; Singh et al., 2008; Wu et al., 2006).
RAD51 has a key role in promoting HR, especially in presynaptic and synaptic phases (Sung and Klein, 2006; Sung et al., 2003). It is known that amino acid residues in the N- and C-terminal region of BLM can independently interact with RAD51 (Wu et al., 2001). We and others have also demonstrated that BLM and RAD51 colocalize during stalling of the replication forks and reside in a matrix-bound complex (Bischof et al., 2001; Sengupta et al., 2003). Using genetic and cell biology approaches, we recently provided in vivo evidence that BLM can abrogate endogenous RAD51 foci formation and disrupt RAD51 polymerization (Tripathi et al., 2007). Phosphorylation of BLM at Thr99 by ATR is required for the disruption of RAD51 filaments (Tripathi et al., 2008). In vitro biochemical evidence confirmed that BLM can dislodge RAD51 from ssDNA in an ATPase-dependent manner (Bugreev et al., 2007).
One of the most studied interacting partners of RAD51 during HR is RAD54. RAD54 is a member of the snf2 family of DNA-dependent ATPases in the SF2 family of DNA helicases. Both ATP hydrolysis and DNA supercoiling activities of RAD54 are stimulated in the presence of RAD51 ssDNA or dsDNA filaments, thereby suggesting that RAD51 and RAD54 function together in vivo (Heyer et al., 2006; Tan et al., 2003). The functional interaction between RAD51 and RAD54 happens during presynaptic, synaptic and postsynaptic phases of HR. Using in vitro chromatin reconstitution systems, it was observed that RAD51-ssDNA stimulates RAD54-dependent chromatin remodeling in a homology-dependent or -independent manner (Alexeev et al., 2003; Zhang et al., 2007).
Although the parameters of the functional interaction between BLM and RAD51 are comparatively well established, little is known about whether or how BLM affects RAD54 function, especially in human cells. RAD54 and BLM double-knockout DT40 cells (chicken B-lymphocyte line) demonstrated increased chromosome breaks and gaps than either single gene mutant alone. Hence, the defects due to lack of BLM are repaired by RAD54-mediated HR (Wang et al., 2000). In this report, we examined how these two proteins functionally interact in human cells. We found that BLM can disrupt the RAD51-RAD54 complex formed on chromatin via the N-terminal region of the helicase, which can directly bind to RAD54. Binding of BLM to RAD54 enhances the ATPase function and chromatin-remodeling activities of RAD54. Based on the above results, we propose a novel transient pro-recombinogenic function of the helicase.
Results
BLM and RAD54 physically and functionally interact during HR
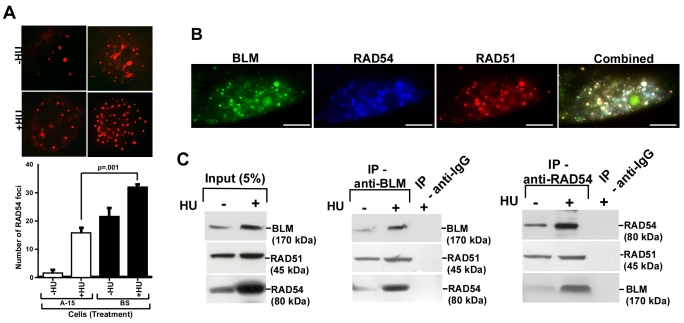
Initially, we wanted to determine whether the anti-recombinogenic function of BLM was dependent on RAD54. For this purpose, we carried out spontaneous SCE analysis, which is mediated by HR in vertebrate cells (Sonoda et al., 1999). shRNA against RAD54 stably transfected into isogenic BS (which do not express BLM) and A-15 (BS cells complemented with mini-chromosome 15 encoding BLM) cell lines, resulted in BS shRNA-RAD54 and A-15 shRNA-RAD54 cells, both of which exhibited acute depletion in endogenous RAD54 levels (supplementary material Fig. S1A). Compared with normal cells (A-15), loss of RAD54 (A-15 shRNA-RAD54 cells) led to a significant 60% decrease in the rate of SCEs (supplementary material Fig. S1B). The 800% increase in the rate of SCE observed upon loss of BLM, was abrogated in BS shRNA-RAD54 cells, thereby indicating that in human cells in vivo, BLM has a predominant anti-recombinogenic function. Consistent with this role, we found that the number of foci for RAD54 increased in BS compared with A-15 cells, irrespective of the absence or presence of replicative stress (Fig. 1A).
Fig. 1.
BLM and RAD54 have in vivo functional interaction. (A) Loss of BLM enhances the number of RAD54 foci. BS/A-15 cells were either left untreated or treated with HU for 12 hours. Extent of RAD54 foci formation was determined by immunofluorescence (top) and quantified (bottom). (B) BLM, RAD51 and RAD54 colocalize in vivo. IF with RAD51, BLM and RAD54 antibodies was carried out in HU-treated NHFs. Scale bars: 5 μm. (C) BLM and RAD54 interact in vivo. Immunoprecipitation was carried out with nuclear extracts from NHFs with antibodies against either BLM or RAD54 and probed with indicated antibodies.
Next, we wanted to determine whether BLM, RAD51 and RAD54 were present in the same complex either in absence or presence of DNA damage. Using immunofluorescence studies, we found that the three proteins colocalized extensively during hydroxyurea (HU) treatment (as visualized by white foci in merged images) in hTERT-immortalized normal human fibroblasts (NHF) cells (Fig. 1B). Using reciprocal coimmunoprecipitation, we found that the three proteins physically interacted with each other, irrespective of replicative stress (Fig. 1C). The enhanced BLM-RAD54 interaction due to HU-treatment was probably a reflection of the enhanced protein levels during replication arrest.
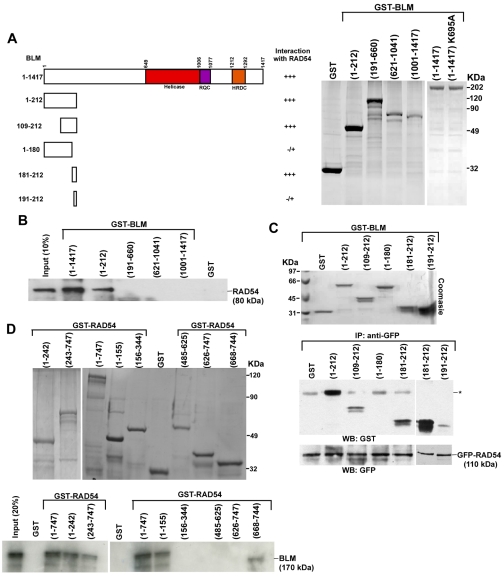
The above experiment indicated that RAD54 and BLM were part of the RAD51 complex at the site of DNA lesions. We hypothesized that BLM and RAD54 directly interact. To determine the region(s) of BLM which governed the interaction between the helicase and RAD54, we cloned, expressed and purified full-length BLM with a GST tag in E. coli (Fig. 2A, right). Using an in vitro GST pull-down assay, we found that full-length BLM and RAD54 directly interacted. Using purified BLM fragments (Tripathi et al., 2008), we further demonstrated that the N-terminal 212 amino acids of BLM mediated this interaction (Fig. 2B).
Fig. 2.
Internal residues of BLM interact with N- and C-termini of RAD54. (A) Summary of BLM-RAD54 interactions (left). Coomassie-blue-stained gel showing the expression of purified GST or GST-tagged BLM fragments BLM(1-212), BLM(191-660), BLM(621-1041), BLM(1001-1417), BLM(1-1417) and BLM(1-1417) K695A (right). (B) N-terminal region of bound GST-tagged BLM interacts with 35S-labeled full-length RAD54. After pull-down, bound radioactive RAD54 was determined by autoradiography. (C) A 10 amino acid stretch in the N-terminus of BLM interacts with RAD54. Bound GFP-RAD54 was incubated with soluble BLM fragments. After pull-down, the blots were probed with anti-GST and anti-GFP antibody. Asterisk indicates a crossreactive band migrating at the same molecular mass as BLM 1-212. (D) Coomassie-blue-stained gel showing the expression of purified GST or GST-tagged RAD54 fragments. Bottom gel shows that N- and C-terminal domains of RAD54 are required for its interaction with BLM. 35S-labeled full-length BLM was incubated with wild-type RAD54 or its various fragments.
Next, we wanted to determine the minimum interacting region within the N-terminal 212 amino acids of BLM that could bind with RAD54. Hence, we initially cloned, expressed and purified in soluble form four GST-tagged fragments within the first 212 amino acids of BLM. The soluble GST-bound BLM fragments were interacted with affinity-purified bead-bound GFP-RAD54. We found that that apart from the 1-212 fragment, 109-212 amino acids of BLM also interacted with RAD54 (Fig. 2C, bottom). The 1-180 fragment of BLM did not interact with RAD54, indicating that that a stretch of 32 amino acids (i.e. 181-212) of BLM mediates its interaction with RAD54. Hence, we cloned expressed and purified GST-tagged BLM(181-212) and BLM(191-212) fragments. Indeed we found that that GST BLM(181-212) but not BLM(191-212) interacted with RAD54 (Fig. 2C), indicating that ten amino acids (181-190) in the N-terminal region of BLM mediate its interaction with RAD54.
RAD54 contains a N-terminal domain (NTD), a C-terminal domain (CTD) and Snf2-specific helical domains (HD1 and HD2) (Heyer et al., 2006). RAD54(1-747), N-terminal region of RAD54(1-242) and C-terminal region of the protein, RAD54(243-747) interacted with full-length BLM (Fig. 2D, bottom left). To determine the minimal region of interaction, we cloned, expressed and purified the fragments of RAD54 encoding different domains (Fig. 2D, top). It was found that extreme N- and C-terminal regions of RAD54 independently interacted with full-length BLM (Fig. 2D, bottom right).
BLM prevents RAD54-RAD51 complex formation
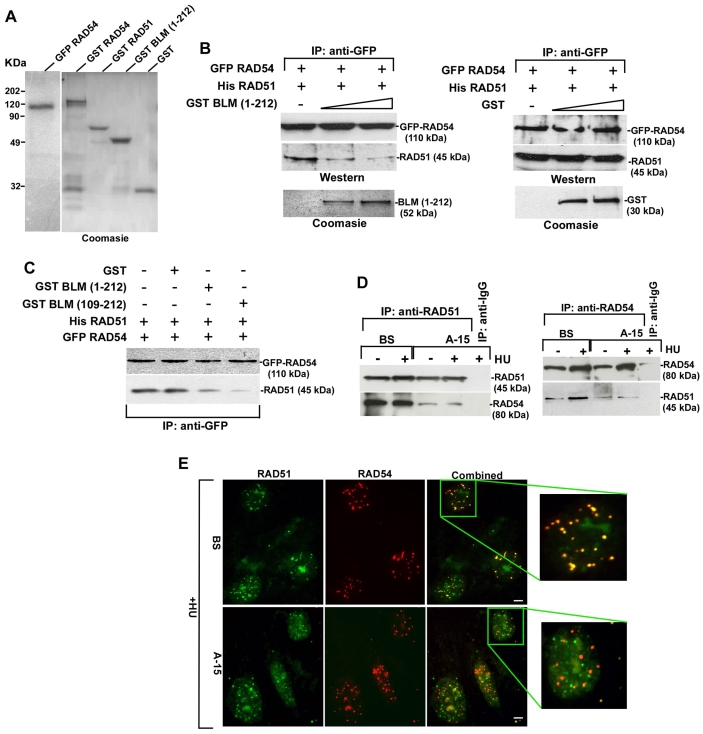
The N-terminal region of BLM binds to both RAD51 (Wu et al., 2001) and RAD54 (Fig. 2B). Incidentally, the N-terminal region of human RAD54 is involved in its interaction with both RAD51 (Golub et al., 1997) and BLM (Fig. 2D). To test the possibility that BLM prevents the RAD51-RAD54 interaction, we carried out in vitro pull-down assay with bead-bound purified GFP-RAD54 and soluble His-RAD51 (Fig. 3A) in the absence or presence of increasing amounts of soluble GST-BLM(1-212) or GST (Fig. 3B). RAD54 and RAD51 formed a readily detectable complex. Increasing amount of BLM(1-212), but not GST, prevented the RAD51-RAD54 interaction (Fig. 3B). Prevention of complex formation was even more enhanced with the BLM(109-212) fragment (Fig. 3C). These results were confirmed in vivo by carrying out reciprocal immunoprecipitations in BS and A-15 cells. Interaction between RAD51 and RAD54 decreased in A-15 cells (compared with BS) both with and without HU (Fig. 3D). At a single-cell level, RAD51 and RAD54 proteins colocalized well in BS cells in the presence of HU (98% of the cells demonstrated 87±3% foci colocalization) (Fig. 3E). Under similar conditions, in 99% of A-15 cells, only 68±4% of RAD51 foci colocalized with RAD54. The nucleolus has been postulated to be the site of RAD51 degradation (Orre et al., 2006). In both BS and A-15 cells, accumulation of RAD51 was also observed in the nucleolus, as revealed by co-staining with a nucleoli marker (data not shown). Altogether, the above results indicated that BLM prevented RAD51-RAD54 complex formation in vitro and in vivo.
Fig. 3.
BLM prevents RAD54-RAD51 complex formation in vitro and in vivo. (A) Purified proteins used to test the BLM(1-212)-mediated prevention of RAD51-RAD54 complex formation. (B,C) BLM prevents RAD54-RAD51 interaction. Interaction between bound GFP-RAD54 and soluble His-RAD51 was determined in the absence or presence of soluble BLM(1-212) or GST. The levels of the respective proteins after interaction were assessed by western blotting (B). Soluble BLM(109-212) was also used (C). (D) BLM prevents RAD51-RAD54 complex formation in vivo. Immunoprecipitation was carried out on the nuclear extract from BS/A-15 cells with antibodies against RAD51 or RAD54. Western blots were then performed with same antibodies. (E) BLM decreases colocalization between RAD51 and RAD54. Immunofluorescence of BS/A-15 cells with antibodies against RAD51 and RAD54. Scale bars: 5 μm.
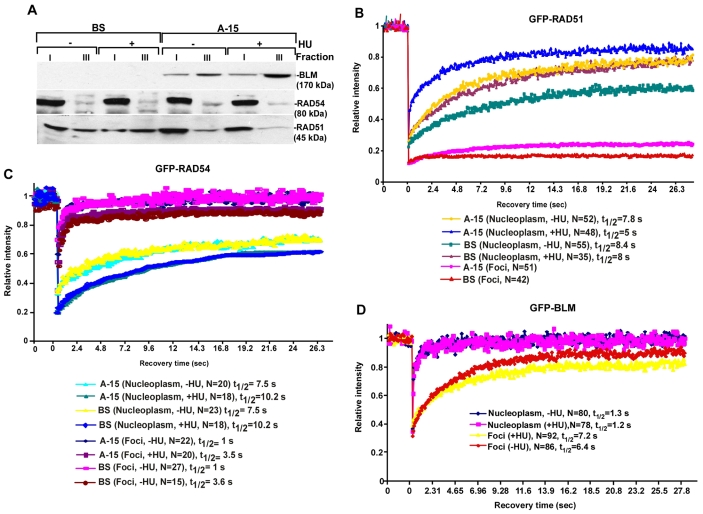
Based on these results, we hypothesized that the extent of accumulation of RAD51 and RAD54 in the chromatinized foci and the nucleoplasm should be different in the absence or presence of BLM. We carried out ribonucleotide (RNP) fractionation assays in BS/A-15 cells. BLM was present only in the A-15 cells and it accumulated predominantly in the ribonucleoprotein (RNP)-enriched fraction (fraction III), especially in the presence of HU (Fig. 4A). RAD54 was predominantly present in fraction I in both BS/A-15 cells, indicating that RAD54 was mostly in a soluble form. The small fraction of RAD54 at the sites of spontaneous or damage-induced foci, was not dependent on BLM. However RAD51 exhibited a BLM-dependent difference in accumulation between fractions I and III. In the absence of BLM, more RAD51 was found in the RNP-enriched fraction III and the amount of soluble RAD51 in fraction I was lower.
Fig. 4.
Lack of BLM leads to a decrease in the dynamic mobility of RAD51. (A) RAD51 is present in the soluble fraction in the presence of BLM. Nuclei of BS/A-15 cells were subfractionated into RNP-enriched (fraction III) and soluble (fraction I). Levels of BLM, RAD54 and RAD51 were determined by western blotting. (B) FRAP recovery curves of GFP-RAD51 in BS/A-15 cells. The recovery curves were obtained for GFP-RAD51 present in nucleoplasm or foci. Each data point represented the mean of a number of cells (indicated on right). t1/2 represents half maximal recovery time. (C) FRAP recovery curves of BS/A-15 cells transfected with GFP-RAD54. (D) FRAP recovery curves on of SV40-immortalized GM08505 cells stably expressing GFP-BLM (GFP-BLM).
We hypothesized that BLM might prevent RAD51-RAD54 complex formation by altering the dynamic relocalization of RAD51 between the nucleoplasm and the foci. To test this hypothesis, we transfected low levels of GFP-tagged RAD51 into BS/A-15 cells either with or without 2 hours of HU treatment (supplementary material Fig. S2). The relative levels of GFP-RAD51 and endogenous RAD51 were similar between A-15 and BS cells. Compared with A-15, GFP-RAD51 levels were higher in BS cells and were induced after HU treatment. Fluorescent recovery after photobleaching (FRAP) was measured to determine the dynamics of GFP-RAD51 in BS/A-15 cells (Fig. 4B; supplementary material Table S1). We found that in cells expressing BLM (i.e. A-15), 75±1.8% of RAD51 in the nucleoplasm was mobile (A-15, nucleoplasm, –HU). The half maximal recovery time (t1/2) for nucleoplasmic RAD51 in A-15 cells was 7.8 seconds. These values compare well with the reported value for the mobility of GFP-RAD51 in nucleoplasm of wild-type cells expressing BLM (Essers et al., 2002; Yu et al., 2003). By comparison, in BS cells, 55±2.2% of nucleoplasmic GFP-RAD51 was mobile and it manifested a slower recovery, with a t1/2 of 8.4 seconds (BS, nucleoplasm, –HU). The BLM-dependent difference in the dynamic behavior of nucleoplasmic GFP-RAD51 existed even after 2 hours of HU treatment (A-15, nucleoplasm, +HU vs BS, nucleoplasm, +HU). In both BS and A-15, the mobility of the GFP-RAD51 in the nucleoplasm increased after 2 hours of HU treatment, confirming earlier results that replication arrest enhances the dynamic mobility of nucleoplasmic RAD51 (Yu et al., 2003). This enhanced mobility of RAD51 after 2 hours of HU treatment probably indicates a unique case of transient mobilization of the protein during the process of the stalling of the replication forks. This might in turn help in the generation of new interactions during the process. The difference in the mobility of RAD51 between A-15 and BS was not limited to nucleoplasmic GFP-RAD51. As reported earlier (Essers et al., 2002; Yu et al., 2003), a slow RAD51 fluorescence recovery was observed in the foci of wild-type (A-15) cells. However, the recovery was completely absent when FRAP was carried out on the foci of BS cells (A-15 foci vs BS foci). Together, the above data indicate that RAD51 is more dynamic and exchanged at a faster rate in the presence of BLM. Conversely, in the absence of BLM, RAD51 seems to be more stable both in the foci and in the nucleoplasm.
The above experiments raised the possibility that the mobility of RAD54 is also altered in the presence or absence of BLM. We carried out FRAP after transiently transfecting BS/A-15 cells with GFP-RAD54, to levels similar to its endogenous counterpart (data not shown). As reported earlier (Essers et al., 2002), RAD54 foci recovered very fast (t1/2 of 1 second) indicating that the RAD54 in the foci is highly dynamic (Fig. 4C; supplementary material Table S1). RAD54 in the nucleoplasm was mobile showing a t1/2 of 7.5-10 seconds, depending on the absence or presence of stalled replication. Presence of HU led to a slight decrease in the mobility of both foci-bound and nucleoplasmic GFP-RAD54. The relative dynamic behavior of foci-bound and nucleoplasmic GFP-RAD54 is in contrast to that of GFP-RAD51 (Fig. 4B) and GFP-BLM (see below). The fast recovery of the foci-bound RAD54 appears to be counter-intuitive to the relatively slower recovery of the nucleoplasmic fraction. However, foci-bound RAD54 could never be completely bleached, and about 50% of the fluorescence recovered very fast (Fig. 4C, see extent of fluorescence intensity after laser pulse on foci). This probably indicates the distribution of RAD54 in the nucleoplasm into two pools, each having different mobility: the highly mobile nucleoplasmic fraction, which recovered very fast (as indicated by the fraction which could not be bleached due to laser pulse on the foci) and the slower-exchanging fraction with a residence time of 7.5-10 seconds. RAD54 in the foci probably rapidly exchanges with the highly mobile fraction of the nucleoplasm. This might be the reason why the less-mobile nucleoplasmic RAD54 does not affect the recovery seen for the highly mobile foci bound counterpart during the FRAP experiments. Importantly, the rate of movement of fluorescent RAD54 was not altered in either the nucleoplasm or the foci in the absence or presence of BLM (even in the absence or presence of HU).
BLM undergoes dynamic intranuclear exchange
The above biochemical and cell biology experiments hinted at a temporal regulation governing the interaction between RAD54, RAD51 and BLM in the nucleoplasm and foci. BLM might regulate the dynamic exchange of RAD51, but not RAD54, between the chromatinized foci and the nucleoplasm (Fig. 4A-C). Hence, it is possible that BLM also concurrently undergoes exchange at the foci and the nucleoplasm, thereby regulating its own interaction with RAD54 and RAD51. To investigate this, we used a stable line expressing GFP-BLM in SV40-immortalized GM08505 cells (Hu et al., 2001). The expression levels of BLM, RAD51 and RAD54 were very similar in GFP-BLM and A-15 cells, thereby allowing extrapolation of results between the cell lines (data not shown). The formation of GFP-BLM foci was observed in only 10-15% of cells in the absence of HU, indicating that they formed during spontaneous DNA damage in S-phase. After 2 hours of HU treatment, approximately 45-55% of the cells developed detectable GFP-BLM foci.
FRAP experiments indicated that after photobleaching there was 80-85% recovery of BLM present in the foci at a fast rate (t1/2 of 7 seconds), irrespective of the presence or absence of induced stalled forks (Fig. 4D; supplementary material Table S1). Thus BLM resides transiently at the chromatinized foci and dynamically exchanges with its counterpart in the nucleoplasm, which itself is present in an extremely mobile form (100% recovery with a t1/2 of 1 second). The markedly slower kinetics of recovery for GFP-BLM in the foci, in comparison to the nucleoplasm, indicated that a fraction of the helicase is transiently immobilized at the site of either spontaneous or HU-induced DNA damage. The residence time of BLM in the HU-induced foci (7.2 seconds) is similar to that of MDC1 and 53BP1 at the double-strand breaks (Bekker-Jensen et al., 2005; Lukas et al., 2004).
BLM stimulates RAD54-mediated ATPase and chromatin remodeling activities in a RAD51-independent manner
The above results (Figs 2, 3, 4) indicated that the RAD51-independent binding of BLM to RAD54 occurs in cells. To investigate this possibility, we expressed in NHFs a dominant-negative form of RAD51, the yeast-mouse chimera SMRAD51, which downregulates HR without affecting cell viability (Lambert and Lopez, 2000). Expression of SMRAD51 in HU-treated NHFs, led to stabilization of both endogenous RAD54 and BLM and decreased the rate of HR as measured by host-cell reactivation assay (supplementary material Fig. S3A). However, BLM and RAD54 colocalized to the same extent irrespective of the presence or absence of functional RAD51. In both cases, in 95% of the cells, around 82±3% BLM and RAD54 colocalization was observed (supplementary material Fig. S3B). These results demonstrated that the direct interactions detected between BLM and RAD54 (Fig. 2) could also occur in vivo, even in the absence of functional RAD51.
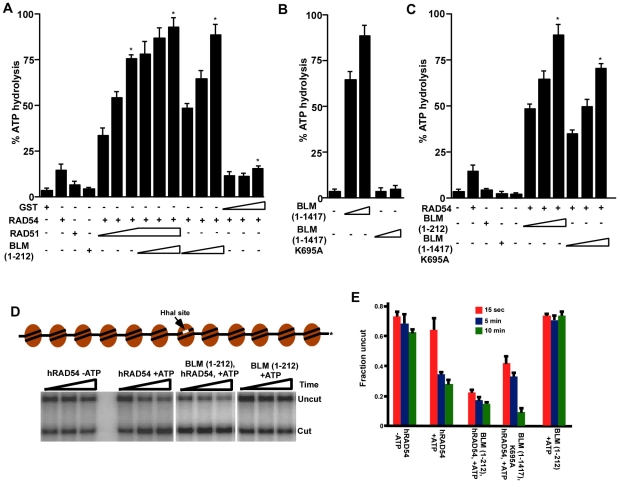
Many of the enzymatic functions of RAD54, including the ATPase and the homology-driven chromatin-remodeling function, are upregulated by RAD51 (Heyer et al., 2006; Zhang et al., 2007). To determine a functional role for the BLM-RAD54 interaction, we wanted to elucidate whether the N-terminal 1-212 amino acids of BLM (which interacts with RAD54, Fig. 2B) could affect RAD54-mediated ATPase activity. Based on our results from pull-down assays (Fig. 3B), we hypothesized that BLM(1-212), similarly to the Saccharomyces cerevisiae meiosis-specific gene Hed1 (Busygina et al., 2008), would ablate the enhancement of RAD54 ATPase activity by RAD51. Instead, we found that addition of the BLM(1-212) fragment led to a further increase in the rate of RAD54-mediated ATP hydrolysis. This result indicated that the N-terminal region of BLM might enhance the ATPase function of RAD54. Indeed, BLM(1-212), but not GST, could enhance RAD54 ATPase hydrolysis in a dose-dependent manner, to an extent similar to that seen with RAD51 (Fig. 5A).
Fig. 5.
BLM stimulates RAD54 function. (A) The N-terminal 1-212 amino acids of BLM enhance RAD54-mediated ATPase activity. ATPase activity was assessed with different combinations of proteins, as indicated. The concentrations of each protein are indicated in the Materials and Methods. *P<0.05 compared with the ATPase activity obtained by RAD54 alone. (B) Loss of ATPase activity in BLM(1-1417) K695A mutant. ssDNA-dependent ATPase activity was assessed using BLM(1-1417) or BLM(1-1417) K695A. (C) ATPase-dead mutant of BLM stimulates RAD54-mediated ATPase activity. BLM(1-1417) K695A was used to determine its effect on RAD54-mediated ATPase activity. (D) Schematic diagram of G5E4 array (Zhang et al., 2007). Representative REA assays with RAD54 alone (±ATP), RAD54+BLM(1-212) (+ATP) and BLM(1-212) alone (+ATP). (E) Quantification of results in D and of REA assays involving RAD54 and BLM(1-1417) K695A (+ATP).
We also wanted to know whether stimulation by the 1-212 fragment of BLM was also observed for the full-length helicase. We did not use wild-type BLM because the helicase has a strong ssDNA-dependent and a mild dsDNA-dependent ATPase activity (Bugreev et al., 2007). We generated and purified an ATPase-dead mutant of the full-length BLM, BLM(1-1417) K695A, which did not have any detectable ATPase activity (Fig. 2A, right; Fig. 5B). BLM(1-1417) K695A stimulated RAD54 ATPase activity to a similar extent as the BLM(1-212) fragment (Fig. 5C).
Since BLM could stimulate the ATPase activity of RAD54 (Fig. 5A,C), it might also stimulate the chromatin-remodeling activity of RAD54. It has been reported that RAD51 ssDNA filaments stimulated RAD54-dependent chromatin remodeling in a homology-dependent or -independent manner (Alexeev et al., 2003; Zhang et al., 2007). We verified ATP- and time-dependent RAD54 chromatin-remodeling activity by using restriction enzyme accessibility (REA) assay on the G5E4 array, containing 12 nucleosomes with a centrally located HhaI site occluded by one nucleosome (Fig. 5D,E). Although BLM(1-212) alone did not have any effect on the REA assay, it enhanced RAD54-dependent chromatin-remodeling activity, especially at the early (15 second) time point. The effect was also seen for the BLM(1-1417) K695A mutant. At the early time point, the BLM(1-212) fragment enhanced the RAD54 chromatin-remodeling activity to 300% more than that observed for RAD54 alone, thereby indicating that stimulation of RAD54 remodeling by BLM is an early event in HR.
Discussion
BLM is a multifunctional protein that takes an active part in the regulation of HR. This is manifested in vivo by its colocalization and physical interaction with proteins involved in HR, such as RAD51 and RAD54 (Fig. 1B,C). There is increasing evidence that BLM stimulates HR (Adams et al., 2003; Bugreev et al., 2007). Human exonuclease 1 and BLM interact to resect DNA and initiate the process of DNA repair (Nimonkar et al., 2008). However, much of the evidence regarding the pro-recombinogenic role of BLM is either from in vitro experiments or from studies involving BLM mutants in Drosophila. Using a set of four isogenic human cells, which differ only in the status of two genes, BLM and RAD54, we have provided evidence that though the predominant function of BLM is anti-recombinogenic (supplementary material Fig. S1), it also has a transient pro-recombinogenic role at the chromatin-remodeling stage of HR (Fig. 5).
In vitro biochemical analysis provided the evidence that an internal stretch of 10 amino acids in the N-terminal region of BLM is sufficient to interact with RAD54 (Fig. 2C). Apart from RAD54, the N-terminal region of BLM binds to a number of proteins involved in the control of HR, such as RAD51 (Wu et al., 2001) and TopoIIIα (Hu et al., 2001). One reason for this could be the presence of distinct and specific epitopes in the N-terminal region of BLM for each of the above interactions. However, the possibility also exists that there are overlapping epitopes, and consequently, the interactions of BLM with the different HR regulators are transient and/or dynamic. We found that the N-terminal 109-212 amino acids of BLM were sufficient to prevent the interaction between RAD51 and RAD54 in vitro (Fig. 3C). Immunoprecipitation, immunofluorescence, RNP fractionation studies and FRAP assays (Fig. 3D,E; Fig. 4A,B) verified that the presence of BLM prevented the interaction between RAD51 and RAD54 in vivo and resulted in RAD51 being released from the chromatin-bound fraction into the soluble nucleoplasm.
It is known that RAD54 and RAD51 mutually stimulate the other's function during chromatin remodeling (by RAD54) and strand invasion (by RAD51). It has been demonstrated that RAD51-ssDNA can stimulate RAD54-dependent chromatin remodeling either in a homology-dependent polarity-independent or homology-independent (Alexeev et al., 2003; Zhang et al., 2007) manner. It has been proposed that the RAD51-RAD54-ssDNA complex initially recognizes a homologous site on chromatin, remodeling it and thereby promoting strand invasion. Using FRAP studies. we aimed to determine the relative mobility of RAD51 and RAD54 with respect to BLM during these early steps of HR. Nucleoplasmic BLM and foci-bound RAD54 displayed very fast, yet similar, kinetics (Fig. 4C,D) which might allow these two proteins to accumulate on the chromatinized foci. Once bound, BLM probably continues to exchange between the foci and nucleoplasm. Given the time scales involved, the BLM-RAD54 interaction on the foci takes place very early during HR, possibly during the initial chromatin remodeling step. Instead of the RAD51-ssDNA complex, BLM helps RAD54 (by enhancing its ATPase activity) to initially remodel the chromatin (Fig. 5). Hence, at this stage, BLM acts as a transient pro-recombinogenic protein. This activity of BLM is not sequence driven, indicating that it might be the initial repair response upon DNA damage. Subsequently, BLM probably dissociates from the N-terminal region of RAD54, allowing the same region to instead bind to RAD51. The next stage of RAD54-driven chromatin remodeling is possibly homology driven and stimulated by RAD51-ssDNA. RAD54 then promotes the nucleation of RAD51 on the RPA-coated ssDNA, thereby initiating the presynaptic phase of HR. Later, at the synaptic phase, BLM again interacts with RAD51 and disrupts RAD51 filaments (Fig. 6) (Bugreev et al., 2007; Tripathi et al., 2007). Hence, the question arises: how does BLM switch its function from a pro-recombinogenic to an anti-recombinogenic protein? We have recently demonstrated that lack of ATR-mediated BLM phosphorylation on Thr99 prevents the helicase from disrupting RAD51 polymerization (Tripathi et al., 2008). Additional cellular and molecular event(s) including additional post-translational modification(s) on BLM and/or interaction with other anti-recombinogenic proteins, such as 53BP1 and p53 (Sengupta et al., 2003; Tripathi et al., 2007) might also be required during this transition process. We cannot exclude the idea that BLM does not stimulate the ATPase activity of RAD54 in the other steps of HR. We have proposed the present hypothesis (Fig. 6) taking into consideration the predominant anti-recombinogenic role of BLM (supplementary material Fig. S1B), the time scales involved in the interaction between BLM, RAD51 and RAD54 (Fig. 4B-D), the disruption of the RAD51-RAD54 complex by BLM (Fig. 3) and the stimulation of RAD54 functions by BLM (Fig. 5). Hence, our results do not in any way exclude the additional mechanistic processes which BLM uses in the late phase of HR.
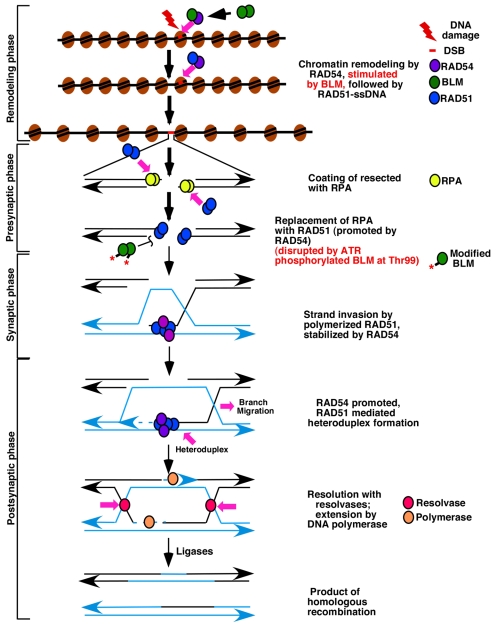
Fig. 6.
Model depicting the regulation of homologous recombination by BLM as a result of alteration of RAD51 and RAD54 function. Double-strand breaks (DSBs) are recognized by the HR machinery after the chromatin-remodeling phase. BLM enhances the ATPase activity of the chromatin remodeler RAD54, thereby enhancing its remodeling activity in a homology-independent manner. Subsequently, BLM dissociates from RAD54, allowing the same region to instead bind to RAD51. The next stage of RAD54-driven chromatin remodeling is possibly homology driven and stimulated by RAD51 bound to single-stranded DNA (RAD51-ssDNA). The result of chromatin remodeling allows the sequential accumulation of proteins during subsequent stages of HR. After detection of DSB and resection of the DNA in the 5′-3′ direction, RAD51 binds to ssDNA and displaces replication protein A (RPA), which leads to RAD51 polymerization (this phase is referred to as the presynaptic phase). RAD54 promotes the nucleation of RAD51 on the RPA-coated ssDNA, thereby initiating the presynaptic phase of HR. Once the homology search is successful, the duplex is captured and the RAD51 filament invades it to form the heteroduplex structure (synaptic phase). RAD54 stabilizes the RAD51-ssDNA complex, thereby promoting this process. At synaptic phase BLM(possibly phosphorylated by ATR at Thr99) interacts with RAD51 and disrupts RAD51 filaments. Heteroduplex DNA extension and branch migration normally occurs during the postsynaptic phase of HR. DNA polymerases use the intact copy to re-synthesize the deleted DNA sequences, DNA ligases join the newly synthesized fragments and the Holliday junctions are resolved by specific endonucleases, known as resolvases. As a result of the above two activities, BLM can accurately control HR. Additional mechanistic processes, which BLM is known to use at late stages of HR, are not shown.
In conclusion, we have characterized a RAD51-independent interaction between BLM and RAD54. Using live-cell dynamics and biochemical analysis, we have provided evidence on how RAD51 mobility could be altered as a result of the BLM-RAD54 interaction. BLM stimulated the ATPase and chromatin remodeling activities of RAD54. In fact, this is the first report of any other protein apart from RAD51 that can stimulate these functions of RAD54. Finally, we have proposed an integrated model of BLM function, which might help to explain its dual, pro- and anti-recombinogenic roles, and to understand how this helicase integrates with and controls the HR machinery.
Materials and Methods
Plasmids and shRNA
p2085S-G5E4 (gift from Jerry Workman, Stowers Institute for Medical Research, Kansas City, MO), pcDNA3.1.puro SMRAD51 (gifted by Bernard Lopez, Institut de Radiobiologie Cellulaire et Moléculaire, Fontenay-aux-Roses, France), pcDNA 3.1 Flag RAD54 (gift from Kiyoshi Miyagawa, The University of Tokyo, Tokyo, Japan). pcDNA Flag BLM, pGEX4T-1 BLM(1-212) (gifts from Ian Hickson, University of Oxford, Oxford, UK), EGFP-RAD51, EGFP-RAD54 (gift from Ronald Kanaar, Erasmus Medical Center, Rotterdam, The Netherlands), pGEX4T-1 RAD54 (1-242) (gift from Kiyoshi Miyagawa). pGEX4T-1 BLM(191-660), pGEX4T-1 BLM(621-1041), pGEX4T-1 BLM(1001-1417) (Tripathi et al., 2008). pGEX4T-1 BLM(109-212), pGEX4T-1 BLM(1-180) were obtained by cloning the PCR products into the EcoR1-XhoI sites of the vector. pGEX4T-1 BLM(181-212), pGEX4T-1 BLM(191-212) were obtained by cloning the PCR products into the BamHI-XhoI site of the vector. pGEX4T-1 BLM(1-1417) was obtained by a two-step cloning process: (1) cloning the PCR product into BamH1-XhoI site of pGEX4T-1 to generate pGEX4T-1 BLM(1-1041); (2) cloning the PCR product in the correct orientation into XhoI site of pGEX4T-1 BLM(1-1041) to generate pGEX4T-1 BLM(1-1417). pGEX4T-1 BLM(1-1417) K695A mutant was obtained by site-directed mutagenesis (Stratagene). His-tagged RAD51 was purchased from Calbiochem. pGEX4T-1 RAD54 constructs were generated by cloning the respective PCR products into: (1) NotI site for pGEX4T-1 RAD54(1-747), (2) BamH1-XhoI sites for pGEX4T-1 RAD54(243-747), pGEX4T-1 RAD54(485-625), pGEX4T-1 RAD54(626-748), pGEX4T-1 RAD54(668-748); (3) BamH1-EcoRI sites for pGEX4T-1 RAD54(1-155) and pGEX4T-1 RAD54(156-344). pRS-shRNA RAD54 constructs (TR309964) were purchased from Origene.
Antibodies
Anti-BLM: rabbit polyclonals NB 100-214 (Novus); DR1034 (Calbiochem) and mouse monoclonal BFL-103 (Novus). Anti-RAD54: rabbit polyclonal ab10705 and mouse monoclonal (4E3/1) ab11055 (Novus). Anti-RAD51: rabbit polyclonal Ab-1 (PC-130) and mouse monoclonal Ab-2 (NA71) (Calbiochem). Anti-GFP: rabbit polyclonal 632460 and mouse monoclonal 632375 (Clontech).
Cell culture, treatment and transfection
hTERT-immortalized Bloom Syndrome (BS) fibroblasts, chromosome 15 minochromosome corrected BS fibroblasts (referred to as A-15), hTERT-immortalized NHF strain GM07532 (referred to as NHF), SV40-immortalized GFP BLM complemented GM08505 (referred to as GFP-BLM) were maintained as described (Hu et al., 2001; Sengupta et al., 2003). For HU experiments during immunoprecipitation and immunofluorescence, cells were either left untreated (–HU) or treated (+HU) for 12 hours. All transfections were done using Lipofactamine 2000 (Invitrogen).
Immunoprecipitation and immunofluorescence
Cytoplasmic and nuclear extracts from cells were made using NE-PER Nuclear and Cytoplasmic Extraction reagent (Pierce). Immunoprecipitation was done as described previously (Sengupta et al., 2003) using 1 mg nuclear extracts. The experiments were repeated at least twice and representative blots shown. Immunofluorescence experiments were carried out as described previously (Sengupta et al., 2003). Briefly the cells were washed and either directly fixed in 100% ice cold ethanol or were subjected to a hypotonic lysis buffer (10 mM Tris-HCl, pH 7.5, 2.5 mM MgCl2, 1 mM PMSF, 0.5% NP-40) on ice and subsequently fixed with ethanol. After staining, cells were visualized in an Upright Axioimager M1 motorized epifluorescence microscope equipped with a high resolution AxioCam MRm Rev. 2 camera. The images were taken with Plan Apochromate ×100/1.40 NA oil-immersion objective using FITC, Texas Red or Cy5 fluorophores (Jackson ImmunoResearch). At least 100 cells were analyzed for all immunofluorescence experiments, which were repeated twice.
Expression, purification and interaction of GST-tagged proteins
GST-tagged proteins were expressed according to standard protocols in E. coli at 16°C and subsequently purified by binding to Glutathione-S-Sepharose (GE Healthcare) for use in interaction studies. Soluble proteins were obtained by eluting the bound proteins with reduced glutathione. The proteins subsequently dialyzed in Slide-A-Lyzer Dialysis Cassettes (Pierce) and used for ATPase and chromatin-remodeling assays.
pcDNA FLAG-BLM and pcDNA3.1 FLAG-RAD54 were used for coupled in vitro transcription or translation of BLM and RAD54, respectively. Reactions were carried out with T7 Quick coupled Transcription/Translation System kit (Promega) using 1 μg of the respective cDNAs, 5 μCi of [35S]methionine for 90 minutes at 30°C. GST-bound target proteins were incubated with the in vitro translated interacting partner for 4 hours at 4°C with constant inversion. Interaction was assayed by determining the radioactivity bound to GST beads after washing, SDS-PAGE and fluorography. The experiments were repeated three times and representative blots shown.
Overexpressed GFP-RAD54 (in HEK293T cells) was used for interaction with soluble GST proteins to facilitate purification. The lysates, obtained in UTB lysis buffer (8M Urea, 50 mM Tris-HCl pH 7.5, 150 mM β-mercaptoethanol) were sonicated twice for 10 seconds with a 5 minute cool down period on ice between sonications. Samples were centrifuged at 4°C for 20 minutes at 33,000 g and the clarified supernatant was bound to GFP-antibody-bound Protein G beads. Bound GFP-RAD54 was checked by SDS-PAGE and Coomassie blue staining and for the lack of RAD51 and BLM by co-immunoprecipitation followed by western analysis. Bound GFP-RAD54 was incubated with soluble GST or GST-tagged BLM fragments for 6 hours at 4°C with constant inversion. Interaction was assayed by determining the GST proteins bound to GFP-RAD54 beads after washing, SDS-PAGE and western blotting with anti-GST antibodies. The experiments were repeated three times and representative blots shown.
Affinity pull-down assays
To show RAD51-RAD54 interaction, Protein-G-Sepharose (GE Healthcare) bound immunoprecipitated GFP-tagged RAD54 (3 μg, as estimated from Coomassie-blue-stained gels) and soluble His-tagged RAD51 (1 μg) were used. To determine whether BLM fragments can prevent of RAD51-RAD54 complex formation, increasing amount of soluble BLM fragment (1 μg, 3 μg) were added to the RAD51-RAD54 reaction. The reaction was carried out in PBS + 0.1% NP40 for 6 hours at 4°C. After incubation, GFP-RAD54 bound to the beads and the interacting proteins (if any) was pelleted down by centrifugation, washed three times with binding buffer and subjected to SDS-PAGE and western blotting.
Host cell reactivation assay
The host cell reactivation assay to determine HR rate was carried out as described (Slebos and Taylor, 2001). Transfection was done using either HR substrate (pBHRF) alone or pBHRF combination with RAD51 or SMRAD51. Cells transfected with pBHRF were grown for 24 hours. Each experiment was done in triplicate and repeated three times. The substrate (pBHRF), encoding an intact, emission shifted `blue' variant of GFP (BFP), with a 300 nucleotide stretch of homology to a nonfunctional copy of GFP. In the absence of HR, only BFP is present whereas HR can also create a functional GFP. Green and blue fluorescence were simultaneously examined by exciting the cells using a 488 nm Argon laser (GFP) and a UV (350-360 nm) laser (BFP). After transfection, incubation was continued for 36 hours in complete medium, after which the cells were harvested and flow cytomentry carried out. Each experiment was carried out at least three times. Data analysis was done using Cell Quest Pro.
Sister chromatid exchange
SCE analysis for was carried out according to standard protocols (Sengupta et al., 2003). 60 metaphase spreads for each cell line were imaged at ×100 in a Zeiss Axio Imager M1 microscope. The whole experiment was carried out twice, and for each cell line, SCE was scored blind. P-values were obtained by Student's t-test with two-tailed, unpaired data with unequal variance.
FRAP and time-lapse microscopy
Transfection with GFP-RAD51 and GFP-RAD54 was carried out in A-15/BS cells grown on Lab-Tek chambered coverslips (Nalge Nunc International). Cells were grown in normal growth medium for 2 days before the transfection. 6 hours after transfection, cells were washed and culture continued. After 20 hours, cells were either treated with vehicle or with HU for 2 hours. At this stage, normal cell growth medium was replaced with phenol-red-free DMEM containing 10% charcoal-stripped serum. Cells were transferred to a computer-controlled incubation chamber with active CO2, temperature and humidity control. FRAP analysis was carried out by using a Zeiss 510 Meta system with a 100×/1.4 NA oil-immersion objective and a 40 mW argon laser. Cells expressing abnormally high or low levels of the GFP-tagged proteins were not included in the analysis. 30 single imaging scans were acquired before bleaching with a bleach pulse of 160 mseconds using the 458, 488 and 512 nm laser lines at 100% laser power and 70% laser output without attenuation. Defined areas (2 μm in diameter) were photoblached either in the foci or in the nucleoplasm. Images of single z-sections were collected every 0.08 seconds up to 30 seconds post bleach using 488 nm laser line with laser power attenuated to 0.2%. FRAP recovery curves were generated by using LSM software and Microsoft Excel as described (Phair and Misteli, 2001). FRAP of cells stably expressing GFP-BLM were done as above. Student's t-test was used to determine the statistical significance of the results. All quantitative data represent averages ± s.d. from the total number of indicated cells imaged in three independent experiments.
Ribonucleoprotein fractionation assay
RNP fractionation assay was carried out as described (Lou et al., 2006). Briefly, the cells were lysed in buffer I (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.05% NP40, and protease and phosphatase inhibitors) for 5 minutes on ice. Cell lysates were centrifuged at 1575 g for 5 minutes at 4°C. The supernatants were collected (fraction I). The precipitates were washed once with buffer I (fraction II), then extracted with buffer II (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS and protease and phosphatase inhibitors) on ice for 20 minutes. The extracts were centrifuged at 25,200 g for 20 minutes at 4°C and supernatants were collected as the RNP-enriched fraction (fraction III).
ATPase assay
Assay for RAD54 ATPase activity was carried out as described (Busygina et al., 2008). During the ATPase reactions RAD54 (23 nM) was incubated with the designated proteins, i.e. GST-BLM(1-212), GST-RAD51 (Tripathi et al., 2008) or GST itself (133, 266, 532 nM). φX174RF1 DNA (22 μM base pairs) was added to initiate the ATP hydrolysis reaction at 30°C for 15 minutes. 1 μl of the sample was spotted on the polyethyleneimine-coated TLC plate (Merck), resolved in 1.5 M KH2PO4 (pH 3.4) buffer and visualized by phosphoimaging. Assay for BLM ATPase activity using 15 nM of purified protein was carried out as described (Bugreev et al., 2007).
Remodeling substrate and restriction enzyme accessibility assays
The purified p2085S-G5E4 substrate (Ikeda et al., 1999), was digested with Asp718 and ClaI to excise the 2.5 kb fragment and 3′-end labelled with 32P. Nucleosomal reconstitution on the purified fragment was carried out using an in vitro chromatin assembly kit (Vaxron) according to the manufacturer's protocol and checked by micrococcal nuclease digestion. The reconstituted array, referred to as the G5E4 array, was used in restriction enzyme accessibility (REA) assays, which were done as described (Zhang et al., 2007), using RAD54 (500 nM) and radiolabeled substrate (G5E4 array, 1 nM) with BLM(1-212) or BLM(1-1417) K695A (500 nM) as indicated. The assays were done in 20 mM HEPES (pH 7.9) and 40 mM KCl in the presence of 0.4 U/μl of HhaI, 2 mM ATP (where required), 4 mM MgCl2 at 30°C. The reactions were incubated for 15 seconds, 5 minutes or 10 minutes. The reactions were stopped by addition of 50 mM EDTA, 1.2% SDS, digested with proteinase K (final concentration 1 mg/ml, at 37°C for 30 minutes). Proteins were removed by phenol-chloroform extraction, DNA was ethanol precipitated, washed and analyzed on 1% agarose gels. The fraction of uncut products was determined from Phoshoimager scans.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/17/3093/DC1
We thank Ian Hickson, Jerry Shay, Bernard Lopez, Ronald Kanaar, Kiyoshi Miyagawa, Nathan Ellis, Jerry Workman for recombinants and cells; Geethavani Rayasam for initial help in FRAP experiments, National Institute of Immunology core funds, Department of Biotechnology, India (BT/PR9598/Med/30/33/2007), Department of Science and Technology, India (SR/SO/HS-24/2005), Council of Scientific and Industrial Research [37(1348)/08/EMR-II] and National Institutes of Health, USA (1 R01 TW007302-01A1) for financial assistance. Deposited in PMC for release after 12 months.
References
- Adams, M. D., McVey, M. and Sekelsky, J. J. (2003). Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299, 265-267. [DOI] [PubMed] [Google Scholar]
- Alexeev, A., Mazin, A. and Kowalczykowski, S. C. (2003). Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 10, 182-186. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen, S., Lukas, C., Melander, F., Bartek, J. and Lukas, J. (2005). Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 170, 201-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof, O., Kim, S. H., Irving, J., Beresten, S., Ellis, N. A. and Campisi, J. (2001). Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 153, 367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev, D. V., Yu, X., Egelman, E. H. and Mazin, A. V. (2007). Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 21, 3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busygina, V., Sehorn, M. G., Shi, I. Y., Tsubouchi, H., Roeder, G. S. and Sung, P. (2008). Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 22, 786-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers, J., Houtsmuller, A. B., van Veelen, L., Paulusma, C., Nigg, A. L., Pastink, A., Vermeulen, W., Hoeijmakers, J. H. and Kanaar, R. (2002). Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 21, 2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub, E. I., Kovalenko, O. V., Gupta, R. C., Ward, D. C. and Radding, C. M. (1997). Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res. 25, 4106-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer, W. D., Li, X., Rolfsmeier, M. and Zhang, X. P. (2006). Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 34, 4115-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, P., Beresten, S. F., van Brabant, A. J., Ye, T. Z., Pandolfi, P. P., Johnson, F. B., Guarente, L. and Ellis, N. A. (2001). Evidence for BLM and Topoisomerase IIIalpha interaction in genomic stability. Hum. Mol. Genet. 10, 1287-1298. [DOI] [PubMed] [Google Scholar]
- Ikeda, K., Steger, D. J., Eberharter, A. and Workman, J. L. (1999). Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol. 19, 855-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, S. and Lopez, B. S. (2000). Characterization of mammalian RAD51 double strand break repair using non-lethal dominant-negative forms. EMBO J. 19, 3090-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, Z., Minter-Dykhouse, K., Franco, S., Gostissa, M., Rivera, M. A., Celeste, A., Manis, J. P., van Deursen, J., Nussenzweig, A., Paull, T. T. et al. (2006). MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 21, 187-200. [DOI] [PubMed] [Google Scholar]
- Lukas, C., Melander, F., Stucki, M., Falck, J., Bekker-Jensen, S., Goldberg, M., Lerenthal, Y., Jackson, S. P., Bartek, J. and Lukas, J. (2004). Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 23, 2674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar, A. V., Ozsoy, A. Z., Genschel, J., Modrich, P. and Kowalczykowski, S. C. (2008). Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. USA 105, 16906-16911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orre, L. M., Stenerlow, B., Dhar, S., Larsson, R., Lewensohn, R. and Lehtio, J. (2006). p53 is involved in clearance of ionizing radiation-induced RAD51 foci in a human colon cancer cell line. Biochem. Biophys. Res. Commun. 342, 1211-1217. [DOI] [PubMed] [Google Scholar]
- Ouyang, K. J., Woo, L. L. and Ellis, N. A. (2008). Homologous recombination and maintenance of genome integrity: cancer and aging through the prism of human RecQ helicases. Mech. Ageing Dev. 129, 425-440. [DOI] [PubMed] [Google Scholar]
- Phair, R. D. and Misteli, T. (2001). Kinetic modelling approaches to in vivo imaging. Nat. Rev. Mol. Cell Biol. 2, 898-907. [DOI] [PubMed] [Google Scholar]
- Raynard, S., Bussen, W. and Sung, P. (2006). A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J. Biol. Chem. 281, 13861-13864. [DOI] [PubMed] [Google Scholar]
- Sengupta, S., Linke, S. P., Pedeux, R., Yang, Q., Farnsworth, J., Garfield, S. H., Valerie, K., Shay, J. W., Ellis, N. A., Wasylyk, B. et al. (2003). BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 22, 1210-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S., Doherty, K. M. and Brosh, R. M., Jr (2006). Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 398, 319-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, T. R., Ali, A. M., Busygina, V., Raynard, S., Fan, Q., Du, C. H., Andreassen, P. R., Sung, P. and Meetei, A. R. (2008). BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 22, 2856-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slebos, R. J. and Taylor, J. A. (2001). A novel host cell reactivation assay to assess homologous recombination capacity in human cancer cell lines. Biochem. Biophys. Res. Commun. 281, 212-219. [DOI] [PubMed] [Google Scholar]
- Sonoda, E., Sasaki, M. S., Morrison, C., Yamaguchi-Iwai, Y., Takata, M. and Takeda, S. (1999). Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 19, 5166-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, P. and Klein, H. (2006). Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 7, 739-750. [DOI] [PubMed] [Google Scholar]
- Sung, P., Krejci, L., Van Komen, S. and Sehorn, M. G. (2003). Rad51 recombinase and recombination mediators. J. Biol. Chem. 278, 42729-42732. [DOI] [PubMed] [Google Scholar]
- Tan, T. L., Kanaar, R. and Wyman, C. (2003). Rad54, a Jack of all trades in homologous recombination. DNA Repair (Amst.) 2, 787-794. [DOI] [PubMed] [Google Scholar]
- Tripathi, V., Nagarjuna, T. and Sengupta, S. (2007). BLM helicase-dependent and -independent roles of 53BP1 during replication stress-mediated homologous recombination. J. Cell Biol. 178, 9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, V., Kaur, S. and Sengupta, S. (2008). Phosphorylation-dependent interactions of BLM and 53BP1 are required for their anti-recombinogenic roles during homologous recombination. Carcinogenesis 29, 52-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Seki, M., Narita, Y., Sonoda, E., Takeda, S., Yamada, K., Masuko, T., Katada, T. and Enomoto, T. (2000). Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J. 19, 3428-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L., Davies, S. L., Levitt, N. C. and Hickson, I. D. (2001). Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 276, 19375-19381. [DOI] [PubMed] [Google Scholar]
- Wu, L., Bachrati, C. Z., Ou, J., Xu, C., Yin, J., Chang, M., Wang, W., Li, L., Brown, G. W. and Hickson, I. D. (2006). BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl. Acad. Sci. USA 103, 4068-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D. S., Sonoda, E., Takeda, S., Huang, C. L., Pellegrini, L., Blundell, T. L. and Venkitaraman, A. R. (2003). Dynamic control of Rad51 recombinase by self-association and interaction with BRCA2. Mol. Cell 12, 1029-1041. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Fan, H. Y., Goldman, J. A. and Kingston, R. E. (2007). Homology-driven chromatin remodeling by human RAD54. Nat. Struct. Mol. Biol. 14, 397-405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.