Summary
The TonB system of Escherichia coli (TonB/ExbB/ExbD) transduces the protonmotive force (pmf) of the cytoplasmic membrane to drive active transport by high affinity outer membrane transporters. In this study, chromosomally encoded ExbD formed formaldehyde-linked complexes with TonB, ExbB, and itself (homodimers) in vivo. Pmf was required for detectable crosslinking between TonB-ExbD periplasmic domains. Consistent with that observation, the presence of inactivating transmembrane domain mutations ExbD(D25N) or TonB(H20A) also prevented efficient formaldehyde crosslinking between ExbD and TonB. A specific site of periplasmic interaction occurred between ExbD(A92C) and TonB(A150C) and required functional transmembrane domains in both proteins. Conversely, neither TonB, ExbB, nor pmf were required for ExbD dimer formation. These data suggest two possible models where either dynamic complex formation occurred through transmembrane domains or the transmembrane domains of ExbD and TonB configure their respective periplasmic domains. Analysis of T7-tagged ExbD with anti-ExbD antibodies revealed that a T7 tag was responsible both for our previous failure to detect T7-ExbD-ExbB and T7-ExbD-TonB formaldehyde-linked complexes and for the concomitant artifactual appearance of T7-ExbD trimers.
Keywords: Escherichia coli, ExbD, TonB, transmembrane domains, protein conformation, iron transport
Introduction
The Gram-negative bacterial cell envelope serves the dual roles of protective barrier for the cell and a gateway for the entry of essential elements and nutrients. The cell envelope consists of the concentric cytoplasmic (CM) and outer (OM) membranes, separated by an aqueous periplasmic space. The OM protects the cell from hydrophobic antibiotics, degradative enzymes, and detergents while allowing small (< 600 Da) hydrophilic nutrients entry by passive diffusion through OM porin proteins (Nikaido, 2003). Nutrients that are too large, too scarce or too important to pass through porins—iron siderophore complexes, vitamin B12, nickel, sucrose and possibly sulfate—are actively transported across the OM. Energy for transport across the unenergized OM is supplied by the protonmotive force (pmf) of the CM, with the TonB system acting as the energy-coupling agent between the two membranes (recently reviewed in (Postle and Larsen, 2007)). In the presence of protonophores such as dinitrophenol (DNP) or carbonylcyanide m-chlorophenylhydrazone (CCCP), ligands can bind to the outer membrane transporters but are not transported across the outer membrane. The conformation of TonB changes depending on whether pmf is present or absent (Larsen et al., 1999). The pmf-dependent mechanisms of this system, however, remain largely unknown.
The TonB system of Escherichia coli consists of a complex of the CM proteins TonB, ExbB, and ExbD. ExbD is topologically similar to TonB, with each containing a single transmembrane domain (TMD) and the majority of the soluble domain occupying the periplasm (Hannavy et al., 1990; Kampfenkel and Braun, 1992; Roof et al., 1991). In contrast, ExbB has three transmembrane domains, with the majority of its soluble domains occupying the cytoplasm (Kampfenkel and Braun, 1993). TonB is known to form homodimers in the CM, and both ExbB and ExbD form homomultimers in vivo (Ghosh and Postle, 2005; Higgs et al., 1998; Sauter et al., 2003). The cellular ratio of TonB:ExbB:ExbD is 1:7:2, but the stoichiometry within an energy transduction complex is unknown (Held and Postle, 2002; Higgs et al., 2002b). Paralogues of ExbB and ExbD have been proposed to form complexes with a 4:2 MotA:MotB or 6-4:2 TolQ:TolR stoichiometry (Braun et al., 2004; Kojima and Blair, 2004; Cascales et al., 2001).
ExbD is an essential component of the TonB system, required for TonB activity (Brinkman and Larsen, 2008). Little is known about the precise role of ExbD, though it was recently proposed to have a chaperone-like function in regulating the dynamics of TonB conformation (Ghosh and Postle, 2005; Larsen et al., 2007). Only two essential residues, aspartate 25 in the TMD and periplasmic residue leucine 132, have been identified, such that D25N or L132Q substitutions render ExbD inactive (Braun et al., 1996). The functional significance of these residues, however, remains obscure. The conserved corresponding TMD residues in TolR and MotB, D23 and D32, are also essential for activity within their respective systems. It has been proposed that these essential acidic residues are part of proton pathways through the putative TolQR or MotAB proton channels (Cascales et al., 2001; Zhou et al., 1998).
The sole functionally significant side chain in the TonB TMD is histidine 20. The remainder of the TonB TMD residues can be replaced by alanine without significant effect (Larsen et al., 2007). The TonB amino terminal TMD serves as a signal anchor, a means by which TonB dimerizes in vivo, a means of contact with ExbB, and to regulate the conformation of the TonB carboxy terminus (Ghosh and Postle, 2005; Jaskula et al., 1994; Karlsson et al., 1993; Larsen et al., 1994; Larsen et al., 1999; Larsen and Postle, 2001; Larsen et al., 2007; Postle and Skare, 1988; Sauter et al., 2003; Skare et al., 1989).
Although many details remain unclear, current data suggest a mechanism for energy transduction whereby ExbB/ExbD harvest the pmf and transmit it to TonB, allowing the TonB carboxy terminus to transduce energy to a ligand-loaded OM transporter. Following the energy transduction event, TonB is recycled back to an energizable state by ExbB/ExbD, undergoing conformational changes both prior to and following interaction with OM transporters (Ghosh and Postle, 2005; Larsen et al., 1999; Postle and Kadner, 2003). These conformations require a functional TonB transmembrane domain, the pmf, and ExbB/ExbD.
In this study, we demonstrated for the first time that pmf was required for the interaction of ExbD and TonB periplasmic domains trapped by formaldehyde crosslinking. Consistent with that, transmembrane domain mutations proposed to be on the proton pathway across the cytoplasmic membrane in either ExbD or TonB prevented formaldehyde and disulfide-directed crosslinking of the periplasmic domains. ExbD also efficiently crosslinked to ExbB and formed homo-dimers, but not homo-trimers in vivo. Our earlier study showed that T7-tag-ExbD formed crosslinked homo-dimers and homo-trimers, but did not crosslink to TonB or ExbB (Higgs et al., 1998). We show here that those artifactual results were due to the presence of the T7 tag at the ExbD amino terminus.
Results
Wild-type ExbD crosslinks to ExbB and TonB and forms homodimers in vivo
To examine in vivo interactions of wild-type, chromosomally-encoded ExbD with itself or other proteins, whole cells were treated with monomeric formaldehyde, processed for immunoblot analysis, and characterized using polyclonal anti-ExbD antibody (Higgs et al., 2002a). Along with the ExbD monomer, migrating at an apparent molecular mass of 15 kDa, three higher molecular mass complexes, at approximately 30, 41, and 52 kDa, were detected (Fig. 1, wild-type lane). An active plasmid-encoded ExbD size variant, ExbDΔ2-11, which lacked ten residues in the cytoplasmic domain, was used to determine which, if any, of the complexes represented ExbD homo-multimers. This size variant migrated with an apparent molecular mass of 13 kDa. Accordingly, for the formaldehyde crosslinking profile of ExbDΔ2-11, complexes containing homomultimers of ExbDΔ2-11 were expected to show a shift in migration equal to a multiple of this difference. The 30 kDa complex obtained with wild-type ExbD was replaced by a 26 kDa complex when the ExbDΔ2-11 size variant was crosslinked (Fig. 1). This shift of twice the difference in monomeric masses identified the 30 kDa complex as a homodimer of ExbD. Both of the remaining complexes showed a shift in migration of approximately 2 kDa, suggesting each contained monomeric ExbD in complex with other proteins. Previous work using T7 epitope tag-specific antibody had demonstrated the ability of a T7 epitope-tagged ExbD to form homodimers and homotrimers in vivo (Higgs et al., 1998). This work confirmed the ability of wild-type ExbD to form homodimers in vivo. The ExbD trimer previously observed with T7 epitope-tagged ExbD at ∼48 kDa was not observed for wild-type, chromosomally-encoded ExbD or plasmid-encoded ExbD expressed near chromosomal levels. An explanation for this discrepancy will be addressed below.
Fig. 1.
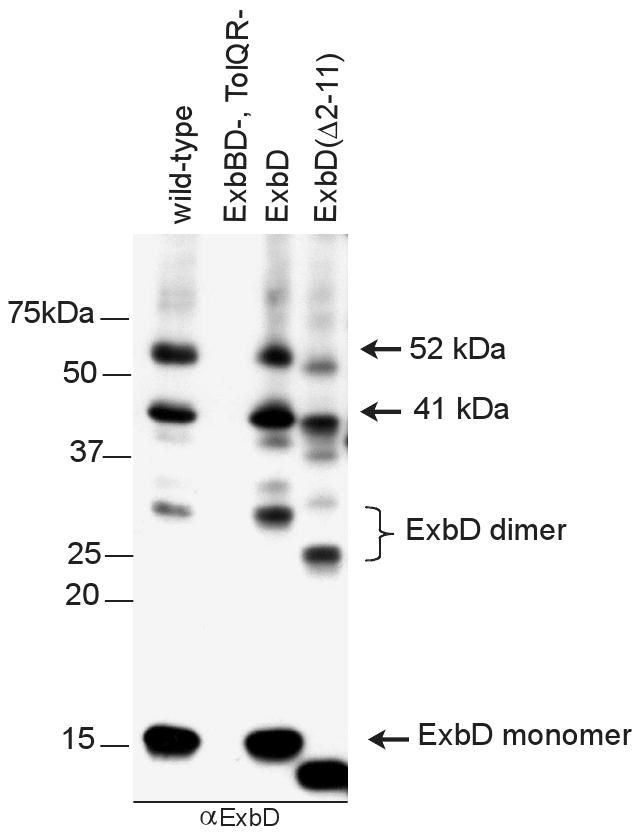
Wild-type ExbD forms homo-dimers in vivo. Strains expressing chromosomally encoded (W3110) or plasmid-encoded wild-type ExbD (RA1017/pKP660), and ExbD(Δ2-11) (RA1017/pKP761) were crosslinked with formaldehyde as described in Materials and Methods. Plasmids encoding ExbD also encoded wild-type ExbB. Levels of L-arabinose for induction were 0.0002% (w/v) for pKP660 and 0.001% for pKP761. Samples were resolved on a 13% SDS-polyacrylamide gel and immunoblotted. ExbD was visualized with ExbD-specific polyclonal antibodies. Positions of molecular mass standards are indicated on the left. Identities or apparent molecular masses of ExbD-specific crosslinked complexes and the ExbD monomer are indicated on the right.
Other proteins likely to interact with ExbD include TonB and ExbB. Based on a theoretical mass of 41.6 kDa for an ExbB-ExbD heterodimer, the 41 kDa ExbD-specific complex had the potential to be a complex between ExbB and ExbD. When ExbD was expressed in a strain lacking ExbB, both the 41 kDa and 52 kDa complexes were absent, demonstrating the dependence of both of these complexes on the presence of ExbB (Fig. 2).
Fig. 2.
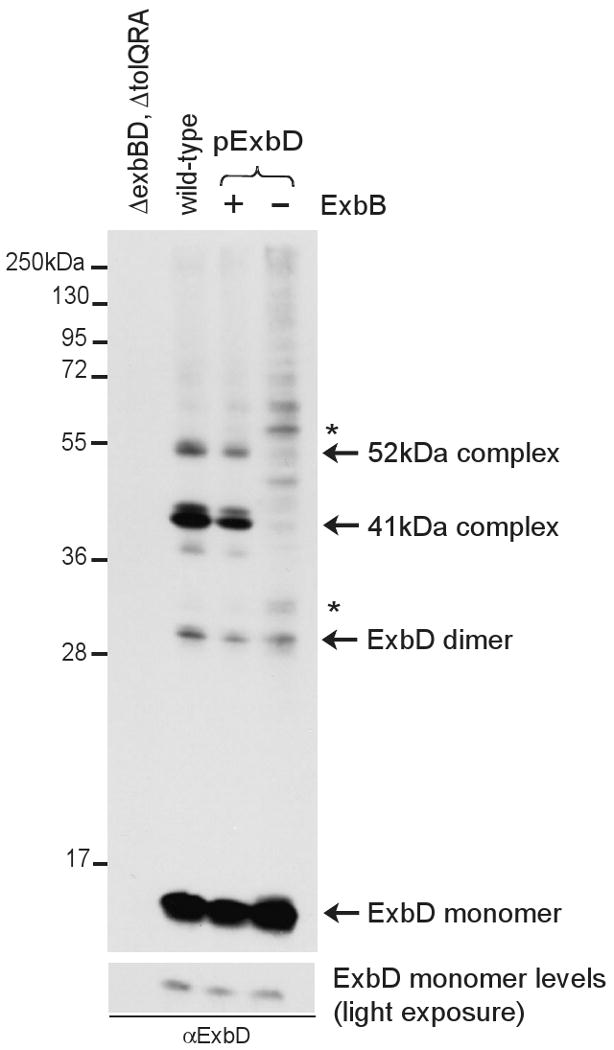
The 41 and 52 kDa ExbD-specific complexes are dependent on the presence of ExbB. Strains expressing chromosomally-encoded (W3110) or plasmid-encoded wild-type ExbD (pKP999) in ExbB+ (RA1045) or ExbB- (RA1017) backgrounds were crosslinked with formaldehyde as described in Materials and Methods. pKP999 was induced with 3mM sodium propionate, pH 8 in RA1045 and 20mM sodium propionate, pH 8 in RA1017. Samples were resolved on a 13% SDS-polyacrylamide gel and immunoblotted. Approximately 40% more of the ExbB- sample (right lane) was loaded to achieve ExbD monomer levels of equal intensity to wild-type. ExbD was visualized with ExbD-specific polyclonal antibodies. “+” or “-” indicates the presence or absence, respectively, of ExbB in the sample resolved in the lane below the symbol. Positions of molecular mass standards are indicated on the left. Identities of ExbD-specific crosslinked complexes and monomers are indicated on the right. (*) indicates an unidentified complex.
To determine if either complex contained ExbB protein, exbBD cells (KP1392) expressing wild-type ExbD and ExbB fused to the fluorescent protein Venus were crosslinked with formaldehyde. When expressed at normal chromosomal levels, ExbB-Venus (52.3 kDa) was approximately 90% active compared to wild-type ExbB (Bulathsinghala and Postle, unpublished results). The presence of ExbB-Venus resulted in the loss of the 41 kDa ExbD-specific complex and the appearance of two novel higher molecular mass complexes at ∼68 kDa and ∼84 kDa. The 68 kDa complex corresponded to the theoretical mass of an ExbD-(ExbB-Venus) complex (Fig. 3). The 41 kDa complex therefore represented a complex between wild-type ExbB and ExbD. While previous work identified the potential for ExbB-ExbD interaction through in vitro binding of ExbD to ExbB (Braun et al., 1996), this is the first direct evidence of in vivo ExbB-ExbD complex formation. A less intense band migrating slightly above the ExbB-ExbD complex, at approximately 44 kDa, was also dependent on the presence of ExbB. However, a corresponding band was not detected for wt ExbD in the presence of ExbB-Venus and its identity remains unknown. The identity of the 84 kDa complex observed for ExbD in experiments with ExbB-Venus also was not determined. The 52 kDa complex was still detected in the presence of ExbB-Venus, indicating it did not contain ExbB. But based on its absence in the exbB strain, it clearly required ExbB for its assembly (Fig. 2, 3).
Fig. 3.
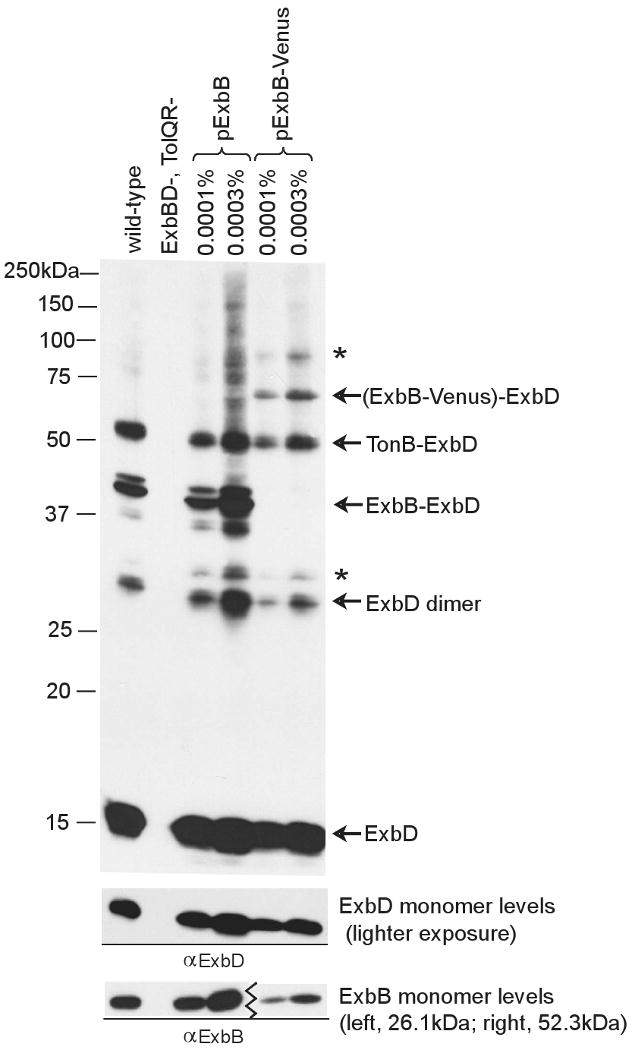
The 41 kDa complex contains one ExbD and one ExbB. Strains expressing chromosomally-encoded (GM1) or plasmid-encoded (KP1392/pKP660) ExbB and ExbB-Venus fusion protein (KP1392/pKP944) were crosslinked with formaldehyde as described in Materials and Methods. All plasmids also encoded wild-type ExbD. Proteins were expressed using two different percentages of arabinose, as indicated above each lane. ExbB monomer levels were determined from culture samples that were TCA precipitated immediately after harvesting. Samples were resolved on a 13% SDS-polyacrylamide gel and immunoblotted. ExbD and ExbB were visualized with ExbD- or ExbB-specific polyclonal antibodies, respectively. Positions of molecular mass standards are indicated on the left. Identities of ExbD-specific crosslinked complexes and the ExbD monomer are indicated on the right. (*) indicates an unidentified complex.
The mass of the 52 kDa ExbD-containing complex closely matched the predicted mass of a complex between ExbD (15.5 kDa) and TonB (with a calculated molecular mass of 26 kDa, but an apparent molecular mass of 36 kDa in SDS polyacrylamide gels) (Eick-Helmerich and Braun, 1989; Postle and Reznikoff, 1979). To determine if the 52 kDa complex was dependent on the presence of TonB, the formaldehyde crosslinking profile of ExbD was examined in a strain lacking TonB (KP1503). In this strain, the 52 kDa ExbD-specific complex was not detected (Fig. 4, A), suggesting this complex consisted of a heterodimer of TonB and ExbD. Using anti-TonB antibody, a TonB-ExbD complex was also identified by the absence of the 52 kDa complex in a strain lacking ExbD (RA1045) (Fig. 4, B). Taken together these data indicated that the 52 kDa complex detected with ExbD-specific antibody was a TonB-ExbD complex that required ExbB to form. A TonB-ExbD-ExbB complex was not detected, most likely due to inefficiency of trimolecular crosslinking.
Fig. 4.
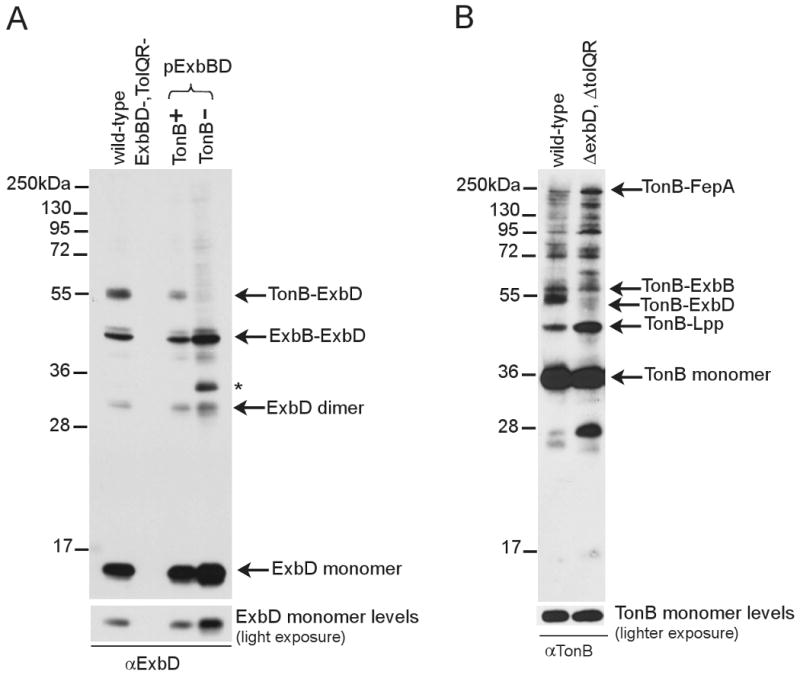
The 52 kDa complex contains one ExbD and one TonB. Strains expressing chromosomally-encoded (GM1) or plasmid-encoded wild-type ExbD in the presence (KP1392/pKP660) or absence (KP1503/pKP660) of TonB were crosslinked with formaldehyde as described in Materials and Methods. Plasmids encoding ExbD also encoded wild-type ExbB and were induced with 0.0004% (w/v) L-arabinose. Samples were resolved on a 13% SDS-polyacrylamide gel and immunoblotted. A. ExbD visualized with ExbD-specific polyclonal antibodies. B. TonB visualized with TonB-specific monoclonal antibodies. Positions of molecular mass standards are indicated on the left. Identities of ExbD- or TonB-specific crosslinked complexes and monomers are indicated on the right. Light exposures for comparison of monomer levels are present at the bottom of each figure. (*) indicates an unidentified complex.
Formaldehyde-specific crosslinks between TonB and ExbD almost certainly occurred between their periplasmic domains rather than their transmembrane domains. Formaldehyde crosslinking is initiated by formation of methylol derivatives at 1° amino groups or 1° thiol groups which then undergo a condensation to form a Schiff-base that can subsequently crosslink to a variety of amino acids (Means and Feeney, 1971; Metz et al., 2004). However, under the conditions of rapid in vivo crosslinking with formaldehyde, the spectrum of subsequent interactions is limited primarily to lysyl, tryptophanyl, and cysteinyl residues (Toews et al., 2008). Given the sequences of the ExbD and TonB cytoplasmic and transmembrane domains and their identical topologies, the only likely crosslink would form between the amino terminus of ExbD and Trp 11 in TonB; however, deletion of Trp 11 does not prevent ExbD-TonB crosslinking (data not shown). In contrast, the periplasmic domains of each protein contain numerous crosslinkable residues; the TonB periplasmic domain has 1 tryptophanyl and 18 lysyl residues and the ExbD periplasmic domain has 10 lysyl residues. The predicted pIs of the periplasmic domains [TonB residues 33-239 (9.6) and ExbD residues 44-141 (5.3)] are also consistent with their interactions.
Pmf is required for in vivo formation of TonB-ExbD formaldehyde crosslinks
The role of ExbB in formation of TonB-ExbD crosslinks suggested that proposed proton translocation through ExbB might also be important (Braun and Herrmann, 2004). Although the pmf is clearly required for TonB-dependent transport across the OM, there is little known about its mechanistic role (Bradbeer, 1993). To determine if pmf was required for any ExbD interactions, we treated cells with various amounts of DNP and with CCCP prior to and during formaldehyde crosslinking (Fig. 5). In the presence of the protonophores, TonB-ExbD crosslinks were undetectable while levels of ExbD dimer and ExbB-ExbD crosslinks remained unchanged. A similar decrease in TonB-ExbD crosslinking was also observed with anti-TonB antibodies with no other detectable changes to the normal crosslinking profile other than slight increases in the TonB-FepA and TonB-Lpp complexes (data not shown), consistent with the fact that these complexes occur with unenergized TonB (Ghosh and Postle, 2005). Comparison with control immunoblots indicated that monomeric TonB and ExbD levels were unaffected by protonophore treatments.
Fig. 5.
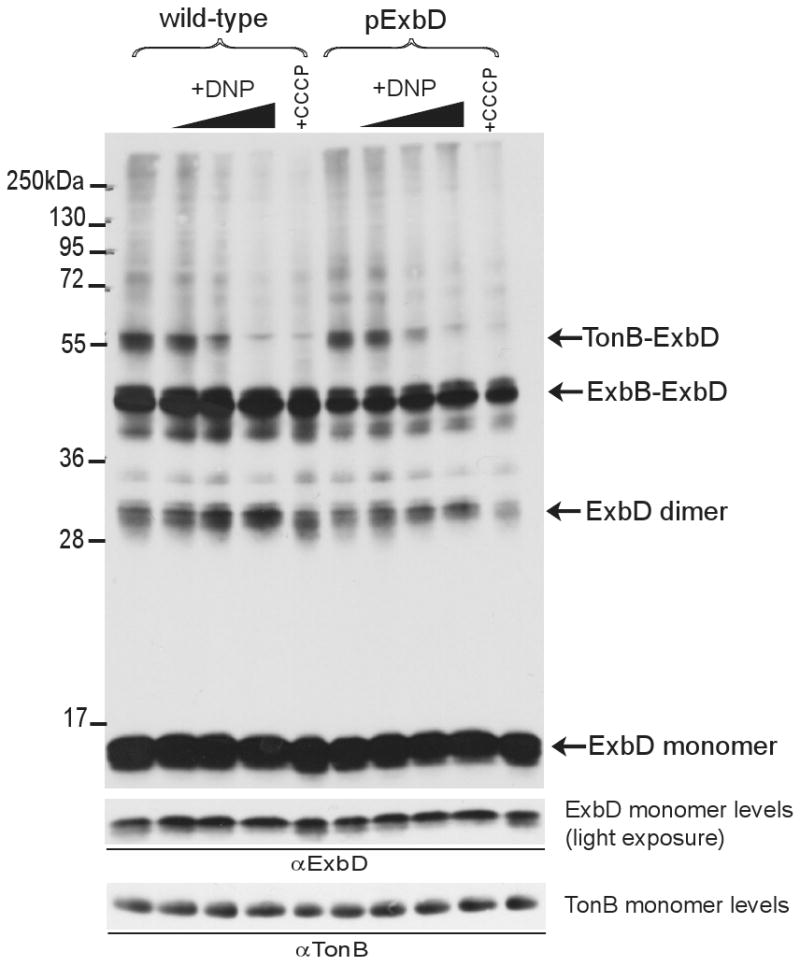
Pmf regulates TonB-ExbD complex formation. Strains expressing chromosomally-encoded (W3110) or plasmid-encoded (RA1045/pKP999) ExbD were crosslinked with formaldehyde in the presence of protonophores that collapse the pmf as described in Materials and Methods. The expanding triangle above the +DNP lanes indicates the presence of 1, 5, or 10 mM DNP. +CCCP indicates the presence of 50 μM CCCP. Solvent only (DMSO) was added to samples lacking protonophore. Plasmid-encoded ExbD was expressed with 3 mM sodium propionate, pH 8. Samples were resolved on 13% SDS-polyacrylamide gels and immunoblotted. ExbD was visualized with ExbD-specific polyclonal antibodies. Positions of molecular mass standards are indicated on the left. Identities of ExbD-specific crosslinked complexes and monomers are indicated on the right. Light exposures for comparison of monomer levels are present at the bottom of each figure. TonB monomer levels were visualized with TonB-specific monoclonal antibodies.
A D25N substitution in the ExbD transmembrane domain disrupts TonB-ExbD periplasmic domain interaction
The transmembrane domain of ExbD might be on the proton translocation pathway. Asp 25, a residue in the transmembrane domain of ExbD, is essential for ExbD activity (Braun et al., 1996). We confirmed that D25N inactivates ExbD and also showed that ExbD(D25A) is inactive (Table 1), which ruled out the possibility that D25N was inactive due to steric hindrance. The D25N substitution also did not prevent proper localization of ExbD to the cytoplasmic membrane (Fig. 6).
Table 1. Spot titer assay results.
TonB system activity of strains expressing variants of ExbD and TonB was evaluated using spot titer assays. Cultures of the mutants expressed to near-chromosomal levels (verified through Western blot—not shown) were plated on T-plates and spotted with fivefold serial dilutions of colicins and tenfold serial dilutions of bacteriophage ϕ80. Values were recorded as the reciprocal of the highest dilution at which clearing of the bacterial lawn was evident after 18 hours of incubation at 37°C. “T” indicates tolerance (no sensitivity).
| Strain | Phenotype | Sensitivitya | |||
|---|---|---|---|---|---|
| Colicin B | Colicin Ia | Colicin M | ϕ80 | ||
| W3110 | WT | 8,8,8 | 7,7,7 | 6,6,6 | 8,8,8 |
| RA1045 | ExbD-,TolQR- | T,T,T | T,T,T | T,T,T | T,T,T |
| KP1509 | ExbD- TonB- | T,T,T | T,T,T | T,T,T | T,T,T |
| KP1344/pKP381 | TonB(H20A) | T,T,T | T,T,T | T,T,T | T,T,T |
| KP1344/pKP1054 | TonB(H20D) | T,T,T | T,T,T | T,T,T | T,T,T |
| RA1045/pKP1055 | ExbD(D25H) | T,T,T | T,T,T | T,T,T | T,T,T |
| KP1509/pKP1054/pKP1055 | ExbD(D25H)/TonB(H20D) | T,T,T | T,T,T | T,T,T | T,T,T |
| RA1045/pKP999 | ExbD | 8,8,7 | 7,7,7 | 6,6,6 | 8,8,8 |
| RA1045/pKP1064 | ExbD(D25N) | T,T,T | T,T,T | T,T,T | T,T,T |
| RA1045/pKP1191 | ExbD(D25A) | T,T,T | T,T,T | T,T,T | T,T,T |
Scored as the highest fivefold dilution of a standard colicin preparation, or tenfold dilution of bacteriophage ϕ80, that provided an evident zone of clearing on a cell lawn. “T” indicates tolerance (i.e. no clearing of the lawn) to undiluted colicin or phage. The values of three platings are presented for each strain/plasmid and colicin or phage pairing.
Fig. 6.
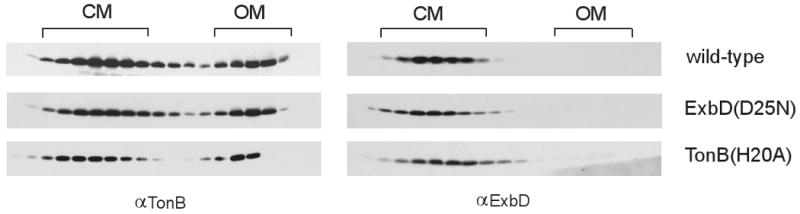
ExbD(D25N) and TonB(H20A) fractionate identically to the wild-type forms of each protein. Strains expressing chromosomally-encoded ExbD and TonB (W3110), ExbD(D25N) (RA1021/pKP1064), and TonB(H20A) (KP1344/pKP381) were fractionated using sucrose density gradient fractionation, as described in Materials and Methods. No inducer was needed for ExbD(D25N). TonB(H20A) was induced with .00025% (w/v) arabinose. Samples were resolved on 13% SDS-polyacrylamide gels and immunoblotted. ExbD and TonB were visualized with ExbD-specific polyclonal or TonB-specific monoclonal antibodies, respectively.
Interestingly, the (D25N) or (D25A) substitutions in the ExbD transmembrane domain both prevented formaldehyde crosslinking to TonB, as detected by either anti-ExbD or anti-TonB immunoblots (Fig. 7 and data not shown). The presence of ExbD(D25N)-ExbB complexes also confirmed that ExbD(D25N) was localized properly to the cytoplasmic membrane (Fig. 7). ExbD(D25N) also formed homodimers, although the apparent molecular mass of the complex was slightly less than that observed for the wild-type ExbD dimer, even though ExbD(D25N) monomer has an apparent molecular mass similar to wildtype. The identity of the ExbD(D25N) dimer was confirmed using size variants (data not shown). An unidentified complex containing ExbD(D25N) migrated slightly above the dimer and was more abundant compared to wild-type ExbD. The increased intensity for this band was also observed for the crosslinking of ExbD in the absence of TonB (Fig. 4, A), suggesting that it increases when ExbD does not interact with TonB.
Fig. 7.
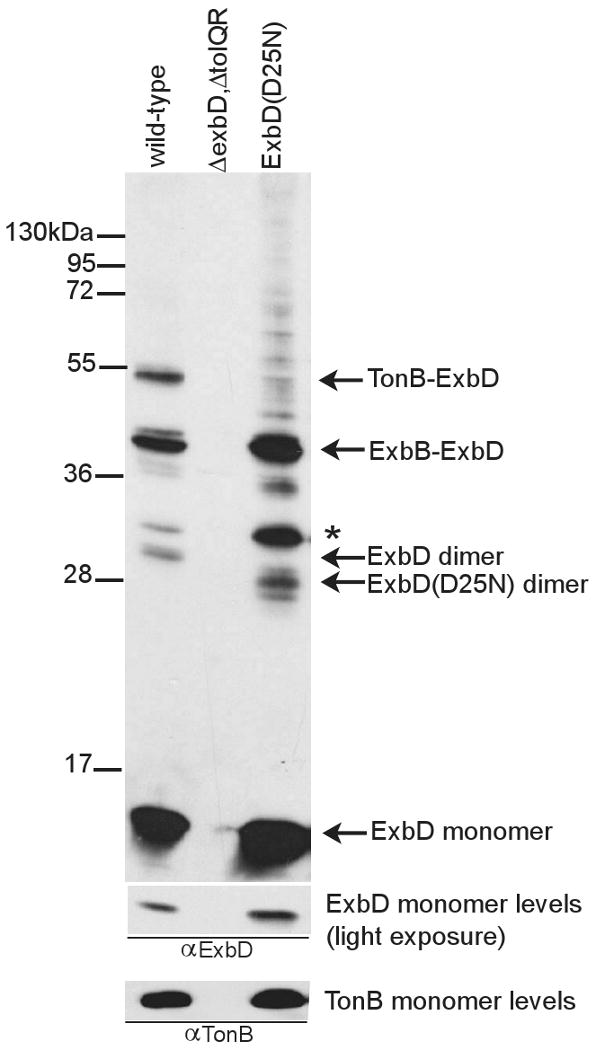
ExbD(D25N) does not crosslink to TonB in vivo. Strains expressing chromosomally-encoded ExbD (W3110) and ExbD(D25N) (RA1045/pKP1064) were crosslinked with formaldehyde as described in Materials and Methods. ExbD(D25N) was induced with 0.05mM sodium propionate, pH 8. Samples were resolved on a 13% SDS-polyacrylamide gel and immunoblotted. ExbD was visualized with ExbD-specific polyclonal antibodies. To verify that TonB levels were unchanged, TonB monomer was visualized with TonB-specific monoclonal antibodies. Positions of molecular mass standards are indicated on the left. Identities of ExbD-specific crosslinked complexes and the ExbD monomer are indicated on the right. (*) indicates an unidentified complex.
The H20A substitution in the TonB transmembrane domain also disrupts ExbD-TonB interaction
A complementary approach was used to examine the effect of a TonB transmembrane domain mutation on TonB-ExbD complex formation. Histidine 20 in TonB is the only functionally important side chain in its transmembrane domain, with a H20A substitution leading to inactivity of chromosomally encoded TonB in all assays (Larsen et al., 1999; Larsen et al., 2007), (Table 1). Like ExbD (D25N), the TonB(H20A) was properly localized to the CM. (Fig. 6). The formation of a formaldehyde-crosslinked complex between TonB and ExbB was unaffected by the H20A substitution, also confirming that TonB(H20A) was correctly assembled in the cytoplasmic membrane. Like ExbD(D25N), the TonB(H20A) mutation eliminated detection of the TonB-ExbD formaldehyde-crosslinked complex by either anti-TonB or anti-ExbD antibodies (Fig. 8). The formaldehyde crosslinked complexes of TonB(H20A) with Lpp and FepA increased in intensity relative to wild-type TonB, consistent with previous observations that those complexes arise from inactive TonB and do not require a functional TonB TMD to form (Ghosh and Postle, 2005; Jaskula et al., 1994). In an attempt to determine whether TonB(H20) and ExbD(D25) form a salt bridge, TonB(H20D) and ExbD(D25H) were constructed. Each was individually inactive, and they were inactive (and present at chromosomal levels) when co-expressed—a negative, and thus uninterpretable, result (Table 1).
Fig. 8.
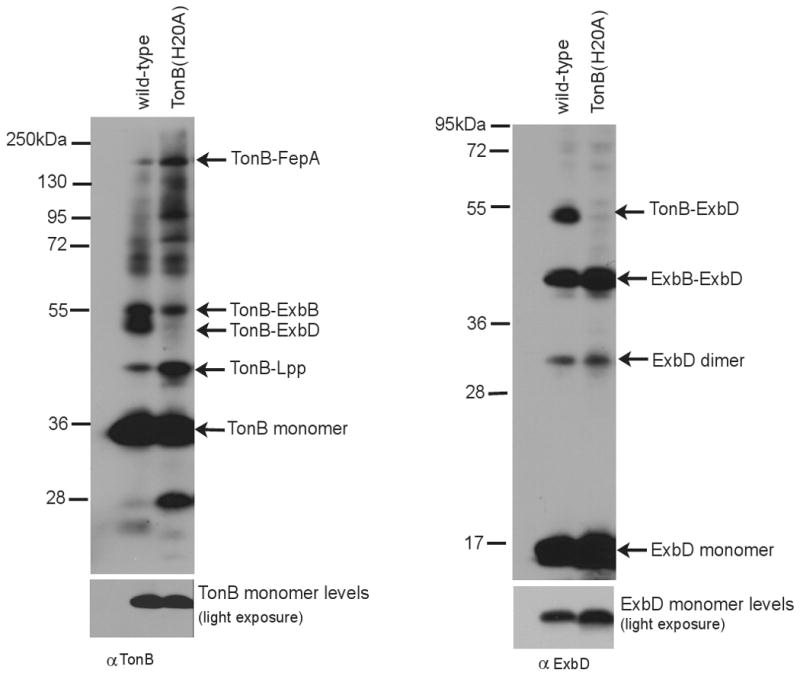
TonB(H20A) does not crosslink to ExbD in vivo. Strains expressing chromosomally-encoded wildtype TonB (W3110) and TonB(H20A) (KP1344/pKP381) were crosslinked using formaldehyde. L-arabinose at a final concentration of 0.001% (wt/vol) was used to induce pKP381. Samples were resolved on an 11% SDS-polyacrylamide gel and analyzed using immunobloting with ExbD-specific polyclonal antibodies and TonB-specific monoclonal antibodies. Positions of molecular mass standards are indicated on the left. Identities of crosslinked complexes and the protein monomers are indicated on the right.
T7-tagged ExbD crosslinks artifactually
As noted above, we had previously observed that T7-tagged ExbD complemented an exbD mutation and could be formaldehyde-crosslinked into dimers and trimers, but did not detectably crosslink to TonB or ExbB (Higgs et al., 1998). To determine the source of this difference with chromosomally encoded ExbD, the formaldehyde crosslinking of T7-ExbD was revisited, this time using ExbD-specific antibody for detection. T7-ExbD was recloned into propionate and arabinose expression vectors, expressed by induction to chromosomal levels or overexpressed, crosslinked in vivo with formaldehyde, and analyzed by immunoblot with ExbD- or T7-epitope-tag-specific antibodies.
In contrast to the previous results, the formaldehyde crosslinked T7-ExbD detected by anti-ExbD antisera matched the profile of wild-type ExbD when expressed at chromosomal levels (Fig. 9A). This difference was explained, however, by comparison to identical samples detected with T7 epitope tag-specific antibody, which unexpectedly revealed that the vast majority of the T7-ExbD had been proteolytically processed to remove the T7-tag. Thus the normal ExbD crosslinking profile of the “T7-ExbD” originated from ExbD lacking the T7 tag. In this experiment the level of intact T7-ExbD was so low as to make detection of formaldehyde cross-linked complexes impossible.
Fig. 9.
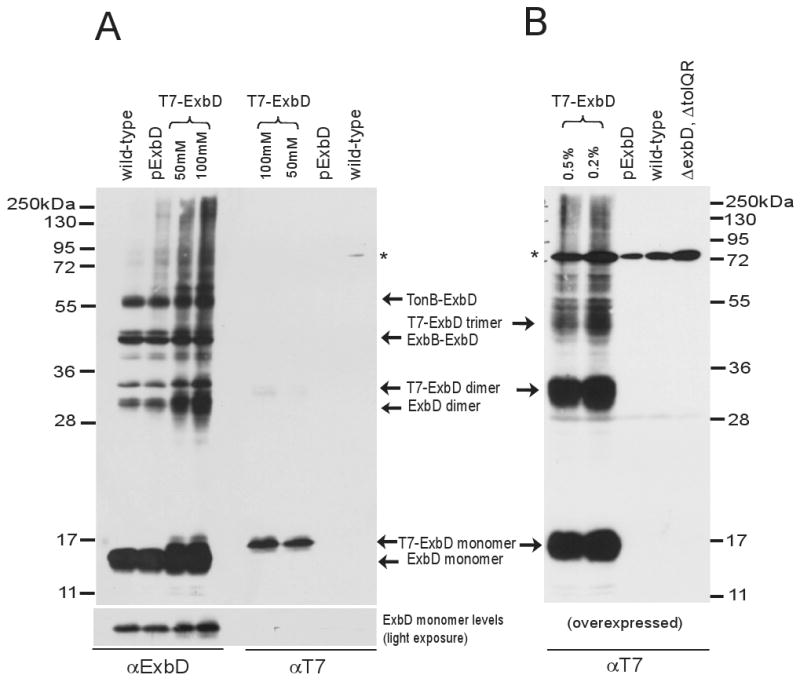
ExbD with an amino terminal T7-epitope tag crosslinks artifactually. Strains expressing chromosomally encoded ExbD (W3110), plasmid-encoded wild-type ExbD (RA1045/pKP999), and T7-epitope tagged ExbD (RA1045/pKP1186 or RA1045/pKP1195 for overexpression) were crosslinked with formaldehyde as described in Materials and Methods. Plasmid-encoded ExbD was expressed with 3mM sodium propionate, pH 8. T7-ExbD was induced with two different concentrations of sodium propionate for pKP1186 (A) or two different percentages of arabinose for overexpression from pKP1195 (B) as indicated above each lane. Samples were resolved on a 13% SDS-polyacrylamide gel and immunoblotted. ExbD was visualized with ExbD-specific polyclonal antibodies or T7 epitope tag-specific monoclonal antibodies. Positions of molecular mass standards are indicated on the side. Identities of ExbD-specific crosslinked complexes and monomers are indicated in the middle. (*) indicates a non-specific cross-reactive band.
To detect formaldehyde crosslinked complexes specific to the tagged population of ExbD, T7-ExbD was overexpressed from the arabinose promoter, crosslinked with formaldehyde and detected with anti-T7-tag antibody. Like the 1998 study that this replicated, the anti-T7 crosslinking profile contained complexes at the molecular masses of a T7-ExbD dimer (33.4 kDa) and trimer (50.1 kDa), and did not contain TonB-ExbD or ExbB-ExbD complexes (Fig. 9B and data not shown). This crosslinking result was not due to overexpression, since overexpressed ExbD had the same crosslinking profile as chromosomally expressed ExbD (data not shown).
Taken together, these results indicated that 1) 1998 immunoblots with T7 tag-specific antibody were detecting only the minor subpopulation of ExbD protein that retained the T7 epitope tag and 2) that the artifactual formation of formaldehyde crosslinked trimeric T7-ExbD and lack of TonB-ExbD and ExbB-ExbD complexes was due to the presence of the T7 tag and did not reflect the normal behavior of ExbD. Even though the activity of T7-ExbD could not be determined against a background containing a preponderance of full-length active ExbD, based on its abnormal crosslinking behavior the T7-ExbD was almost certainly inactive.
Active cysteine substitutions ExbD(A92C) and TonB(C18G, A150C) demonstrate specific TonB-ExbD periplasmic domain contact in vivo
While the periplasmic domain of ExbD was identified as the site of formaldehyde-mediated crosslinking to TonB, we did not identify specific residues through which it occurred. To begin to map regions of interaction between ExbD and TonB, cysteine substitutions were engineered in their respective periplasmic domains, and the existence of disulfide-linked heterodimers was monitored on non-reducing SDS-polyacrylamide gels. ExbD has no native cysteinyl residues whereas TonB carries a single cysteinyl residue at position 18. As an example of this approach, ExbD(A92C) and TonB(C18G, A150C), which were fully active when expressed at near chromosomal levels (Ollis, Kastead, and Postle, unpublished results), were analyzed. The appearance of identical novel complexes at 52 kDa on immunoblots developed with either TonB- or ExbD-specific antibodies indicated that the periplasmic domains of these two proteins were indeed interacting in vivo (Fig. 10). The 52 kDa complex was specific to the presence of the introduced cysteine in each protein (data not shown).
Fig. 10.
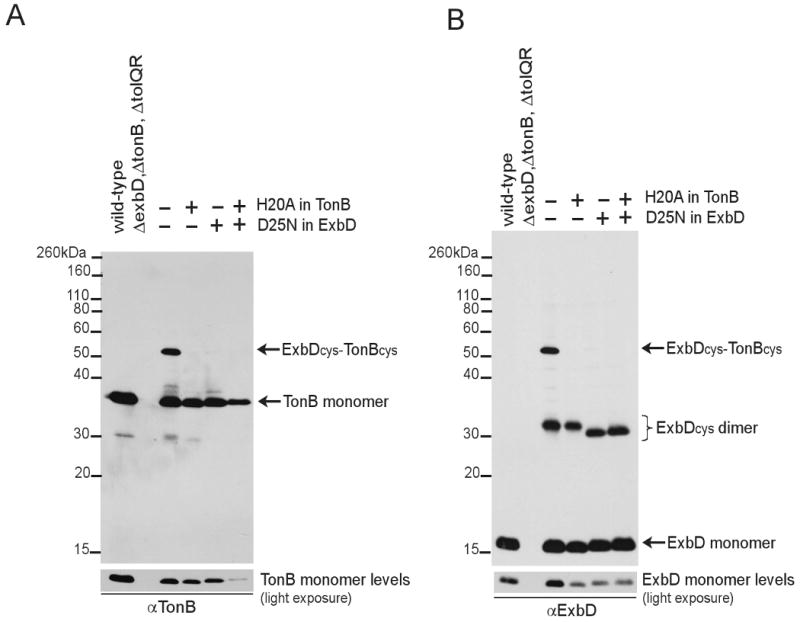
Active TonB and ExbD cysteine substitutions form specific periplasmic domain contacts. Strains expressing wild-type ExbD and TonB (W3110), ExbD(A92C) with TonB(C18G, A150) [KP1509/pKP1000, pKP945], ExbD(A92C) with TonB(C18G, H20A, A150C) [KP1509/pKP1000, pKP1031], ExbD(D25N, A92C) with TonB(C18G, A150C) [KP1509/pKP1049, pKP945] were processed in non-reducing sample buffer containing iodoacetamide as described in Materials and Methods. Samples were resolved on a 13% non-reducing SDS-polyacrylamide gel and immunoblotted. ExbD (A) or TonB (B) was visualized with ExbD-specific polyclonal antibodies or TonB-specific monoclonal antibodies. Positions of molecular mass standards are indicated on the left. Lanes 3 through 6 contained strain KP1509 expressing derivatives of ExbD(A92C) coexpressed with TonB(C18G, A150C). Derivatives contained (+) or lacked (-) the residue substitutions listed to the right.
To determine if the 52 kDa disulfide-linked complex represented a biologically relevant interaction, the effect of ExbD(D25N) or TonB(H20A) transmembrane domain substitutions was also examined using either anti-TonB or anti-ExbD antibodies (Fig. 10 A, B respectively). The inactivating H20A substitution in the transmembrane domain of TonB(C18G, A150) essentially eliminated TonB-ExbD disulfide-linked complex formation detected with either antibody. The inactivating D25N substitution likewise prevented TonB-ExbD complex formation. Coexpression of the inactive mutants did not restore detection of a disulfide-linked complex. Taken together these results indicate that interactions of the TonB and ExbD periplasmic domains require activities attributable to their transmembrane domains. It was not possible to assess the effects of protonophores on formation of disulfide crosslinks because 1) disulfide crosslinks are pre-existing in the population and 2) synthesis of the two proteins in a pulse requires pmf for their export to the periplasm.
Similar to the formation of ExbD dimers through formaldehyde crosslinking, ExbD(A92C) formed disulfide-linked dimers through its periplasmic domain (Fig. 8, B). ExbD(D25N, A92C) monomer ran, if anything, slightly slower than the ExbD(A92C) monomer. Interestingly, the D25N transmembrane domain substitution resulted in an apparently smaller molecular mass, suggesting that a novel conformational change had been trapped.
Discussion
The TonB/ExbB/ExbD proteins of E. coli couple the cytoplasmic membrane ion electrochemical potential (most likely a proton potential) to active transport of iron-siderophore and vitamin B12 nutrients across the outer membrane. In other Gram-negative bacteria, the TonB system energizes outer membrane transport of iron-binding proteins, sucrose, Ni(II), and potentially sulfate, suggesting that it serves as the general means by which the limiting porosity of the outer membrane can be overcome (Blanvillain et al., 2007; Cescau et al., 2007; Schauer et al., 2007; Tralau et al., 2007). TonB undergoes cyclic energization, transduction of that energy to a TonB-gated transporter, and recharging to allow re-energization (Fischer et al., 1989; Larsen et al., 1999). ExbB and ExbD appear to have roles in both harvesting the protonmotive force, allowing TonB to then transduce this energy to TonB-gated transporters, and in recycling TonB after it has transduced energy. If TonB is not energized, it is not recycled (Larsen et al., 1999; Letain and Postle, 1997). ExbD has been proposed to chaperone the conformation of the TonB carboxy terminus and is specifically involved in the recycling of TonB following energy transduction (Brinkman and Larsen, 2008; Larsen et al., 2007). Its role in energization of TonB has not been directly determined.
A new role for the pmf in TonB-dependent energy transduction
Our results here indicate for the first time a definitive role for the cytoplasmic membrane pmf in promoting functionally important interaction of TonB and ExbD through their periplasmic domains. First, two different protonophores that collapse the proton gradient of the cytoplasmic membrane prevent formation of ExbD-TonB formaldehyde crosslinks in vivo. Second, the ExbD(D25N) transmembrane domain mutation, which inactivates ExbD, also prevents ExbD-TonB formaldehyde crosslinks and disulfide-directed crosslinks between their periplasmic domains. The ExbD(D25N) mutation occurs at a conserved residue that is equally important in ExbD paralogues TolR and MotA, considered to be on the proton pathway, and responsible for conformational changes in those proteins (Cascales et al., 2001; Goemaere et al., 2007; Kojima and Blair, 2001). Third, the TonB(H20A) transmembrane domain mutation, which inactivates TonB, also prevents ExbD-TonB formaldehyde crosslinks and disulfide crosslinks between their periplasmic domains. The His 20 is conserved among most TonB genes and also conserved in the analogous TolA protein of the Tol system (Germon et al., 1998). His20 is required for pmf-dependent conformational changes in the TonB carboxy terminus and is the sole functionally significant side-chain in the entire transmembrane domain (Larsen et al., 1999; Larsen et al., 2007). Fourth, the L132Q mutation in the periplasmic domain of ExbD knocks out ExbD function (Braun et al., 1996). ExbD(L132Q) does not crosslink in vivo to TonB although it can still crosslink into dimers and crosslink to ExbB, indicating that the periplasmic interaction between TonB and ExbD is a functionally important one (data not shown). These data support the idea that ExbD manages the conformational changes in the carboxy terminus of TonB.
We previously observed that the TonB energy transduction cycle is functionally divided into events that occur prior to energy transduction and those that occur following energy transduction, by performing the experiments in an aroB strain that cannot synthesize enterochelin or any of its precursors. In the absence of ligand, TonB does not transduce energy to the TonB-gated transporter FepA, thus interrupting the cycle (Larsen et al., 1999). Because the ExbD-TonB crosslinked complex (as well as the ExbD dimer and ExbD-ExbB complex described below) was detected equally well in a wild-type or aroB strain, it indicated that the TonB-ExbD interaction detected by formaldehyde crosslinking occurred prior to the energy transduction step (data not shown). This was also consistent with the requirement for pmf and intact transmembrane domains, and indicated that ExbD plays a role in the energization step on the front half of the energy transduction cycle. Since a role for ExbD in recycling TonB has been identified, ExbD appears to play critical roles both before and after energy transduction by TonB.
Two models for the TonB-ExbD interaction
These results suggest two possible models for TonB-ExbD interaction. In the first model, the TonB-ExbD complex is formed dynamically, and only in response to the presence of the pmf. Thus the pmf would be responsible for allowing TonB and ExbD transmembrane domains to move close enough for interactions between their periplasmic domains to be captured through crosslinking. There is evidence to support the idea of dynamic complexes: the ratios of the total numbers per cell for ExbB and ExbD proteins are, at 7:2, significantly higher than the ratios of the total numbers per active complex for paralogues MotA:MotB, TolQ:TolR, or PomA:PomB at 4:2 (Cascales et al., 2001; Guihard et al., 1994; Kojima and Blair, 2004; Sato and Homma, 2000). It thus may be that the TonB/ExbB/ExbD complex is in equilibrium with pools of uncomplexed ExbB, assembling in response to cellular signals to transduce energy. Consistent with that idea, it has been recently shown that MotB moves in and out of the flagellar rotor complex (Leake et al., 2006).
In the second model, the stably assembled transmembrane domains of TonB and ExbD in association with ExbB would be somehow responsible for directly transmitting conformational information to their periplasmic domains. The TonB transmembrane domain is known to play a role in regulating the conformation of its carboxy terminus (Ghosh and Postle, 2005; Larsen et al., 1999; Larsen et al., 2007). Because the TonB amino terminus and carboxy terminus are separated by a non-essential proline-rich region, it seems unlikely that the regulation occurs via a proton-wire (Larsen et al., 1993; Seliger et al., 2001). However, different types of transmembrane helix motions have been proposed to propagate conformational changes to adjacent domains including a piston motion between helices, pivoting of helices and rotation of helices (Matthews et al., 2006). For the ExbD paralogue, MotB, Asp32 is required for conformational changes in MotA, the ExbB paralogue (Kojima and Blair, 2001). It will be important to distinguish between the two models.
The nature of ExbD dimerization
ExbD dimerization occurred in the absence of ExbB and in the presence of the ExbD(D25N) substitution believed to render ExbD unresponsive to pmf, consistent with previous observations that ExbD(D25N) is dominant negative (Braun et al., 1996). Also consistent with these observations, formaldehyde crosslinking of ExbD dimers did not require pmf. In spite of the fact that the ExbD transmembrane domains appear to interact closely, the formaldehyde-specific ExbD dimers were mediated through the periplasmic domain and almost certainly required interaction of many residues. Indeed, deletion of the periplasmic domain of ExbD(D25N) relieved its dominant negativity (Braun et al., 1996). Perhaps ExbD(D25N) is blocked in the ability to transition from homodimeric periplasmic domain interactions to functionally important heterodimeric interactions with TonB. If so, deletion of periplasmic domain residues, which eliminated the dominant negative effect of ExbD(D25N), would then have freed the periplasmic domain of wild-type ExbD to transition to its normal interactions.
At least one of the residues in the dimerization region was A92, which when substituted with a cysteinyl residue, was capable of trapping a disulfide-linked ExbD dimer. A92C was also a residue through which ExbD contacted the periplasmic domain of TonB at residue A150C. It may be that the ExbD dimer was maintained through its transmembrane domain while the periplasmic domain cycled between interactions with another ExbD periplasmic domain or a TonB periplasmic domain. In our hands, ToxR-ExbD fusion proteins can activate a ctx∷cat fusion (Russ and Engelman, 1999), indicating that the transmembrane domains of ExbD are sufficiently close that that they allow functional dimerization of ToxR, whether or not ExbB is present (Vakharia-Rao and Postle, unpublished results). Movement of the dimeric ExbD transmembrane domains relative to one another could drive changes in interactions between periplasmic domains. Rotation of dimeric paralogue TolR transmembrane helices relative to each other has been documented recently (Zhang et al., 2009). The physiological role of the ExbD dimer is currently unknown.
In contrast to results seen here, in the Tol system formaldehyde crosslinking of TonB paralogue TolA with ExbD paralogue TolR is not pmf-dependent; however, TolA interaction with lipoprotein Pal is (Cascales et al., 2000). TolA-TolR interaction is mediated through the last 25 amino acids of TolR (Journet et al., 1999). Structural changes in the periplasmic carboxy terminus of TolR are, however, dependent on the presence of pmf and residues in the predicted TolQR ion pathway, including TolR Asp23, the residue analogous to ExbD Asp25 (Goemaere et al., 2007). The differences in pmf-dependent interaction partners may reflect the divergent functions identified for the periplasmic domains of ExbD and TolR (Brinkman and Larsen, 2008).
Comparison of in vitro and in vivo structural predictions
The importance of TonB and ExbD transmembrane domains in determining the conformations and interactions of their periplasmic domains is underscored by comparison to the structures of the soluble periplasmic domains of these two proteins lacking their transmembrane domains (Chang et al., 2001; Garcia-Herrero et al., 2007; Kodding et al., 2005; Peacock et al., 2005). In the recently solved NMR structure of the ExbD soluble domain 5-7 copies of the ExbD monomer formed a multimeric complex at pH 7.0. Residue A92, through which ExbD can efficiently form dimers in vivo, was far from this multimeric interface (Fig. 11). Additionally in that paper, no significant interactions between purified ExbD and TonB periplasmic domains were detected in vitro, leading the authors to conclude that the functional interactions between TonB and ExbD likely occurred primarily through their transmembrane domains in vivo. In contrast, we propose here that the lack of detectable interaction in vitro was almost certainly due to the absence of the transmembrane domains of TonB and ExbD as well as ExbB and the pmf.
Fig. 11.
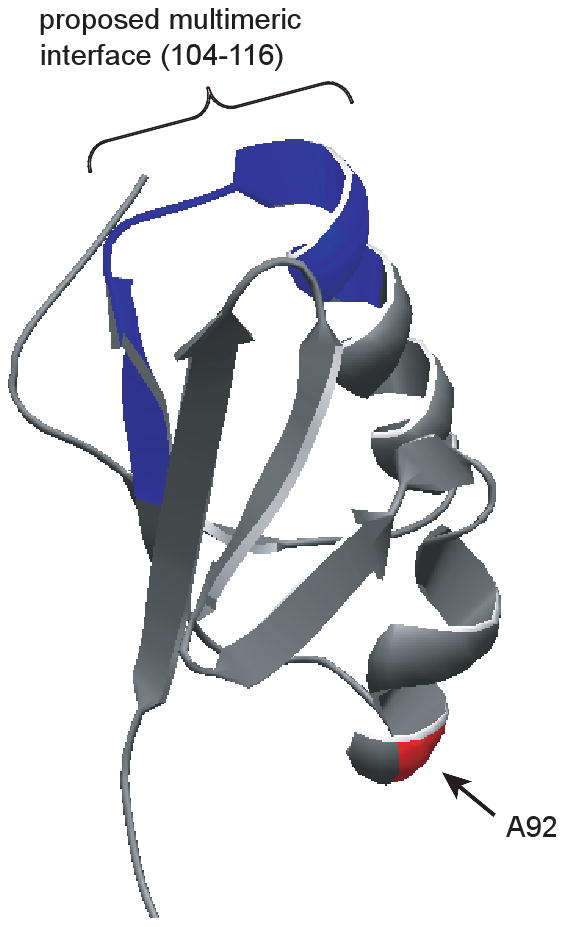
ExbD A92 is distantly located from the proposed multimeric interface of ExbD. The NMR structure of the carboxy-terminal domain (amino acids 44-141) of ExbD is shown (pdb code: 2pfu). The proposed multimeric interface (amino acids 104-116) (Garcia-Herrero et al., 2007) is highlighted in blue. Residue A92 is highlighted in red. ExbD(A92C) spontaneously formed dimers through this residue in vivo (Fig.10).
With respect to the TonB transmembrane domain, the in vivo data on full-length TonB also diverge from major aspects of the solved structures for the periplasmic carboxy terminus of TonB protein. In particular, while the 5 aromatic residues of the carboxy terminus are buried in the crystal and NMR structures, the in vivo data indicate that they are surface exposed, accessible for homodimeric interactions, and virtually the only functionally important residues in the carboxy terminus [(Ghosh and Postle, 2004; Ghosh and Postle, 2005); Kastead and Postle, unpublished observations]. A mutant TonB transmembrane domain that inactivates TonB also prevents TonB homodimer formation through cysteine substitutions at the aromatic residues (Ghosh and Postle, 2005). Thus for both ExbD and TonB, the structural results obtained in vitro without transmembrane domains and access to the pmf are significantly different than those obtained in vivo where all needed components are present. It is thus not clear if the structures of the soluble domains of ExbD and TonB represent in vivo conformations.
Artifactual crosslinking results arising from the use of a T7 epitope tag
The acquisition of anti-ExbD antibodies allowed the characterization of wild-type ExbD interactions at chromosomally encoded levels (Higgs et al., 2002a). ExbD could be crosslinked by formaldehyde to itself (homo-dimers), to ExbB, and to TonB. This was the first time that crosslinks to either ExbB or TonB had been observed in vivo. Consistent with the requirement for pmf, TonB-ExbD crosslinks did not form in the absence of ExbB. Both TonB and ExbD are proteolytically unstable in the absence of ExbB [(Fischer et al., 1989) and data not shown]. ExbB has three transmembrane domains and appears to be the glue that holds the complex together since when expressed in the absence of TonB or ExbD, ExbB is proteolytically stable [(Fischer et al., 1989) and Higgs and Postle, unpublished results].
Before anti-ExbD antibodies were available, we had characterized plasmid-encoded ExbD tagged with a T7-epitope and observed only ExbD dimers and trimers in vivo (Higgs et al., 1998). The data presented here show that the ability to detect artifactual ExbD trimers, as well as the artifactual inability to detect interactions with ExbB and TonB, was due to the presence of the T7-tag. The fact that the T7-ExbD could complement an exbD mutation was meant to provide confidence in the results from an overexpressed epitope tagged protein study. Instead, the observed complementation was almost certainly due to proteolytic cleavage of the majority of the T7 tag, leaving behind full-length ExbD. Thus we have no evidence that the ExbD trimers are biologically relevant. Without the use of anti-ExbD antibodies, for which absence the T7-tag was originally meant to compensate, these discrepancies would not have been apparent. These results provide a direct and important demonstration of the hazards involved in relying on interpretations of data from tagged or fused proteins.
In summary, TonB and ExbD interact through their periplasmic domains in vivo, with that interaction guided by not-well-understood aspects of their transmembrane domains. The study of the periplasmic domains of TonB and ExbD in vitro has greatly enhanced our knowledge of what their structures and behaviors are in the absence of transmembrane domains and protonmotive force, and provided an important basis for comparison with in vivo results. Could it be that the in vitro structures of soluble domains of TonB and ExbD represent a default non-energized state of these proteins and that the protonmotive force is somehow used to perturb these conformations? As one of the central questions in membrane protein signal transduction biology, it will be important to understand how transmembrane domains regulate conformations and interactions of their soluble domains.
Experimental Procedures
Bacterial strains and plasmids
Bacterial strains and plasmids used in this study are listed in Table 2. KP1484 was constructed by P1vir transduction of ΔtonB, P14∷kan from KP1477 into GM1. KP1503 was constructed by P1vir transduction of ΔtonB, P14∷kan from KP1484 into KP1038. KP1509 was constructed by P1vir transduction of ΔtonB, P14∷kan from KP1484 into RA1045. To create pKP660, the exbB, exbD operon was amplified by polymerase chain reaction (PCR) and cloned into the SmaI site of plasmid pBAD24.
Table 2.
Strains and Plasmids used in this study.
| Strain or Plasmid | Genotype or Phenotype | Reference |
|---|---|---|
| Strains | ||
| W3110 | F− IN(rrnD-rrnE)1 | (Hill and Harnish, 1981) |
| GM1 | ara, Δ(pro-lac), thi, F′ pro lac | (Sun and Webster, 1987) |
| KP1038 | GM1 exbB∷Tn10, tolQ(am) | |
| KP1344 | W3110 tonB∷blaM | (Larsen et al., 1999) |
| KP1392 | GM1 exbB∷Tn10, tolQ(am), recA∷cat | (Held and Postle, 2002) |
| KP1477 | W3110 ΔtonB∷kan | (Devanathan and Postle, 2007) |
| KP1484 | GM1 ΔtonB∷kan | Present study |
| KP1503 | GM1 exbB∷Tn10, tolQ(am), ΔtonB∷kan | Present study |
| KP1509 | W3110 ΔexbD, ΔtolQR, ΔtonB∷kan | Present study |
| RA1017 | W3110 ΔexbBD∷kan, ΔtolQRA | (Larsen et al., 2007) |
| RA1021 | W3110 ΔexbD | Ray Larsen |
| RA1045 | W3110 ΔexbD, ΔtolQR | (Brinkman and Larsen, 2008) |
| Plasmids | ||
| pKP325 | pBAD-regulated TonB | (Larsen et al., 1999) |
| pKP381 | TonB(H20A) | (Larsen et al., 2007) |
| pKP568 | TonB(C18G) | (Ghosh and Postle, 2005) |
| pKP879 | TonB(C18G, H20A) | Present study |
| pKP945 | TonB(C18G, A150C) | Present study |
| pKP1054 | TonB(H20D) | Present study |
| pKP1031 | TonB(C18G, H20A, A150C) | Present study |
| pBAD24 | L-arabinose-inducible, pBR322 ori | (Guzman et al., 1995) |
| pKP660 | pBAD24 expressing exbBD from the pBAD promoter | Present study |
| pKP761 | ExbB, ExbDΔ2-11 | Present study |
| pKP880 | ExbB, ExbD(A92C) | Present study |
| pKP930 | ExbB with 7 residue C-terminal insertion | Present study |
| pKP944 | ExbB-Venus, ExbD | Present study |
| pET21a-Venus | Venus fluorescent protein | (Anderson and Yang, 2008) |
| pKP1186 | pPro24-(T7-ExbD) | Present study |
| pKP1195 | pBAD24-(T7-ExbD) | Present study |
| pPro24 | propionate-inducible, pBR322 ori | (Lee and Keasling, 2005) |
| pKP999 | pPro24 expressing exbD | Present study |
| pKP1000 | ExbD(A92C) | Present study |
| pKP1049 | ExbD(D25N, A92C) | Present study |
| pKP1055 | ExbD(D25H) | Present study |
| pKP1064 | ExbD(D25N) | Present study |
| pKP1191 | ExbD(D25A) | Present study |
pKP1186 was constructed by extra-long PCR on pKP999, using forward and reverse primers each encoding one half of the T7 epitope tag, placed at the extreme amino-terminus of ExbD. The PCR products were recircularized and ligated, joining the halves of the T7 tag sequence. The correct T7 epitope tagged ExbD was confirmed by DNA sequencing of the T7-exbD gene. pKP1195 was constructed by digestion of pKP1186 and pBAD24 with NcoI. Fragments were separated by gel electrophoresis. The 4542 bp fragment of pBAD24 and 539 bp fragment of pKP1186 were purified by gel extraction and ligated together after treatment of the vector fragment with Antarctic Phosphatase (New England Biolabs). Proper orientation of the insert was verified by FspI digestion. The correct T7 epitope tagged ExbD in pBAD24 was confirmed by DNA sequencing.
pKP761 was constructed by in-frame deletion of ten exbD codons using extra-long PCR, as previously described (Higgs et al., 1998). The resulting construct, ExbD(Δ2-11), was determined to be active by standard Fe transport and spot titer assays performed as previously described (data not shown) (Larsen et al., 2003; Postle, 2007). To construct pKP999 and pKP1000, forward and reverse primers were designed to amplify the last 22 codons of exbB through the stop codon of exbD from a pKP660 or pKP880 template, respectively, introducing flanking NcoI sites. The PCR-amplified, NcoI digested fragment was cloned into the unique NcoI site in pPro24. Proper orientation was determined by FspI digestion. Sequences of the exbB segment and exbD gene were confirmed by DNA sequencing.
TonB and ExbD single residue substitutions are derivatives of pKP325 and pKP999, respectively, unless otherwise stated. pKP879 and pKP945 are derivatives of pKP568. pKP1049 is a derivative of pKP1000. Substitutions were generated using 30-cycle extra-long PCR using Pfu Ultra Hotstart DNA Polymerase from Stratagene or Phusion Hotstart DNA Polymerase from Finnzymes. Forward and reverse primers were designed with the desired base change flanked on both sides by 12-15 homologous bases (primer sequences available upon request). DpnI digestion was used to remove the template plasmid. Substitutions were verified by DNA sequencing to avoid unintended base changes.
pKP944 was constructed by directional cloning. First, using a pKP660 template, KpnI and XhoI sites were introduced to the 3′ end of exbB, adding 7 residues (Ala, Gly, Thr, Gly, Gly Leu, Glu) before the stop codon, creating pKP930. The gene encoding the fluorescent GFP derivative Venus was amplified from a pET21a background, introducing a 5′ KpnI site and 3′ XhoI site. The KpnI, XhoI venus fragment was ligated in frame into the corresponding sites in the exbB gene of pKP930 to create pKP944. The resulting ExbB-Venus fusion has three introduced residues, Ala Gly Thr, linking the cytoplasmic carboxy terminal domain of ExbB to Venus. Sequences of exbB and exbD genes were confirmed by DNA sequencing to rule out unintended base changes. To construct pKP1031, plasmids pKP879 and pKP945 were digested with BstEII, resulting in 2 fragments for each. Fragments were separated by gel electrophoresis. The large fragment of pKP945 and small fragment of pKP879 were purified by gel extraction and ligated together after treatment of the large fragment with antarctic phosphatase (New England Biolabs). Proper orientation was determined by BamHI digestion. All DNA sequencing occurred at The Pennsylvania State University Nucleic Acid Facility, University Park, PA.
Media and culture conditions
Luria-Bertani (LB), tryptone (T), and M9 minimal salts were prepared as previously described (Miller, 1972). Liquid cultures, agar plates, and T-top agar were supplemented with 34 μg ml-1 chloramphenicol and/or 100 μg ml-1 ampicillin and plasmid-specific levels of L-arabinose and/or sodium propionate, pH 8, as needed for expression of TonB and ExbD proteins from plasmids. M9 salts were supplemented with 0.5% glycerol (w/v), 0.4 μg ml-1 thiamine, 1 mM MgSO4, 0.5 mM CaCl2, 0.2% casamino acids (w/v), 40 μg ml-1 tryptophan, and 1.85 μM FeCl3. Cultures were grown with aeration at 37°C.
Spot titer activity assays
Assays were performed essentially as previously described (Larsen et al., 2003; Postle, 2007).
Sucrose density gradient fractionation
Mid -exponential phase cultures were grown in M9 medium as described above, harvested, lysed by French pressure cell and fractionated on a 25%-56% (w/w) sucrose gradient as described previously (Letain and Postle, 1997).
In vivo formaldehyde crosslinking
Saturated overnight cultures were subcultured 1:100 into M9 minimal media (above) supplemented arabinose and/or propionate concentrations as needed to achieve chromosomal levels of plasmid expression, and at mid -exponential phase treated with formaldehyde as previously described (Higgs et al., 1998). Crosslinked complexes were detected by immunoblotting with ExbD-specific polyclonal antibodies(Higgs et al., 2002a), TonB-specific monoclonal antibodies (Larsen et al., 1996), or T7-epitope tag-specific monoclonal antibodies (Novagen). For crosslinking in the presence of protonophores, 1, 5, or 10mM 2, 4 dinitrophenol (DNP) or 50μM carbonylcyanide-m-chlorophenylhydrazone (CCCP) were added following resuspension of cell pellets in phosphate buffer. An equal volume of dimethyl sulfoxide (DMSO) was added to wild-type samples as a solvent control. Cells were incubated 5 min at 37°C. Formaldehyde was then added and procedure continued as referenced above.
In vivo disulfide crosslinking assay
Saturated overnight cultures of strains carrying plasmids encoding combinations of TonB and ExbD cysteine substitutions were subcultured 1:100 in T broth containing chloramphenicol and ampicillin and supplemented with L-arabinose and sodium propionate, pH 8, as described below. Cultures were harvested in mid-exponential phase and precipitated with trichloroacetic acid (TCA). Cell pellets were resuspended in non-reducing Laemmli sample buffer containing 50mM iodoacetamide, as previously described (Ghosh and Postle, 2005). Samples were resolved on 13% non-reducing SDS-polyacrylamide gels and evaluated by immunoblot analysis. Levels of inducers for coexpression of the TonB and ExbD cysteine variants were as follows:
pKP1000, pKP945 = 1mM sodium propionate, 0.0005% (w/v) L-arabinose;
pKP1000, pKP1031 = 0.5mM sodium propionate, 0.0005% (w/v) L-arabinose;
pKP1049, pKP945 = 0.5mM sodium propionate, 0.0003% (w/v) L-arabinose;
pKP1049, pKP1031 = 0.3mM sodium propionate, 0.0003% (w/v) L-arabinose
Acknowledgments
We thank Ray Larsen for critical reading of the manuscript, for generously providing strain RA1021, and for early observations on crosslinking of TonBΔTrp11. The generous gifts of plasmids pPro24 from Jay Keasling, and pET21a-Venus from Roger Tsien are gratefully acknowledged. We thank Charlie Bulathsinghala for construction of KP1484, pKP660, pKP930, and pKP944. We thank Loretta Tu for construction of pKP879. We thank Aruna Kumar for construction of pKP761. We thank Kyle Kastead for construction of KP1509 and pKP945. We thank Mary Huber for construction of pKP880. We thank Bryant Schultz for construction of pKP1031. We thank Christine Dubowy for construction of pKP1191. We wish to acknowledge Qian Zhang for early studies on ExbD crosslinking. This work was supported by a grant from the National Institute of General Medical Sciences to K.P.
References
- Anderson LM, Yang H. DNA looping can enhance lysogenic CI transcription in phage lambda. Proc Natl Acad Sci U S A. 2008;105:5827–5832. doi: 10.1073/pnas.0705570105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denance N, Vasse J, Lauber E, Arlat M. Plant Carbohydrate Scavenging through TonB-Dependent Receptors: A Feature Shared by Phytopathogenic and Aquatic Bacteria. PLoS ONE. 2007;2:e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbeer C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J Bacteriol. 1993;175:3146–3150. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Gaisser S, Herrmann C, Kampfenkel K, Killman H, Traub I. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J Bacteriol. 1996;178:2836–2845. doi: 10.1128/jb.178.10.2836-2845.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Herrmann C. Point mutations in transmembrane helices 2 and 3 of ExbB and TolQ affect their activities in Escherichia coli K-12. J Bacteriol. 2004;186:4402–4406. doi: 10.1128/JB.186.13.4402-4406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman KK, Larsen RA. Interactions of the energy transducer TonB with noncognate energy-harvesting complexes. J Bacteriol. 2008;190:421–427. doi: 10.1128/JB.01093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Gavioli M, Sturgis JN, Lloubes R. Proton motive force drives the interaction of the inner membrane TolA and outer membrane pal proteins in Escherichia coli. Mol Microbiol. 2000;38:904–915. doi: 10.1046/j.1365-2958.2000.02190.x. [DOI] [PubMed] [Google Scholar]
- Cascales E, Lloubes R, Sturgis JN. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol Microbiol. 2001;42:795–807. doi: 10.1046/j.1365-2958.2001.02673.x. [DOI] [PubMed] [Google Scholar]
- Cescau S, Cwerman H, Letoffe S, Delepelaire P, Wandersman C, Biville F. Heme acquisition by hemophores. Biometals. 2007;20:603–613. doi: 10.1007/s10534-006-9050-y. [DOI] [PubMed] [Google Scholar]
- Chang C, Mooser A, Pluckthun A, Wlodawer A. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J Biol Chem. 2001;276:27535–27540. doi: 10.1074/jbc.M102778200. [DOI] [PubMed] [Google Scholar]
- Devanathan S, Postle K. Studies on colicin B translocation: FepA is gated by TonB. Mol Microbiol. 2007;65:441–453. doi: 10.1111/j.1365-2958.2007.05808.x. [DOI] [PubMed] [Google Scholar]
- Eick-Helmerich K, Braun V. Import of biopolymers into Escherichia coli: nucleotide sequences of the exbB and exbD genes are homologous to those of the tolQ and tolR genes, respectively. J Bacteriol. 1989;171:5117–5126. doi: 10.1128/jb.171.9.5117-5126.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E, Günter K, Braun V. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J Bacteriol. 1989;171:5127–5134. doi: 10.1128/jb.171.9.5127-5134.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Herrero A, Peacock RS, Howard SP, Vogel HJ. The solution structure of the periplasmic domain of the TonB system ExbD protein reveals an unexpected structural homology with siderophore-binding proteins. Mol Microbiol. 2007;66:872–889. doi: 10.1111/j.1365-2958.2007.05957.x. [DOI] [PubMed] [Google Scholar]
- Germon P, Clavel T, Vianney A, Portalier R, Lazzaroni JC. Mutational analysis of the Escherichia coli K-12 TolA N-terminal region and characterization of its TolQ-interacting domain by genetic suppression. J Bacteriol. 1998;180:6433–6439. doi: 10.1128/jb.180.24.6433-6439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh J, Postle K. Evidence for dynamic clustering of carboxy-terminal aromatic amino acids in TonB-dependent energy transduction. Mol Microbiol. 2004;51:203–213. doi: 10.1046/j.1365-2958.2003.03816.x. [DOI] [PubMed] [Google Scholar]
- Ghosh J, Postle K. Disulphide trapping of an in vivo energy-dependent conformation of Escherichia coli TonB protein. Mol Microbiol. 2005;55:276–288. doi: 10.1111/j.1365-2958.2004.04384.x. [DOI] [PubMed] [Google Scholar]
- Goemaere EL, Devert A, Lloubes R, Cascales E. Movements of the TolR C-terminal domain depend on TolQR ionizable key residues and regulate activity of the Tol complex. J Biol Chem. 2007;282:17749–17757. doi: 10.1074/jbc.M701002200. [DOI] [PubMed] [Google Scholar]
- Guihard G, Boulanger P, Benedetti H, Lloubes R, Besnard M, Letellier L. Colicin A and the Tol proteins involved in its translocation are preferentially located in the contact sites between the inner and outer membranes of Escherichia coli cells. J Biol Chem. 1994;269:5874–5880. [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose P-BAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannavy K, Barr GC, Dorman CJ, Adamson J, Mazengera LR, Gallagher MP, Evans JS, Levine BA, Trayer IP, Higgins CF. TonB protein of Salmonella typhimurium. A model for signal transduction between membranes. J Mol Biol. 1990;216:897–910. doi: 10.1016/S0022-2836(99)80009-6. [DOI] [PubMed] [Google Scholar]
- Held KG, Postle K. ExbB and ExbD do not function independently in TonB-dependent energy transduction. J Bacteriol. 2002;184:5170–5173. doi: 10.1128/JB.184.18.5170-5173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs PI, Myers PS, Postle K. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J Bacteriol. 1998;180:6031–6038. doi: 10.1128/jb.180.22.6031-6038.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs PI, Larsen RA, Postle K. Quantitation of known components of the Escherichia coli TonB-dependent energy transduction system: TonB, ExbB, ExbD, and FepA. Mol Microbiol. 2002a;44:271–281. doi: 10.1046/j.1365-2958.2002.02880.x. [DOI] [PubMed] [Google Scholar]
- Higgs PI, Letain TE, Merriam KK, Burke NS, Park H, Kang C, Postle K. TonB interacts with nonreceptor proteins in the outer membrane of Escherichia coli. J Bacteriol. 2002b;184:1640–1648. doi: 10.1128/JB.184.6.1640-1648.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CW, Harnish BW. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:7069–7072. doi: 10.1073/pnas.78.11.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskula JC, Letain TE, Roof SK, Skare JT, Postle K. Role of the TonB amino terminus in energy transduction between membranes. J Bacteriol. 1994;176:2326–2338. doi: 10.1128/jb.176.8.2326-2338.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet L, Rigal A, Lazdunski C, Benedetti H. Role of TolR N-terminal, central, and C-terminal domains in dimerization and interaction with TolA and TolQ. J Bacteriol. 1999;181:4476–4484. doi: 10.1128/jb.181.15.4476-4484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampfenkel K, Braun V. Membrane topology of the Escherichia coli ExbD protein. J Bacteriol. 1992;174:5485–5487. doi: 10.1128/jb.174.16.5485-5487.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampfenkel K, Braun V. Topology of the ExbB protein in the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1993;268:6050–6057. [PubMed] [Google Scholar]
- Karlsson M, Hannavy K, Higgins CF. A sequence-specific function for the N-terminal signal-like sequence of the TonB protein. Mol Microbiol. 1993;8:379–388. doi: 10.1111/j.1365-2958.1993.tb01581.x. [DOI] [PubMed] [Google Scholar]
- Kodding J, Killig F, Polzer P, Howard SP, Diederichs K, Welte W. Crystal structure of a 92-residue C-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments. J Biol Chem. 2005;280:3022–3028. doi: 10.1074/jbc.M411155200. [DOI] [PubMed] [Google Scholar]
- Kojima S, Blair DF. Conformational change in the stator of the bacterial flagellar motor. Biochemistry. 2001;40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- Kojima S, Blair DF. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry. 2004;43:26–34. doi: 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- Larsen RA, Wood GE, Postle K. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol Microbiol. 1993;10:943–953. doi: 10.1111/j.1365-2958.1993.tb00966.x. [DOI] [PubMed] [Google Scholar]
- Larsen RA, Thomas MT, Wood GE, Postle K. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (ΔV17) by a missense mutation in ExbB. Mol Microbiol. 1994;13:627–640. doi: 10.1111/j.1365-2958.1994.tb00457.x. [DOI] [PubMed] [Google Scholar]
- Larsen RA, Myers PS, Skare JT, Seachord CL, Darveau RP, Postle K. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J Bacteriol. 1996;178:1363–1373. doi: 10.1128/jb.178.5.1363-1373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen RA, Thomas MG, Postle K. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol Microbiol. 1999;31:1809–1824. doi: 10.1046/j.1365-2958.1999.01317.x. [DOI] [PubMed] [Google Scholar]
- Larsen RA, Postle K. Conserved residues Ser(16) and His(20) and their relative positioning are essential for TonB activity, cross-linking of TonB with ExbB, and the ability of TonB to respond to proton motive force. J Biol Chem. 2001;276:8111–8117. doi: 10.1074/jbc.M007479200. [DOI] [PubMed] [Google Scholar]
- Larsen RA, Chen GJ, Postle K. Performance of standard phenotypic assays for TonB activity, as evaluated by varying the level of functional, wild-type TonB. J Bacteriol. 2003;185:4699–4706. doi: 10.1128/JB.185.16.4699-4706.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen RA, Deckert GE, Kastead KA, Devanathan S, Keller KL, Postle K. His20 provides the sole functionally significant side chain in the essential TonB transmembrane domain. J Bacteriology. 2007;189:2825–2833. doi: 10.1128/JB.01925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- Lee SK, Keasling JD. A propionate-inducible expression system for enteric bacteria. Appl Environ Microbiol. 2005;71:6856–6862. doi: 10.1128/AEM.71.11.6856-6862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letain TE, Postle K. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Gram-negative bacteria. Mol Microbiol. 1997;24:271–283. doi: 10.1046/j.1365-2958.1997.3331703.x. [DOI] [PubMed] [Google Scholar]
- Matthews EE, Zoonens M, Engelman DM. Dynamic helix interactions in transmembrane signaling. Cell. 2006;127:447–450. doi: 10.1016/j.cell.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Means GE, Feeney RE. Chemical modification of proteins. San Francisco, Calif.: Holden-Day Inc.; 1971. [Google Scholar]
- Metz B, Kersten GF, Hoogerhout P, Brugghe HF, Timmermans HA, de Jong A, Meiring H, ten Hove J, Hennink WE, Crommelin DJ, Jiskoot W. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J Biol Chem. 2004;279:6235–6243. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, N. Y.: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock SR, Weljie AM, Peter Howard S, Price FD, Vogel HJ. The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J Mol Biol. 2005;345:1185–1197. doi: 10.1016/j.jmb.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Postle K, Reznikoff WS. Identification of the Escherichia coli tonB gene product in minicells containing tonB hybrid plasmids. J Mol Biol. 1979;131:619–636. doi: 10.1016/0022-2836(79)90011-1. [DOI] [PubMed] [Google Scholar]
- Postle K, Skare JT. Escherichia coli TonB protein is exported from the cytoplasm without proteolytic cleavage of its amino terminus. J Biol Chem. 1988;263:11000–11007. [PubMed] [Google Scholar]
- Postle K, Kadner RJ. Touch and go: tying TonB to transport. Mol Microbiol. 2003;49:869–882. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- Postle K. TonB system, in vivo assays and characterization. Methods in Enzymology. 2007;422:245–269. doi: 10.1016/S0076-6879(06)22012-3. [DOI] [PubMed] [Google Scholar]
- Postle K, Larsen RA. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals. 2007;20:453–465. doi: 10.1007/s10534-006-9071-6. [DOI] [PubMed] [Google Scholar]
- Roof SK, Allard JD, Bertrand KP, Postle K. Analysis of Escherichia coli TonB membrane topology by use of PhoA fusions. J Bacteriol. 1991;173:5554–5557. doi: 10.1128/jb.173.17.5554-5557.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ WP, Engelman DM. TOXCAT: a measure of transmembrane helix association in a biological membrane. Proc Natl Acad Sci U S A. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Homma M. Multimeric structure of PomA, a component of the Na+-driven polar flagellar motor of vibrio alginolyticus. J Biol Chem. 2000;275:20223–20228. doi: 10.1074/jbc.M002236200. [DOI] [PubMed] [Google Scholar]
- Sauter A, Howard SP, Braun V. In vivo evidence for TonB dimerization. J Bacteriol. 2003;185:5747–5754. doi: 10.1128/JB.185.19.5747-5754.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer K, Gouget B, Carriere M, Labigne A, de Reuse H. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol Microbiol. 2007;63:1054–1068. doi: 10.1111/j.1365-2958.2006.05578.x. [DOI] [PubMed] [Google Scholar]
- Seliger SS, Mey AR, Valle AM, Payne SM. The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol Microbiol. 2001;39:801–812. doi: 10.1046/j.1365-2958.2001.02273.x. [DOI] [PubMed] [Google Scholar]
- Skare JT, Roof SK, Postle K. A mutation in the amino terminus of a hybrid TrpC-TonB protein relieves overproduction lethality and results in cytoplasmic accumulation. J Bacteriol. 1989;171:4442–4447. doi: 10.1128/jb.171.8.4442-4447.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Webster RE. Nucleotide sequence of a gene cluster involved in entry of E colicins and single-stranded DNA of infecting filamentous bacteriophages into Escherichia coli. J Bacteriol. 1987;169:2667–2674. doi: 10.1128/jb.169.6.2667-2674.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toews J, Rogalski JC, Clark TJ, Kast J. Mass spectrometric identification of formaldehyde-induced peptide modifications under in vivo protein cross-linking conditions. Anal Chim Acta. 2008;618:168–183. doi: 10.1016/j.aca.2008.04.049. [DOI] [PubMed] [Google Scholar]
- Tralau T, Vuilleumier S, Thibault C, Campbell BJ, Hart CA, Kertesz MA. Transcriptomic analysis of the sulfate starvation response of Pseudomonas aeruginosa. J Bacteriol. 2007;189:6743–6750. doi: 10.1128/JB.00889-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Goemaere EL, Thome R, Gavioli M, Cascales E, Lloubes R. Mapping the Interactions between Escherichia coli Tol Subunits: Rotation of the TolR Transmembrane Helix. J Biol Chem. 2009;284:4275–4282. doi: 10.1074/jbc.M805257200. [DOI] [PubMed] [Google Scholar]
- Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol. 1998;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]


