Abstract
In many naturally occurring habitats, bacteria live in micrometer-size confined spaces. Although bacterial growth and motility in such constrictions is of great interest to fields as varied as soil microbiology, water purification, and biomedical research, quantitative studies of the effects of confinement on bacteria have been limited. Here, we establish how Gram-negative Escherichia coli and Gram-positive Bacillus subtilis bacteria can grow, move, and penetrate very narrow constrictions with a size comparable to or even smaller than their diameter. We show that peritrichously flagellated E. coli and B. subtilis are still motile in microfabricated channels where the width of the channel exceeds their diameters only marginally (∼30%). For smaller widths, the motility vanishes but bacteria can still pass through these channels by growth and division. We observe E. coli, but not B. subtilis, to penetrate channels with a width that is smaller than their diameter by a factor of approximately 2. Within these channels, bacteria are considerably squeezed but they still grow and divide. After exiting the channels, E. coli bacteria obtain a variety of anomalous cell shapes. Our results reveal that sub-micron size pores and cavities are unexpectedly prolific bacterial habitats where bacteria exhibit morphological adaptations.
Keywords: biophysics, confinement, microbiology, microfluidics
Bacterial growth and movement in confined spaces is ubiquitous in nature and plays an important role in diverse fields ranging from soil microbiology, water purification, to microbial pathogenesis. The majority of bacteria in soil and bedrock live in pores of size 6 micrometer and smaller (1). These bacteria constitute a large portion of the Earth's biomass (2) and are essential for the functioning of soil. Although distributions of bacteria in soil and Earth's subsurfaces have been studied, it is largely unknown how bacteria grow, move, and penetrate pores of very small size. The latter is also an important question for water treatment and purification. Whereas microbiology textbooks consider output from 0.2-μm pore size filters sterile, it has recently been found that numerous bacteria can pass through these membranes and grow thereafter (3, 4). It is unclear what mechanism bacteria employ to penetrate such membranes. Also, in microbial pathogenesis, bacterial growth and penetration is a problem, for example in dental implants (5), but likely also in soft tissues and bones of a host organism where confined spaces are relevant to bacterial propagation through the extracellular matrix.
Some experimental (6–10) and theoretical (11, 12) studies have discussed the effects of restricting geometry on bacterial motility. It has been established that Escherichia coli bacteria can swim in 2.0 μm and wider channels without appreciable slowdown (8) and that bacteria regularly swim in close proximity to surfaces (9), preferring some types of surfaces to others (10). It has been shown that these behaviors can be used to guide bacterial movement in microfluidic structures (10, 13). The effects of confinement on bacterial growth have received very limited attention. Growth of E. coli bacteria has been studied by Takeuchi et al. in microfabricated structures (14). In these experiments, confinement affected bacterial growth in the direction of its elongation. The experiments showed that the filamentous bacteria can bend during growth and conform to the shapes of the microfabricated structures. All previous research on bacterial motility and growth have been carried out in relatively large channels and constrictions where the critical dimension is larger than one micrometer, that is, larger than bacterial diameter. So far, no one has addressed questions such as: How narrow channels can bacteria penetrate using their own motility? How do bacterial movement and growth change in very narrow channels compared with that in unbound medium?
Here, we establish how E. coli and Bacillus subtilis bacteria can grow and move in very narrow constrictions with sub-μm width, and we determine the lower limits for the constriction size which these bacteria are able to penetrate. We show that E. coli and B. subtilis retain their motility in microfabricated channels with a width that exceeds their diameters by only approximately 30%. We also show that bacteria can penetrate even narrower channels. To achieve this, bacteria initiate growth into the channels. In this process, elongation and division pushes bacteria forward until they fill the whole channel. Whereas B. subtilis bacteria can grow in such a way in channels as narrow as their diameter, E. coli bacteria are even able to penetrate channels with a width that is much smaller than their diameter. Our work demonstrates that growth in channels which are narrower than the bacterial diameter can drastically change the shape of E. coli and lead to a morphological phenotype which has not been described previously.
Results
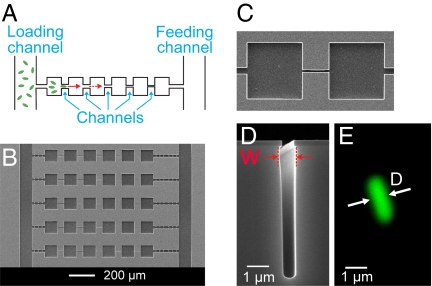
To carry out this study, we design and fabricate microfluidic channels (constrictions) which connect chambers (small bacterial wells) on a silicon chip and image individual GFP-labeled bacteria in these structures using fluorescent time-lapse microscopy. The advantages of using silicon chips for these experiments are the possibility to define sub-micrometer size channels and the capability to carry out long-term measurements with these bacteria. Whereas the bacteria can pass wide channels within seconds, it can take several days before the bacteria are able to cross the narrowest channels. The microfluidic chips that we use make it possible to maintain the necessary conditions for bacterial growth and motility for such periods of time. The schematic of the microfluidic chip used in this experiment is presented in Fig. 1A. The motile bacteria enter the structure of chambers and channels from the left vertical channel. The observations start when the bacteria reach to the left-most chamber of the structure and start to move through the channels toward the chambers on the right end of the arrays. Bacterial movement in channels is partially driven by chemotaxis toward the nutrient source which consists of growth media in the ‘feeding channel’ on the right end of the arrays. From left to right, the channels are fabricated progressively narrower. This allows monitoring how the population of bacteria is able to negotiate increasingly narrower channels and adapt to life in such a confined environment. On the same silicon chip, a large number of array structures are made, each of which consists of series of chambers and channels (Fig. 1B). Most channels that we fabricate and report here are straight channels of 50 μm length connecting two near-by chambers (Fig. 1C). These narrow and deep channels in silicon are made using electron-beam lithography and dry etching (see Materials and Methods). The typical depth of the channels is between 5–7 μm. The widths of the channels (W) reported here vary from 5 μm to ≈0.3 μm (Fig. 1D). Besides these channels, we have also fabricated channels of 0.3-μm width and narrower using a different fabrication process which will be discussed later. The widths of the channels on our chips are comparable to or even smaller than the diameters (D) of the E. coli and B. subtilis bacteria used in these experiments (Fig. 1E). The typical diameters of these two bacteria range from D = 0.7—1.1 μm, with a mean value of 0.76 μm for E. coli and of 0.86 μm for B. subtilis.
Fig. 1.
Setup for studying bacterial movement in small constrictions. (A) Schematic of the experiment. Using time-lapse fluorescent microscopy, bacterial movement from left to right is observed in array structures consisting of multiple channels and chambers in series. (B) SEM image of a section of a microfluidic chip where five of these array structures can be seen. (C) Top view of a 3-μm wide and 50-μm long channel connecting two chambers. (D) SEM side view image of the cross-section of a 0.8-μm wide channel. The width of the channel (W) is measured at the widest cross-section. For small W, bacteria are confined to the slightly wider location near the top. (E) Fluorescent image of a typical E. coli bacterium shown at the same scale as the 0.8-μm wide channel on panel (D). The intensity profile across its cross-section is used to determine the bacterial diameter D, see SI Text.
Bacterial Motility in Channels.
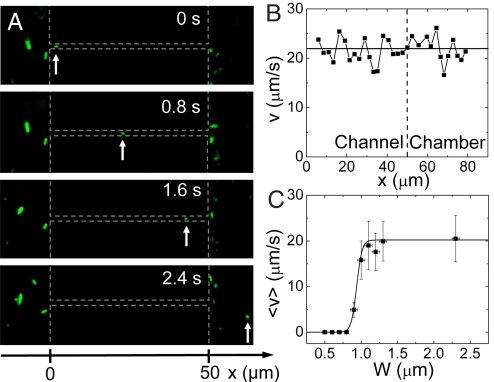
In this section, we study the motility of E. coli in channels of ≈2-μm width and narrower, and determine how the swimming motion of these bacteria comes to a halt as the channel width decreases. An example of the bacterial motility in a 1.2 μm wide channel is shown in Fig. 2A (see also Movie S1). Qualitatively, bacteria in these channels show the typical bulk motility pattern, where ‘tumbling’ events interrupt periods of straight ‘runs’ (15). Because the bacterial movement is essentially one-dimensional in our case because of shallowness of the channel and its cross-sectional profile, tumbling leads to only two possible outcomes: reversing of the direction or continuation of the motion in the same direction. From the time-lapse movies, the speed of the bacterium can be determined in the channel as well as in the chamber area. In the example shown on Fig. 2B, it can be seen that the bacterium swims at the same average speed within the 1.2 μm wide channel and in the chamber. This is generally true in wide channels (>1.1 μm). We have verified that forward and backward moving bacteria have on average the same speed and that bacteria which reverse their direction of motion in channels, display the same speed after reversal. We measure an average swimming speed of 20 ± 5 μm/s in chambers and wide channels, which is close to the values cited for this E. coli strain (16, 17). As Fig. 2C shows, the swimming speed is essentially unhindered down to a channel width of 1.1 μm. This is remarkable because 1.1 μm is only slightly larger than the diameter of E. coli (∼0.8 μm). For channels narrower than 1.1 μm, the average speed of bacterial swimming starts to decrease, and the movement completely stalls for channels with a width of 0.8 μm and narrower. Along with the decreasing bacterial swimming speed, also the frequency and the duration of tumbling events increase in these increasingly narrow channels.
Fig. 2.
Bacterial motility in channels. (A) Time-lapse images of an E. coli bacterium that swims through a 1.2-μm wide channel (three topmost images) and into the chamber area (Bottom image). The arrow points at the swimming bacterium. The vertical dashed lines mark the boundaries of the two chambers and the horizontal lines indicate the location of the channel. (B) Velocity of the bacterium v as a function of its coordinate along a 1.2-μm wide channel (left from the vertical dashed line) and in the chamber area (right from the vertical dashed line). The x-coordinate is measured along the channel with x = 0 at the channel entrance from the left chamber. (C) Average velocity <v> versus the width of the channel. The time-averaged velocity is first calculated from traces such as shown in panel B, excluding tumbling events which last >0.2 s. Subsequently, averaging over the population of bacteria in the same-size channel is carried out to yield <v>. The error bars correspond to the standard deviation of velocities among the population of bacteria in a given-size channel. Solid line represents a sigmoidial fit to the data, with a midpoint W = 0.95 μm.
Bacterial Growth in Narrow Channels.
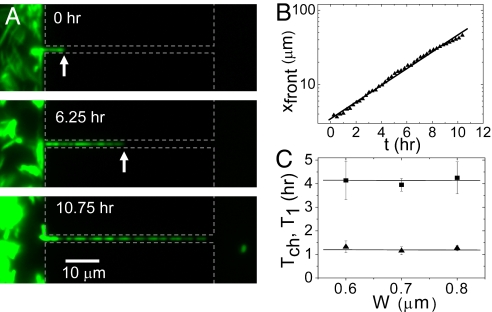
In channels with a width <0.8 μm, E. coli lose their ability to swim. Unexpectedly, however, they are still able to penetrate such channels. In these narrow constrictions, bacterial dispersal is not driven by motility but by growth through the channels (Fig. 3A and Movie S2). We have observed such penetration by growth through channels as narrow as 0.4 μm. Typically, a single ancestor bacterium lodges itself near the entrance of the channel or is pressed to the channel entrance by other bacteria. This ancestor cell gives rise to a population of daughter cells that push themselves through the channel by growth and division. This process moves approximately half of the bacteria forward along the channel and the other half backwards toward the chamber from where they originated. As Fig. 3B demonstrates, the bacterial populations in these channels undergo an exponential growth: the front of bacterial filament extends approximately as xfront(t) ≈ 2t/Tch (dashed line in Fig. 3B), where Tch is average doubling time of the bacteria in the chain.
Fig. 3.
Bacterial growth through narrow channels. (A) Time-lapse fluorescent images of bacterial growth in a 0.6-μm wide channel. Dashed gray lines show the approximate boundaries of the chambers and channels. The arrows point to the position of the bacterial front. (B) Position of the bacterial front vs. time for the growth process on panel A. The line presents a fit of the function xfront(t) = L0 + L1 2t/Tch. (C) Doubling time of the chain length Tch (squares) and division time of the first bacterium in the chain T1 (triangles) vs. channel width. Each Tch and T1 value in the Inset of B represents an average over several populations in channels of the given width on the same chip. Error bars represent standard deviations among different populations.
It is noteworthy that the measured doubling times Tch do not show a slowdown when the channels get narrower, i.e., when larger stresses to the bacteria should occur. The Inset of Fig. 3B shows that the Tch values stay constant for bacterial populations in channels with a width from 0.6 to 0.8 μm. Although 0.2 μm variation in channel width may seem small, it does represent a significant fraction (25%) of the bacterial diameter. The constancy of the measured doubling times is further verified by plotting the doubling times of the first bacterium at the leading position in the chain (T1). These doubling times are determined directly from time-lapse movies as time difference between consecutive division events of the bacterium. As shown in the Inset of Fig. 3B (triangles), this time is factor of 3.5 shorter and independent of the channel width as well. The average doubling time of the first bacterium (T1 = 73 ± 10 min) compares well to the division time of bacteria in the chambers (70 ± 26 min) at low cell density.
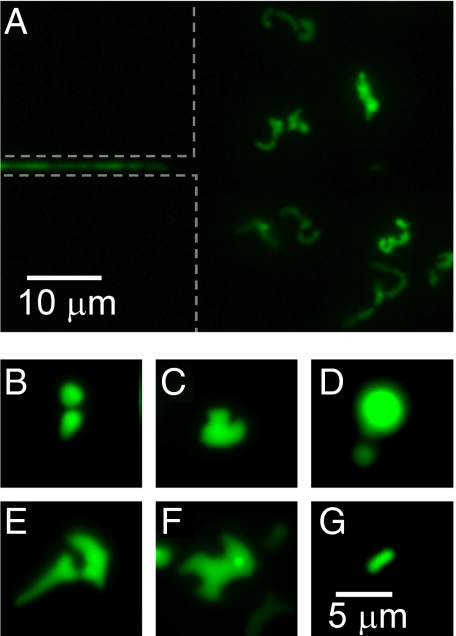
After bacteria exit from the narrow channels (W ≤ 0.8 μm) to the chambers, they further proliferate there. These bacteria, which usually can be traced back to a single ancestor in the narrow channel, show a wide variety of shapes and sizes that can differ substantially from the regular rod-shape of E. coli. As an example, Fig. 4A shows an image of the chamber, 5 hours after the first bacterium has reached it through a 0.6 μm constriction. This image follows the sequence shown in Fig. 3A. Most bacteria in this image show aberrant shapes and cross-sectional sizes that considerably exceed the size of regular E. coli. The lateral dimensions of aberrant bacteria (Fig. 4 B and C) exceed the diameters of regular E. coli (Fig. 4G) by factor of two or more. In some extreme cases, the bacteria are completely round shaped (Fig. 4D), reminiscent to the well-studied L-form morphology (18–20). The aberrant bacteria which we observe frequently show bulges and protuberances on their sidewalls as well as bent cell shapes (Fig. 4 E and F). No bacteria with such widths and shape irregularities are observed in chambers which are connected to the inlet through wide channels where motility is possible. Although treatment with cell-wall acting antibiotics has been used in the past to produce the L-form morphology (18, 19), these shapes appear in our experiments where none of such antibiotics were present. The appearance of aberrantly shaped bacteria correlates with the width of the channel: In narrower channels, the ratio of occurrence of aberrant versus regular bacteria increases. Based on these observations, we associate the aberrant phenotype with the passage through narrow channels.
Fig. 4.
Aberrant bacteria exit narrow channels. (A) Fluorescence image of 0.6-μm wide channel and a chamber 5 h after the first bacterium appeared in this chamber. Variety of aberrantly shaped bacteria can be seen to populate the chamber. The image is sequence to the series shown in Fig. 4A. (B–F) Different aberrant bacterial shapes at higher magnification. (G) For comparison, fluorescence image of a regularly shaped and sized bacterium which has emerged from the same channel as the bacteria shown in panels E and F. The same scale bar applies for all of the panels from B to G.
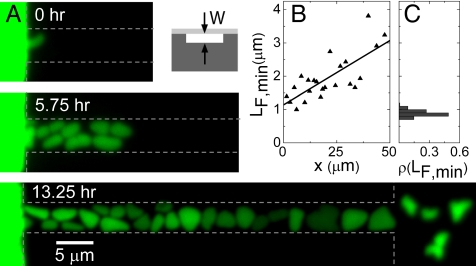
To follow the formation of aberrantly shaped bacteria in real time, we fabricate channels with a different geometry (Fig. 5A Inset on the right and Fig. S1 and Fig. S2). Whereas previously we discussed bacterial growth in channels which were etched deep and narrow into silicon (vertical channels), the channels which are used here are etched shallow into silicon nitride (horizontal channels) (for details of fabrication see Materials and Methods). In this case, the depth of the trench in silicon nitride determines the effective width of the channel. Such geometry allows a detailed imaging of the bacterial shapes during the growth. Fig. 5A shows an example of the development of bacteria in a 0.3-μm wide horizontal channel. The bacteria are pressed into the channel by the neighboring bacteria in a densely packed chamber (Fig. S3). Upon entering the channel, the bacteria are strongly squeezed by the channel walls and they flatten. Despite the significant flattening, these bacteria are still able to grow and divide (Movie S3 and Movie S4). During growth in the channel, the bacteria widen further, becoming almost round shaped near the exit from the 50-μm long channel. The broadening of bacteria is quantitatively analyzed in Fig. 5B where the minimum Feret's diameter LF,min is plotted as a function of the bacterial position from the channel entrance. Here, LF,min is defined as the minimum distance between parallel tangents at opposing borders of the bacterium. The borders of the object are defined by zero crossings of the second derivative in the direction of intensity gradient. For comparison, Fig. 5C shows the distribution of LF,min in batch cultures. Note that LF,min overestimates the actual diameter of the rod-shaped bacterium by ≈17%. Fig. 5B shows that bacteria considerably widen as they move within the channel. At the exit of the 50-μm long channel they have become wider by about factor of 3 than their width at the entrance. The stress imposed by the channel walls thus not only deforms the bacteria, but also promotes bacterial growth in the sideways direction. Further changes in bacterial shape occur when bacteria exit the channels. Here, bacteria contract and frequently obtain rugged shapes with bulges and protuberances as described above (Movie S5).
Fig. 5.
Morphogenesis of E. coli in shallow horizontal channels. (A) Time lapse fluorescence images of bacteria in channel of 0.3 μm width (and other dimensions 5 and 50 μm). Dashed lines show approximate boundaries of chambers and channels. Inset: schematic side-view cross-section of such channel. Darker gray corresponds to silicon and lighter gray to PDMS. (B) Minimum Feret's diameter vs. bacterial position in the channel (right vertical axes). The position is measured from the channel entrance. The data points correspond to the bacteria on the Bottom image of panel A. (C) For comparison, distribution of minimum Feret's diameters in batch culture. Batch culture has grown to optical density OD600 = 2.3. The vertical axes of this plot is the same as in B.
Essentially all of the aberrant bacteria that exit narrow channels (W < 0.8 μm) are nonmotile. Most of these bacteria are able to grow and divide. Exceptions are the round-shaped bacteria which we do not observe to divide. After 1–2 days, a population with regular shape and size recovers from the aberrantly shaped bacteria that seeded the chamber in the beginning (Fig. S4 and Movie S5). These bacteria have also restored their motility. Interestingly, on the chip, where several chambers and narrow channels are connected in row, repeated transitions from aberrant to regular cells thus take place multiple times as the bacterial population advances from one chamber to the next.
Comparison of the Channel Widths to the Bacterial Diameters.
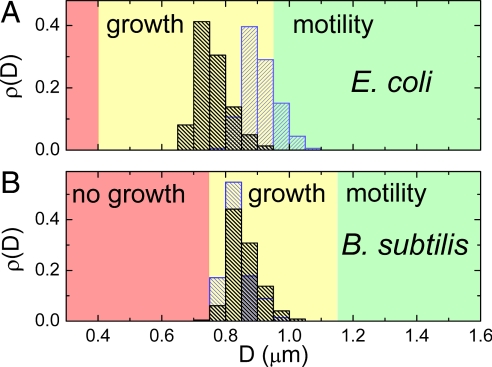
Next we make a detailed comparison of the diameter of our E. coli strain to the width of the channels that these bacteria penetrate by either motility or growth. We measure the diameter distributions of bacteria in batch cultures using fluorescence images. These measurements are summarized in Fig. 6A. As Fig. 4A shows, the diameters of E. coli in stationary-phase cultures (black, D̄ = 0.76 ± 0.05 μm) are ≈18% smaller than in mid-log phase (blue, D̄ = 0.91 ± 0.05 μm), in agreement with previous observations (21–23). Because the bacterial density in the chambers is comparable with the density of stationary culture, we consider the average diameter D̄ = 0.76 μm to be a good estimate for the typical bacterial diameter in our experiments. The ratio of the channel width to the bacterial diameter, W/D̄, is a relevant parameter for the hydrodynamics of bacterial swimming as well as for comparisons to measurements performed with other organisms. We find the channel width that is characteristic to the transition between motility and no motility to be W = 0.95 ± 0.05 μm (Fig. 2C), which results in W/D̄ = 1.25 ± 0.07. However, it is likely that the bacteria that are observed moving in channels where W/D̄ ≈ 1.25, are those whose diameters are smaller than the mean diameter of the population D̄. The smallest bacterial diameter that we measure in a population is 12% lower than the mean diameter. If we assume that the bacteria with this smallest diameter are responsible for the observed motility then W/Dsmallest = 1.4. We thus conclude that flagellar motility is still possible in channels that are only 25–40% wider than the cell diameter.
Fig. 6.
Comparison of channel widths to bacterial diameters. (A) Distribution of bacterial diameters in log (OD600 = 0.6, blue) and in stationary phase (OD600 = 2.3, black) for E. coli as determined from fluorescence intensity profiles of individual bacteria from batch cultures. Details of the size determination are given in SI Text. The average diameters are D̄ = 0.91 ± 0.05 μm in log and D̄ = 0.76 ± 0.05 μm in stationary phase. The range of channel widths for which bacteria are motile or grow are indicated by the green or yellow colored background, respectively. The range of channel widths where no penetration is observed is shown in red. (B) The same for B. subtilis strain for the log (OD600 = 0.4, blue) and for stationary phase distributions (OD600 = 2.0, black) where the average diameters are D̄ = 0.84 ± 0.04 μm and D̄ = 0.86 ± 0.04 μm, respectively.
The narrowest vertical channel which we observe E. coli to penetrate by growth and division is 0.4-μm wide. Fig. 5 shows that E. coli can penetrate horizontal channels of 0.3-μm width and we have observed E. coli to enter and grow in even narrower horizontal channels. In these channels, however, we expect that bacteria are able to deform to some extent the ceiling of the channel which is made of elastic PDMS. Because of such a deformation, the actual width of the channel may exceed its defined value of 0.3 μm once the bacteria enter it. Even for an observation period of 1 week, we have not observed bacteria to enter and grow in 0.3-μm wide vertical channels, which are much less deformable because of their rigid silicon walls. The limiting width of the channel is independent on nutrient conditions in which we carried out the experiments. In separate experiments we varied the nutrient concentration by allowing or stopping flow of growth medium in the “feeding channel” (see Fig. 1 A and B), and we obtained the same results. Based on these observations, we consider 0.4 μm the limit to the channel width which E. coli can penetrate by growth and division. If we use the average diameter D̄ = 0.76 μm, we obtain a ratio W/D̄ ≈ 0.5. Thus, E. coli can penetrate constrictions with a size twice narrower than their typical sizes in stationary-phase batch cultures.
We also investigate growth and motility of B. subtilis in the same on-chip setting as for E. coli. The diameters of the B. subtilis strain used in this study (24) (D̄ = 0.86 ± 0.04 μm, see Fig. 4B) are comparable to those of E. coli. Unlike E. coli, the diameter of B. subtilis is essentially constant during the various growth phases, indicating that B. subtilis exhibits less morphological plasticity than E. coli. Swimming of B. subtilis is similar to the swimming of E. coli in wider constrictions. The transition from swimming motility to no motility takes place at W = 1.15 ± 0.05 μm, yielding a ratio W/D̄ = 1.3 ± 0.06 and W/Dsmallest = 1.5 (the diameter of the narrowest bacterium was 13% smaller than the mean), comparable to the values found for E. coli. In the range of channel widths from 1.05 to 0.75 μm, B. subtilis is able to penetrate the channels through growth, similar to what was observed for E. coli (Fig. S5 and Movie S6). However, we do not see B. subtilis growing through channels narrower than 0.75 μm even after a week-long observation. This yields a lower limit for growth of W/D̄ = 0.9. The ratio for the smallest measured bacterium in the distribution is W/Dsmallest = 1.0. In contrast to E. coli, B. subtilis thus are not able to penetrate channels which are smaller than their width. This statement holds only for rigid vertical channels and not for the horizontal channels of Fig. 5 that are somewhat deformable. Thus, whereas the ability of B. subtilis to swim through constrictions is comparable with that of E. coli, its ability to grow through narrow constrictions is much more limited. Although B. subtilis shows tendency to grow filamentous in channels and right after exiting it, we do not observe aberrant morphologies similar to E. coli in B. subtilis in narrow channels and after exiting the channels. Because E. coli and B. subtilis are typical representatives of Gram-negative and Gram-positive bacteria, it is likely that the ability to squeeze through small constrictions is different in general for these two classes of bacteria.
Discussion
Limits of Bacterial Motility in Channels.
These observations trigger questions about what sets the minimum size for bacterial propagation through constrictions. How much free space exists between the cell wall and the channel wall in the limiting case when bacterial swimming stalls? In narrowest channels where we observed motility, the two walls of the channel are each ≈100–150 nm away from the cell if bacterium swims in the middle of the channel. This estimation is in line with the observation that E. coli can swim in a very close proximity (∼40 nm) to a planar surface (9). Our data show that E. coli and B. subtilis can swim not only at close proximity to a single surface but also to two parallel surfaces in the channel.
What sets the lower limit for the channel width where these peritrichously flagellated bacteria can still swim? We discuss hydrodynamic drag, constraints to movement of flagella, and adhesion forces as three possible physical constraints which limit bacterial movement in the channel: (i) Hydrodynamic drag. Whereas hydrodynamics of bacterial swimming near surfaces presents a complicated mathematical problem (11, 12) Stokes' law straightforwardly shows that hydrodynamic drag does not lead to a complete stalling of bacteria in channels. Although hydrodynamic drag may slow down bacterial swimming, it will not stop the bacteria swimming through narrow channels. (ii) Constraints to the movement of bacterial flagella. Whereas flagellar filaments of bacteria are long (10–20 μm), they form a compact bundle which has helical diameter of ≈0.5 μm (25). This is smaller than the width of the channel where the bacterial swimming stalls (0.8 μm for E. coli). It thus seems unlikely that the flagellar filaments are severely hindered in channels where the bacterial motility stops. (iii) Adhesive and friction forces acting between the cell and the channel walls. Once the diameter of bacterium becomes comparable to the width of the channel, the bacterium will be in contact with the walls of the channel and experience adhesive and friction forces. These forces have been reported to be in the range from 1 pN to 10 nN (26). They easily exceed the force provided by flagellar motors of bacteria which is ≈0.5 pN (27). Adhesive and friction forces thus clearly set severe constraints to bacterial motility in narrow constrictions.
Growth in Narrow Channels.
The near equal division times of bacteria in the chambers and that of the first bacterium in the growing chain in narrow channels indicates that the channel width is not limiting bacterial growth. This observation holds in vertical as well as in horizontal channels. Thus, somewhat surprisingly, the lateral confinement does not limit the growth rate of E. coli even when considerable squeezing takes place. However, as Fig. 3C shows, the majority of bacteria in the channel have division times which are longer than the division time of the first bacterium in the chain. The slower division times of bacteria closer to the entrance of the channel can be explained by the gradient of habitat quality along the channel. Habitat quality, which combines the concentration of nutrients and the level of metabolic waste products to characterize the local habitat (28), is high near the first bacterium and low near the chamber from where the bacteria originated.
According to our observations, the Gram-positive B. subtilis possesses a far inferior ability to enter and grow in small pores than Gram-negative E. coli. These two types of bacteria have almost the same diameter and shape, but they have significantly different cell walls. Whereas the cell wall of B. subtilis is 30–40 nm thick (29), the cell wall of E. coli has been measured to be only approximately 3 nm (30). The much thicker cell wall of B. subtilis allows maintaining a high osmotic pressure of 20–50 atm (31) which significantly exceeds the pressure of 2–3 atm in E. coli cell (32). This pressure difference, and to a lesser extent the stiffer cell wall (33), makes B. subtilis significantly less compressible than E. coli. As a consequence, the ability of B. subtilis to enter rigid channels which are narrower than its diameter is very limited. Besides the difference in thickness, the cell walls of Gram-positive and -negative bacteria are also thought to grow and regenerate differently (21). These differences lead to different plastic properties of the cell, that is, ability to change in response to changing environmental conditions such as mechanical stress and nutrient availability. The dynamic and plastic nature of cell wall of E. coli have been observed in earlier experiments where filamentous E. coli cells bent by their guiding structures as they grew (14). The plastic and dynamic nature of the cell wall reveals itself also in ability of E. coli unlike B. subtilis cells to shrink and expand in response to nutrient availability (Fig. 6). Besides being more elastic, the larger plasticity thus also facilitates passage of E. coli through narrow constrictions compared with B. subtilis.
Although it is easier to squeeze E. coli through narrow constrictions, its thin cell wall is more vulnerable to damage. Experiments with cell-wall acting antibiotics and theoretical modeling of bacterial shapes (34) show that aberrant morphologies such as shown in Fig. 4 can result from defects in the cell wall. The cell-wall defects likely form in the channel where E. coli undergoes dramatic change in its morphology. Additional defects may form upon exiting the channel when a sudden change in stress can lead to the rupture of bonds in peptidoglycan. Besides aberrant growth of the cell wall, mechanical stress may disrupt the functioning of cytoskeletal proteins of E. coli in narrow channels. It is an interesting topic for further studies to understand how the mechanical stress affects the functioning of these proteins.
To summarize, our findings provide a microscopic description of how bacteria disperse in environments with small pores and constrictions with a size comparable to their diameters. We show that both E. coli and B. subtilis are motile in channels which only marginally exceed their diameters (30%). Both bacteria can penetrate even smaller channels by growth and division. Whereas entrance and growth of Gram-positive B. subtilis to rigid channels requires the width of channel to exceed its diameter, Gram-negative E. coli can flatten upon entering, and grow even in narrower channels. In these channels, E. coli bacteria undergo significant deformations and lose their regular shapes, but they still have growth rates that are not significantly different than in unbound medium. These findings help to understand how bacteria move in soil ecosystems, penetrate water filtering systems where small pores are present, and move in tissues and biofilms. The results of this work indicate that sub-micrometer-size pores and cavities can be much more prolific bacterial habitats than previously assumed. In these habitats bacteria are likely morphologically very diverse.
Materials and Methods
Microfluidic Chip Fabrication.
Microfluidic chips were fabricated using Si microfabrication techniques. The channels and chambers on the chip were defined by using e-beam lithography. Two types of channels with different geometry were fabricated. Following their geometry they were referred to as vertical or horizontal channels. The vertical channels were etched deep (5–7 μm) and narrow (from 2 to 0.3 μm) whereas horizontal channels were etched shallow (300 nm and less) but extended more in lateral direction (from 1 to 10 μm). To create vertical channels a cryo-etch process with SF6 and O2 gases at −120°C was used to etch silicon. For horizontal channels a standard reactive ion etching process with CHF3 and Ar gases was used to etch silicon nitride layer which was deposited on the top of the silicon wafer. In the chip with horizontal channels, the chambers and connecting flow channels were created using additional cryo-etching step which allowed making these structures deeper (1.7–1.8 μm). The channels and chambers were closed using PDMS (polydimethylsiloxane) covered glass coverslips of 150-μm thickness. The approximately 30-μm thick PDMS layer was treated with oxygen plasma just before bonding the glass slide to the silicon chip. Access holes to the microfluidic channels were created by a KOH wet etch through the silicon chip beforehand. The widths of the vertical channels were determined by cutting channels across and imaging the cross-sections of channels in a SEM. The widths of horizontal channels were determined by Tencor Alpha Step 500 profilometer.
Bacterial Strains.
E. coli used in the experiments were RP437 strain which was transformed with two different plasmids. The original was a high copy number plasmid expressing ampicillin resistance and GFP (35). The second plasmid used in these bacteria was derived from the original plasmid in our laboratory. In this plasmid, ampicillin-resistance gene was excised and a kanamycin resistance gene inserted into this location. The B. subtilis strain used in this study carries GFP fused to the abrB promoter (PabrB-gfp) stably integrated in the chromosome along with chloroamphenicol resistance (24). All bacteria were grown in standard Luria-Bertani media (1% wt peptone from casein, 0.5% wt yeast extract, and 1% wt NaCl) complemented with respective antibiotics for each strain of bacteria (100 μg/mL ampicillin, 50 μg/mL kanamycin, and 5 μg/mL chloramphenicol). The measurements with bacteria were done at 25–26°C.
Fluorescence Microscopy.
An Olympus IX81 inverted fluorescence microscope with a 100× NA 1.3 oil immersion and a 60× dry objective was used for imaging the bacteria. Fluorescence from GFP was excited by 100 W Hg lamp through a 0.25 or 0.5 neutral density filter and Chroma EN GFP filtercube. Hamamatsu 3CCD color camera was used for imaging at maximum frame rate of 10 Hz. A Mad City Labs MicroStage 20E stage was used for automated positioning over multiple locations on the chip. For image analysis, MatLab Image Analysis Toolbox and DipImage Toolbox were used. A detailed description of the measurements of the bacterial diameters using fluorescent images can be found from SI Text and Fig. S6–S10.
Supplementary Material
Acknowledgments.
We thank S. Donkers, S. Hage, and M. Zuiddam for technical assistance; J.-W. Veening for the B. subtilis strain; and B. Rieger, K.C. Huang, and C. Woldringh for valuable discussions. This work has been supported in part by research grants from Stichting voor Fundamenteel Onderzoek der Materie (FOM), de Nederlandse Organisatie voor Watenschappelijk Onderzoek (NWO), and Nanoned. J.E.K. acknowledges support from Delft University of Technology Start-up Fund.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907542106/DCSupplemental.
References
- 1.Ranjard L, Richaume AS. Quantitative and qualitative microscale distribution of bacteria in soil. Res Microbiol. 2001;152:707–716. doi: 10.1016/s0923-2508(01)01251-7. [DOI] [PubMed] [Google Scholar]
- 2.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hahn MW. Broad diversity of viable bacteria in ‘sterile’ (0.2 μm) filtered water. Res Microbiol. 2004;155:688–691. doi: 10.1016/j.resmic.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Hammes F, Boon N, Egli T. Quantification of the filterability of freshwater bacteria through 0.45, 0.22, and 0.1 μm pore size filters and shape-dependent enrichment of filterable bacterial communities. Environ Sci Technol. 2007;41:7080–7086. doi: 10.1021/es0707198. [DOI] [PubMed] [Google Scholar]
- 5.do Nascimento C, et al. Bacterial leakage along the implant-abutment interface of premachined or cast components. Int J Oral Maxillofac Surg. 2008;37:177–180. doi: 10.1016/j.ijom.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 6.Frymier PD, Ford RM. Analysis of bacterial swimming speed approaching a solid-liquid interface. AICHE J. 1997;43:1341–1347. [Google Scholar]
- 7.Frymier PD, Ford RM, Berg HC, Cummings PT. 3-Dimensional tracking of motile bacteria near a solid planar surface. Proc Natl Acad Sci USA. 1995;92:6195–6199. doi: 10.1073/pnas.92.13.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biondi SA, Quinn JA, Goldfine H. Random motility of swimming bacteria in restricted geometries. AICHE J. 1998;44:1923–1929. [Google Scholar]
- 9.Vigeant MAS, Ford RM, Wagner M, Tamm LK. Reversible and irreversible adhesion of motile Escherichia coli cells analyzed by total internal reflection aqueous fluorescence microscopy. Appl Environ Microbiol. 2002;68:2794–2801. doi: 10.1128/AEM.68.6.2794-2801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiLuzio WR, et al. Escherichia coli swim on the right-hand side. Nature. 2005;435:1271–1274. doi: 10.1038/nature03660. [DOI] [PubMed] [Google Scholar]
- 11.Ramia M, Tullock DL, Phanthien N. The role of hydrodynamic interaction in the locomotion of microorganisms. Biophys J. 1993;65:755–778. doi: 10.1016/S0006-3495(93)81129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauga E, DiLuzio WR, Whitesides GM, Stone HA. Swimming in circles: Motion of bacteria near solid boundaries. Biophys J. 2006;90:400–412. doi: 10.1529/biophysj.105.069401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galajda P, Keymer J, Chaikin P, Austin R. A wall of funnels concentrates swimming bacteria. J Bacteriol. 2007;189:8704–8707. doi: 10.1128/JB.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeuchi S, DiLuzio WR, Weibel DB, Whitesides GM. Controlling the shape of filamentous cells of Escherichia coli. Nano Lett. 2005;5:1819–1823. doi: 10.1021/nl0507360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berg HC. Motile behavior of bacteria. Phys Today. 2000;53:24–29. [Google Scholar]
- 16.Alon U, et al. Response regulator output in bacterial chemotaxis. EMBO J. 1998;17:4238–4248. doi: 10.1093/emboj/17.15.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amsler CD, Cho MS, Matsumura P. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter postexponential growth. J Bacteriol. 1993;175:6238–6244. doi: 10.1128/jb.175.19.6238-6244.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleineberger E. The natural occurence of pleuropneumonia-like organisms in apparent symbiosis with Streptobacillus moniliformis and other bacteria. J Pathol Bacteriol. 1935;40:93. [Google Scholar]
- 19.Joseleau-Petit D, Liebart JC, Ayala JA, D'Ari R. Unstable Escherichia coli L forms revisited: Growth requires peptidoglycan synthesis. J Bacteriol. 2007;189:6512–6520. doi: 10.1128/JB.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leaver M, Dominguez-Cuevas P, Coxhead JM, Daniel RA, Errington J. Life without a wall or division machine in Bacillus subtilis. Nature. 2009;457:849–854. doi: 10.1038/nature07742. [DOI] [PubMed] [Google Scholar]
- 21.Koch AL. Bacterial Growth and Form. Dordrecht: Kluwer; 2001. [Google Scholar]
- 22.Bronk BV, Vandemerwe WP, Stanley M. In vivo measure of average bacterial-cell size from a polarized-light scattering function. Cytometry. 1992;13:155–162. doi: 10.1002/cyto.990130208. [DOI] [PubMed] [Google Scholar]
- 23.Woldringh CL, Grover NB, Rosenberger RF, Zaritsky A. Dimensional rearrangement of rod-shaped bacteria following nutritional shift-up. 2. Experiments with Escherichia coli B-R. J Theor Biol. 1980;86:441–454. doi: 10.1016/0022-5193(80)90344-6. [DOI] [PubMed] [Google Scholar]
- 24.Veening JW, Kuipers OP, Brul S, Hellingwerf KJ, Kort R. Effects of phosphorelay perturbations on architecture, sporulation, and spore resistance in biofilms of Bacillus subtilis. J Bacteriol. 2006;188:3099–3109. doi: 10.1128/JB.188.8.3099-3109.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner L, Ryu WS, Berg HC. Real-time imaging of fluorescent flagellar filaments. J Bacteriol. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao X, et al. Atomic force microscopy and theoretical considerations of surface properties and turgor pressures of bacteria. Colloid Surf B-Biointerfaces. 2002;23:213–230. [Google Scholar]
- 27.Chattopadhyay S, Moldovan R, Yeung C, Wu XL. Swimming efficiency of bacterium Escherichia coli. Proc Natl Acad Sci USA. 2006;103:13712–13717. doi: 10.1073/pnas.0602043103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keymer JE, Galajda P, Muldoon C, Park S, Austin RH. Bacterial metapopulations in nanofabricated landscapes. Proc Natl Acad Sci USA. 2006;103:17290–17295. doi: 10.1073/pnas.0607971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matias VRF, Beveridge TJ. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol Microbiol. 2005;56:240–251. doi: 10.1111/j.1365-2958.2005.04535.x. [DOI] [PubMed] [Google Scholar]
- 30.Yao X, Jericho M, Pink D, Beveridge T. Thickness and elasticity of Gram-negative Murein sacculi measured by atomic force microscopy. J Bacteriol. 1999;181:6865–6875. doi: 10.1128/jb.181.22.6865-6875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnoldi M, et al. Bacterial turgor pressure can be measured by atomic force microscopy. Phys Rev E. 2000;62:1034–1044. doi: 10.1103/physreve.62.1034. [DOI] [PubMed] [Google Scholar]
- 32.Koch AL. The surface stress theory of microbial morphogenesis. Adv Microb Physiol. 1983;24:301–366. doi: 10.1016/s0065-2911(08)60388-4. [DOI] [PubMed] [Google Scholar]
- 33.Boulbitch A, Quinn B, Pink D. Elasticity of the rod-shaped Gram-negative eubacteria. Phys Rev Lett. 2000;85:5246–5249. doi: 10.1103/PhysRevLett.85.5246. [DOI] [PubMed] [Google Scholar]
- 34.Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci USA. 2008;105:19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.