Abstract
Regulation of gene activity by microRNAs is critical to myriad aspects of eukaryotic development and physiology. Amidst an extensive regulatory web that is predicted to involve thousands of transcripts, emergent themes are now beginning to illustrate how microRNAs have been incorporated into diverse settings. These include potent inhibition of individual key targets, fine-tuning of target activity, the coordinated regulation of target batteries, and the reversibility of some aspects of microRNA-mediated repression. Such themes may reflect some of the inherent advantages of exploiting microRNA control in biological circuits, and provide insight into the consequences of microRNA dysfunction in disease.
MicroRNAs (miRNAs) are ~21–24 nucleotide regulatory RNAs derived from hairpin transcripts that are abundant in animals, plants and even viruses. Although miRNAs were formally codified as a gene class in late 2001 (REFS 1–3), their study was rooted in genetic analyses of worm4–7 and fly8,9 development during the 1990s. These early works revealed key features of miRNA transcripts, some of the significant biological uses of miRNAs and insights into how ~7-nucleotide 3′ UTR elements mediate miRNA function through target deadenylation and destabilization, as well as translational inhibition. The subsequent explosion of miRNA research in the current decade has yielded breathtaking advances in our understanding of the mechanism and biology of miRNA control10,11. Our goal in this Review is to rationalize how and why miRNAs have been incorporated into biological networks. In particular, some of the emerging principles that are shared across unrelated developmental or physiological settings seem to reflect the particular utilities of miRNAs in gene regulation.
Overview of miRNA biogenesis and activity
miRNAs derive from longer primary transcripts bearing one or more local hairpins, which are cleaved by RNase III enzymes to yield ~21–24 nucleotide RNA duplexes. Single strands from the duplexes are selected for association with Argonaute proteins, which guide them to complementary targets for regulation10.
In plants, most miRNAs exhibit extended complementarity to one or a few predicted targets12, and directed tests have validated ~100 mRNAs that are cleaved by an endogenous miRNA and an Argonaute protein with Slicer activity13–15. High-throughput sequencing of RNAs with 5′ monophosphates (which excludes capped mRNAs) provides an opportunity to identify miRNA-cleaved transcripts genome-wide16,17. Application of this unbiased approach to Arabidopsis thaliana uncovered some novel targets, but most of the high-confidence targets had been previously identified or predicted. This suggested that plant miRNAs indeed have only a small number of targets. In addition to cleaving their targets, plant miRNAs can direct the translational inhibition of highly complementary targets, raising the possibility of reversible regulation18.
In animals, few mRNAs seem to be sufficiently complementary to miRNAs to be cleaved by a Slicer mechanism. Instead, most miRNA target sites contain seven nucleotides of Watson–Crick base-pairing to positions 2–8 of the miRNA (numbering from the 5′ end), also known as the miRNA ‘seed’19. However, there is still mechanistic diversity in animal target regulation, which can take place through target degradation and/or through translational repression20. Target predictions based primarily on conserved seed pairing and local sequence or structural features suggest that individual animal miRNAs often have >100 targets, and that at least 20–30% of animal transcripts bear one or more conserved miRNA binding sites in their 3′ UTR21–23. Additional targets may potentially be regulated through miRNA binding to atypical sites with imperfect seeds6,24–26 or non-conserved sites27–29. Therefore, the direct target network of animal miRNAs is inferred to be quite substantial.
Classifying miRNA–target relationships
Because individual organisms can have hundreds of miRNAs and many thousands of miRNA targets, it is unlikely that the biological consequences of miRNA-mediated regulation will be the same in all cases. Some relevant parameters to consider are how much a given target is repressed by a miRNA, and how much the target repression matters in a given biological setting. One can also ask whether the miRNA operates through one target or many targets, each of which might behave differently with respect to the quantitative and qualitative consequence of miRNA control.
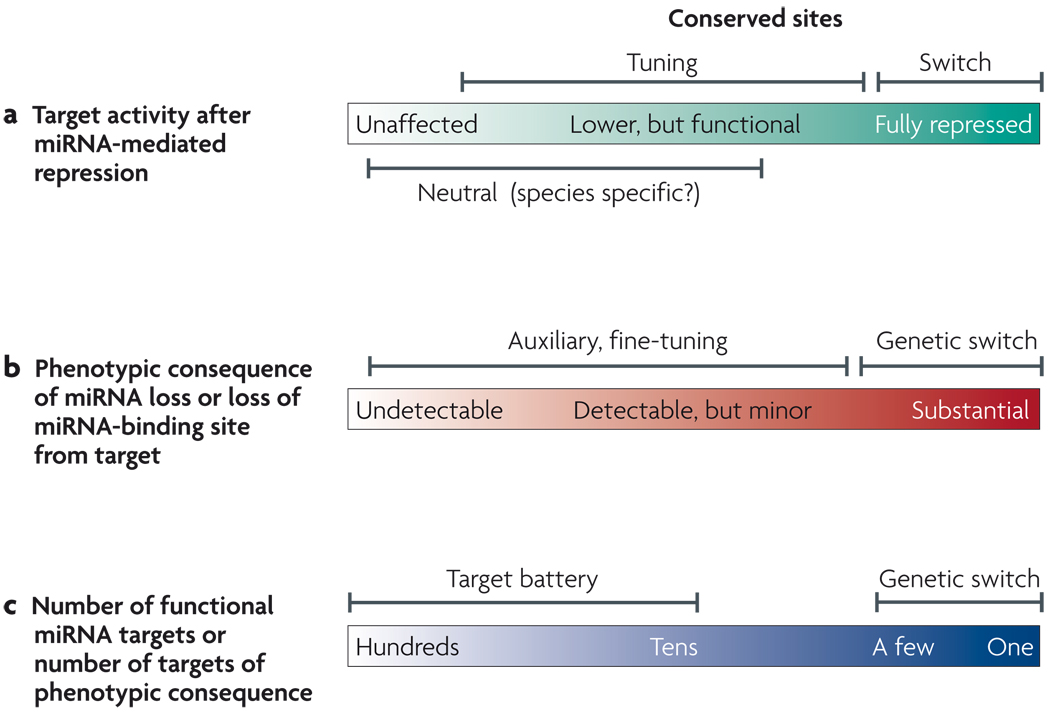
In one of the earliest proposals for how to classify miRNA activities, their targets were categorized as ‘switch’, ‘tuning’ and ‘neutral’30 (FIG. 1a). In this scheme, switch targets are ones with activity that is reduced by miRNAs to inconsequential levels — that is, ones that are essentially turned off by miRNAs. Tuning targets are ones with activity that is not eliminated by miRNAs — that is, targets that remain functional in miRNA-expressing cells. The regulation that is mediated by switch and tuning sites is useful enough to be positively selected during evolution. Neutral targets are involved in species-specific regulatory interactions that are of no particular consequence to the cell. As they are neither advantageous nor adverse, neutral sites are expected to readily appear and disappear through genetic drift during evolution. Presumably only in rare instances do evolutionarily novel miRNA target sites provide beneficial regulation that is species-specific. It was also suggested that many genes would be negatively affected by the acquisition of miRNA-binding sites, and ‘anti-targets’ that are depleted for miRNA-binding sites indeed exist28,29,31.
Figure 1. Classifying microRNA (miRNA) activities and functions.
a The activity of a given target can be evaluated in the presence of a cognate miRNA. ‘Switch’ targets are essentially inactive following miRNA-mediated repression, whereas ‘tuning’ targets produce functional protein in the domain of miRNA activity. Both of these terms apply to evolutionarily conserved target relationships. Non-conserved but functional sites may mediate repression that is incidental or otherwise compensated for by other regulatory mechanisms; these are termed ‘neutral’ targets. Non-conserved sites may occasionally mediate beneficial, species-specific regulation. Not shown are ‘anti-targets’, transcripts for which miRNA-mediated repression is deterimental, such that the acquistion of target sites is avoided. b Another means of classification is to evaluate whether loss of function of a specific miRNA is associated with a phenotype, or whether the removal of target sites from a specific target causes a phenotype. For many conserved miRNAs and their conserved targets, the repression is likely to be beneficial enough to be selected during evolution but, overall, to be auxiliary to other means of gene regulation. Only a subset of miRNAs, with a small number of targets, mediate regulation with genetically defined consequences; we call these ‘miRNA genetic switches’, c One may consider the total number of targets whose repression by a miRNA in a given setting can be detected by quantitative means. Alternatively, a more stringent classification would be to consider only those targets whose repression by a miRNA is of detectable phenotypic consequence.
Another way to classify miRNA activities regards whether they make intrinsic detectable contributions to morphology, physiology or behaviour. We call these miRNA activities genetic switches', to reflect the fact that the importance of target regulation can be detected by organismal phenotype (FIG. 1b). Importantly, genetic switch targets do not wholly overlap with the general switch category described above32. For example, a miRNA can reduce a target to an inconsequential level, but this might be essentially auxiliary if transcriptional repression is sufficient to silence the target. Conversely, a miRNA need not eliminate its target to exert an important phenotypic effect. For instance, a pathway might be finely balanced at a tipping point, and the quantitatively modest influence of a tuning miRNA site might flip the switch.
A third way to classify miRNA activities depends on whether their major effect is mediated through one or a few targets or through many targets (tens or hundreds) (FIG. 1c). All known genetic switches concern cases of one or a few important miRNA targets, although it is theoretically possible for one miRNA to have many genetically important targets. In a setting in which hundreds of targets are simultaneously repressed by a given miRNA, it may be that none of the individual regulatory events is particularly important, but that the system collapses when all of the regulatory links are broken. In such a setting, a miRNA might serve as a global enforcer of cell or organ identity
Any of these categorizations depends on a good understanding of direct miRNA targets, and on the phenotypic consequence of failing to repress each of these targets. In this regard, our knowledge of most miRNAs and their targets is arguably shallow Nevertheless, the extant research provides compelling illustrations for each of these rationales to incorporate miRNAs into specific regulatory networks. We consider selected examples in the following sections, with emphasis on principles derived from genetic studies that often provide the clearest evidence for the biological significance of miRNA target regulation.
miRNAs as developmental genetic switches
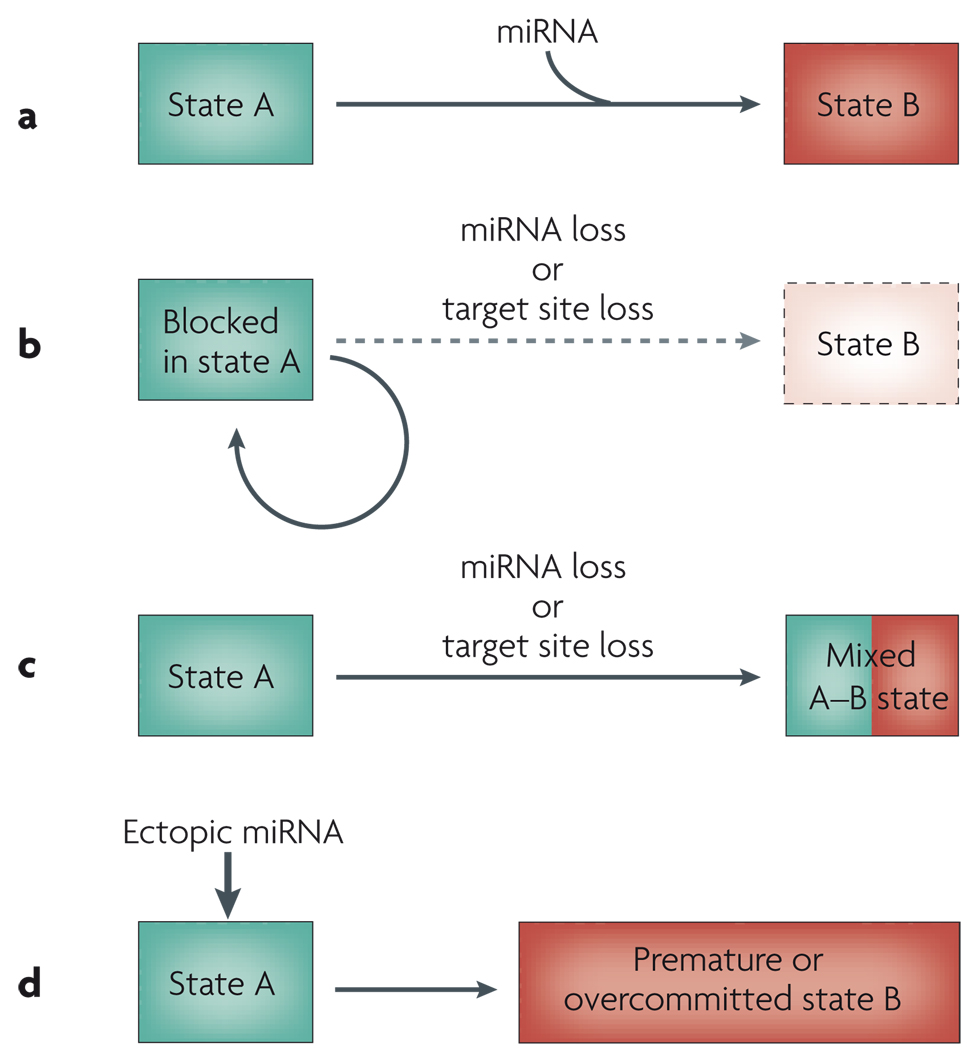
Classical genetics analysis and epistasis analysis have successfully distilled several biological processes into core components that function in orderly pathways. In a few known cases, one of the components is a miRNA. Such ‘miRNA genetic switches’ often share the property of reciprocal gain- and loss-of-function phenotypes. Suppose that activation of a miRNA switches the cell state from A to B (FIG. 2a). Loss of the miRNA, or perhaps even the loss of miRNA-binding sites from a particular gene, could lead to a cell becoming stuck in state A (FIG. 2b). Alternatively, it could lead to the adoption of a mixed A–B fate (FIG. 2c). On the other hand, ectopic activation of the miRNA could induce temporally premature adoption of, or spatial overcommitment to, state B (FIG. 2d). We consider here a few examples of miRNA genetic switches that govern developmental programmes.
Figure 2. Consequences of microRNA (miRNA) loss- or gain-of-function for cell state.
The activation of a miRNA can direct or aid a transition from state A to state B (a). Loss of a miRNA, or of its binding sites in a target, can lead to the cell becoming stuck in state A (b) or can result in a mixed state with characteristics of both state A and state B (c). An ectopic miRNA can prematurely induce state B or can cause cells to overcommit to this state (d).
Control of developmental timing by miRNAs
The miRNA field emerged from studies that screened for genes that mediate developmental transitions in nematodes (FIG. 3). For example, the lin-14 gene encodes a transcription factor that is crucial for the completion of the first larval stage (L1)33. Worms that lack lin-14 skip L1 and go straight to L2-specific lineages, whereas lin-14 gain-of-function mutants lacking 3′ UTR control sequences reiterate L1-specific lineages34,35. The lin-4 gene was genetically identified as a repressor of lin-14 that becomes active during the transition to L2, and the phenotypes of lin-4 mutants are entirely reciprocal to those of lin-14 mutants36,37. The surprise, of course, was that lin-4 did not encode a protein as expected, but instead a small regulatory RNA (lin-4) that binds lin-14 3′ UTR control sequences6,7. A second key heterochronic target of lin-4 is an RNA-binding protein encoded by lin-28. Like lin-14, its miRNA control is so important that mutation of the lin-4-binding site in a lin-28 rescuing transgene induces developmental delays, in this case reiteration of L2 lineages4 (FIG. 2b). Conversely, the genetic switch can be thrown by overexpressing lin-4 during L1; this induces early loss of LIN-14 and precocious entry into L2 (REF. 38) (FIG. 2d).
Figure 3. Multiple microRNA (miRNA) genetic switches in the nematode heterochronic pathway.
The lin-4 miRNA promotes the transition between the first and second larval stages (L1 and L2, respectively) by repressing the transcription factor LIN-14 and the RNA-binding protein LIN-28. Similarly, three related miRNAs with homology to let-7 (miR-48, miR-84 and miR-241) act in concert to repress the transcription factor HBL-1, promoting the transition from L2 to L3. Finally, let-7 represses the TRIM protein LIN-41, and possibly HBL-1, to promote the transition from L4 to the adult.
Subsequent studies of the worm heterochronic pathway revealed it to be rich with miRNA components (FIG. 3). The miRNA let-7 controls the L4–adult transition, primarily by repressing the TRIM protein that is encoded by lin-41 (REF. 24). A cohort of miRNAs produced from let-7 ‘sister genes’ (miR-48, miR-84 and miR-241) control the L2–L3 transition, by repressing the zinc finger transcription factor that is encoded by hbl-1 (REF. 39). Although many genes might in principle be derepressed in the absence of these miRNAs, the phenotypes of let-7 mutants and those of let-7 sister-gene mutants were substantially suppressed by reducing the activity of lin-41 and hbl-1, respectively. This powerful genetic evidence argues that the repression of a handful of targets accounts for the major functions of these heterochronic miRNAs, even though they clearly have many other conserved targets40–42.
Potentially analogous temporal initiation of let-7 expression in various animals was noticed from early studies43, and some of the lin-4 and let-7 targets seem to be conserved outside nematodes44,45. Recent studies of the Drosophila melanogaster let-7 gene cluster (which includes mir-100 and the lin-4 homologue mir-125) showed that the heterochronic function of let-7, and possibly that of miR-125, is conserved among invertebrates46,47. Flies that carry deletions of the let-7 gene cluster are subviable, and adult survivors exhibit various behavioural defects and shortened lifespan. Closer examination revealed that let-7 and/or miR-125 mediate terminal cell-cycle exit in wing imaginal discs, and that let-7 is specifically required for remodelling neuromuscular junctions (NMJs) from the larval pattern to the adult pattern46,47. These functions are analogous to that of the nematode let-7 in promoting cell-cycle exit of hypodermal cells during the L4–adult transition.
The D. melanogaster abrupt gene (which encodes a BTB zinc finger transcription factor) is an important heterochronic target of let-7 at the NMJ, as reduced abrupt dosage could suppress the temporal remodelling defect of let-7-mutant NMJs47. However, abrupt activity does not seem to account for the cell-cycle defect or shortened lifespan of let-7 mutants, so other important targets might be deregulated in these settings. These studies demonstrate that conserved miRNAs can have conserved functions, but the specific targets of conserved miRNAs are not necessarily fixed (for example, nematodes do not have an abrupt homologue). In general, although many miRNAs are highly conserved in sequence and expression pattern across animal genomes, vanishingly few miRNA–target interactions have been similarly well conserved48,49. This suggests that extensive rewiring of the miRNA regulatory network has occurred during animal evolution.
Widespread role for plant miRNAs in directing developmental patterning
The plant miRNA target network encompasses various gene functions, but it is heavily biased towards developmental transcription factors12. A provocative early hint of the in vivo importance of these targets came with the realization that gain-of-function alleles of PHABULOSA and PHAVOLUTA, genes that encode HD-ZIP transcription factors that regulate the radial polarity of A. thaliana shoots50, contain mutations that disrupt binding sites for miR-165 (REF. 12). The significance of this was initially confounded by amino-acid changes resulting from these mutations. However, subsequent work on the paralogous gene REVOLUTA showed that translationally silent mutations in the miR-165-binding site of a REVOLUTA transgene caused it to induce characteristic gain-of-function polarity defects51. Therefore, HD-ZIP repression is mediated at the mRNA level by miR-165.
Many additional studies have shown that transcription factor alleles or transgenes bearing miRNA target-site mutations induce gain-of-function patterning defects15. Complementary evidence from loss-of-function mutations of miRNA alleles has also emerged. Single or compound mutations in A. thaliana mir-164 genes, which regulate CUP-SHAPED COTYLEDON (CUC)-family NAC-domain transcription factors, revealed defects in flower development and spatial arrangement that were indicative of target deregulation52,53. In addition, the floral homeotic defects of petunia BLIND and snapdragon FISTULATA mutants were both found to be caused by inactivation of mir-169-family genes54. miR-169 miRNAs repress NF-YA-class transcription factors that enhance the expression of floral patterning genes. Although some plant miRNAs are also deployed during specific stress conditions, the preponderance of highly conserved transcription factor miRNA targets regulate developmental patterning, suggesting that this is the major function of miRNAs in plants12,55.
These animal and plant genes serve as paradigms of miRNA genetic switches: the strong repression of one or a few targets has a major impact on a specific biological process (FIG. 4a). Especially in animals, it seems likely that these miRNA switches simultaneously repress multiple transcripts; nevertheless, the loss-of-function phenotypes of these miRNAs mostly reflect the deregulation of specific targets (FIG. 4c).
Figure 4. Configurations of microRNA (miRNA)–target networks.
a Potent inhibition by a miRNA can have a substantial phenotypic effect in some cases, but can lead to only minor phenotypic consequences in other cases. b Relatively weak repression by a miRNA might often have minor consequences, but could have substantial effects if the biological system is sensitive to minor fluctuations in target activity. c Although most animal miRNAs are likely to influence a battery of targets in a given cell, the phenotype caused by loss of a miRNA might demonstrably be due to one or a few targets. d In some cases a miRNA might exert relatively equivalent effects across a large number of targets, the collective deregulation of which might lead to a detectable phenotype. However, the loss of miRNA-mediated repression of an entire target battery can also be fully compensated for by other regulatory mechanisms. RISC, RNA-induced silencing complex.
MicroRNAs that modulate specific targets
A thematic variation on the use of miRNAs is to modulate target activity. So-called ‘tuning”, ‘thresholding’ or ‘de-noising’ targets are functionally active within the domain of miRNA expression. In these cases, the role of the miRNA is to precisely set a limit on target mRNA and/or protein level (FIG. 4b). In contrast to genetic switches, in which the miRNA and its target protein are perhaps transiently co-expressed as the miRNA takes over (FIG. 3), miRNAs that are modulatory might be expected to be more continuously co-expressed with their targets.
Multiple miRNAs set thresholds on neural precursor selection
D. melanogaster studies from the 1990s provided early evidence that Notch target genes are thresholding miRNA targets during sensory bristle and eye development. Although Notch target genes are expressed in specific cells that actively engage Notch signalling, they are repressed by spatially broad regulatory systems mediated by Brd box- and K box-class miRNAs8,9,56,57. A key demonstration was that genomic rescue transgenes for the Notch target genes Bearded and E(spl)m8 (also known as E(spl)) induced gain-of-function bristle and eye phenotypes when specifically mutated for their miRNA-binding sites, whereas corresponding wild-type transgenes did not. This suggested that miRNAs set a threshold activity level for Notch target gene transcription, possibly to ensure that Notch pathway activity is not deployed inappropriately by transcriptional noise58. Similar functions were ascribed to miR-9a, which regulates sensory-bristle development by repressing Senseless, a zinc finger transcription factor that is sufficient to induce sensory-bristle specification59. mir-9a mutants exhibit stochastic and modest numbers of ectopic sensory bristles, perhaps reflecting the accidental triggering of the cascade for sensory-bristle specification60.
miR-8 regulates the tuning target atrophin
A recent study of D. melanogaster miR-8 revealed that the transcriptional corepressor atrophin (also known as Gug) is a tuning target61. mir-8 deletion mutants exhibit various morphological and behavioural defects, but most of these could be attributed as downstream consequences of aberrant brain apoptosis induced by ectopic Atrophin61. Impressively, most of these overt phenotypes could be dominantly suppressed by removing one copy of the atrophin gene, implying that a major function of miR-8 is to regulate this single target. Expression of an atrophin-knockdown transgene under the control of miR-8 regulatory sequences resulted in a mutant phenotype, suggesting that the Atrophin that is present in miR-8-expressing cells is necessary for development. These tests therefore qualify atrophin as a tuning target.
Plant tuning targets
Some plant miRNAs are not spatially co-expressed with their targets, consistent with their ability to slice and eliminate target mRNAs. In other cases, however, miRNA and target transcripts are co-expressed. This raises the possibility that plant miRNAs, like animal miRNAs, can tune their targets. Indeed, the A. thaliana mir-164abc triple mutant exhibits seemingly random enlargements in target CUC expression domains, which correspondingly define variable flower positions, suggesting that miRNAs provide an essential thresholding function that delimits CUC activity53. Similarly, BLIND, FISTULATA and miR-169 were interpreted to lend a dynamic homeostatic control to the transcriptional regulation of homeotic gene activity during flower development54.
In these examples the importance of miRNA-mediated tuning is substantial because minor changes in these targets have phenotypic consequences (FIG. 4b). Presumably, the subtle control of many other conserved targets will not underlie significant mutant phenotypes, even though such regulation is beneficial enough to be selected for during evolution (FIG. 1b). In general, the fact that many miRNAs fine-tune targets or lend robustness to pathway output has led to the hypothesis that miRNAs are often exploited to canalize developmental traits62 (BOX 1).
Box 1 Canalization of regulatory programmes by microRNAs (miRNAs).
Although miRNAs can have essential roles in directing some biological processes, it seems likely that the action of many miRNAs with highly conserved targets has no discernable consequence, at least under assayable laboratory conditions. For example, systematic deletions of nearly all Caenorhabditis elegans miRNA genes revealed unexpectedly few developmental or behavioural phenotypes86. By extension, few miRNA–target relationships seem likely to be required for substantial observable traits. What is the meaning of regulatory networks that can be computationally inferred but not experimentally visualized?
One possibility is that the incorporation of miRNA-mediated control raises the fidelity of stereotyped processes that are already under exacting control by other mechanisms, such as transcriptional regulation or post-translational control62. By buffering inherent genetic noise, leaky transcription, or decreasing variability incurred by ever-changing environmental conditions, many miRNA-target regulatory interactions might be beneficial enough to be selected during evolution. Overall, however, the regulation of many individual miRNA targets might still be responsible for minute improvements to individual fitness.
In such cases, it will be extremely challenging to attain phenotypic evidence for the endogenous utility of highly conserved miRNA regulatory targets. In search of effects beyond purely molecular phenotypes (for example, the slight increase in the levels of a target transcript or protein in a miRNA mutant), perhaps a useful approach will be to subject miRNA mutants lacking obvious phenotypes to sensitized genetic backgrounds or to environmental stresses. With the current lack of phenotypic evidence from such approaches, however, bioinformatic evidence from evolutionary conservation, functional enrichments of targets or spatiotemporal expression of miRNA target cohorts might be our best indication of the usefulness of many miRNA–target interactions.
Target batteries control spatial or temporal state
Thus far, we have mostly considered the control of single targets by miRNAs. Because animal miRNAs can regulate targets with as few as seven nucleotides of complementarity, their potential to regulate large numbers of targets is obvious (FIG. 4d). How have animals capitalized on such expansive regulatory potential? Computational and experimental studies support models in which miRNAs are used to broadly influence spatial or temporal identity.
Expression profiling reveals miRNA control of gene batteries
Microarray expression profiling provided evidence for broad enforcement of tissue identity by tissue-specific miRNAs. When the heart- and muscle-specific miR-1 or the brain-specific miR-124 was introduced into HeLa cells, hundreds of genes containing miR-1 or miR-124 seed matches, respectively, were downregulated63. The inferred direct targets of miR-1 were normally expressed at their lowest levels in heart and muscle tissue, relative to all other tissues in the body, and the inferred direct targets of miR-124 were expressed at their lowest relative levels in the brain. Thus, the introduction of tissue-specific miRNAs into a naive cell type detectably shifts their gene-expression profile towards that of the endogenous miRNA tissue of origin.
Analysis of cell lines and whole organs provided further evidence that highly expressed miRNAs impose an mRNA expression signature in which their direct targets are depressed relative to their levels in other cells that do not express the miRNA28,29. These approaches were recently complemented by large-scale proteomics studies that similarly show that individual miRNAs can subtly, but detectably, repress the protein output of hundreds of seed-bearing targets64,65. This supports the notion that miRNAs direct global changes in gene activity that are precisely appropriate for a given cell type or organ.
Mutual exclusion model for miRNA–target expression
The aforementioned studies concluded that miRNA targets are frequently ‘lowered, but not off when they are co-expressed with their cognate miRNAs. A variation of this concept is ‘mutual exclusion’, which proposes that many miRNAs are not substantially expressed in the same cells as their targets. This idea came from considering developmental lineages such as the neuroectoderm, a multipotent tissue that gives rise to the epidermal ectoderm as well as to specialized neural cells. The central nervous system (CNS)-specific miR-124 preferentially targets differentiated epidermis genes while generally avoiding CNS genes31. This suggests a model in which the potential ‘noise’ of epidermal gene expression in neurons, a possibility because of the common prior developmental history of the two tissue types, might be excluded by the activation of the tissue-specific miR-124. In support of this, in situ hybridization experiments in D. melanogaster embryos illustrated several cases in which miRNAs are expressed in cells that are adjacent to, but spatially excluded from, their targets. In this role, miRNAs serve as a backup to silence genes that are already mostly transcriptionally inactive.
Control of maternal–zygotic transition by fish and fly miRNAs
The examples in the previous subsection concern spatial identity. The propensity of miRNAs to regulate large target batteries may also confer temporal identity. Although the first few nematode miRNAs studied had small sets of temporal targets, the study of zebrafish development showed how a single miRNA family controls temporal identity during the maternal–zygotic transition (MZT)27. As with many animals, the rapid initial development of the fertilized fish embryo is fuelled by maternally provided stores of mRNAs and proteins. Eventually the pace of cell division slows down and begins to depend on zygotic transcription; during this time (namely the MZT) maternal gene products are cleared away.
The early fish zygote expresses tremendous amounts of miR-430-family miRNAs, and thus the early phenotype of embryos that are maternally and zygotically deficient for Dicer (MZ-Dicer embryos) is largely attributable to the loss of miR-430 miRNAs. MZ-Dicer embryos accumulate hundreds of maternal transcripts that normally decline after wild-type MZT, and many of these are direct miR-430 targets27. This has been interpreted as a situation in which the loss of miR-430 miRNAs yields an inappropriately mixed state (FIG. 2c). Importantly, these maternal gene expression defects can be substantially suppressed by reintroducing mature miR-430 duplexes into MZ-Dicer embryos27. These findings supported a model in which miR-430 facilitates a temporally sharp MZT by clearing away a battery of maternally provided transcripts (FIG. 4d, upper pathway).
Recently it was shown that D. melanogaster MZT uses miRNAs in an analogous fashion. The situation is not entirely the same as in the zebrafish, as fly MZT is heavily dependent on Smaug, an RNA-binding protein that coordinates the degradation of hundreds of maternal transcripts during MZT66. Nevertheless, among the earliest zygotically active loci is a cluster of eight miRNA genes that encodes five different mature miRNAs (the miR-309 cluster). Although mutants lacking this miRNA locus have only moderately reduced viability and normal morphology, they exhibit an unmistakable molecular signature in that a battery of maternally expressed genes is maintained following MZT; many of which are predicted to be direct targets of the various miR-309-cluster miRNAs67.
As none of the D. melanogaster miR-309-cluster miRNAs is related to zebrafish miR-430, their respective use in MZT is presumably evolutionarily convergent. It is worth noting that MZT involves the regulation of many targets that are divorced from transcriptional control, a property that makes them well suited to regulation by miRNAs. This may conceivably underlie the similar use of miRNAs to effect a temporally sharp MZT in different organisms.
Distinguishing ‘more’ and ‘less’ important targets
It should now be evident that miRNA targets do not comprise a homogenous class. Therefore, the existence of thousands of highly conserved miRNA–target relationships should not be taken to imply that there are thousands of regulatory interactions with the phenotypic importance of, for example, the lin-4–lin-14 interaction. A pressing challenge for the miRNA field is to clarify the extent to which the major activities of miRNAs with many predicted conserved targets might be largely attributable to a few specific targets (FIG. 4c,d), as with the field-founding nematode miRNAs (FIG. 3). Many examples have recently emerged from invertebrate and vertebrate studies, indicating that the early worm genetic studies were not unique in this regard.
miR-150 and haematopoiesis
miR-150 is a well conserved miRNA that is specifically expressed in the haematopoietic system in mature T and B cells, and its misexpression in immature B cells blocks their differentiation68. As with other well conserved miRNAs, there are hundreds of genes with well conserved miR-150-binding sites. But out of this large target battery, careful analysis of mice lacking the sole murine mir-150 gene revealed alterations in B cell numbers that could be attributed to deregulation of the transcription factor MYB alone69. Importantly, subtle alterations in MYB level — in the range of 25–35% — have profound effects on B cell development. This provides a rationale for why regulation of this target by a miRNA might have a particularly substantial impact on the phenotype.
miR-279 and olfactory neurons
miR-279 is one of a small group of miRNAs that were identified through a loss-of-function mutation in a forward genetic screen, in this case for genes that are required for the development of the D. melanogaster olfactory system. Loss of miR-279 causes CO2-sensing neurons, which are normally restricted to the antennae, to be generated ectopically in the maxillary palps70. miR-279 has many predicted targets that are neural transcription factors71; however, the zinc finger transcription factor that is encoded by nerfin stands out because it has multiple conserved target sites. Indeed, loss of one copy of nerfin significantly corrected the mir-279-null phenotype70. On the other hand, misexpression of nerfin was insufficient to recapitulate the mir-279-null phenotype. Therefore, this seems to be a mixed situation in which multiple targets probably contribute to the miRNA phenotype, but nerfin is amongst the most important of these targets.
Many miRNAs regulate key individual targets
A growing number of other miRNA mutants can be phenotypically suppressed by partially reducing the expression of single target genes. For example, D. melanogaster mir-14 mutants exhibit a defective metamorphosis that is suppressed by heterozygous deletion of miR-14’s target gene, Ecdysone receptor72. Mice with mutations in the myeloid-lineage-specific gene mir-223 exhibit increased numbers of neutrophils and granulocytes and upregulation of MEF2C, an enhancer of proliferation in the myeloid lineage. This was inferred to be a key target because removal of Mef2c from the mir-223-mutant mouse rescued its neutrophil defect73. Deletion mutants of the murine miR-17-92 cluster arrest at the pro-B cell to pre-B cell transition and exhibit elevated levels of the product of the pro-apoptotic target Bim (also known as Bcl2l11)74, and the loss of Bim can partially suppress the phenotypic effect of the loss of Dicer in lymphocyte progenitors75. All of these miRNAs have tens to hundreds of highly conserved targets, and the preservation of these targets over tens of millions of years of evolution demonstrates their functional utility. Nevertheless, genetics provides evidence that the deregulation of individual targets can often underlie substantial aspects of miRNA-knockout animals (FIG. 4c).
Many miRNA genetic switches involve signalling genes or transcription factors, the activity modulation of which can potentially have broad consequences. It is also worth noting that several miRNA genetic switches involve targets that encode regulators of miRNA expression and/or activity (BOX 2). Bistable loops offer a means to amplify the effects of miRNA-regulatory interactions, and understanding such circuits can be essential for understanding the consequences of successful or unsuccessful miRNA-mediated target repression.
Box 2 Bistable loops involving microRNAs (miRNAs).
Regulation by miRNAs does not have to be a one-way street. In many cases in which miRNAs function as genetic switches, miRNA targets encode regulatory factors. In several cases, the function of these factors feeds back to repress the miRNA. This regulation therefore constitutes a bistable loop — that is, a regulatory interaction that leads to robust commitment to one of two possible outcomes.
In one simple bistable loop that has a role in the developing Drosophila melanogaster eye (see figure, part a), miR-7 inhibits the transcriptional repressor Yan, and Yan reciprocally represses transcription of mir-7 (REF. 103). This mutually inhibitory relationship helps to partition the expression of Yan into eye progenitor cells and that of miR-7 into differentiating photoreceptors, contributing to these two alternative fates.
A more complicated bistable loop, involving multiple miRNAs and their transcription factor targets, operates to assign left and right fates to the pair of nematode chemosensory neuronal cells termed ASE left (ASEL) and ASE right (ASER)104–106 (see figure, part b). In this system, the transcription factors DIE-1 and COG-1 specify the left and right ASE cells, respectively, in a cell-autonomous manner. The left–right-specific expression of these factors requires their post-transcriptional repression by miRNAs. In ASEL, the miRNA lsy-6 is activated downstream of DIE-1, and it directly represses cog-1. In ASER, miRNAs of the miR-273 family are activated downstream of COG-1, and they directly repress die-1. The importance of miRNAs in directing ASE asymmetry is seen, for example, in the double-ASER phenotype of either lsy-6 loss-of-function animals or cog-1 gain-of-function mutants lacking 3′ UTR sequences.
Bistable loops can also involve miRNA-processing factors. Mammalian LIN-28 was recently demonstrated to bind directly and specifically to the terminal loops of several let-7-miRNA-family precursors, and to inhibit their processing by Drosha and Dicer81–83 (see figure, part c). This seems to be a bistable loop, as let-7 reciprocally represses lin-28 directly44,107. The loop may be relevant during neural differentiation of stem cells, with LIN-28 present and let-7 activity off in pluripotent embryonic stem (ES) cells, and with LIN-28 absent and let-7 activity on following these cells' commitment into neural stem cells. Interestingly, lin-28 was originally identified as a nematode heterochronic gene that resides downstream of the miRNA lin-4 and upstream of the let-7 miRNA4,108. Nematode lin-28 is not only a genetic-switch target of lin-4, it contains also a conserved site for let-7 family members. As lin-28 mutants exhibit precocious expression of let-7, whereas lin-4 mutants (which have elevated LIN-28) have delayed expression of let-7 (REF. 108), it is conceivable that worm development incorporates a LIN-28-let-7 bistable loop.
Shown are examples of bistable loops that incorporate miRNAs. Note that the linkage between these factors and their associated cell states is correlative; in most of these cases there are additional factors not depicted that help determine these cell identities.
miRNAs as reversible regulators
miRNAs can have substantial effects on target transcript levels, but they also often function as translational repressors. In contrast to irreversible target cleavage or degradation, translational regulation offers the possibility for target mRNA reactivation. Such temporal responsiveness may make translational repression a favoured use of miRNAs, as target mRNA reactivation should be quicker than activating the transcription of a repressed genomic locus. In addition, miRNAs have the theoretical utility of spatially compartmentalized regulation. This might be particularly advantageous in polarized cells, in which local translation of certain proteins is desirable for characteristic cell behaviours.
HuR opposes miR-122 function in liver cells
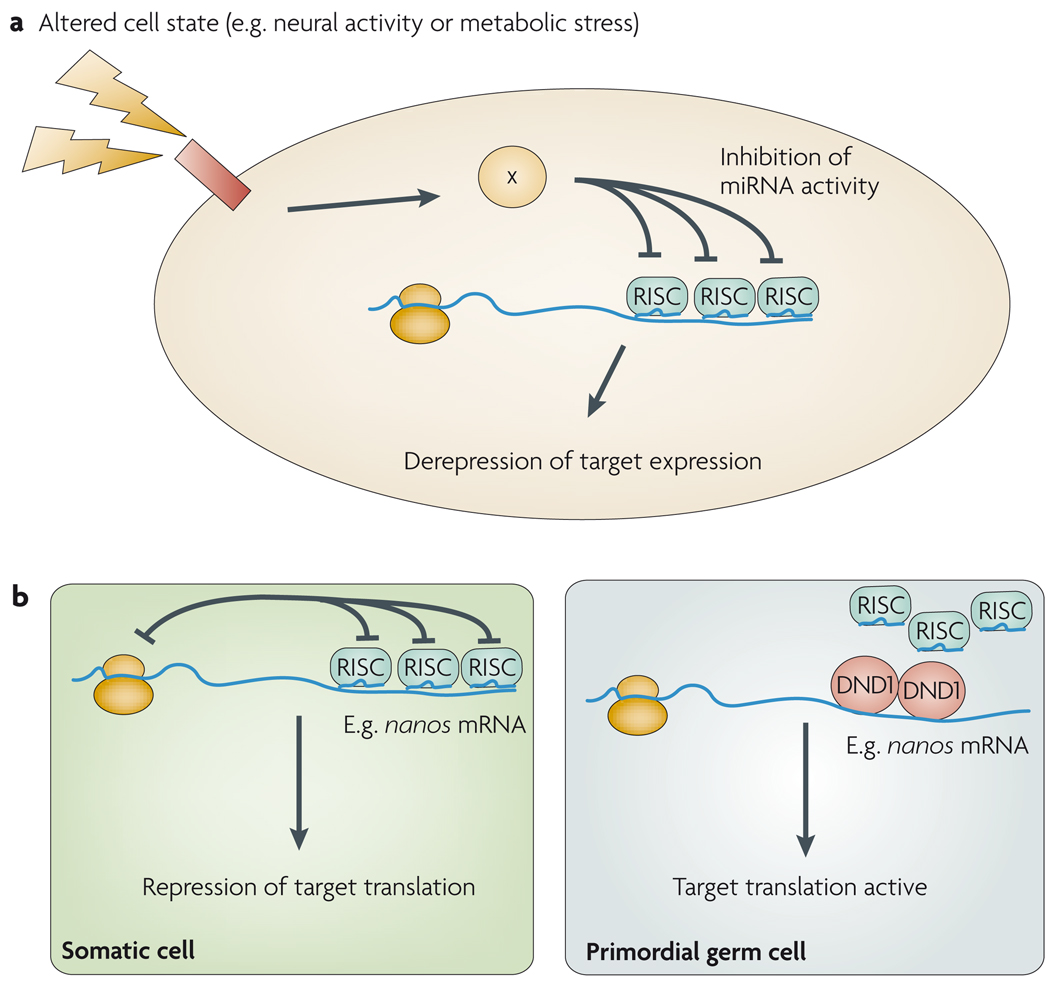
Metabolic pathways are prime candidates for settings in which reversible regulation might be exploited. Rapid responses to changing physiological states might be enabled by the ability to store mRNAs and release them for translation at a moment’s notice. For this to work, there need to be mechanisms that can compete with miRNA-mediated control. The best characterized example of this applies to the liver-specific miR-122. Amongst its targets is the CAT1 amino-acid transporter, the activity of which is generally kept low to minimize arginine hydrolysis by arginase, which is abundant in hepatocytes. However, it is desirable to upregulate CAT1 activity during liver regeneration, to ensure that there are sufficient amino acids for protein synthesis. In this context, the inhibition of CAT1 (also known as SLC7A1) by miR-122 is opposed by the RNA-binding protein HuR during amino-acid deprivation and other stresses, allowing for de novo translation of CAT1 from pre-existing mRNAs76 (FIG. 5a).
Figure 5. Antagonism of microRNA (miRNA)-mediated repression.
a Signal-dependent alleviation of miRNA-mediated repression — for example, at synapses following neural activity, or in response to intracellular signals — might also trigger target re-activation, b Cell-type-specific RNA-binding proteins can antagonize miRNA activity For example, in the germ line, dead end 1 (DND1) can prevent the interaction between a miRNA and targets such as the nanos mRNA. RISC, RNA-induced silencing complex.
Reversal of miRNA repression in neurons
The ability to regulate gene activity at synapses is vital, owing to the tremendous length of the axons that separate these active sites of cell communication from nuclear transcriptional regulation. Local translational regulation at the synapse might allow for rapid activity-dependent changes in gene activity. A study in cultured rat neurons indicated that exposure to brain-derived neurotrophic factors can relieve the repression of Lim domain kinase 1 mRNA by miR-134 (REF. 77). Also, studies in D. melanogaster brains suggest that olfactory stimulation leads to the derepression of CaM kinase II by antagonizing small-RNA-mediated silencing78. Although the mechanisms of miRNA target derepression are not fully understood in these examples, these findings hint at the biological exploitation of reversible miRNA control in neurons (FIG. 5a).
Dead end 1 opposes miRNA–target interactions in germ cells
As mentioned earlier, members of the miR-430 family are ubiquitously active in the early fish embryo and can repress targets in both the early soma and the germ line. It is therefore curious that selected targets, including the translational repressor that is encoded by nanos are repressed in the soma but manage to evade miR-430-mediated repression in primordial germ cells (PGCs)79. How does the animal rescue selected maternal messages from miRNA-mediated silencing? It seems that a major mechanism involves the opposition of germ-cell-expressed miRNAs by the RNA-binding protein Dead end 1 (DND1) (FIG. 5b). Dndl mutation causes germ cell loss in various vertebrates, and in certain mouse backgrounds can lead to testicular germ cell tumours that arise from the few surviving germ cells. DND1 not only opposes miR-430 repression in zebrafish PGCs, it also allows the miR-372 target LATS2 to escape repression in a human germ cell tumour line80.
RNA-binding proteins are notable as one of the largest classes of proteins with biological functions that are generally poorly understood. It seems likely that some of these factors with currently unknown functions will prove to modulate the regulatory capacity of miRNAs, either in particular cells or in response to specific stimuli. miRNA biogenesis itself can also be regulated. For example, the RNA-binding protein LIN-28 was recently shown to inhibit the maturation of let-7-family miRNAs81–83. Finally, although miRNAs are nearly always thought of as repressors, a potentially general role for miRNAs as direct gene activators was defined in cell-cycle-arrested cells84,85. These different strategies and the reversibility of miRNA-mediated control endow miRNAs with regulatory flexibility that has been exploited in diverse biological settings.
Implications of miRNA activities for disease
The principles discussed above have implications for the study of miRNAs in human disease. This is a burgeoning research topic, and we note here only a few recent examples.
The loss of individual miRNAs will derepress targets. At present, it is difficult to predict what consequences that will have for the organism. Systematic analysis of a nematode deletion collection that covers nearly all nematode miRNAs revealed that surprisingly few were obviously required for normal viability, fertility, morphology or behaviour86. Undoubtedly, many of these mutants will eventually prove to be less fit than their wild-type counterparts in some respect. Taken the other way around, the fact that so many miRNA mutants seem to be viable may have substantial implications for disease. For example, mice carrying mutations in haematopoietic-system-expressed miRNAs such as miR-155 (REFS 87,88), miR-150 (REF. 69) or miR-223 (REF. 73) are all viable and fertile, but they have abnormalities in immune cell numbers that can be of substantial consequence following immune challenge. Similarly, mice with mutations in cardiac miRNAs such as miR-1-2 (REF. 89) or miR-208 (REF. 90) are viable and fertile, but they exhibit profound stress-dependent defects in cardiac function and/or remodelling. Although some mammalian miRNA loci are essential (for example, the miR-17-92 cluster74), it seems likely that loss of many individual human miRNA genes will prove to be compatible with life and reproduction but will underlie postnatal cardiac, immune, neurological or metabolic disorders.
As discussed, miRNAs might conceivably exert their major activities through handfuls of key targets or through the sum regulation of large batteries of targets with subtle individual regulation. In cases of the former type, the escape of individual targets from miRNA-mediated regulation may be of particular consequence. A long history of model-organism genetics indicates that very few genes are likely to cause a substantial gain-of-function phenotype simply by losing their 3′ UTRs. On the other hand, it is well worth knowing these few, as they are potentially attractive therapeutic targets. For example, HMGA2 encodes a chromatinassociated protein and contains seven let-7 sites in its 3′ UTR. The specific loss of these miRNA-binding sites strongly potentiated its ability to induce tumours91,92. This correlates well with the observation that various malignant HMGA2 translocation alleles similarly delete the 3′ UTR and let-7 sites. Thus, HMGA2 may represent a type of miRNA genetic switch, especially in cancer cells. Generally speaking, a small number of targets maintain multiple conserved sites for a given miRNA, and many of these have proved to be of substantial genetic importance (BOX 3).
Box 3 Frequent occurrence of multiple target sites in microRNA (miRNA) genetic switches.
It is a challenge to discern individual predicted targets of particular consequence from large numbers of conserved predicted sites. However, it is striking that many of the animal miRNAs that have been identified through the discovery of loss-of-function alleles in forward genetic screens have genetic-switch functions that are substantially mediated by individual targets with multiple conserved sites. These include lin-4–lin-14 (REFS 6,7), lsy-6-cog-1 (REF. 104) and miR-279-nerfin (REF. 70). In fact, these particular targets have the most conserved octomeric sites for the cognate miRNA, according to TargetScan21. Bearded109 and E(spl)m8 (REF. 110) emerged from forward genetic screens as key miRNA targets that have multiple Brd-box miRNA sites or K-box miRNA sites8,9, respectively, and similar trends apply to several miRNAs that have been analysed by reverse genetic screens, such as let-7 sister miRNAs: hbl-1 (REFS 41,42), miR-150-myb (REF. 69) and miR-223–Dmef2c73. This is not to say that individual targets with single binding sites are not important or cannot be genetic switches. A classic example is lin-28, which mediates a genetic-switch function through a single lin-4 target site4. On the other hand, the existence of target genes that maintain exceptional pairing to individual miRNAs may reflect regulatory linkages with biology that is worth investigating.
In the other direction, the overexpression of miRNAs could lead to the potentially detrimental over-repression of targets (FIG. 2d). This is probably of broad consequence, as apparently both genetic-switch and fine-tuning targets are susceptible to strong repression by ectopic miRNAs, yielding target loss-of-function phenotypes32. As miRNAs can operate through as little as seven nucleotides of complementarity, one must also consider the idea that repression through illegitimate sites created by mutations may have phenotypic consequence. For example, the muscular phenotype of the Texel sheep strain is due to a mutation in the myostatin 3′ UTR that creates a binding site for miR-1 and miR-206 — miRNAs that are highly expressed in skeletal muscle93. As a key negative regulator of muscle mass, even slight decreases in myostatin activity yield muscle overgrowth.
A potentially broad connection to human disease was inferred from the location of many miRNA loci near fragile chromosomal sites94. If such aberrations are causally due to miRNA dysfunction in the heterozygous state, it seems likely that many of them either remove repressive cis-regulatory elements or bring into proximity novel transcriptional activation elements that deregulate miRNA expression. The appropriate transcriptional regulation of miRNAs is therefore of great importance. This is well illustrated by Myc, a transcription factor with both activator and repressor function. It was recently reported that there is widespread direct repression of miRNAs by Myc. This may be functionally linked to the oncogenic activity of Myc, as the enforced expression of Myc-repressed miRNAs could suppress Myc-induced tumours95. Conversely, some miRNAs, including the six miRNAs that are encoded by the mir-17-92 Operon, are directly activated by Myc96. This also has clinical consequences, as the mir-17-92 locus is highly amplified in various solid tumours and B cell lymphomas97, and enforced expression of this miRNA Operon accelerates Myc-induced tumorigenesis98. The cis-regulatory control of few miRNAs is known in detail, but these observations indicate that it will be crucial to elucidate both the transcriptional activation and the transcriptional repression of miRNAs.
Conclusions
miRNAs have roles in diverse aspects of plant and animal biology, and careful consideration of how miRNAs are used suggests many parallels with transcription-factor-mediated gene regulation99. There is a fundamental hierarchy to both of these systems. As miRNAs require an RNA substrate to repress, miRNA-mediated regulation acts downstream of transcriptional control. This does not necessarily mean that miRNAs are ‘less important’ than transcription factors. We discussed many examples in which transcriptional regulation is poorly suited or not applicable — for example, at synapses, for maternally deposited transcripts and for messages that require rapid reactivation. Nevertheless, even in these systems miRNAs are not the only solution: proteins can also direct the necessary regulation to great effect. We suggest that it is not constructive to focus on whether transcriptional, post-transcriptional or even post-translational strategies are ‘best’ for any particular system. Instead, the optimal operation of biological systems probably involves the coordinated use of multiple regulatory systems, and many of these will include a miRNA-mediated aspect. But one should bear in mind that the requirement for the miRNA input will vary widely among different settings.
Regarding the commonalities of plant and animal miRNAs, it is clear that both are used to direct developmental processes and physiological responses. Although an early dogma was that animal miRNAs were translational repressors and plant miRNAs were slicers, it is now evident that animal miRNAs can also degrade their targets and plant miRNAs can reciprocally effect translational inhibition. These versatile modes of repression — one reversible, the other not — may underlie the flexible incorporation of miRNAs into a variety of settings.
One difference between plant and animal miRNA seems to have held up regards the general age of miRNA–target interactions. Although many miRNAs have been broadly conserved across the plant or animal kingdoms, only in plants have many miRNA–target interactions been similarly preserved over large evolutionary distances100,101. This may be correlated with the fact that plant miRNAs generally have limited numbers of predicted targets, and with the proposition that plant miRNAs can derive from inverted duplications of their prospective target genes102. Thus, most plant miRNAs that survived the evolutionary test of time seem to have been selected because of their inherent function as dedicated genetic switches, and most of these seem to involve developmental transcription factors.
On the other hand, the targets of highly conserved animal miRNAs seem to evolve comparatively rapidly18. This may reflect a greater exploitation of animal miRNAs relative to plant miRNAs to mediate subtle regulation of many targets. Animal targets might easily be acquired through random mutations that fortuitously create sites. If these ever prove beneficial, their regulatory influence could be positively selected during evolution30,49. Animal miRNA targets are also quite heterogeneous in terms of the quantitative amount and the phenotypic consequence of their regulation by miRNAs (FIG. 4). Thus, although a handful of animal miRNA–target interactions are of the life-or-death variety, most will have only minor effects on the organism.
A pressing challenge for the future will therefore be to determine the extent to which miRNAs directly influence the activity of target genes with predicted conserved sites (or non-conserved sites), and to elucidate the extent to which this has detectable phenotypic consequences. A lesson from both plants and animals is that a miRNA may not need to eliminate a target in order to have a substantial effect on phenotype, if the target itself is highly dose-sensitive. From the disease perspective, knowledge of these genetic-switch targets will prove to be especially valuable, and at least some of them are likely to be genes that have been well studied in the context of development, metabolism and oncogenesis.
Additional genetic studies will be needed to understand whether miRNAs typically regulate only a handful of key targets or coordinately regulate many targets that are simultaneously each of importance. In addition, much remains to be understood about the extent to which co-expressed miRNA–target pairs might be nonfunctional owing to the influence of competing secondary structures or trans-acting factors. Answering these questions will require the ongoing marriage of genetics, molecular and structural biology, biochemistry and bioinformatics, a combination that has provided, and that will undoubtedly continue to provide, fascinating advances in our knowledge of the roles of small regulatory RNAs.
DATABASES.
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
abrupt | atrophin | Bearded | Bim | CAT1 | cog-1 | die-1 | Dndl | Ecdysone receptor | E(spl)m8 | hbl-1 | HMGA2 | LATS2 | Lim domain kinase 1 mRNA | lin-14 | lin-28 | lin-41 | Mej2c | mir-14 | mir-100 | mir-125 | mir-223 | nanos | nerfin | PHABULOSA | PHAVOLUTA | REVOLUTA
FURTHER INFORMATION
TargetScan: http://www.targetscan.org
Acknowledgements
We apologize to the many researchers whose findings were not covered in this Review owing to space limitations. This work was supported by grants from the V Foundation for Cancer Research, the Sidney Kimmel Cancer Foundation, the Alfred Bressler Scholars Fund and the US National Institutes of Health (R01-GM083300).
Glossary
- Argonaute proteins
A family of proteins with PAZ and PIWI domains that directly bind specific small RNAs (including miRNAs, siRNAs and piRNAs). After binding, the small RNA guides the Argonaute, along with its associated protein complex, to regulatory targets.
- Genetic drift
Changes in allele frequency or nucleotide identity that occur entirely by chance.
- Epistasis analysis
A genetic-interaction test that is applied when mutations in two genes yield different phenotypes with respect to a common biological process. One of the genes is said to be epistastic if the phenotype of the double mutant resembles that of the single mutant, as opposed to being intermediate.
- Heterochronic gene
A gene that determines an organism's stage-specific temporal state during its development.
- Imaginal discs
Tissues that, in holometabolous insects such as D. melanogaster, will give rise to the adult structures during metamorphosis.
- Notch
A transmembrane receptor that coordinates a cell–cell signalling cascade that is fundamental to the specification of diverse animal cell fates.
- Canalization
The capacity of individuals within a species to yield similar phenotypic traits despite genetic and/or environmental variability.
- Drosha and Dicer
RNAse III enzymes that catalyse nuclear and cytoplasmic cleavage, respectively of animal miRNA precursors. Plants lack Drosha, and a Dicer homologue called Dicer-like 1 seems to execute both cleavage events in the nucleus during miRNA maturation. Various animal and plant Dicers are also involved in small interfering RNA biogenesis.
References
- 1.Lau N, Lim L, Weinstein E, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 2.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 4.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 5.Wightman B, Burglin TR, Gatto J, Arasu P, Ruvkun G. Negative regulatory sequences in the lin-14 3′-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 1991;5:1813–1824. doi: 10.1101/gad.5.10.1813. [DOI] [PubMed] [Google Scholar]
- 6.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 7. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. References 6 and 7 provide the first formal description of a miRNA (lin-4) and a phenotypically important target (lin-14), which together control developmental timing in nematodes.
- 8.Lai EC, Burks C, Posakony JW. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development. 1998;125:4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- 9. Lai EC, Posakony JW. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–4856. doi: 10.1242/dev.124.23.4847. References 8 and 9 revealed the dominant heptameric logic of animal miRNA binding sites, showed that miRNA binding sites confer both transcript destabilization/de-adenylation and translational inhibition, and demonstrated animal phenotypes that are due to the mutation of miRNA binding sites in single gene targets.
- 10.Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 11.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nature Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 12. Rhoades MW, et al. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. Reference 12 showed that most plant miRNAs identify targets with extended complementarity.
- 13.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Fahlgren N, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 16.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.German MA, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nature Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 18.Brodersen P, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 19.Rajewsky N. MicroRNA target predictions in animals. Nature Genet. 2006;38(Suppl 1):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 20.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 21.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 22. Krek A, et al. Combinatorial microRNA target predictions. Nature Genet. 2005;37:495–500. doi: 10.1038/ng1536. References 21 and 22 are representative analyses that revealed the breadth of miRNA targeting in animal genomes.
- 23.Ruby JG, et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 25.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- 27. Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. Reference 27 provides a convincing example of how the coordinated control of hundreds of targets by a single miRNA family is used to control temporal identity.
- 28.Farh KK, et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 29.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl Acad. Sci. USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nature Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 31.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 32.Bushati N, Cohen SM. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 33.Hristova M, Birse D, Hong Y, Ambros V. The Caenorhabditis elegans heterochronic regulator LIN-14 is a novel transcription factor that controls the developmental timing of transcription from the insulin/insulin-like growth factor gene ins-33 by direct DNA binding. Mol. Cell. Biol. 2005;25:11059–11072. doi: 10.1128/MCB.25.24.11059-11072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruvkun G, et al. Molecular genetics of the Caenorhabditis elegans heterochronic gene lin-14. Genetics. 1989;121:501–516. doi: 10.1093/genetics/121.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ambros V, Horvitz HR. The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes Dev. 1987;1:398–414. doi: 10.1101/gad.1.4.398. [DOI] [PubMed] [Google Scholar]
- 36.Chalfie M, Horvitz HR, Sulston JE. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981;24:59–69. doi: 10.1016/0092-8674(81)90501-8. [DOI] [PubMed] [Google Scholar]
- 37.Arasu P, Wightman B, Ruvkun G. Temporal regulation of lin-14 by the antagonistic action of two other heterochronic genes, lin-4 and lin-28. Genes Dev. 1991;5:1825–1833. doi: 10.1101/gad.5.10.1825. [DOI] [PubMed] [Google Scholar]
- 38.Feinbaum R, Ambros V. The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans. Dev. Biol. 1999;210:87–95. doi: 10.1006/dbio.1999.9272. [DOI] [PubMed] [Google Scholar]
- 39.Abbott AL, et al. The let-7 microRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev. Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 41.Lin SY, et al. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 42.Abrahante JE, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 43.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 44.Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- 45.O'Farrell F, Esfahani SS, Engstrom Y, Kylsten P. Regulation of the Drosophila lin-41 homologue dappled by let-7 reveals conservation of a regulatory mechanism within the LIN-41 subclade. Dev. Dyn. 2008;237:196–208. doi: 10.1002/dvdy.21396. [DOI] [PubMed] [Google Scholar]
- 46.Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr. Biol. 2008;18:943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen K, Rajewsky N. Deep conservation of microRNA-target relationships and 3′UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb. Symp. Quant. Biol. 2006;71:149–156. doi: 10.1101/sqb.2006.71.039. [DOI] [PubMed] [Google Scholar]
- 49.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nature Rev. Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 50.McConnell JR, et al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 51.Emery JF, et al. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 52.Baker CC, Sieber P, Wellmer F, Meyerowitz EM. The early extra petals 1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 2005;15:303–315. doi: 10.1016/j.cub.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 53.Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development. 2007;134:1051–1060. doi: 10.1242/dev.02817. [DOI] [PubMed] [Google Scholar]
- 54.Cartolano M, et al. A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nature Genet. 2007;39:901–905. doi: 10.1038/ng2056. [DOI] [PubMed] [Google Scholar]
- 55.Axtell MJ. Evolution of microRNAs and their targets: are all microRNAs biologically relevant? Biochim. Biophys. Acta. 2008 Mar 10; doi: 10.1016/j.bbagrm.2008.02.007. (doi:10.1016/j.bbagrm.2008.02.007). [DOI] [PubMed] [Google Scholar]
- 56.Lai EC. microRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nature Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 57.Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 59.Nolo R, Abbott L, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- 60.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. Reference 61 provides evidence for a miRNA tuning target — continuing function of the target in a cognate miRNA-expressing domain was demonstrated phenotypically.
- 62.Hornstein E, Shomron N. Canalization of development by microRNAs. Nature Genet. 2006;38(Suppl 1):S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 63.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 64.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 66.Tadros W, et al. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev. Cell. 2007;12:143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 67.Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 68.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc. Natl Acad. Sci. USA. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 70.Cayirlioglu P, et al. Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science. 2008;319:1256–1260. doi: 10.1126/science.1149483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Varghese J, Cohen SM. MicroRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 2007;21:2277–2282. doi: 10.1101/gad.439807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnnidis JB, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 74.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koralov SB, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 76. Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. Reference 76 demonstrates stress-dependent reversibility of miRNA-mediated repression.
- 77.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 78.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 79.Mishima Y, et al. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kedde M, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 81.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 84.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 86.Miska EA, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 89.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 90.van Rooij E, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 91.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Clop A, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nature Genet. 2006;38:813–818. doi: 10.1038/ng1810. Reference 93 provides convincing evidence that the de novo acquisition of a miRNA binding site can have phenotypc consequences.
- 94.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chang TC, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nature Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 97.Ota A, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 98. He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. References 96, 97 and 98 show that a mammalian miRNA cluster exhibits oncogenic activity.
- 99.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 100.Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Floyd SK, Bowman JL. Gene regulation: ancient microRNA target sequences in plants. Nature. 2004;428:485–486. doi: 10.1038/428485a. [DOI] [PubMed] [Google Scholar]
- 102.Allen E, et al. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nature Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- 103.Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 104.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 105.Chang S, Johnston RJ, Jr, Frokjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- 106.Johnston RJ, Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl Acad. Sci. USA. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johnson SM, Lin SY, Slack FJ. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- 109.Leviten MW, Lai EC, Posakony JW. The Drosophila gene Bearded encodes a novel small protein and shares 3′ UTR sequence motifs with multiple Enhancer of split complex genes. Development. 1997;124:4039–4051. doi: 10.1242/dev.124.20.4039. [DOI] [PubMed] [Google Scholar]
- 110.Klämbt C, Knust E, Tietze K, Campos-Ortega J. Closely related transcripts encoded by the neurogenic gene complex Enhancer of split of Drosophila melanogaster. EMBO J. 1989;8:203–210. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]