Abstract
Methadone (MD) is the most established substance abuse pharmacotherapy of choice for the management of heroin dependence. To date, drug-drug interactions involving MD have been characterized asymmetrically among existing reports, which describe how other drugs affect the metabolic or pharmacokinetic profiles of MD; however, limited information is available regarding the potential for MD to influence similar fates of coadministered drugs. Moreover, little to no mechanistic evidence has been explored. Here, we show that MD induces hepatic drug-metabolizing enzymes (DMEs) through the activation of pregnane X receptor (PXR) and constitutive androstane receptor (CAR). Real-time polymerase chain reaction analysis of human hepatocyte cultures revealed that MD induces the mRNA expression of CYP2B6, CYP3A4, UGT1A1, and multidrug resistance 1 in a concentration-related manner, with the maximal induction of CYP2B6 challenging that of the induction by rifampicin. Furthermore, MD-mediated induction of CYP2B6 and CYP3A4 proteins was observed in Western blot analysis. In cell-based reporter assays, MD significantly increased human (h) PXR-mediated CYP2B6 reporter activities but exhibited minimal effect on hCAR activation as a result of the constitutive activity of hCAR in HepG2 cells. Further studies revealed that treatment with MD resulted in significant nuclear accumulation of adenovirus/enhanced yellow fluorescent protein tagged-hCAR in human hepatocytes, which has been regarded as the initial step of CAR activation. Additional analysis of the two enantiomers of MD, R-(–)-MD (active) and S-(+)-MD (inactive), indicates the lack of stereoselectivity pertaining to MD-mediated DME induction. Overall, our results show that MD induces the hepatic expression of multiple DMEs by activating PXR- and CAR-mediated pathways.
Methadone (MD), a synthetic opioid that possesses enduring effects as a result of its long half-life, is a critical clinical drug therapy that continues to be used for the management of heroin addiction and the treatment of chronic pain. Approximately 20% of an estimated 810,000 addicts in the United States receive long-term MD maintenance treatment (American Methadone Treatment Association, 1999), and often MD maintenance patients consume a myriad of concomitant medication as part of an aggressive polypharmacy approach of therapy. For example, MD users are likely also treated with pain medications, such as oxycodone and codeine, or antiretroviral therapies, such as efavirenz and ritonavir (Fornataro, 1999; Ferrari et al., 2004). Thus, the potential for drug-drug interactions (DDIs) involving MD is high.
MD is almost exclusively metabolized in the liver (Nilsson et al., 1982). The primary metabolic route involves hepatic N-demethylation and cyclization to its stable metabolite, 2-ethyl-1,5-dimethyl-3,3-diphenylpyrrolidine, which is pharmacologically inactive (Ferrari et al., 2004). Some controversy exists in the literature regarding which cytochrome P450 (P450) isoform(s) plays the primary role in hepatic metabolism and clearance of MD. In previous studies, the predominant mediator of MD metabolism was thought to be CYP3A4, and numerous in vitro and in vivo studies have shown CYP3A4 involvement in the metabolism of MD in human liver or intestines (Wang and DeVane, 2003; Gerber et al., 2004; Kharasch et al., 2004). However, recent studies suggest that CYP2B6 is also involved in the metabolism of MD and may possess a higher affinity for MD metabolism compared with that of CYP3A4 (Gerber et al., 2004; Kharasch et al., 2008; Weschules et al., 2008). Dramatic variations in the pharmacokinetic profile of MD are speculated to be largely the result of the variable expression and activity of CYP2B6 and CYP3A4, given that both of these P450s are highly inducible isozymes that exhibit significant interindividual variations (Ingelman-Sundberg et al., 2007).
Notably, although MD has been used extensively for several decades for the management of narcotic dependence, characterization of its interaction with other coadministered or coabused drugs is largely incomplete and heavily one-sided. To date, mounting literature, which heavily focuses on describing how other drugs affect the metabolic and pharmacokinetic profiles of MD, has been published (Kharasch et al., 2008; Linderbeck, 2008; Weschules et al., 2008). For example, a number of antiretroviral drugs, such as efavirenz and nevirapine, have been identified as potent inducers of CYP2B6 and CYP3A4, whereas ritonavir and indinavir are efficacious inhibitors of CYP3A4. Subsequently, cotreatment of these antiretroviral drugs with MD has resulted in decrease or increase of the plasma concentrations of MD in HIV-positive heroin addicts (Clarke et al., 2001; Ernest et al., 2005; Faucette et al., 2007). Conversely, only limited data exist regarding the effects of MD on the metabolic fate of other coadministered drugs, and little to no mechanistic evidence has been explored.
Metabolism-based DDIs have received increasing attention during the past 2 decades, in large part because of the increased incidence of multidrug therapy necessitated by overlapping disease states. The majority of clinically significant DDIs occur via induction or inhibition of drug-metabolizing enzymes (DMEs). In the case of enzyme induction, increased expression and activity of DMEs such as P450s can lead to accelerated drug clearance and premature termination of drug actions. Furthermore, enzyme induction is often governed by the activation of several nuclear receptors (NRs), such as pregnane X receptor (PXR) and constitutive androstane receptor (CAR), which coordinately control the drug-induced expression of multiple DMEs and drug transporters, including the highly inducible CYP2B6 and CYP3A4 (Sueyoshi et al., 1999; Moore et al., 2000).
The primary objective of the current study was to characterize the effects of MD on the expression of hepatic DMEs and transporters, as well as the activation of the xenobiotic sensors PXR and CAR. MD-mediated induction of CYP2B6, CYP3A4, UGT1A1, and multidrug resistance 1 (MDR1) was assessed in human primary hepatocytes, which express physiologically relevant liver-enriched transcription factors. Cell-based reporter assays in HepG2 cells were used to determine the differential activation of NRs. A newly generated adenovirus/enhanced yellow fluorescent protein tagged-human CAR (Ad/EYFP-hCAR) was used to assess drug-induced hCAR nuclear accumulation in human primary hepatocytes (Li et al., 2009a). Overall, our results show that MD induces the hepatic expression of multiple DMEs through the activation of PXR- and CAR-mediated pathways.
Materials and Methods
Chemicals and Reagents. MD, PK11195, and rifampicin (RIF) were purchased from Sigma-Aldrich (St. Louis, MO), and 6-(4-chlorophenyl)imidazo-[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichloro-benzyl)-oxime (CITCO) was obtained from BIOMOL Research Laboratories (Plymouth Meeting, PA). Methadone enantiomers, R-(–)-MD and S-(+)-MD, were provided by the National Institute on Drug Abuse Drug Supply Program (Bethesda, MD). Oligonucleotide primers and TaqMan fluorescent probes were synthesized by Sigma-Genosys (The Woodlands, TX) and Applied Biosystems (Foster City, CA), respectively. The Dual-Luciferase Reporter Assay System was purchased from Promega (Madison, WI). Matrigel, insulin, and ITS+ were obtained from BD Biosciences Discovery Labware (Bedford, MA). Horseradish peroxidase-labeled anti-rabbit antibody was purchased from GE Healthcare (Little Chalfont, Buckinghamshire, UK). Other cell culture reagents were purchased from Invitrogen (Carlsbad, CA) or Sigma-Aldrich.
Plasmid Constructs and Ad/EYFP-hCAR. The CYP2B6 reporter constructs, containing both PBREM and the distal XREM [CYP2B6-2.2 kilobases (kb)], were generated as described previously (Wang et al., 2003). The pCR3-hCAR and the EYFP-hCAR expression plasmids were provided by Dr. Masahiko Negishi (National Institute of Environmental and Health Sciences, National Institutes of Health, Research Triangle Park, NC). The pSG5-hPXR and CMV2-hCAR3 expression plasmids were acquired from Dr. Steve Kliewer (University of Texas, Southwestern Medical Center, Dallas, TX) and Dr. Curtis Omiecinski (The Pennsylvania State University, University Park, PA), respectively. The Ad/EYFP-hCAR was generated as described previously (Li et al., 2009a). The pRL-TK Renilla luciferase plasmid used to normalize firefly luciferase activities was purchased from Promega.
Induction Studies in Human Primary Hepatocyte Cultures. Human liver tissues were obtained following surgical resection by qualified pathology staff after diagnostic criteria were met, and approval was obtained previously from the Institutional Review Board at the University of Maryland at Baltimore. Hepatocytes were isolated by a modification of the two-step collagenase digestion method as described previously (LeCluyse et al., 2005). Hepatocytes were seeded at 1.5 × 106 cells/well in six-well BioCoat (BD Biosciences, San Jose, CA) plates in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 4 μg/ml insulin, and 1 μM dexamethasone. After attachment at 37°C in a humidified atmosphere of 5% CO2, hepatocytes were cultured in complete Williams' E medium and then overlaid with Matrigel (0.25 mg/ml). Hepatocytes were maintained for 36 h before treatment with different compounds.
Real-Time Polymerase Chain Reaction Analysis. Total RNA was isolated using the RNeasy Mini Kit (QIAGEN, Valencia, CA) and reverse-transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems) per manufacturers' instructions. Primers and probes for CYP2B6, CYP3A4, UGT1A1, and MDR1 genes (Table 1) were designed using Primer Express version 2.0 (Applied Biosystems) and entered into the National Center for Biotechnology Information BLAST to ensure specificity as described previously (Maglich et al., 2002; Smith et al., 2005; Faucette et al., 2007; Li et al., 2008). The mRNA expression of CYP2B6, CYP3A4, UGT1A1, and MDR1 was normalized against that of human β-actin, which was detected using a predeveloped primer/probe mixture (Applied Biosystems). TaqMan polymerase chain reaction (PCR) assays were performed in 96-well optical plates on an ABI Prism 7000 Sequence Detection System (Applied Biosystems). -Fold induction values were calculated according to the equation 2ΔΔCT, where ΔCT represents the differences in cycle threshold numbers between the target gene and β-actin, and ΔΔCT represents the relative change in these differences between control and treatment groups.
TABLE 1.
Primer and probe sequences for real-time PCR assays
| Gene | Sequence | Reference |
|---|---|---|
| CYP2B6 | Faucette et al., 2007 | |
| Forward primer | 5-AAGCGGATTTGTCTTGGTGAA-3 | |
| Reverse primer | 5-TGGAGGATGGTGGTGAAGAAG-3 | |
| Probe | 6-FAM-CATCGCCCGTGCGGAATTGTTC-TAMRA | |
| CYP3A4 | Faucette et al., 2007 | |
| Forward primer | 5-TCAGCCTGGTGCTCCTCTATCTAT-3 | |
| Reverse primer | 5-AAGCCCTTATGGTAGGACAAAATATTT-3 | |
| Probe | 6-FAM-TCCAGGGCCCACACCTCTGCCT-TAMRA | |
| UGT1A1 | Smith et al., 2005 | |
| Forward primer | 5-GGCCCATCATGCCCAATAT-3 | |
| Reverse primer | 5-TTCAAATTCCTGGGATAGTGGATT-3 | |
| Probe | 6-FAM-TTTTTGTTGGTGGAATCAACTGCCTTCAC-TAMRA | |
| MDR1 | Maglich et al., 2002 | |
| Forward primer | 5-GTCCCAGGAGCCCATCCT-3 | |
| Reverse primer | 5-CCCGGCTGTTGTCTCCAT-3 | |
| Probe | 6-FAM-TGACTGCAGCATTGCTGAGAACATTGC-TAMRA |
Transient Transfection in HepG2 Cells. HepG2 cells seeded in 24-well plates were transfected with CYP2B6-2.2 kb reporter construct in the presence of hPXR, hCAR1, or hCAR3 expression vector using FuGENE 6 Transfection Kit (Roche Applied Science, Indianapolis, IN) according to the manufacturer's protocol. Twenty-four hours post-transfection, cells were treated with solvent [0.1% dimethyl sulfoxide (DMSO)] or test compounds at the concentrations of RIF, 10 μM; CITCO, 1 μM; or MD, 10, 25, or 50 μM for another 24 h. Subsequently, cell lysates were assayed for firefly activities normalized against the activities of Renilla luciferase using the Dual-Luciferase Kit (Promega). Data are represented as mean ± S.D. of three individual transfections.
Translocation of Ad/EYFP-hCAR in Human Primary Hepatocyte Cultures. Human hepatocytes were seeded at 3.75 × 105 cells/well in 24-well BioCoat plates (BD Biosciences) and cultured as described previously (Wang et al., 2003). Twenty-four hours later, hepatocyte cultures were infected with Ad/EYFP-hCAR for 12 h before treatment with vehicle control (0.1% DMSO) or test compounds for another 12 h. Confocal laser scanning microscopy was performed with a Nikon (Melville, NY) C1-LU3 instrument based on an inverted Nikon Eclipse TE2000 microscope. The subcellular localization of Ad/EYFP-hCAR was visualized and quantitatively characterized as nuclear (N), cytosolic (C), and mixed (N + C) by counting 100 Ad/EYFP-hCAR-expressing hepatocytes from each group.
Western Immunoblot Analysis. Homogenate proteins (40 μg each) from treated human hepatocytes were separated on a NuPAGE 4–12% Bis-Tris Gel (Invitrogen) and transferred onto polyvinylidene difluoride Transfer Membrane (Pierce, Rockford, IL). Subsequently, membranes were incubated with specific antibodies against CYP2B6 or CYP3A4 (Millipore Bioscience Research Reagents, Temecula, CA) diluted 1:4000 and 1:5000, respectively. β-Actin (Sigma-Aldrich) was used as internal control. Blots were washed and incubated with horseradish peroxidase goat anti-rabbit IgG antibody diluted 1:4000. Films were developed using enhanced chemiluminescence Western blot detection reagent (GE Healthcare, Little Chalfont, Buckinghamshire, UK).
Statistical Analysis. All the data represent three independent measurements and are expressed as the mean ± S.D. Statistical comparisons were made using the Student's t test, and statistical significance was assessed at three levels (***, p ≤ 0.001; **, p ≤ 0.01, and *, p ≤ 0.05).
Results
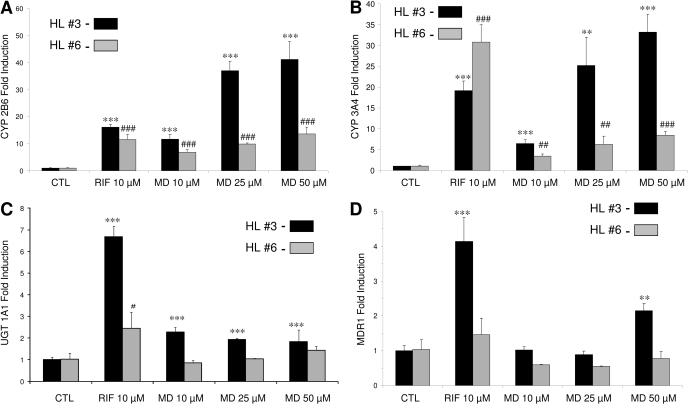
Induction of DMEs and Transporter mRNA Expression in Human Primary Hepatocytes. In the current study, the effects of MD on the expression of CYP2B6, CYP3A4, UGT1A1, and MDR1 genes were evaluated in human primary hepatocyte cultures using real-time reverse transcription-PCR analysis. In human hepatocytes prepared from two donors (HL3, HL6), the mRNA expression of CYP2B6, CYP3A4, UGT1A1, and MDR1 was increased significantly after treatment with MD at 10, 25, and 50 μM (Fig. 1). It is noteworthy that the maximal induction of CYP2B6 at higher concentrations of MD treatment equals or exceeds that induced by the positive control RIF (10 μM). Potent induction of CYP3A4 mRNA was also observed in HL3, where 6-, 25-, and 33-fold CYP3A4 mRNA increases resulted from 10, 25, and 50 μM MD exposures, respectively. Compared with the highly inducible CYP2B6 and CYP3A4 genes, the induction of UGT1A1 and MDR1 was relatively moderate (Fig. 1, C and D). As expected, the positive control RIF efficiently induced CYP2B6, CYP3A4, UGT1A1, and MDR1 expression in both hepatocyte preparations, despite obvious interindividual variations.
Fig. 1.
Induction of DMEs and drug transporter in human primary hepatocytes. Human hepatocytes (HL3 and HL6) cultured in Williams' E medium were treated for 24 h with vehicle, RIF (10 μM), CITCO (1 μM), or MD (10, 25, and 50 μM). Total RNA was isolated, reverse-transcribed, and subjected to TaqMan real-time PCR. The expression levels of mRNA for CYP2B6 (A), CYP3A4 (B), UGT1A1 (C), and MDR1 (D) were determined and normalized against β-actin. Induction of these enzymes relative to vehicle control was calculated as described under Materials and Methods. All the data are expressed as mean ± S.D. (n = 3). *** and ###, p ≤ 0.001; ** and ##, p ≤ 0.01; * and #, p ≤ 0.05.
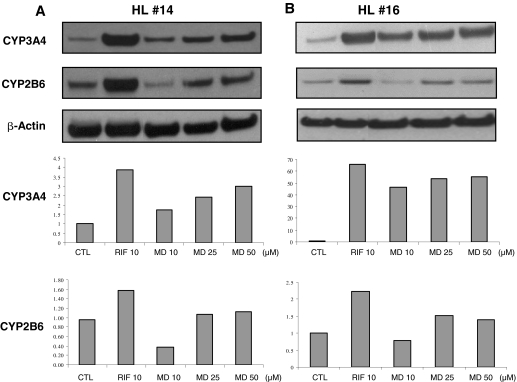
Induction of CYP2B6 and CYP3A4 Protein in Human Primary Hepatocytes. To assess whether MD could induce CYP2B6 and CYP3A4 expression at the protein level, whole-cell homogenate isolated from two preparations of human primary hepatocytes (HL14, HL16) treated with different concentrations of MD was analyzed for CYP2B6 and CYP3A4 protein content by Western blot analysis. As shown in Fig. 2, MD robustly induced the protein expression of CYP3A4 in a dose-dependent manner, whereas the CYP2B6 protein was only induced at higher concentrations of MD (25 and 50 μM) to a relatively minor extent (Fig. 2, A and B). In both hepatocyte preparations, RIF displayed the greatest extent of CYP2B6 and CYP3A4 protein induction.
Fig. 2.
Effects of MD on the expression of CYP3A4 and CYP2B6 immunoreactive proteins. Human hepatocytes, HL14 (A) and HL16 (B), were treated for 72 h with vehicle control, RIF (10 μM), or MD (10, 25, and 50 μM). Whole-cell homogenates were subjected to Western blot and densitometric analyses as described under Materials and Methods.
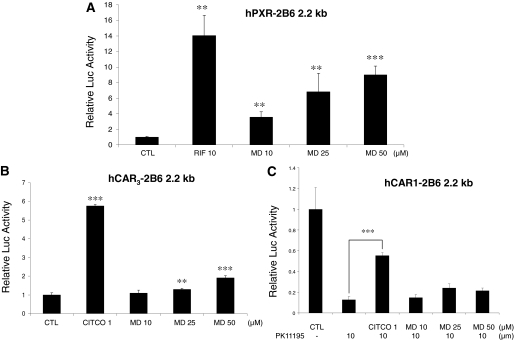
Activation of hPXR and hCAR by Racemic MD. Drug-induced expression of CYP2B6 and CYP3A4 is predominantly controlled at the transcriptional level by the two NRs hCAR and hPXR. Here, we investigated the ability of racemic MD to activate these receptors in cell-based reporter assays conducted in HepG2 cells. As shown in Fig. 3A, MD showed significant increases of hPXR-mediated CYP2B6 reporter activities over the concentration range of 10 to 50 μM, where significant CYP2B6 and CYP3A4 gene induction was observed in human primary hepatocytes. In contrast to hPXR, in vitro assessment of hCAR activation has been problematic because of the constitutive activation nature of CAR in all the immortalized cell lines. More recently, however, several reports indicate that an hCAR splicing variant (hCAR3) displays low basal activity while still retaining chemical-mediated activation in several cell lines (Auerbach et al., 2005; Faucette et al., 2007). Thus, in the current study, we also evaluated hCAR3 activation in HepG2 cells. As expected, the hCAR-selective agonist CITCO strongly enhanced hCAR3 activity, whereas MD resulted in moderate but statistically significant activation of hCAR3 (Fig. 3B). In addition, earlier work conducted in this laboratory showed that the constitutive activity of the wild-type hCAR in HepG2 cells could be substantially repressed by PK11195, a typical ligand for peripheral benzodiazepine receptor, and this inhibitory effect was only recovered by cotreatment with direct hCAR activator CITCO but not by indirect activators such as phenobarbital (PB) (Li et al., 2008). Using this system, our current results showed that MD has no effect on the PK11195-mediated deactivation of hCAR (Fig. 3C).
Fig. 3.
Effects of MD on hPXR-, hCAR3-, and hCAR-mediated CYP2B6 reporter gene activation. HepG2 cells were transfected with hPXR (A), hCAR3 (B), or hCAR (C) expression vectors in the presence of CYP2B6-2.2 kb reporter construct. Transfected cells were then treated with vehicle or MD (10, 25, or 50 μM) for 24 h. RIF (10 μM) and CITCO (1 μM) were used as positive controls for hPXR and hCAR, respectively. Luciferase activities were determined and expressed relative to vehicle control. Data represent the mean ± S.D. (n = 3). ***, p ≤ 0.001; **, p ≤ 0.01.
Both R-(–)-MD and S-(+)-MD Contribute to the Activation of hPXR in HepG2 Cells. MD is clinically administered as a mixture of two stereoisomers, R-(–)-MD and S-(+)-MD, with opioid activity residing in the R-enantiomer. To assess the contribution of each enantiomer to the observed increases of hPXR-mediated CYP2B6 reporter activities, similar reporter assays in HepG2 cells were conducted as described above using R-(–)-MD (active) and S-(+)-MD (inactive). As shown in Fig. 4, both the active R-enantiomer and the inactive S-enantiomer achieved potent and concentration-related activation of hPXR, with maximal activity occurring at 50 μM. These results suggest that although the metabolism and clearance of racemic MD are highly variable, the potential for each enantiomer to achieve DME induction is most likely nonstereoselective.
Fig. 4.
MD enantiomers increase the activities of hPXR in HepG2 cells. HepG2 cells were transfected with hPXR expression vector in the presence of CYP2B6-2.2 kb reporter construct, then treated with vehicle control, R-(–)-MD (A) or S-(+)-MD (B) (10, 25, or 50 μM) for 24 h. RIF (10 μM) was used as positive control. Luciferase activities were determined and expressed relative to vehicle control. Data represent the mean ± S.D. (n = 3). ***, p ≤ 0.001; **, p ≤ 0.01.
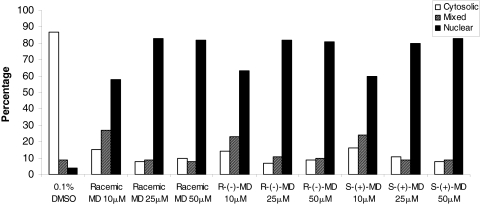
Translocation of Ad/EYFP-hCAR by MD in Human Primary Hepatocytes. The high constitutive activation of hCAR in immortalized cell lines can be attributed to the spontaneous nuclear accumulation of hCAR in these cells. In contrast, in primary hepatocytes and in vivo, CAR is sequestered predominantly in the cytoplasm and translocated to the nucleus only after exposure to xenobiotics. More recently, our laboratory has generated an Ad/EYFP-hCAR, which showed exceptional efficiency in transducing human primary hepatocytes (Li et al., 2009a). Using this system, we sought to determine whether MD can translocate hCAR as the initial step of hCAR activation. Cultured hepatocytes were infected with Ad/EYFP-hCAR and treated with vehicle control, known hCAR indirect activator PB, or 50 μM MD. Fluorescence microscopy analysis showed that both PB and MD treatment result in abundant nuclear accumulation of hCAR (Fig. 5A). Of the hepatocytes expressing hCAR prepared from two liver donors (HL8, HL9), 87 to 93% displayed cytoplasmic localization and 7 to 13% exhibited mixed (cytoplasm and nucleus) localization, whereas cells in the vehicle the control group showed pure nuclear localization (Fig. 5B). The Ad/EYFP-hCAR expression was predominantly accumulated inside the nucleus after the treatment with known indirect hCAR activator PB, where 63 to 80% exhibited nuclear localization, 7 to 38% exhibited a mixed distribution pattern, and only 0 to 13% displayed cytoplasmic localization. It is noteworthy that MD (50 μM) treatment also resulted in 62 to 81% nuclear, 11 to 13% mixed, and 8 to 25% cytoplasmic localizations (Fig. 5B). Subsequently, a parallel experiment was conducted to expand the scope of MD-mediated hCAR translocation by including both racemic MD and its constituent isomers at 10, 25, and 50 μM concentrations. As shown in Fig. 6, all the isomers of MD translocated hCAR efficiently from the inactive cytoplasmic localization to the nucleus at all three concentrations. The maximal translocation extent mimicked that of the positive control PB. Overall, these results indicate that MD and its constituent isomers are capable of accumulating hCAR inside the nucleus of human hepatocytes in a nonstereoselective manner.
Fig. 5.
Methadone translocates Ad/EYFP-hCAR in human primary hepatocytes. Human hepatocytes (HL8 and HL9) were infected with Ad/EYFP-hCAR as described under Materials and Methods and treated with vehicle, PB (1 mM), or MD (50 μM). After 24 h of treatment, hepatocytes were subjected to confocal microscopy. A, representative Ad/EYFP-hCAR localizations from vehicle control, PB- (1 mM), and MD- (50 μM) treated hepatocytes. B, for each treatment, more than 60 hCAR-expressing cells were counted and classified based on cytosolic, nuclear, or mixed (cytosolic + nuclear) hCAR localizations.
Fig. 6.
MD and constituent isomers promote nuclear translocation of Ad/EYPF-hCAR in human hepatocytes. Hepatocytes were infected with Ad/EYFP-hCAR and treated with vehicle (0.1% DMSO) or racemic MD and its constituent isomers at 10, 25, and 50 μM for 24 h. For each treatment group, 100 hCAR-expressing cells were counted and classified based on cytosolic, nuclear, or mixed (cytosolic + nuclear) hCAR distributions.
Discussion
MD is widely prescribed for the management of heroin dependence and different types of chronic pain. Because of frequent coadministration with other therapeutics, DDIs involving MD resulting from a polypharmacy approach to therapy are common. Nevertheless, mounting evidence shows thus far that MD drug interactions have been characterized asymmetrically among existing literature, where focus has been placed heavily on describing how other drugs affect the metabolic or pharmacokinetic profiles of MD. However, limited data exist regarding the potential for MD to influence similar fates of coadministered drugs, and little to no mechanistic evidence has been provided. Although MD is not a new medication, to our knowledge, the current study is the first to show that MD can induce the expression of multiple hepatic DMEs through the activation of PXR- and CAR-dependent pathways.
Sandwich cultures of human primary hepatocytes have been used extensively for evaluating the ability of drugs to induce the expression and activity of DMEs and transporters in humans (LeCluyse, 2001). Enzyme induction in human primary hepatocytes possesses the distinct advantage of mimicking physiological in vivo conditions and exhibiting species-specific induction patterns. Using human primary hepatocytes as a model, our data revealed that the expression of CYP2B6 and CYP3A4 was potently and dose-dependently induced after treatment with MD, whereas the expression of phase II enzyme UGT1A1 and efflux transporter MDR1 was only moderately induced. Given that the clinical use or abuse of MD is usually chronic, and prescribed dosages could range from approximately 30 to 180 mg/day up to as much as 1300 mg/day under certain circumstances (Ali and Woods, 2008), exposure levels could vary dramatically. The current in vitro studies have tested MD over a concentration range of 10 to 50 μM. Although it is difficult to quantitatively correlate in vitro data with in vivo conditions, our findings lead to the speculation that MD may also induce CYP2B6 and CYP3A4 expression to a clinically significant level in vivo. Because the occurrence of HIV disease is common among injection drug users and opioid dependants who often require MD maintenance therapy, DDIs occurring between MD and anti-HIV agents have been investigated previously. However, the particular DDI between MD and antiretroviral drugs has been commonly examined in a unilateral manner, where anti-HIV agents have been reported to affect MD disposition (Clarke et al., 2001; Kharasch et al., 2009), but little has been written about how MD can affect antiretroviral drugs. A number of anti-HIV agents such as efavirenz and nevirapine are primarily metabolized in the liver by CYP3A4 and CYP2B6 (Desta et al., 2007). Thus, MD induction of CYP2B6 and CYP3A4 may result in altered pharmacokinetics of such agents and may contribute to the frequently observed efavirenz adaptation in clinical settings. Likewise, MD itself is metabolized predominantly by CYP2B6, CYP3A4, and CYP2D6, with CYP2B6 exhibiting the highest affinity and efficacy (Gerber et al., 2004; Kharasch et al., 2004). Therefore, MD-mediated autoinduction of P450s may enhance the clearance of MD itself. Overall, the current in vitro observations warrant further in vivo and clinical investigations, with a focus on how the long-term administration of MD may affect the efficacy and toxicity of concomitantly administered medications.
Transcriptional regulation of CYP2B6 and CYP3A4, as well as a number of other DMEs and drug transporters, has been attributed to the cross-talk of the two major xenobiotic receptors, PXR and CAR. In response to structurally diverse xenobiotics, hPXR displays promiscuous ligand binding ability and indiscriminant induction of CYP2B6 and CYP3A4, whereas hCAR exhibits limited ligand binding capacity with preferential induction of CYP2B6 over CYP3A4 (Faucette et al., 2006, 2007). In delineating the molecular mechanisms underlying MD-mediated induction of CYP2B6 and CYP3A4, our cell-based reporter assays in HepG2 cells showed that treatment of MD resulted in potent and concentration-related activation of hPXR-mediated luciferase reporter gene expression. Given that activation of PXR coordinately induces the expression of a plethora of DMEs and drug transporters besides CYP2B6 and CYP3A4, MD treatment holds great potential for causing DDIs by interacting with a broader spectrum of DMEs and transporters.
In contrast to PXR, CAR is spontaneously accumulated in the nucleus and constantly activated in immortalized cells before xenobiotic activation, which makes the in vitro assessment of hCAR activation extremely difficult. More recently, one of the hCAR splicing variants (hCAR3) has been reported as ligand-responsive hCAR with low constitutive activity in several cell lines (Auerbach et al., 2005; Faucette et al., 2007). In addition, recent work conducted in this laboratory showed that the constitutive activity of wild-type hCAR in HepG2 cells was sufficiently repressed by hCAR antagonist PK11195, and this repression was reactivated only by the hCAR agonist CITCO but not by indirect activators such as PB (Li et al., 2008). Using the hCAR3 reporter and PK11195-based hCAR reactivation assays, our data showed that MD mediated a moderate but concentration-related activation of hCAR3 while failing to reactivate PK11195-suppressed hCAR activity in HepG2 cells, indicating that MD may not function as a direct agonist of hCAR. Nevertheless, it is noteworthy that the majority of known hCAR activators are PB-like compounds that activate hCAR through indirect mechanisms without direct ligand binding. As a matter of fact, CITCO and artemisinin are the only two hCAR agonists identified thus far (Maglich et al., 2003; Simonsson et al., 2006). Compared with the cell-based hCAR reporter assays, chemical-mediated hCAR nuclear translocation in hepatocytes of primary culture or in vivo appears to correlate well with hCAR activation and target gene induction, regardless of the distinction between direct or indirect mechanisms (Wang et al., 2004; Faucette et al., 2007). However, in vitro transfection of human primary hepatocytes has been extremely challenging mostly because of the quiescent nature of hepatocytes in cultures. Most recently, we have generated an Ad/EYFP-hCAR that transduces human primary hepatocytes with high efficiency and exhibits a physiologically relevant hCAR distribution pattern (Li et al., 2009a). Further evaluation of MD in Ad/EYFP-hCAR-infected human primary hepatocytes revealed that MD treatment resulted in remarkable nuclear accumulation of hCAR at all the tested concentrations, which achieves an extent similar to the positive control PB. Combined with the reporter assays, these results indicate that MD actives hCAR most likely through indirect, PB-like mechanisms.
MD is clinically administered as a racemic mixture of two stereoisomers with only the R-enantiomer possessing opioid activity. Several lines of evidence indicate that MD metabolism and clearance are stereoselective, where CYP2B6, but not CYP3A4, contributes significantly to the highly variable plasma R/S-MD ratios (Totah et al., 2007). To gain insight into the potential stereoselectivity between R-(–)-MD and S-(+)-MD in the induction of DMEs, the current study further evaluated the active R-(–)-MD and the inactive S-(+)-MD isomers for their activation of hPXR and hCAR. Intriguingly, both isomers robustly activated hPXR in cell-based reporter assays in HepG2 cells and translocated Ad/EYFP-hCAR in human primary hepatocytes. These results suggest that the two enantiomers of MD may equally induce hepatic DMEs through similar molecular mechanisms, even though their own disposition is considerably stereoselective.
In conclusion, the current study shows that MD can induce the expression of multiple hepatic DMEs and drug transporters, including CYP2B6, CYP3A4, UGT1A1, and MDR1, through the activation of xenobiotic receptors PXR and CAR. Furthermore, each constituent isomer, R-(–)-MD and S-(+)-MD, contributed significantly and equivalently to the activation of both hPXR and hCAR. These findings indicate that racemic MD may induce multiple hepatic DMEs and drug transporters in a nonstereoselective manner. Given that opioid drug abuse is a rapidly escalating problem, which has the potential to lead to clinically significant DDIs and adverse effects, the in vitro discoveries from the current studies warrant more systematic future in vivo and clinical studies that focus on how MD can mediate changes in the metabolic and pharmacokinetic profiles of other drugs.
Acknowledgments
We thank Drs. Masahiko Negishi (National Institute of Environmental Health Sciences, National Institute of Health, Research Triangle Park, NC), Steve Kliewer (University of Texas, Southwestern Medical Center, Dallas, TX), and Curtis Omiecinski (The Pennsylvania State University, University Park, PA) for providing pCR3-hCAR, pSG5-hPXR, and CMV2-hCAR3 expression vectors, respectively. We also thank the Drug Supply Program at the National Institute on Drug Abuse for providing the methadone enantiomers and Dr. Andrew Coop (Chair, Department of Pharmaceutical Sciences, University of Maryland at Baltimore, Baltimore, MD) for assistance with obtaining methadone. Human liver tissues were procured with the aid of John Cottrell from the University of Maryland at Baltimore Medical Center (Baltimore, MD).
This work was supported in part by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant DK061651]; and the National Institutes of Health National Institute on Drug Abuse [Grant R01-DA16715].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.109.027854.
ABBREVIATIONS: MD, methadone; DDI, drug-drug interaction; P450, cytochrome P450; DME, drug-metabolizing enzyme; NR, nuclear receptor; PXR, pregnane X receptor; CAR, constitutive androstane receptor; MDR1, multidrug resistance 1; Ad/EYFP-hCAR, adenovirus/enhanced yellow fluorescent protein tagged-human CAR; PK11195, 1-(2-chlorophenyl-N-methylpropyl)-3-isoquinoline-carboxamide; RIF, rifampicin; CITCO, 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichloro-benzyl)-oxime; kb, kilobase; PCR, polymerase chain reaction; CT, cycle threshold; DMSO, dimethyl sulfoxide; PB, phenobarbital.
References
- Ali J and Woods D (2008) Seizure in a cancer patient on methadone. Can J Anesth 55 (Suppl 1): 475850. [Google Scholar]
- American Methadone Treatment Association (1999) 1998 Methadone Maintenance Program and Patient Census in the U.S., American Methadone Treatment Association, New York.
- Auerbach SS, Stoner MA, Su S, and Omiecinski CJ (2005) Retinoid X receptor-alpha-dependent transactivation by a naturally occurring structural variant of human constitutive androstane receptor (NR1I3). Mol Pharmacol 68 1239–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SM, Mulcahy FM, Tjia J, Reynolds HE, Gibbons SE, Barry MG, and Back DJ (2001) The pharmacokinetics of methadone in HIV-positive patients receiving the non-nucleoside reverse transcriptase inhibitor efavirenz. Br J Clin Pharmacol 51 213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desta Z, Saussele T, Ward B, Blievernicht J, Li L, Klein K, Flockhart DA, and Zanger UM (2007) Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics 8 547–558. [DOI] [PubMed] [Google Scholar]
- Ernest CS 2nd, Hall SD, and Jones DR (2005) Mechanism-based inactivation of CYP3A by HIV protease inhibitors. J Pharmacol Exp Ther 312 583–591. [DOI] [PubMed] [Google Scholar]
- Faucette SR, Sueyoshi T, Smith CM, Negishi M, Lecluyse EL, and Wang H (2006) Differential regulation of hepatic CYP2B6 and CYP3A4 genes by constitutive androstane receptor but not pregnane X receptor. J Pharmacol Exp Ther 317 1200–1209. [DOI] [PubMed] [Google Scholar]
- Faucette SR, Zhang TC, Moore R, Sueyoshi T, Omiecinski CJ, LeCluyse EL, Negishi M, and Wang H (2007) Relative activation of human pregnane X receptor versus constitutive androstane receptor defines distinct classes of CYP2B6 and CYP3A4 inducers. J Pharmacol Exp Ther 320 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari A, Coccia CP, Bertolini A, and Sternieri E (2004) Methadone—metabolism, pharmacokinetics, and interactions. Pharmacol Res 50 551–559. [DOI] [PubMed] [Google Scholar]
- Fornataro K (1999) Methadone and anti-HIV drugs. Body Posit 12 13. [PubMed] [Google Scholar]
- Gerber JG, Rhodes RJ, and Gal J (2004) Stereoselective metabolism of methadone N-demethylation by cytochrome P4502B6 and 2C19. Chirality 16 36–44. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Sim SC, Gomez A, and Rodriguez-Antona C (2007) Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther 116 496–526. [DOI] [PubMed] [Google Scholar]
- Kharasch ED, Bedynek PS, Park S, Whittington D, Walker A, and Hoffer C (2008) Mechanism of ritonavir changes in methadone pharmacokinetics and pharmacodynamics: I. Evidence against CYP3A mediation of methadone clearance. Clin Pharmacol Ther 84 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharasch ED, Hoffer C, Whittington D, and Sheffels P (2004) Role of hepatic and intestinal cytochrome P450 3A and 2B6 in the metabolism, disposition, and miotic effects of methadone[ast]. Clin Pharmacol Ther 76 250–269. [DOI] [PubMed] [Google Scholar]
- Kharasch ED, Walker A, Whittington D, Hoffer C, and Bedynek PS (2009) Methadone metabolism and clearance are induced by nelfinavir despite inhibition of cytochrome P4503A (CYP3A) activity. Drug Alcohol Depend 101 158–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCluyse EL (2001) Human hepatocyte culture systems for the in vitro evaluation of cytochrome P450 expression and regulation. Eur J Pharm Sci 13 343–368. [DOI] [PubMed] [Google Scholar]
- LeCluyse EL, Alexandre E, Hamilton GA, Viollon-Abadie C, Coon DJ, Jolley S, and Richert L (2005) Isolation and culture of primary human hepatocytes. Methods Mol Biol 290 207–229. [DOI] [PubMed] [Google Scholar]
- Li H, Chen T, Cottrell J, and Wang H (2009a) Nuclear translocation of Ad/EYFP-hCAR: a novel tool for screening human CAR activators in human primary hepatocytes. Drug Metab Dispos 37 1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chen T, Stanton JD, Sueyoshi T, Negishi M, and Wang H (2008) The peripheral benzodiazepine receptor ligand 1-(2-chlorophenyl-methylpropyl)-3-isoquinoline-carboxamide is a novel antagonist of human constitutive androstane receptor. Mol Pharmacol 74 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stanton JD, Tolson AH, Luo Y, and Wang H (2009b) Bioactive terpenoids and flavonoids from ginkgo biloba extract induce the expression of hepatic drug-metabolizing enzymes through pregnane X receptor, constitutive androstane receptor, and aryl hydrocarbon receptor-mediated pathways. Pharm Res 26 872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderbeck LR (2008) Update of the clinical issues regarding methadone (dolophine) therapy in pain management. AACN Adv Crit Care 19 253–257. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, et al. (2003) Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem 278 17277–17283. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, and Kliewer SA (2002) Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol 62 638–646. [DOI] [PubMed] [Google Scholar]
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, et al. (2000) Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem 275 15122–15127. [DOI] [PubMed] [Google Scholar]
- Nilsson MI, Meresaar U, and Anggård E (1982) Clinical pharmacokinetics of methadone. Acta Anaesthesiol Scand Suppl 74 66–69. [DOI] [PubMed] [Google Scholar]
- Simonsson US, Lindell M, Raffalli-Mathieu F, Lannerbro A, Honkakoski P, and Lang MA (2006) In vivo and mechanistic evidence of nuclear receptor CAR induction by artemisinin. Eur J Clin Invest 36 647–653. [DOI] [PubMed] [Google Scholar]
- Smith CM, Graham RA, Krol WL, Silver IS, Negishi M, Wang H, and Lecluyse EL (2005) Differential UGT1A1 induction by chrysin in primary human hepatocytes and HepG2 Cells. J Pharmacol Exp Ther 315 1256–1264. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, and Negishi M (1999) The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem 274 6043–6046. [DOI] [PubMed] [Google Scholar]
- Totah RA, Allen KE, Sheffels P, Whittington D, and Kharasch ED (2007) Enantiomeric metabolic interactions and stereoselective human methadone metabolism. J Pharmacol Exp Ther 321 389–399. [DOI] [PubMed] [Google Scholar]
- Wang H, Faucette S, Moore R, Sueyoshi T, Negishi M, and LeCluyse E (2004) Human constitutive androstane receptor mediates induction of CYP2B6 gene expression by phenytoin. J Biol Chem 279 29295–29301. [DOI] [PubMed] [Google Scholar]
- Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, and LeCluyse EL (2003) A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem 278 14146–14152. [DOI] [PubMed] [Google Scholar]
- Wang JS and DeVane CL (2003) Involvement of CYP3A4, CYP2C8, and CYP2D6 in the metabolism of (R)- and (S)-methadone in vitro. Drug Metab Dispos 31 742–747. [DOI] [PubMed] [Google Scholar]
- Weschules DJ, Bain KT, and Richeimer S (2008) Actual and potential drug interactions associated with methadone. Pain Medicine 9 315–344. [DOI] [PubMed] [Google Scholar]