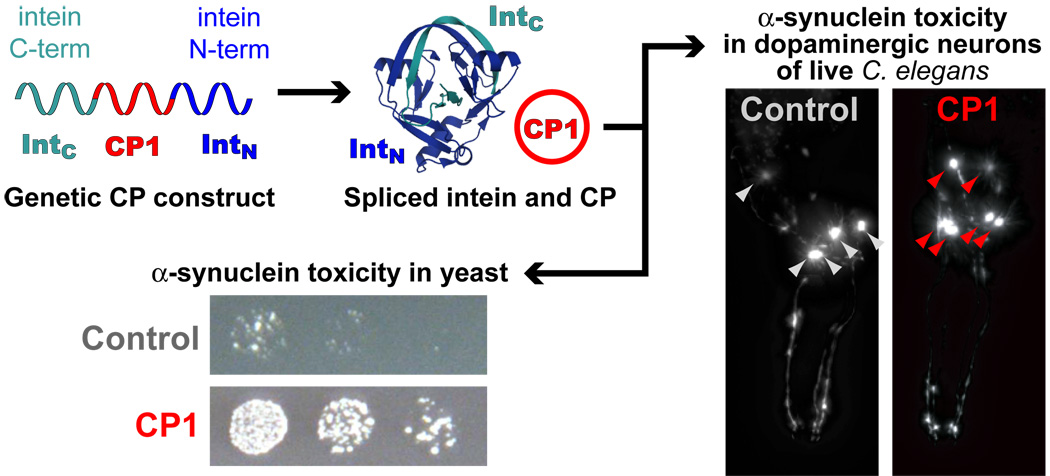
Abstract
Phage display has demonstrated the utility of cyclic peptides as general protein ligands, but cannot access proteins inside eukaryotic cells. Expanding a novel chemical genetics tool, we describe the first expressed library of head-to-tail cyclic peptides in yeast (Saccharomyces cerevisiae). We applied the library to selections in a yeast model of α-synuclein toxicity that recapitulates much of the cellular pathology of Parkinson’s disease. From a pool of five million transformants, we isolated two related cyclic peptide constructs which specifically reduce the toxicity of human α-synuclein. These expressed cyclic peptide constructs also prevent dopaminergic neuron loss in an established Caenorhabditis elegans Parkinson’s model. This work highlights the speed and efficiency of using libraries of expressed cyclic peptides for forward chemical genetics in cellular models of human disease.
Cyclic peptides (CPs) and their derivatives are potent bioactive compounds, and represent an underexplored, natural-product-like chemical space1,2. Phage and RNA display have made it possible to screen large libraries of CPs, cyclized via disulfide bonds or other side chain linkages, to identify high-affinity ligands for nearly any in vitro target3,4. By contrast, to date there are few methods for directly screening large libraries of CPs inside eukaryotic cells. Such methods would provide several advantages over in vitro techniques, ensuring that hits are nontoxic, can bind their target(s) in the appropriate cellular environment, are not rapidly degraded by cellular proteases, and possess at least enough selectivity to function in living cells. In addition, in vivo methods would enable phenotypic selections of CPs, providing a less expensive alternative to traditional high-throughput screening with a greater chance of identifying effectors with non-traditional modes of action such as inhibition of protein-protein interactions.
Recent reports describe a promising method of generating libraries of head-to-tail CPs in vivo using a single genetic construct named SICLOPPS (split-intein-mediated circular ligation of proteins and peptides, Fig. 1a)5–7. This construct uses a cleverly arranged split intein that splices out a linker region as a CP post-translationally; the linker can be as small as four amino acids or as large as a whole protein5,8. SICLOPPS-based CP libraries represent a powerful opportunity for rapid forward and reverse chemical genetics using in vivo selections7. Previous work has interfaced expressed CP libraries with bacterial two-hybrid selections, an elegant strategy for reverse chemical genetics7,9. Phenotypic screening of CP libraries was also performed in bacteria10,11. Despite these successes, to date there has been only one reported attempt to adapt SICLOPPS libraries to a eukaryotic system. A retroviral CP library was applied to a selection for inhibitors of interleukin-4 signaling in human B cells, yielding roughly a dozen CP pentamers with varying activity but no sequence consensus12. The general utility of this library was hampered by its low actual diversity (2.7 × 105 members), a lack of quantitative quality assessment, and the several weeks required just to perform an initial round of selection. Building upon these earlier studies, we sought to apply expressed CP libraries to eukaryotic cells and perform phenotypic selections in cellular models of human disease in a rapid, efficient and generally applicable manner.
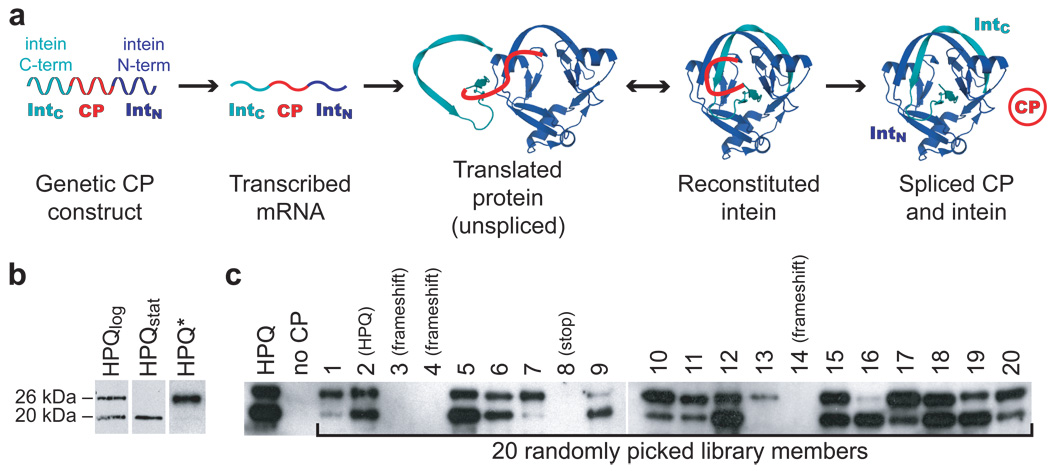
Figure 1. A cyclic peptide library that expresses and processes in yeast.
(a) The SICLOPPS (split-intein mediated circular ligation of peptides and proteins) construct encodes a single protein construct that yields a cyclic peptide (CP) after post-translational splicing5. The dnaE intein C-terminal domain is encoded first, followed by the linker to be cyclized, the intein N-terminal domain, and finally a chitin binding domain affinity tag (not shown). Graphic was generated using the crystal structure of the post-splicing form of the Synechocystis Sp. PCC6803 dnaE intein, PBD ID 1ZD7 26. (b) Western blots against the affinity tag quantitate the expression and processing of the split intein construct. Log-phase yeast cultures expressing the control HPQ construct (HPQlog) showed robust expression and roughly 50% distribution between the unprocessed 26 kDa construct and the processed 20 kDa byproduct. Yeast blotted at stationary phase (HPQstat) showed complete processing, while an HPQ variant with intein-disabling T69A/H72A mutations (HPQ*) showed no processed byproduct in log phase. (c) Blots of log-phase cultures of yeast transformed with 20 randomly picked library members demonstrated that roughly 70% of the library encodes novel CP constructs that express and process in yeast.
We and others have demonstrated that, because protein misfolding often affects highly conserved biological pathways, complex diseases such as Parkinson’s disease (PD) can be modeled in simple organisms such as yeast13–18. The human protein α-synuclein (α-syn) has been linked to PD via genetic evidence and its prominence in the PD-associated intracellular aggregates known as Lewy bodies19–21. α-syn is a small lipid-binding protein that is prone to misfolding and aggregation, and in the yeast Saccharomyces cerevisiae expression of human α-syn over a threshold level leads to ER stress, disruption of ER-Golgi vesicle trafficking, accumulation of lipid droplets, mitochondrial dysfunction, and ultimately cell death16–18. This cellular pathology mirrors many aspects of dysfunction seen in neurons and glia of individuals with PD and other synucleinopathies21. Genetic screening using our yeast synucleinopathy model has yielded suppressors of α-syn toxicity that are also effective in neuronal models17,18,22. Moreover, genetic analyses in these models have directly linked α-syn toxicity to the function of PARK9/ATP13A2, a protein whose mutations lead to an early-onset form of PD but otherwise had no known connection to α-syn22,23. Thus, cellular models have been critical to our understanding of α-syn and its role in the selective degeneration of dopaminergic (DA) neurons in PD. However, despite these and other intensive efforts, there remains a paucity of proven pharmacological targets for PD and other synucleinopathies.
Taking advantage of our established yeast synucleinopathy model, we constructed the first yeast-compatible CP library and used rapid phenotypic selections to isolate CPs that specifically reduce α-syn toxicity. Further, we identified the CP motif responsible for activity and demonstrated that the selected CPs significantly reduce DA neuron loss in a C. elegans PD model. These advances establish in vivo CP selections as an immediately useful tool for a broad range of biologists.
RESULTS
The SICLOPPS construct expresses and splices in yeast
We expressed a SICLOPPS construct using a high-copy 2µ plasmid in Saccharomyces cerevisiae under a constitutive PGK promoter. Lysates from log-phase and stationary-phase cultures were blotted using an antibody to a C-terminal tag (Fig. 1b), which revealed the expression level and processing status of the split intein construct8. The log-phase cultures showed approximately equal amounts of 26 kDa and 20 kDa proteins, which are the expected sizes of the unprocessed and processed constructs, respectively. Cultures harvested in stationary phase showed only the 20 kDa band corresponding to a processed form of the construct that has undergone either complete splicing or hydrolysis. These findings are consistent with a splicing reaction that is somewhat slower than the rate of log-phase PGK-driven protein synthesis.
An intein-disabled version of the gene was constructed by mutating the residues corresponding to T69 and H72 of the dnaE split intein to alanine. Previous work has established that these mutations, which reside in conserved block B, permit association of the two halves of the split intein but prohibit processing at the extein/N-intein junction. This blocks the first obligatory step in the accepted splicing mechanism24–26. When a T69A/H72A mutant of the construct was expressed in yeast, only the full-length, unprocessed form was detected (Fig. 1b). Thus, the lower 20 kDa band indeed represents the expected product of intein processing.
To verify directly that the SICLOPPS construct produces CPs in yeast, we purified the encoded CP from yeast lysates by virtue of its His-Pro-Gln (HPQ) motif. This sequence has well-established streptavidin affinity which was previously used to purify spliced CPs from bacterial cultures27. We used streptavidin agarose to pull down the HPQ CP from lysates of stationary-phase yeast and confirmed its identity by MALDI and electrospray mass spectrometry (MW(M+H)=1330.6, MWobs=1330.5). These data provide the first demonstration that the SICLOPPS construct can be used to produce CPs in yeast.
Construction of a yeast-compatible CP library
Having established that the SICLOPPS construct expresses and splices in yeast, we sought to construct a high-diversity library compatible with yeast-based selections. We used a previously described PCR cloning strategy to generate a randomized insert with a minimum of frameshift errors (see Supplementary Methods, Supplementary Table 1, and Supplementary Table 2 online)28. In contrast to previous SICLOPPS libraries with smaller CP backbones, we chose to construct an octamer library to maximize diversity and minimize effects of side chains on splicing efficiency6,8,12. The library was designed to encode CP octamers with a single cysteine residue followed by seven randomized residues, since a nucleophile at the first position is required for the transesterification step of intein processing24. After transformation into bacterial cells, the total diversity of the library (as measured by counting the number of independent transformants by serial dilution) was 5 × 107 independent members.
Library quality was assessed by picking and analyzing twenty colonies at random (Fig. 1c and Supplementary Table 2 online). Sequencing showed that library constructs encoded CPs with high diversity at the protein sequence level. Sequence data also confirmed that the library possessed low background: only one of twenty clones was the original HPQ-encoding vector, three of twenty possessed frameshift mutations, and one of twenty had a stop codon in the randomized region. Thus, ~75% (15 of 20) of the library is estimated to encode novel CPs.
We next assessed expression and processing of each of the twenty randomly picked constructs. The laboratory yeast strain W303 was individually transformed with each of the twenty constructs, grown to log phase, and lysed. Western blots of these lysates revealed that all fifteen of the novel CP constructs expressed. Fourteen showed evidence of some processing at log phase, most of them quite robustly (Fig. 1c). This quality control analysis indicated that roughly 70% of the library encodes CP constructs that express and undergo intein processing in yeast. This potential diversity of over thirty million CPs represents a vast, diverse, and largely unexplored chemical space ready for screening or selection.
Selection in a yeast synucleinopathy model
With a diverse, high-quality library in hand, we next sought to perform selections in a complex biological system, one which would not be amenable to in vitro methods such as phage display. To this end, the library was applied to our yeast synucleinopathy model using a simple bulk selection protocol. The yeast synucleinopathy model uses the GAL1 promoter to tightly control expression of human α-syn, so that no α-syn is produced until the yeast are grown on galactose media16,17. One liter of yeast culture was transformed with 80 µg library DNA and plated onto large library plates lacking uracil to select for transformed cells. This yielded five million independent transformants, which were scraped, re-plated onto galactose media to induce α-syn expression, and incubated for three days (Fig. 2a). Ninety-six large colonies were picked and the corresponding plasmids were isolated, amplified in bacteria, and individually re-tested in the screening strain. Thirty-one clones demonstrated reproducible suppression of α-syn toxicity in yeast (four are shown in Fig. 2b). False positives likely stemmed from spontaneous genomic suppressor mutations, which are common in most yeast selections17,29.
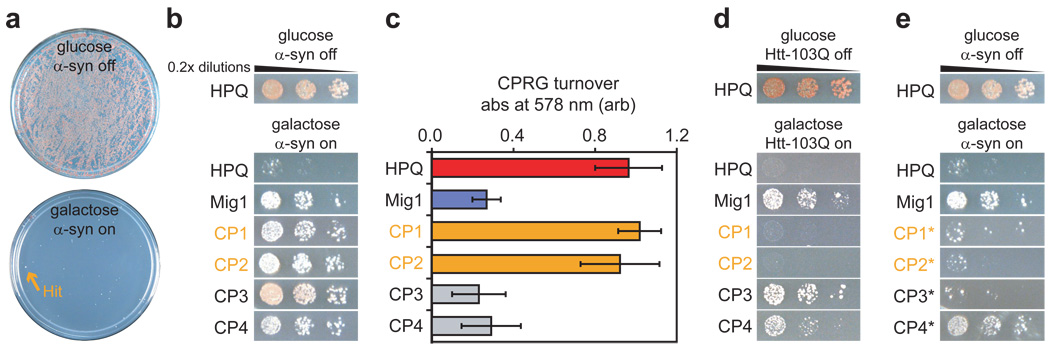
Figure 2. Identification of two CPs that selectively reduce α-synuclein toxicity in yeast.
(a) The α-synuclein (α-syn) selection strain was transformed with the CP library, allowed to recover, and re-plated on galactose media to induce α-syn expression. Glucose and galactose plates are shown to illustrate how a single robust hit clone was isolated from >150,000 transformants on a single plate. (b) Following isolation and amplification in bacteria, hit plasmids were individually transformed into the screening strain. Transformants were allowed to recover, normalized for cell number, serially diluted, and plated on galactose media. Four hits, named CP1-4, are shown along with a negative control CP plasmid (HPQ) and the positive control Mig1, a genetic repressor of the GAL1 promoter. (c) A GAL1-LacZ reporter assay using the soluble β-galactosidase substrate chlorophenol red-beta-D-galactopyranoside (CPRG) demonstrated that CP1 and CP2 do not affect protein expression. Error bars show standard deviation from five independent trials. (d) Spotting assay, similar to (b), demonstrating that CP1 and CP2 do not affect toxicity in a yeast strain expressing multiple copies of Htt-103Q from the GAL1 promoter. (e) Spotting assay, similar to (b), demonstrating that intein-disabled T69A/H72A mutants of CP1-3 (denoted with asterisks) no longer suppress α-syn toxicity. Thus, for these constructs both specific CP sequences and intein processing are required for activity.
Filtering assays
The thirty-one selected CP constructs were further evaluated using two secondary assays to exclude promoter effects and other nonspecific modes of action. First, the constructs were transformed into a reporter yeast strain with the LacZ gene downstream of the GAL1 promoter30. Expression of β-galactosidase from the GAL1 promoter was then quantified using the soluble β-galactosidase substrate chlorophenol red-beta-D-galactopyranoside (CPRG). This enabled us to quantify the effects of each construct on GAL1-mediated expression independently of the presence of α-syn (Fig. 2c). Second, the constructs were transformed into a yeast disease model that uses the GAL1 promoter for the inducible expression of a toxic polypeptide unrelated to α-syn, exon 1 of human huntingtin with 103 glutamine residues (Htt-103Q)14,15. The transformants were then spotted onto galactose media to assess the abilities of selected hits to suppress the toxicity of Htt-103Q in yeast (Fig. 2d). This assay provided a second check on whether the constructs acted by interfering with expression from the GAL1 promoter, and also tested whether the observed effects were specific to α-syn. Of course, selected CP constructs that suppressed both α-syn and Htt-103Q toxicity could be affecting a pathway common to both31. However, all selected constructs that prevented Htt-103Q toxicity also reduced GAL1-mediated expression (for example, CP3 shown in Fig. 2b–c). Two of the thirty-one selected constructs, denoted CP1 and CP2, suppressed α-syn toxicity but had no effects in the GAL1 reporter assay or on Htt-103Q toxicity, and thus act via an α-syn-specific mechanism. Notably, these results are in keeping with our previous results from genetic screens, in which genes that suppress α-syn toxicity possessed little overlap with genes that suppress Htt-103Q toxicity15,17. Thus, suppressors such as CP1 and CP2 affect specific pathways of eukaryotic cell biology involved in the toxicity of these human proteins, rather than nonspecific pathways involved in general protection from misfolded or aggregated proteins.
At this point we sought to ensure that the spliced CPs were responsible for the observed suppression of α-syn toxicity rather than nucleic acid or peptide aptamers encoded by the selected constructs. To this end, we performed three independent tests to ensure complete splicing was required. We first tested T69A/H72A mutants of each construct, which are unable to process at the extein/N-intein junction24,25. T69A/H72A mutants of CP1 and CP2 were unable to suppress α-syn toxicity (Fig. 2e). Next, a different set of mutations at residues H24 and F26 in the dnaE C-intein domain (responsible for promoting asparagine cyclization in the final processing step at the extein/C-intein junction)24,32 were also shown to abolish the activities of CP1 and CP2 (Supplementary Results and Supplementary Fig. 1). Thus, the complete splicing mechanism, including processing at both the N-intein and C-intein junctions, is required for CP1 and CP2 activity. Finally, to completely rule out the possibility that linear peptide byproducts are responsible for suppression of α-syn toxicity, we also directly tested alternative constructs that represent products of an N-intein cleavage event rather than complete cyclization. These constructs also failed to suppress α-syn toxicity, demonstrating that the sequences encoded by CP1 and CP2 are inactive without complete intein splicing (Supplementary Results and Supplementary Fig. 2). Remarkably, one of the strongest inhibitors of expression from the GAL1 promoter, CP3, also required intein processing for function, suggesting that its mode of action is also mediated by a spliced CP.
Structure-activity relationships by point mutagenesis
A critical bottleneck in chemical genetics is the generation of useful structure-activity relationship (SAR) data once hits are identified from large compound libraries. A great advantage of the CP approach is that SAR data can be generated rapidly and cost-effectively using point mutagenesis. We generated a series of alanine-scanning mutants to identify which side-chains of CP1 and CP2 are responsible for function, and to determine whether the two suppressors share a common functional motif (Fig. 3 and Supplementary Figure 3). Each mutant was transformed into yeast and expression and processing were verified by blotting log-phase cultures. Yeast cells expressing each mutant were serially diluted and plated on galactose media to quantify the degree of α-syn toxicity suppression (Fig. 3b). We found that the cysteine in the fourth CP position was absolutely required for activity in both constructs. Mutation of the hydrophobic residue in the third CP position (tryptophan for CP1 and leucine for CP2) resulted in constructs with very weak activity. Mutation of the residue in the fifth CP position (serine for CP1 and glutamate for CP2) resulted in a reproducible 5-fold decrease in growth, indicating a possible role for these side chains as well.
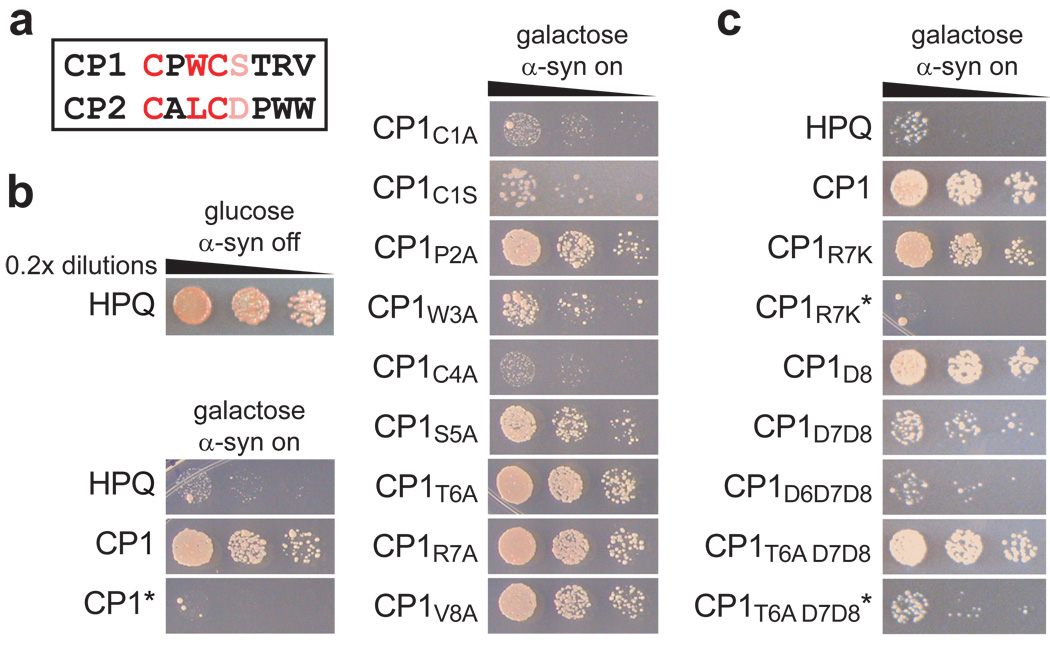
Figure 3. Rapid generation of structure-activity relationship (SAR) data and rapid minimization using point mutagenesis.
(a) Sequences of the CPs encoded by CP1 and CP2. Residues found by SAR analysis to be required for function are shown in red, and residues with minor contributions are shown in pink. (b) Spotting assays of normalized, serially diluted cells of the screening strain expressing the negative control HPQ, the selected construct CP1, indicated point mutants of CP1, and the intein-disabled T69A/H72A mutant CP1*. Each construct was verified to express and process in yeast. Similar results were obtained for CP2 (Supplementary Fig. 3). (c) Spotting assays of yeast expressing HPQ, CP1, and additional CP1 variants. These variants illustrate incorporation of a chemical handle (lysine substitution at position seven) as well as minimization of the encoded CP. Each construct was verified to express and process in yeast. Asterisks denote intein-disabled T69A/H72A mutants, which were verified to express but not process.
Replacement of the cysteine in the first position with alanine resulted in another intein-disabled construct, as expected from the accepted mechanism of intein splicing. To test the requirement for the first cysteine thiol in the spliced CP, we mutated it to serine. These mutants expressed and processed as efficiently as the original CP1 and CP2 constructs as assessed by blotting, but did not suppress α-syn toxicity (Fig. 3b and Supplementary Figure 3). Thus the cysteine in the first position is also required for the biological activities of selected CPs. Overall, CP1 and CP2 share a common CXΦC motif, wherein X is any residue and Φ is a hydrophobic residue.
CP backbone minimization using iterative design
We constructed an octamer CP library to provide diversity, but it was unclear whether selected hits would function only as octamers or might be amenable to minimization. Minimization of CPs would evaluate the extent to which selected functional motifs are dependent on macrocycle size and structure, and would also be desirable for downstream applications as chemical genetics agents. Having selected two functional CP constructs with a common tetrapeptide motif, we had the opportunity to test directly whether the remaining portions were necessary for proper orientation of the motif. Thus, building upon our side chain SAR data, we designed further mutants in order to minimize CP ring size.
CP1 proved especially amenable to minimization (Fig. 3c). Residue 8 could be deleted entirely with no loss of function. However, a construct with both residues 8 and 7 deleted showed only minimal function and a construct with residues 8, 7, and 6 deleted showed no ability to rescue α-syn toxicity. Since the side chains of these residues are not required for function, we hypothesized that conformational effects on the CXΦC motif were responsible for the loss of function. To restore proper orientation in the context of the hexamer, we tested a variety of substitutions in positions 2, 5 and 6, including glycine, alanine, proline, lysine, and tryptophan. Of the twelve mutants tested, substitution of the threonine at position 6 with alanine or glycine had the greatest effect, restoring the ability to suppress α-syn toxicity to an extent comparable to that of the original eight-residue CP1 (Fig. 3c). Intein-disabled mutants of these constructs were also tested to ensure that activity was still dependent on intein processing.
Overall, two rounds of rationally designed mutagenesis generated constructs encoding CP hexamers that suppress α-syn toxicity equally as well as the selected CP octamers CP1 and CP2. Thus, simple iterative testing of designed mutants was used to minimize the originally selected eight-residue CP (predicted MW=933) to a six-residue CP (predicted MW=634) with no observable change in in vivo activity.
CPs are easily incorporated into affinity reagents
Target identification is another notoriously difficult hurdle in forward chemical genetics33. CPs are synthetically accessible in multi-milligram quantities and easily derivatized, making them ideal for physical pull-down experiments. We verified genetically that incorporation of a lysine at position 7 did not affect CP function (Fig. 3c), then synthesized CP1R7K (1) and CP2W7K (2) (Supplementary Figure 4 and Supplementary Methods online). Since the CPs have no free N-termini, the lysine amines represent useful handles for site-specific derivatization or attachment to solid phase. We linked the synthetic CPs to agarose beads and optimized pulldown conditions (Supplementary Methods online). Notably, high lysate protein concentrations (5–10 mg/mL) could be used without extensive nonspecific binding due to the hydrophilic nature of the CPs.
Proteins that selectively bound CP1 or CP2 beads over control CP-linked beads were eluted, separated by SDS-PAGE, and identified by mass spectrometry. Yeast genetics greatly streamlines the process of validating potential targets, and so we were able to rapidly test several target candidates. However, modulation of the levels of these proteins in yeast by genetic overexpression or deletion did not affect α-syn toxicity or CP-mediated suppression of α-syn toxicity. We also easily linked CP1R7K and CP2W7K to biotin and to photoactivatable cross-linking groups, but were unable to selectively isolate additional target candidates. There are three likely explanations for these results. First, CP targets may be transient, unstable, or present at low levels within the cell. Second, selected CPs may possess low affinities for their targets, as CPs selected in bacteria showed high micromolar Ki’s and IC50’s in in vitro assays6,34. Finally, CP1 and CP2 may act via a nontraditional mechanism that is incompatible with physical target identification. Our results demonstrate that the preparation of CP-based affinity reagents can be rapid and straightforward. We are currently exploring further methods to translate this capability into similarly rapid techniques for target identification.
CP1 and CP2 are unique suppressors of α-syn toxicity
Previous work using the yeast model has established that overexpression of human α-syn affects multiple steps in vesicle trafficking18. We wondered whether our selected CPs act upstream or downstream of this defect. The yeast synucleinopathy model is particularly useful for examining the effects of α-syn on vesicle trafficking because α-syn induction can be tightly controlled and α-syn localization within the cell can be monitored in real time using YFP fusions17,18. Shortly after induction, α-syn-YFP accumulates on the plasma membrane. After 2–3 hours of α-syn-YFP induction, small foci are observed peripherally to the membrane, and after 4–6 hours these foci coalesce and begin to migrate inward toward the vacuole. This latter stage is associated with the onset of cell death. Immunoelectron microscopy has revealed that these foci correspond to pools of stalled vesicles that are coated with α-syn-YFP18.
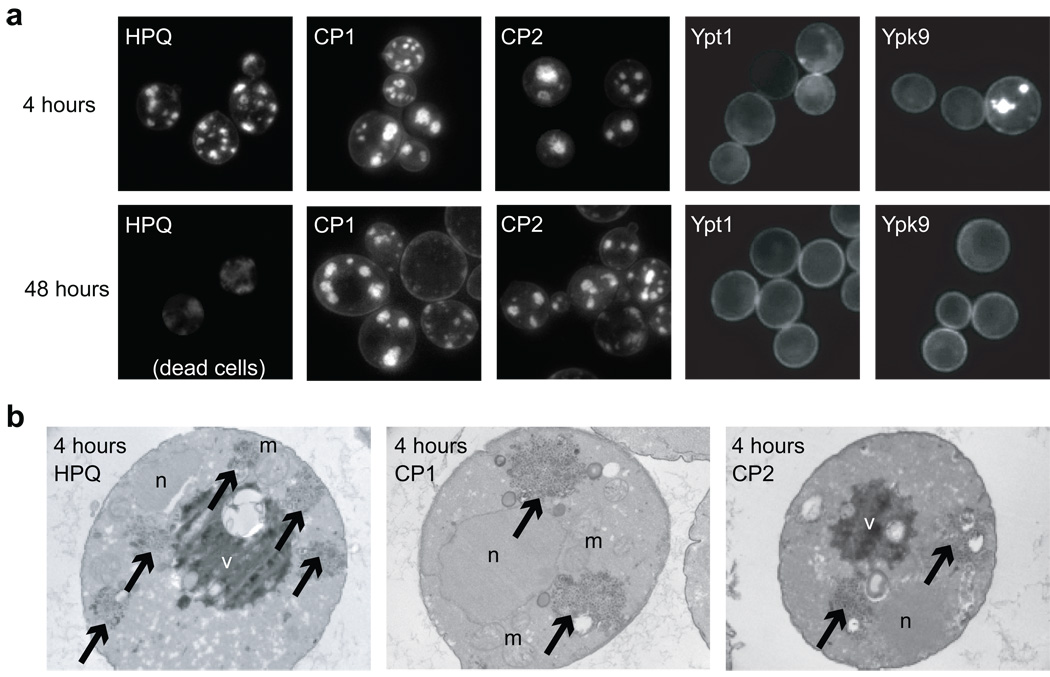
We wondered whether CP1 and CP2 prevent the accumulation of pools of stalled vesicles, since previously described genetic suppressors of α-syn toxicity mitigate the trafficking blockage17. For instance, the expression of a Rab homologue involved in ER-Golgi trafficking (Ypt1) or the yeast homologue of the P-type ATPase PARK9/ATP13A2 (Ypk9) suppresses α-syn-YFP foci 17,18,22. By contrast, CP1 and CP2 do not alter the occurrence or appearance of α-syn-YFP foci after 4 hours (Fig. 4a). Electron microscopy confirms that cells co-expressing CP1 or CP2 along with α-syn still possess pools of stalled vesicles similar to those found in cells expressing a negative control CP along with α-syn (Fig. 4b). Remarkably, even though 8–12 hours of α-syn-YFP induction is lethal to control cells, CP1 and CP2 possess α-syn-YFP foci continuously for over 48 hours after induction, with little change in appearance after 8 hours (Fig. 4a). Thus, CP1 and CP2 permit cells to grow and divide, albeit at a reduced rate, despite the continuous presence of stalled α-syn-coated vesicles. This phenotype is unique among the genetic and small molecule suppressors of α-syn toxicity identified to date, and implies that selected CPs target a pathway downstream of established vesicle trafficking defects17,18,22,31.
Figure 4. Selected CPs operate downstream of known vesicle trafficking defects.
(a) Representative fluorescence micrographs of cells expressing α-syn-YFP after 4 hours (first row) or 48 hours (second row), along with the indicated CP or yeast gene. (b) Representative electron micrographs of cells expressing α-syn-YFP after 4 hours, along with the indicated CP. m = mitochondrion, n = nucleus, v = vacuole, and arrows highlight pools of stalled vesicles. White ovals are lipid droplets.
The previously described genetic suppressors Ypt1 and Ypk9 act in independent pathways, so that the simultaneous expression of both genes results in more potent suppression of α-syn toxicity than the expression of either gene by itself22. To genetically test whether selected CPs act in a novel pathway, we expressed them in combination with Ypt1 or Ypk9. The effects of CP1 and CP2 combine in an additive manner with the effects of either of these genetic suppressors (Supplementary Results and Supplementary Fig. 5). Thus, the CPs likely act in pathways that are distinct from those already linked to α-syn toxicity.
Expression of CPs in a metazoan animal model
We previously demonstrated that human homologues of suppressors of yeast α-syn toxicity identified were able to rescue dopaminergic (DA) neurons in animal models of synucleinopathy17,18,22. In a Caenorhabditis elegans model, expression of human α-syn from the dopamine transporter promoter Pdat-1 results in degeneration of DA neurons18,35,36. This enables direct evaluation of putative α-syn antagonists in DA neurons in the context of whole animals. Nematode development is highly stereotyped and reproducible, so that any alteration in neuron quantity or morphology is highly significant. Nematodes are also transparent, enabling assessment and scoring of DA neurons in live animals by virtue of GFP expression under the control of the Pdat-1 promoter. Using this model, we have found that expression of genes such as Rab1 and PARK9/ATP13A2 (homologues of Ypt1 and Ypk9) ameliorate α-syn toxicity in the DA neurons of live animals18,22,36.
We sought to evaluate whether CP1 and CP2 would similarly rescue DA neurons in the nematode synucleinopathy model. Because the original split intein was isolated from a cyanobacterium, we first codon-optimized and re-synthesized the entire gene to ensure efficient expression in metazoan systems such as C. elegans and mammalian cells. Then Pdat-1::mCP-gfp expression vectors were constructed by recombinational cloning with the pDEST-DAT-1 destination vector35 to generate a construct that would express the mCP gene with a C-terminal GFP fusion in nematode DA neurons.
First, to verify that the mCP construct is expressed in the DA neurons of C. elegans, we injected Pdat-1::mCP-gfp expression vectors into wild-type animals. Because nematodes have only eight DA neurons, detection of gene products expressed exclusively in these few cells by Western blotting is problematic. However, GFP fusions allow direct observation of protein expression in live animals. Expression of mCP-GFP started during embryogenesis, and in adult animals robust GFP fluorescence was observed selectively in DA neurons (Supplementary Fig. 6). Because the GFP is encoded downstream of the mCP gene, we inferred that the mCP gene expresses in C. elegans DA neurons.
CP1 and CP2 suppress α-syn toxicity in DA neurons
We next tested the effects of expressed CP1 and CP2 on α-syn-mediated DA neuron loss in C. elegans. The CP1 and CP2 sequences were introduced into mCP genes without GFP fusions by site-directed mutagenesis and the resulting constructs were cloned into the pDEST-DAT-1 vector35. Transgenic worm lines were generated using these expression vectors as described17,36. We also constructed and tested negative controls, denoted CP1* and CP2*, with the intein-disabling T69A/H72A mutation.
We quantitated the effects of mCP constructs on α-syn toxicity as previously described for human and nematode genes, staging individual worms and inspecting the six anterior DA neurons (Fig. 5)17,18,36. Expression of all mCP constructs had no discernable effect on neuron health or morphology in the absence of α-syn, indicating the mCP gene is well-tolerated by live animals in DA neurons (Fig. 5a–c,g). As previously described, GFP expression also had no effect on DA neurons, while α-syn expression caused striking DA neurodegeneration (Fig. 5d,g)17,18,22,36. Only 15% of 7-day old worms expressing α-syn retained wild-type numbers and morphologies of DA neurons. Expression of mCP constructs encoding either CP1 or CP2 in these animals significantly reduced DA neurodegeneration, raising the proportions of animals devoid of neurodegeneration to 26% and 24%, respectively (Fig. 5e,g). Intein-disabled variants CP1* and CP2* were ineffective (Fig. 5f,g).
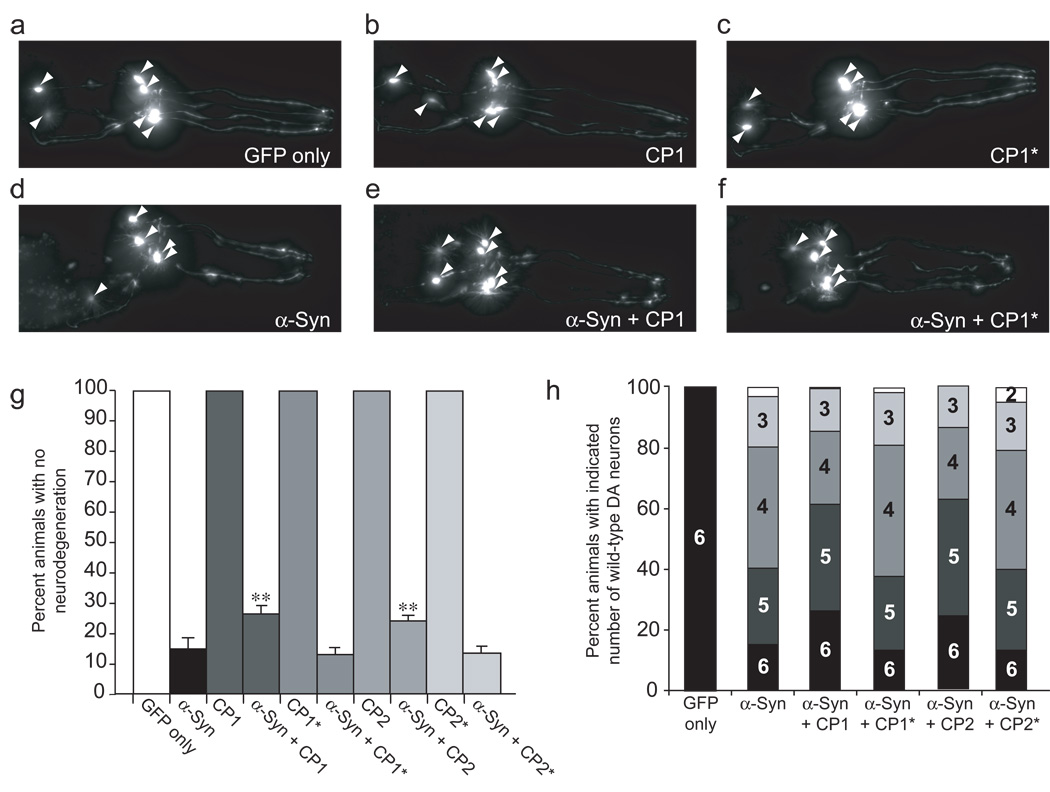
Figure 5. Selected CPs reduce α-syn-mediated toxicity in C. elegans dopaminergic (DA) neurons.
(a–f) Representative fluorescence micrographs of isogenic worm strains with GFP expression in DA neurons. Cell bodies are indicated by arrowheads, with processes visible to the right. (a) At the 7 day-old stage, worms exhibit six intact anterior DA neurons. (b,c) Expression of CP1 and the intein-disabled CP1* constructs in anterior DA neurons had no discernable effect on neuron health or development in the absence of α-syn. (d) Most worms expressing α-syn have missing or degenerated anterior DA neurons. (e) CP1 protected the DA neurons from α-syn-induced neurodegeneration, resulting in a greater proportion of worms with no degeneration in all 4 CEP and 2 ADE neurons. (f) Expression of the intein-disabled CP1* construct does not afford the same protection as CP1. (g) Population analysis revealed that 26% and 24% of worms expressing CP1 and CP2 showed no neurodegeneration, respectively, compared with only ~15% of control worms expressing only α-syn or expressing intein-disabled constructs CP1* and CP2* (**indicates p<0.05, student’s t-test). Error bars show standard deviations of three trials with three independently generated worm lines, n=90 for each line, making a total of 270 animals examined for each transgenic strain. (h) Distributions of worms by number of intact, wild-type DA neurons reveals that selected CPs also reduce the severity of the observed neurodegeneration, preferentially rescuing the two more sensitive ADE neurons.
We also quantitated the severity of the neurodegeneration by counting the number of healthy DA neurons in each animal studied. We observed that 15% of animals expressing α-syn alone retained all six anterior DA neurons, while 40% retained five or more and 80% retained four or more (Fig. 5h). CP1 and CP2 significantly increased the proportion of worms with all six DA neurons intact and the proportion of animals that retained five or more DA neurons, consistent with our observations that worms expressing α-syn are most susceptible to degeneration in their two ADE neurons, and less susceptible in their four anterior CEP neurons18,22,36. Animals expressing intein-disabled variants CP1* and CP2* showed similar distributions to those expressing α-syn alone, again demonstrating that intein processing is required for the observed activities of the CP1 and CP2 constructs.
DISCUSSION
We report a rapid and versatile technique for the in vivo selection of bioactive CPs in yeast. Our CP octamer library encodes diverse macrocycles of molecular weights from ~600 to 1200 daltons and can be effectively introduced into any yeast strain, including models of human disease. Hits can be rapidly identified using selections or screens that sort millions of CPs in a single day without expensive robotics. In selections using our yeast synucleinopathy model, two hits were isolated from an original pool of five million after only a single round of selection. While additional rounds of selection were not required for selections in the synucleinopathy model, we note that multiple rounds of selection could be performed by pooling colonies and amplifying their plasmids en masse. Stringency of the selection could be increased in subsequent rounds by using more robust selection conditions or by transferring selected CP genes to expression vectors with weaker promoters.
We also report the first expression of optimized mCP genetic constructs in live metazoan animals and their effectiveness in suppressing α-syn toxicity in C. elegans DA neurons. Notably, the extents to which CP1 and CP2 reduce the amount and severity of DA neurodegeneration match those of human homologues of hits from our yeast genetic screen17. These include Rab GTPases critical for proper vesicle trafficking and PARK9/ATP13A2, a putative P-type ATPase linked to a genetic, early-onset form of parkinsonism17,18,22,23. Thus, in yeast and C. elegans synucleinopathy models, selected CPs are as potent as endogenous regulators of key cellular pathways implicated in Parkinson’s disease.
The ability to use mutagenesis to generate SAR data and the straightforward synthesis of CPs greatly streamlines the turnaround of hits into affinity reagents for target identification. The time from initial selections to having an affinity reagent in hand was less than two months. Physical pull-downs have not yet revealed a clear target for CP1 and CP2, but various alternatives are being employed to identify their modes of action. For instance, we are currently developing CPs with higher potency by designing more stringent selections and applying second-generation libraries that incorporate the CXΦC motif. We are also exploring whether CP1 and CP2 reduce α-syn toxicity by buffering the thiol redox state of the cytosol or by otherwise altering the activity of thioredoxin-fold proteins37. These possibilities are suggested by the striking similarity between the CXΦC motif and the thioredoxin-fold CXXC motif, the critical roles of thiol redox pathways in ER stress (which is a prominent effect of α-syn expression), and previous findings that dithiol small molecules, peptides and small CPs have biologically significant thiol redox potentials that vary greatly depending on neighboring functional groups and backbone conformation37–40. The CXΦC motif may also function by binding metal ions, since metal ion transporters were discovered among the genetic suppressors and enhancers of α-syn toxicity and manganese exposure has long been known as a risk factor for PD-like syndromes22,41. Either of these possibilities would represent novel proteostasis-based mechanisms that would defy physical target identification. Finally, a variety of genetic and genomic tools are available for target identification in yeast, including transcriptional profiling, genetic screening, and the use of libraries of bar-coded overexpression and deletion strains33,42. All of these strategies will be applied to CP1 and CP2.
CPs selected in the yeast synucleinopathy model produce an unexpected and unique phenotype, apparently acting downstream of α-syn-mediated vesicle trafficking defects. Moreover, CP1 and CP2 act independently of known genetic suppressors of α-syn toxicity, and are effective in both yeast and nematode synucleinopathy models. This provides critical evidence that the target pathways are indeed conserved and relevant to the pathobiology of α-syn in neurons. Whether they act by binding a specific protein or by modulating thiol or metal homeostasis, CP1 and CP2 represent a promising new avenue for the exploration of therapeutics for PD and other synucleinopathies. Moving forward, the successful application of the new mCP construct in a metazoan animal opens the door to further validation in other PD models, including viral introduction into cell culture and incorporation in transgenic mice18,22,43.
Overall, the CP approach represents an empowering technology for the rapid development of chemical genetics agents for systems that are difficult to address in vitro. In addition, the abundance of established yeast two-hybrid selection strains, already validated in explorations of the binary interactomes of yeast, C. elegans and man, will enable rapid and low-cost selections for CP inhibitors of specific protein-protein interactions at an unprecedented scale44–46. Our approach provides a powerful alternative to traditional small molecule screening, straying from traditional “drug-like” chemical space but offering higher throughput and lower cost. It also possesses distinct advantages over display technologies, requiring fewer rounds of selection, obviating the need to purify and immobilize the target, and enabling selections in vivo, albeit with lower throughput. These trade-offs will appeal to biologists interested primarily in developing tools for probing specific cellular pathways and identifying key druggable proteins in disease models.
METHODS
Yeast strains, manipulation, selections and blotting
The α-syn-expressing yeast strains used in this study were all haploid W303 strains with the following genotype: MATa can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1. Selections and SAR data generation was performed in the previously described IntTox strain, which has human α-syn integrated using pRS303-αSynWT-YFP and pRS304-αSynWT-YFP vectors17. Co-expression of CPs with genetic suppressors was performed using the related HiTox strain, in which human α-syn is integrated using pRS304-αSynWT-GFP and pRS306-αSynWT-GFP vectors16. The Htt-103Q strain used was similarly constructed from the W303 background using integrated pRS303-Htt103Q-GFP and pRS305-Htt103Q-GFP constructs as described14.
Yeast cultures were transformed using standard lithium acetate heat shock. In all cases, at least three independent transformants were isolated and tested to ensure reproducibility of the resulting phenotype. Library transformations were performed using a scaled-up heat shock procedure. One liter of IntTox yeast were grown to log phase in SC –his –trp media, pelleted and washed several times with water. Pellets were resuspended in 20 mL of 0.1 M lithium acetate and incubated for 20 minutes at 30 °C with shaking. Cells were then spun down and resuspended in 10 mLs of 0.1 M lithium acetate with 10 mM Tris-Cl, pH 7.5 and 1 mM EDTA. 80 µg library DNA was added along with 1.4 mLs of carrier DNA (salmon sperm DNA, 2 mg/mL, previously boiled and kept on ice) and 25 mLs of 50% PEG-4000. After 30 minutes of incubation at 30 °C with shaking, 3 mLs of DMSO was added and the cells were heat shocked at 42 °C for 23 minutes. Cells were pelleted and resuspended in water prior to plating on large 24.5 × 24.5 cm Petri dishes of SC –his –trp –ura solid medium. After two days, all colonies were scraped and pooled, and 150,000–200,000 cells were plated on each of 35 SGal –his –trp –ura plates. Picked colonies were cultured overnight in SC –ura and pelleted. Plasmids were isolated using the PrepEase yeast plasmid isolation kit (USB).
Cultures for blotting were produced by growing cultures until saturation, diluting to an OD600 of 0.1 and growing for 4–5 hours. Lysates were produced by bead-beating pelleted cells in ethanol with 1 mM phenylmethylsulphonyl fluoride (PMSF), removing the ethanol by evaporation in a Speed-Vac (Savant), and resuspending in 20 mM Tris with 2% SDS. These were then normalized by protein concentration using a BCA assay (Pierce), mixed with SDS-free loading buffer and subjected to SDS-PAGE and blotting onto PVDF membranes. Blots were probed with anti-CBD serum (New England Biolabs) or anti-HA purified antibody (Sigma) according to standard protocols.
HPQ cyclic peptide isolation
Log-phase cultures expressing the HPQ-encoding SICLOPPS construct were spun down and resuspended in 50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, with protease inhibitors (Roche) and 1 mM DTT. Cells were lysed by passing through a French press at 10,000 psi twice. Lysates were incubated with streptavidin-agarose beads for 4 hours at 4 °C and washed extensively. 1 mM biotin was used to elute the HPQ cyclic peptide. Eluates were freeze-dried using a Speed-Vac (Savant) and resuspended in 10 µL water/acetonitrile with 0.1% formic acid. These were further cleaned using C18 ZipTips (Millipore) and analyzed by MALDI-MS (MW(M+H)=1330.6, MWobs=1330.5) as well as electrospray mass spectrometry (MW(M+H)=1330.6, MWobs=1330.6).
Colorimetric LacZ assay in yeast
Cells with a freshly integrated GAL1::LacZ construct were used to assess the effects of isolated CP constructs on expression from the GAL1 promoter30. Samples were prepared by growing cells overnight in raffinose media, then diluting to an OD600 of 0.1 and growing for 4–8 hours in galactose media to induce LacZ expression. Alternatively, cells were plated onto galactose plates and individual colonies were picked after 36–48 hours. Both methods produced similar results; data shown (Fig. 2c) are from picked colonies due to the ease with which many independent clones were analyzed. Cells were spun down, washed with water, and normalized to an OD600 of 0.4–0.8 in 110 µL in wells of a 96-well plate. 90 µL of CPRG solution (100 mM HEPES, pH 7.25, 150 mM NaCl, 0.65 mg/mL L-aspartate, 0.01 mg/mL BSA, 0.05% Tween, 0.5% SDS, 0.75 mg/mL chlorophenol red-beta-D-galactopyranoside (Sigma)) was added to each well. Plates were shaken at room temperature for 30–120 minutes. Plates were read at 578 nm and normalized for absorbance due to yeast optical density.
C. elegans strain generation and analysis of DA neuron degeneration
Nematodes were maintained following standard procedures47. The transgenic strains, UA109 {baInl1[Pdat-1:: a-syn; Pdat-1::gfp]; baEx84[Pdat-1::mCP_CP1; rol-6 (su1006)]}, UA110 {baInl1[Pdat-1:: a-syn; Pdat-1::gfp]; baEx85[Pdat-1::mCP_CP1*; rol-6 (su1006)]}, UA111 {baInl1[Pdat-1:: a-syn; Pdat-1::gfp]; baEx86[Pdat-1::mCP_CP2; rol-6 (su1006)]}, UA112 {baInl1[Pdat-1:: a-syn; Pdat-1::gfp]; baEx87[Pdat-1::mCP_CP2*; rol-6 (su1006)]}, UA119 {vtIs1[Pdat-1::gfp; rol-6 (su1006)]; baEx93[Pdat-1::mCP_CP1; Punc-54::mCherry]}, UA120 {vtIs1[Pdat-1::gfp;rol-6 (su1006)]; baEx94[Pdat-1::mCP_CP1*; Punc-54::mCherry]}, UA121 {vtIs1[Pdat-1::gfp; rol-6 (su1006)]; baEx95[Pdat-1::mCP_CP2; Punc-54::mCherry]}, and UA122 {vtIs1[Pdat-1::gfp; rol-6 (su1006)]; baEx96[Pdat-1::mCP_CP2*; Punc-54::mCherry]} were generated by directly microinjecting 50 ug/ml expression plasmids encoding cyclic peptide and 50 ug/ml rol-6 or Punc-54::mCherry marker into either the integrated UA44 {baInl1[Pdat-1:: a-syn; Pdat-1::gfp]} strain or the integrated BY200 {vtls1[Pdat-1::gfp; rol-6 (su1006)]} as a control for toxicity48. Three independent transgenic lines of worms were generated for each strain. Age-synchronized worms were obtained and analyzed at the indicated time as described previously18,36. The six anterior DA neurons (4 CEP and 2 ADE neurons) of 30 animals per trial were examined for neurodegeneration when the animals were 7 days old. After three trials, 90 animals for each line were analyzed, making a total of 270 worms analyzed for each transgenic strain. If a worm displayed at least one degenerative change (dendrite or axon loss, cell body loss), the animal was scored as exhibiting degenerating neurons35,36. For each trial, 30 worms were transferred onto a 2% agarose pad, immobilized with 2 mM levamisole, and analyzed using Nikon Eclipse E800 epifluorescence microscope equipped with Endow GFP HYQ filter cube (Chroma Technology, Rockingham, VT). Images were captured with a Cool Snap CCD camera (Photometrics, Tucson, AZ) driven by MetaMorph software (Universal Imaging, West Chester, PA).
Supplementary Material
Figure 6.
Acknowledgments
We thank S. Benkovic for plasmids, E. Spooner for mass spectrometry assistance, and N. Azubuine for media preparation. This work was supported by an NRSA fellowship from NINDS/NIA (J.A.K.), an R21 grant from NINDS (S.L.L. and J.A.K.), and the Morris K. Udall Centers of Excellence for Parkinson's Disease Research (S.L.L.). Parkinson's disease research in The Caldwell Lab (G.A.C, K.A.C., S.H.) was supported by the American Parkinson Disease Association, Michael J. Fox Foundation, and NIEHS.
REFERENCES
- 1.Driggers EM, Hale SP, Lee J, Terrett NK. The exploration of macrocycles for drug discovery-an underexploited structural class. Nat. Rev. Drug Discov. 2008;7:608–624. doi: 10.1038/nrd2590. [DOI] [PubMed] [Google Scholar]
- 2.Burja AM, Banaigs B, Abou-Mansour E, Burgess JG, Wright PC. Marine cyanobacteria-a prolific source of natural products. Tetrahedron. 2001;57:9347–9377. [Google Scholar]
- 3.Wilson DS, Keefe AD, Szostak JW. The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl. Acad. Sci. USA. 2001;98:3750–3755. doi: 10.1073/pnas.061028198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kehoe JW, Kay BK. Filamentous phage display in the new millennium. Chem. Rev. 2005;105:4056–4072. doi: 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- 5.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Production of cyclic peptides and proteins in vivo. Proc. Natl. Acad. Sci. USA. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horswill AR, Savinov SN, Benkovic SJ. A systematic method for identifying small-molecule modulators of protein-protein interactions. Proc. Natl. Acad. Sci. USA. 2004;101:15591–15596. doi: 10.1073/pnas.0406999101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horswill AR, Benkovic SJ. Cyclic peptides, a chemical genetics tool for biologists. Cell Cycle. 2005;4:552–555. doi: 10.4161/cc.4.4.1585. [DOI] [PubMed] [Google Scholar]
- 8.Scott CP, Abel-Santos E, Jones AD, Benkovic SJ. Structural requirements for the biosynthesis of backbone cyclic peptide libraries. Chem. Biol. 2001;8:801–815. doi: 10.1016/s1074-5521(01)00052-7. [DOI] [PubMed] [Google Scholar]
- 9.Tavassoli A, et al. Inhibition of HIV Budding by a Genetically Selected Cyclic Peptide Targeting the Gag-TSG101 Interaction. ACS Chem. Biol. 2008;3:757–764. doi: 10.1021/cb800193n. [DOI] [PubMed] [Google Scholar]
- 10.Cheng L, et al. Discovery of antibacterial cyclic peptides that inhibit the ClpXP protease. Protein Sci. 2007;16:1535–1542. doi: 10.1110/ps.072933007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nilsson LO, Louassini M, Abel-Santos E. Using siclopps for the discovery of novel antimicrobial peptides and their targets. Protein Pept. Lett. 2005;12:795–799. doi: 10.2174/0929866054864247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinsella TM, et al. Retrovirally delivered random cyclic peptide libraries yield inhibitors of interleukin-4 signaling in human B cells. J. Biol. Chem. 2002;277:37512–37518. doi: 10.1074/jbc.M206162200. [DOI] [PubMed] [Google Scholar]
- 13.Winderickx J, et al. Protein folding diseases and neurodegeneration: Lessons learned from yeast. Biochim. Biophys. Acta Mol. Cell Res. 2008;1783:1381–1395. doi: 10.1016/j.bbamcr.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 14.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc. Natl. Acad. Sci. USA. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willingham S, Outeiro TF, DeVit MJ, Lindquist SL, Muchowski PJ. Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science. 2003;302:1769–1772. doi: 10.1126/science.1090389. [DOI] [PubMed] [Google Scholar]
- 16.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper AA, et al. alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gitler AD, et al. The Parkinson's disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc. Natl. Acad. Sci. USA. 2008;105:145–150. doi: 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 20.Spillantini MG, et al. alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 21.Lee VMY, Trojanowski JQ. Mechanisms of Parkinson's disease linked to pathological alpha-synuclein: New targets for drug discovery. Neuron. 2006;52:33–38. doi: 10.1016/j.neuron.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 22.Gitler AD, et al. alpha-Synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat. Genet. 2009;41:308–315. doi: 10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez A, et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 24.Evans TC, Xu MQ. Mechanistic and kinetic considerations of protein splicing. Chem. Rev. 2002;102:4869–4883. doi: 10.1021/cr9601369. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh I, Sun L, Xu MQ. Zinc inhibition of protein trans-splicing and identification of regions essential for splicing and association of a split intein. J. Biol. Chem. 2001;276:24051–24058. doi: 10.1074/jbc.M011049200. [DOI] [PubMed] [Google Scholar]
- 26.Sun P, et al. Crystal structures of an intein from the split dnaE gene of Synechocystis sp PCC6803 reveal the catalytic model without the penultimate histidine and the mechanism of zinc ion inhibition of protein splicing. J. Mol. Biol. 2005;353:1093–1105. doi: 10.1016/j.jmb.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 27.Naumann TA, Savinov SN, Benkovic SJ. Engineering an affinity tag for genetically encoded cyclic peptides. Biotechnol. Bioeng. 2005;92:820–830. doi: 10.1002/bit.20644. [DOI] [PubMed] [Google Scholar]
- 28.Tavassoli A, Benkovic SJ. Split-intein mediated circular ligation used in the synthesis of cyclic peptide libraries in E-coli. Nat. Protoc. 2007;2:1126–1133. doi: 10.1038/nprot.2007.152. [DOI] [PubMed] [Google Scholar]
- 29.Bickle MBT, Dusserre E, Moncorge O, Bottin H, Colas P. Selection and characterization of large collections of peptide aptamers through optimized yeast two-hybrid procedures. Nat. Protoc. 2006;1:1066–1091. doi: 10.1038/nprot.2006.32. [DOI] [PubMed] [Google Scholar]
- 30.Yocum RR, Hanley S, West R, Ptashne M. Use of Lacz Fusions to Delimit Regulatory Elements of the Inducible Divergent Gal1-Gal10 Promoter in Saccharomyces-Cerevisiae. Mol. Cell Biol. 1984;4:1985–1998. doi: 10.1128/mcb.4.10.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Outeiro TF, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 32.Ding Y, et al. Crystal structure of a mini-intein reveals a conserved catalytic module involved in side chain cyclization of asparagine during protein splicing. J. Biol. Chem. 2003;278:39133–39142. doi: 10.1074/jbc.M306197200. [DOI] [PubMed] [Google Scholar]
- 33.Burdine L, Kodadek T. Target identification in chemical genetics: The (often) missing link. Chem. Biol. 2004;11:593–597. doi: 10.1016/j.chembiol.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Tavassoli A, Benkovic SJ. Genetically selected cyclic-peptide inhibitors of AICAR transformylase homodimerization. Angew. Chem. Int. Ed. 2005;44:2760–2763. doi: 10.1002/anie.200500417. [DOI] [PubMed] [Google Scholar]
- 35.Cao SS, Gelwix CC, Caldwell KA, Caldwell GA. Torsin-mediated protection from cellular stress in the dopaminergic neurons of Caenorhabditis elegans. J. Neurosci. 2005;25:3801–3812. doi: 10.1523/JNEUROSCI.5157-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamamichi S, et al. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson's disease model. Proc. Natl. Acad. Sci. USA. 2008;105:728–733. doi: 10.1073/pnas.0711018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abou-Sleiman PM, Muqit MMK, Wood NW. Expanding insights of mitochondrial dysfunction in Parkinson's disease. Nat. Rev. Neurosci. 2006;7:207–219. doi: 10.1038/nrn1868. [DOI] [PubMed] [Google Scholar]
- 39.Cabrele C, Fiori S, Pegoraro S, Moroder L. Redox-active cyclic bis(cysteinyl)peptides as catalysts for in vitro oxidative protein folding. Chem. Biol. 2002;9:731–740. doi: 10.1016/s1074-5521(02)00152-7. [DOI] [PubMed] [Google Scholar]
- 40.Kersteen EA, Raines RT. Catalysis of protein folding by protein disulfide isomerase and small-molecule mimics. Antioxid. Redox. Signal. 2003;5:413–424. doi: 10.1089/152308603768295159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olanow CW. Manganese-induced parkinsonism and Parkinson's disease. In: LeVine SM, editor. Redox-Active Metals in Neurological Disorders. Vol. 1012. New York: New York Academy of Sciences; 2004. pp. 209–223. [DOI] [PubMed] [Google Scholar]
- 42.Hoon S, et al. An integrated platform of genomic assays reveals small-molecule bioactivities. Nat. Chem. Biol. 2008;4:498–506. doi: 10.1038/nchembio.100. [DOI] [PubMed] [Google Scholar]
- 43.van der Putten H, et al. Neuropathology in mice expressing human alpha-synuclein. J. Neurosci. 2000;20:6021–6029. doi: 10.1523/JNEUROSCI.20-16-06021.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 45.Yu HY, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li SM, et al. A map of the interactome network of the metazoan C-elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brenner S. Genetics of Caenorhabditis-Elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nass R, Hall DH, Miller DM, Blakely RD. Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2002;99:3264–3269. doi: 10.1073/pnas.042497999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.