Abstract
The amino terminal domain of glutamate receptor ion channels, which controls their selective assembly into AMPA, kainate and NMDA receptor subtypes, is also the site of action of NMDA receptor allosteric modulators. Here we report the crystal structure of the ATD from the kainate receptor GluR6. The ATD forms dimers in solution at micromolar protein concentrations and crystallizes as a dimer. Unexpectedly, each subunit adopts an intermediate extent of domain closure compared to the apo and ligand bound complexes of LIVBP and G-Protein coupled glutamate receptors, and the dimer assembly has a strikingly different conformation from that found in mGluRs. This conformation is stabilized by contacts between large hydrophobic patches in the R2 domain which are absent in NMDA receptors, suggesting that the ATDs of individual glutamate receptor ion channels have evolved into functionally distinct families.
Excitatory synaptic transmission in the brain of vertebrates is mediated by a family of 18 glutamate receptor ion channel genes (iGluRs) which exhibit subtype selective assembly, forming three major classes named AMPA, kainate and NMDA receptors1,2. G-protein coupled receptors (mGluRs) also play key roles in the response to glutamate3. Our first insights into the structure of glutamate receptors came from cDNA cloning and bioinformatic analysis4-6. This revealed that iGluRs and mGluRs were multi-domain proteins, which likely evolved by fusion of bacterial periplasmic binding proteins with prokaryotic ion channels or signaling domains. A prototypical iGluR subunit contains an extracellular N-terminal assembly domain with a leucine/isoleucine/valine binding protein-like fold (LIVBP), and a glutamate or glycine binding domain with a glutamine binding protein-like fold (Fig. 1a); by contrast mGluRs contain only a single LIVBP like domain and paradoxically this forms the binding site for glutamate. The assembly of mGluRs as dimers is well established7, and although iGluRs are tetrameric proteins, their extracellular domains are also believed to assemble as pairs of dimers. The 1st crystal structures of iGluR ligand binding domains were solved more than 10 years ago8, followed by structures for mGluR ligand binding domains7, but no high resolution structures for the 45 kDa amino terminal domain of an iGluR have been solved.
Figure 1.
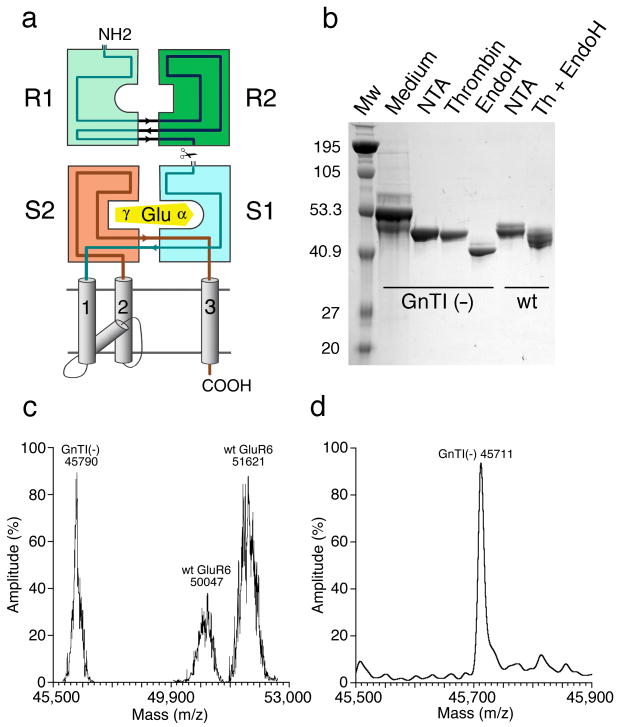
Expression and purification of the GluR6 amino terminal domain. (a) Topology diagram for a glutamate receptor subunit showing the R1 and R2 amino terminal domain clam shells, the S1S2 glutamate binding domain, and the ion channel. (b) SDS PAGE for GluR6 ATD expressed in either wt HEK cells or GnTI- cells; the major band in the Medium lane is residual serum protein; NTA indicates eluate from a Ni2+ affinity column; Thrombin indicates cleavage of the C-terminal His tag; Endo H indicates eluate from an SP ion exchange column following glycosidic cleavage. (c) Mass spectral analysis by MALDI for GluR6 ATD expressed in either wt HEK cells or GnTI- cells; the wt protein has two major peaks, both of which are broader than for Endo H digested protein from GnTI- cells. (d) Mass spectral analysis by ESI for Endo H digested GluR6 ATD expressed in GnTI- cells; the predicted MW of the R6ATD with 5 NAG molecules is 5×203 plus 44,696 which matches the experimental value of 45,711.
The ATDs of iGluRs are generally assumed to show ligand induced domain closure like that observed in LIVBP and mGluR1, and which is a key component in models developed to explain the allosteric modulation of NMDA receptors by Zn2+ and ifenprodil which bind to the ATD9-12. However, the generation of homology models based on crystal structures for LIVBP and mGluR1 is complicated by both insertions in the ATD of iGluRs compared to LIVBP, and insertions in the ligand binding domain of mGluRs compared to the ATD of iGluRs. These homology models have focused on NMDA receptor allosteric modulators, have not addressed the mechanisms underlying subtype selective assembly, and assume that the ATDs of different classes of glutamate receptors are functionally similar9-12.
To address these issues we performed crystallographic and sedimentation experiments. Our results reveal that the ATD of the rat kainate receptor GluR6 can be expressed as a soluble glycoprotein which forms dimers in solution at micromolar concentrations; that it crystallizes as a dimer with a much larger buried surface than found for mGluRs; and that both subunits in the dimer assembly have a partially closed conformation distinct from that observed for mGluRs and LIVBP. The structures reveal novel loop regions, different from those found in mGluRs, which likely play roles in the mechanism for subtype selective assembly. We also find striking amino acid sequence differences in the dimer interface of NMDA receptors versus AMPA and kainate receptors, which suggests unexpectedly that these families are functionally distinct.
Results
The GluR6 ATD has five N-linked glycosylation sites
We focused on the GluR6 ATD as a promising target which could be expressed as a soluble glycoprotein (Fig. 1a), and purified to homogeneity by affinity and ion exchange chromatography (Fig. 1b). N-terminal sequencing established that the 31 amino acid native signal peptide was cleaved from the purified secreted protein at the junction between Gly31 and Thr32. Because expression in wild type HEK293 cells yielded protein with non uniform glycosylation, as assayed by SDS PAGE and mass spectrometry using MALDI (Fig. 1 b,c), we used protein expressed in N‐Acetylglucosaminyltransferase I-negative (GnTI-) cells followed by digestion with Endo H for crystallization trials13. This yielded protein with a uniform molecular weight of 45,711 by mass spectrometry using ESI (Fig. 1d), indicating the presence of five N-linked acetylglucosamine residues. Of relevance to the poor biochemical properties of bacterially expressed iGluR ATDs, complete removal of glycans by digestion with PNGase F led to aggregation and precipitation, indicating a key biological role for glycosylation in preventing non specific interactions between protein surfaces shielded by glycan moieties.
GluR6 ATD dimerizes with micromolar affinity
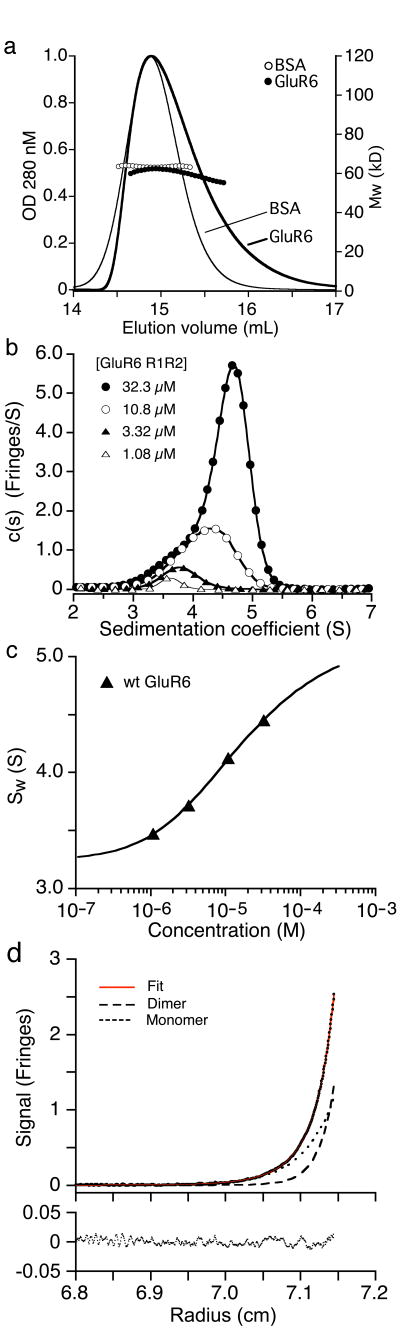
We determined the oligomerization state of GluR6 ATD by analytical size-exclusion chromatography, with multi-angle light scattering, refractive index and UV detectors (SEC/MALS/RI/UV). At a loading concentration of 2 mg/ml GluR6 ATD coeluted with bovine serum albumin (MW 66,429), but with an asymmetric profile (Fig. 2a) suggesting reversible oligomerization. Consistent with this, the molecular weight of the GluR6 ATD peak fraction had a mass of 62,400, intermediate between the monomer and dimer masses of 45,711 and 91,422 (Fig. 2a). To measure the dimer dissociation constant for GluR6 ATD we performed sedimentation velocity (SV) and sedimentation equilibrium (SE) experiments using both wt GluR6 ATD, and Endo H digested protein expressed in GnTI- cells. The c(s) distribution measured by SV with wt GluR6 ATD loading concentrations ranging from 1.1 to 32 μM showed the expected concentration dependent shift in sedimentation coefficient for a monomer dimer system in rapid equilibrium (Fig. 2b); weighted-average sedimentation coefficients sw calculated by integration of the c(s) peaks were well fit by a binding isotherm with a Kd for monomer dimer equilibrium of 15 μM (95% confidence interval 13 – 16 μM), with values of 3.47 S and 5.54 S for monomer and dimer respectively (Fig. 2c); comparable results were obtained by SE (Fig. 2d) with a Kd of 11.3 μM (95% confidence interval 10.4 – 12.2 μM). Similar values were obtained for protein expressed in GnTI- cells (data not shown) indicating that dimer formation by the amino terminal domain is not regulated by complex glycosylation. The oligomerization state of kainate receptor ATDs has not been studied before but our results are consistent with prior studies on AMPA receptors in which sucrose gradient centrifugation followed by SDS PAGE revealed the presence of dimers, together with smaller amounts of trimers and unidentified higher molecular weight species14,15.
Figure 2.
The GluR6 amino terminal domain forms dimers with micromolar affinity. (a) Mass analysis by SEC-MALS/RI/UV for sequential runs with GluR6 ATD and bovine serum albumin. (b) Sedimentation coefficient distributions c(s) obtained from analysis of sedimentation boundaries measured at 50,000 r.p.m. for wt GluR6 ATD at loading concentrations of 32.3, 10.8, 3.32 and 1.08 μM. (c) Isotherm of weighted-average sedimentation coefficients determined from peak integration of the c(s) data shown in (b) fit with a monomer-dimer model. (d) Sedimentation equilibrium interference fringe profiles for wt GluR6 ATD at a rotor speed of 23,000 rpm with a global fit of data from multiple rotor speeds and multiple loading concentrations. Shown are the best-fit estimates for the contributions from monomer and dimer.
Crystal structures of GluR6 ATD dimers
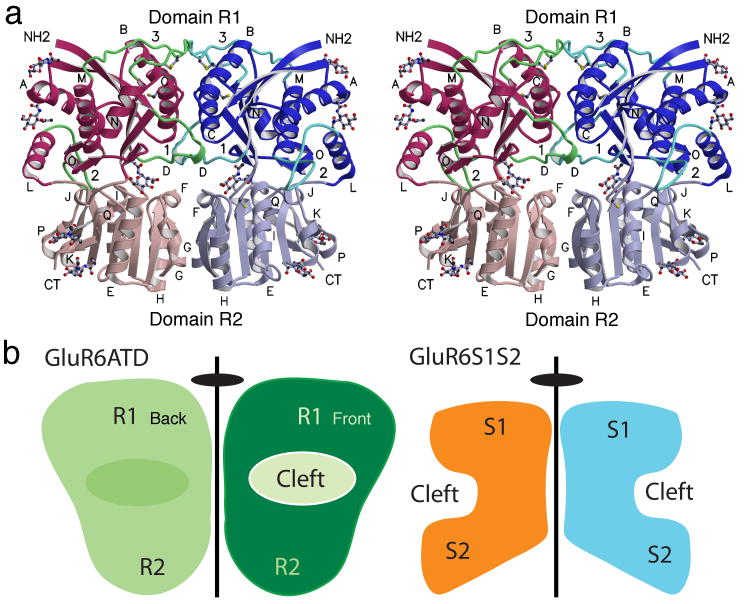
The ATDs of GluR6 and GluR2 share only 25% amino acid sequence identity, substantially more than for LIVBP (12%) and mGluR1 (17%). Molecular replacement trials with LIVBP, mGluR1 or homology models for NR2A and NR2B gave solutions with low Z scores; because a better solution was obtained using a GluR2 ATD monomer solved by the Gouaux lab as the search probe16, we pursued this for model building and refinement. The solution was complicated by the large unit cell and high solvent content. A statistical analysis of Matthews coefficients in the PDB17 suggested 5 molecules in the asymmetric unit as the most probable solution (50.3%), followed by solutions with either six (23.7%) or four copies (21.1%); however, in the solution found there were only two copies, giving a solvent content of 77%. We obtained two different crystal forms at pH 5 and pH 9 which gave essentially identical structures arranged as a dimer with two-fold molecular symmetry (Fig. 3a). The solutions were verified by inspection of crystal packing and electron density maps (Supplementary Fig. 1) and convergence to reasonable Rfree values (Table 1). Each protomer had a clam-shell like two domain αβ core structure, similar to those for LIVBP and the ligand binding domain of mGluR1; following the convention proposed by Masuko et al.9 we label the domains R1 and R2. The N-terminus starts in domain R1 and the C-terminus ends at Lys389 in domain R2, with the peptide chain forming domains R1 and R2 making three interdomain crossings. In domain R2, Thr383, the last amino acid in β-strand 16, is 22 residues upstream of Val404 in the 1st β-strand of the GluR6 ligand binding domain18. In the tartrate form main chain electron density for helix L and loop 2 was complete only for chain B, with a break from Arg269-Pro274 in chain A. For the MPD form the trace was complete for chain A, but with a break from Gln271-Lys275 in chain B; the ribbon diagram shown in Fig. 3 is a composite structure generated after least squares superposition to supply the trace for the 5 missing residues in the MPD form. Using C α coordinates for conserved α helices and β strands, the rmsd values for independent superposition of domains 1 and 2 on GluR6 ATD protomers were 1.16 and 1.22 Å for the glutamate complex of mGluR1, 1.20 and 1.14 Å for the leucine complex of LIVBP and 1.38 and 1.25 Å for the GluR2 ATD, indicating similar core structures. However, the extent of domain closure varies appreciably amongst these structures.
Figure 3.
The GluR6 amino terminal domain crystallizes as a dimer. (a) Stereoview of the dimer assembly with domains R1 and R2 from each subunit shaded in dark and light red and blue respectively; loops 1-3 are colored green; N-linked NAG molecules and Cys side chains are drawn in ball and stick representation; α-helices are labeled for each subunit. The cleft between domains 1 and 2 for the left subunit is on the rear plane of the dimer, while for the right subunit the cleft faces the viewer resulting in a back to front arrangement with respect to the dimer axis of symmetry. (b) Cartoons showing the location of the clefts between domains 1 and 2 relative to the dimer 2-fold axis of symmetry in the GluR6 ATD dimer (left) and the GluR2 S1S2 ligand binding domain dimer (right). The view for the ATD cartoon matches that in (a), for which the interdomain cleft for the left hand subunit is not visible.
Table 1.
Data collection and refinement statistics (molecular replacement)
| Tartrate | MPD | |
|---|---|---|
| Data collection | ||
| Space group | P61 | P61 |
| Cell dimensions | ||
| a, b, c (Å) | 172.1, 172.1, 111.6 | 177.2, 177.2, 81.2 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 50.20 - 2.69 (2.74 - 2.69)* | 50.00 - 2.90 (3.00 - 2.90)* |
| Rmerge | 0.073 (0.626) | 0.089 (0.763) |
| I / σI | 14.0 (2.1) | 21.0 (1.8) |
| Completeness (%) | 99.5 (99.4) | 98.7 (92.6) |
| Redundancy | 4.4 (4.3) | 8.1 (7.3) |
| Refinement | ||
| Resolution (Å) | 46.80 - 2.70 | 44.31 - 2.90 |
| No. reflections | 50,708 | 31,930 |
| Rwork / Rfree | 20.38 / 22.71 | 21.05 / 25.42 |
| No. atoms | ||
| Protein | 6055 | 6072 |
| NAG / Tartrate / Ca2+ | 112 / 20 / 2 | 112 / 0 / 2 |
| Water | 44 | - |
| B-factors | ||
| Protein | 89.77 | 97.95 |
| NAG / Tartrate / Ca2+ | 148.1 / 89.0 / 111.3 | 144.14 / - / 142.9 |
| Water | 70.00 | - |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.009 | 0.004 |
| Bond angles (°) | 0.841 | 0.753 |
Consistent with the reversible monomer dimer equilibrium observed by sedimentation analysis (Fig. 2) there are no disulfide bonds linking the protomers in a dimer assembly, and instead Cys residues at positions 65 and 316 form intramolecular disulfide bonds linking loop 3 with helix B. Amino acid alignments reveal that this disulfide bond is conserved in all iGluR ATDs (Supplementary Fig. 2). The protomers in the GluR6 ATD dimer pack side to side, with the cleft between domains 1 and 2 facing the front and back faces of the dimer (Fig. 3b); in contrast, for GluR6 ligand binding domain dimers the interdomain clefts are on the lateral edges of the dimer assembly19. The GluR6 ATD has 5 consensus N-linked glycosylation sites (NXS/T) the modification of all of which was established by mass spectrometry (Fig. 1d). A total of eight NAG residues were visible in the structure crystallized with tartrate, 5 on chain A and 3 on chain B, while for the MPD structure 4 NAG residues were resolved per protomer. The glycosylation sites are not uniformly distributed over the protein surface, and instead lie in bands decorating the lateral edges of the dimer assembly (Supplementary Fig. 3).
Differences in dimer assemblies of GluR6 ATD and mGluRs
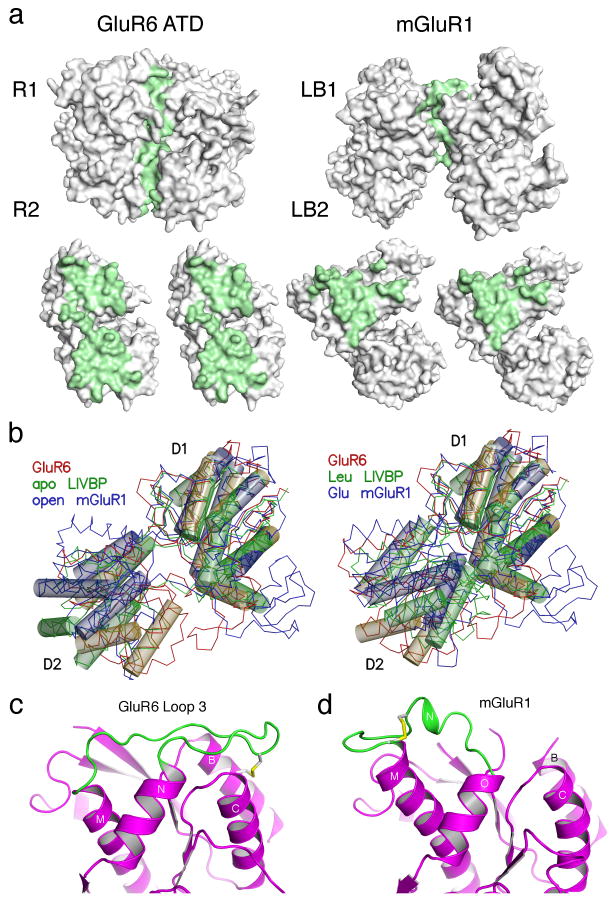
In the GluR6 ATD dimer assembly each protomer has a buried surface area of 1536 Å2, approximately equally distributed between domains R1 and R2. By contrast, the buried surface in mGluR1 dimers has an area of only 880 Å2 per subunit, and the dimer is stabilized by intermolecular disulfide bonds7,20. The involvement of domain R2 in GluR6 ATD dimer assembly, is strikingly different from the packing found for dimers of G-protein coupled glutamate receptors crystallized in a range of conformational states7,20 for which lobe 1 forms the major contact surface (Fig. 4a).
Figure 4.
The GluR6 ATD and mGluR dimers have different conformations. (a) Molecular surfaces for the GluR6 ATD and mGluR1 dimers crystallized in the ligand free open-open/R conformation (1EWT) for the dimer assembly, and after rotation by ± 90° of the partner subunits; residues with solvent inaccessible atoms are colored green. (b) The left panel shows crystal structures for the apo forms of LIVBP and mGluR1 superimposed on a GluR6 ATD protomer. Thin lines show Cα traces and transparent cylinders illustrate conserved α-helices which overlap when either the R1 or R2 domains are used for superposition. The right panel shows the superposition for the leucine and glutamate bound complexes of LIVBP and mGluR1. (c) Ribbon diagram showing the conformation of loop 3 and associated disulfide bridges in GluR6 ATD; the view is the same as in (a) and shows the left subunit in a dimer assembly. (d) The disulfide bridge and loop conformation in the corresponding region of mGluR1.
A second difference between the GluR6 ATD and mGluR crystal structures results from variations in the extent of domain closure. Periplasmic binding proteins and the ligand binding domains of mGluRs undergo large amplitude ligand-induced conformational changes; in the apo state the clam shell formed by domains 1 and 2 is wide open and upon binding leucine or glutamate the clam shell closes. Surprisingly, we found that the ATD of GluR6 adopts an intermediate extent of domain closure between these extremes (Fig. 4b). As a result the lips of the GluR6 ATD clamshell are too far apart to create a closed off cavity in the interior of the protein, as occurs for leucine bound LIVBP, or a narrow solvent filled tunnel leading to the glutamate binding site in mGluR1(Supplementary material).
The final notable difference between the GluR6 ATD and mGluR protomers is the conformation of loop 3 and the location of the disulfide bonds which link it to the rest of the protein. In the GluR6 ATD dimer loop3 projects into the dimer interface (Fig. 4c) while in mGluRs the corresponding loop projects outwards towards the lateral edge of the dimer (Fig. 4d). As a result of this difference it is very unlikely that the ATDs can form intermolecular disulfide bonds as proposed previously for NR1 homodimers 21. Despite these differences we identified an ion binding site in the GluR6 ATD in a location corresponding to the Ca binding site found in mGluR1 (Supplementary Fig. 4); at present, for both receptor species it is not known if this site modulates receptor function.
Loops 1 and 3 likely specify subtype specific assembly signals
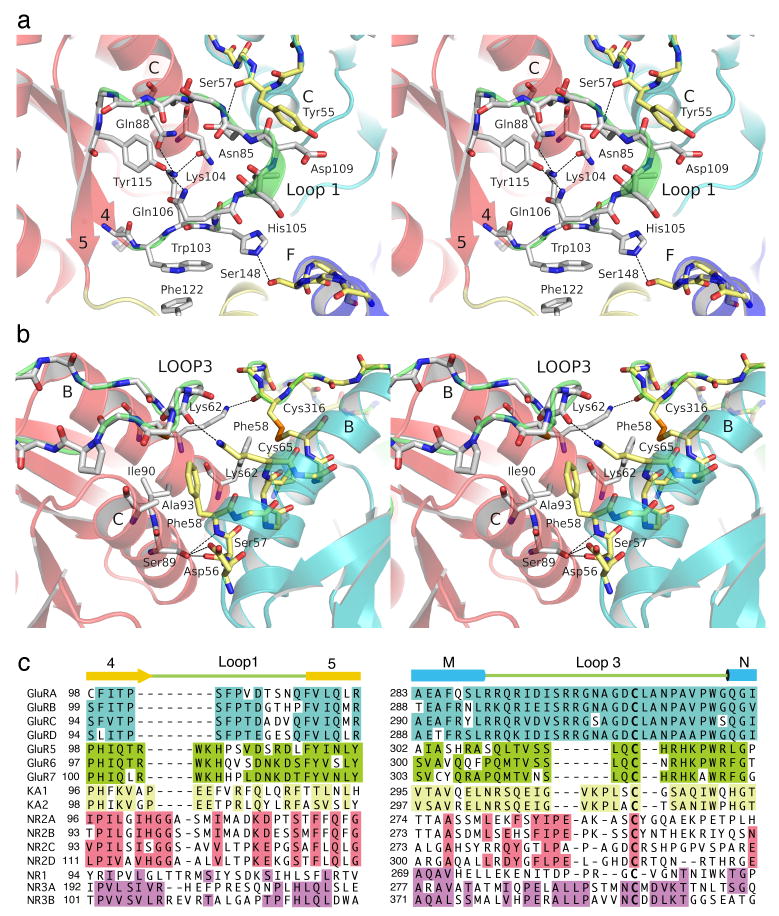
Compared to LIVBP, the core structure of a GluR6 ATD protomer is extended by three loops, which are absent in periplasmic proteins, which have different conformations in mGluRs, and which in the GluR6 ATD mediate either dimer contacts (loops 1 and 3) or contacts between domains R1 and R2 (loop 2). The GluR6 ATD domain R1 dimer interface is formed primarily by interactions between helices B and C. At the base of domain R1 helix C is flanked by loop 1. Because the protomers in the dimer assembly are arranged with side to side packing, loop 1 projects into the mouth of the cleft formed between domains 1 and 2 of the dimer partner subunit, and interacts with residues in both domains (Fig. 5a). The side chain amide of Asn 110 forms an intermolecular hydrogen bond with the main chain carbonyl of Tyr55 in domain R1, while the side chain of His105 makes an intermolecular hydrogen bond with the side chain of Ser148 in domain R2. The conformation of loop 1 in GluR6 is stabilized by polar and hydrophobic interactions made by residues absent in AMPA receptors. The side chain of Arg102 forms a salt bridge with Glu 22 in helix A, while Trp103 makes π stacking interactions with Phe122 in domain R2.
Figure 5.
Dimer interactions in domain R1. (a) Stereoview of the lower segment of domain R1 showing interactions between loop 1 and helices C and F in the dimer partner. (b) Stereoview of the upper segment of domain R1 showing interactions between helices B, C and loop 3. (c) Amino acid sequence alignments for loops 1 and 3 in five iGluR gene families, with sequence identities within each family indicated by colored backgrounds.
In domain R1 helices B and C are capped by loop 3 which is aligned perpendicular to the top surface of each protomer, and held in place by a disulfide bond between Cys65 and Cys316 (Fig. 5b). Despite its close physical proximity to helices B and C, loop 3 forms a link connecting helices M and N, and is separated from helix B by 220 residues in linear sequence. In addition to the disulfide link, the conformation of loop three is stabilized by intermolecular contacts across the dimer interface. The side chain of Lys62 in helix B forms a hydrogen bond with the main chain carbonyl oxygen of Cys316 of the dimer partner. Phe58 at the base of helix B is conserved in AMPA and kainate receptors, and forms intermolecular hydrophobic van der Waals contacts with Cys316 and with Ile90 and Ala93 in helix C. These hydrophobic interactions are supplemented by several hydrogen bond contributions from the main chain and side chains of residues Tyr55, Asp56, Ser57 and Phe58 at the base of helix B, which interact with Ser89 in helix C and Asp109 in loop 1 (Fig. 5b). Within this region kainate and AMPA receptors show conserved differences in the dimer interaction surface which likely play roles in selective assembly. For example, in helix C Ile90 is replaced by Phe, Ala88 by Met or Thr, and Ser84 by Arg, Lys or Met (Supplementary Fig. 2).
The projection of loops 1 and 3 into the dimer interface suggests that they also have a major role in specifying dimer assembly. Within the five major iGluR gene families, an amino acid sequence alignment based on the GluR6 ATD crystal structure reveals that these regions have highly conserved family specific sequences (Fig. 5c and Supplementary Fig. 2). The length of loop1 is shortest in AMPA receptors, and contains inserts of 3, 4, 7 and 6-8 residues in the GluR5-7, KA, glutamate binding NR2 and glycine binding NR1 and NR3 subtypes of iGluRs. Conversely, loop 3 is longest in AMPA and NR3 subtype NMDA receptors, and contains deletions of 8, 5, 2 and 4 residues in GluR5-7, KA, NR2 and NR1, respectively. The variable length and different sequence of these segments would hinder interactions at the domain R1 interface between different receptor species and hence might be a major determinant of subtype specificity in ATDs of iGluRs. However, the buried surfaces in domain R1 of different iGluR subtypes also have family specific sequence conservation, and thus loop swapping experiments are unlikely to switch assembly specificity.
Domain R2 has a large hydrophobic surface absent in NMDA receptors
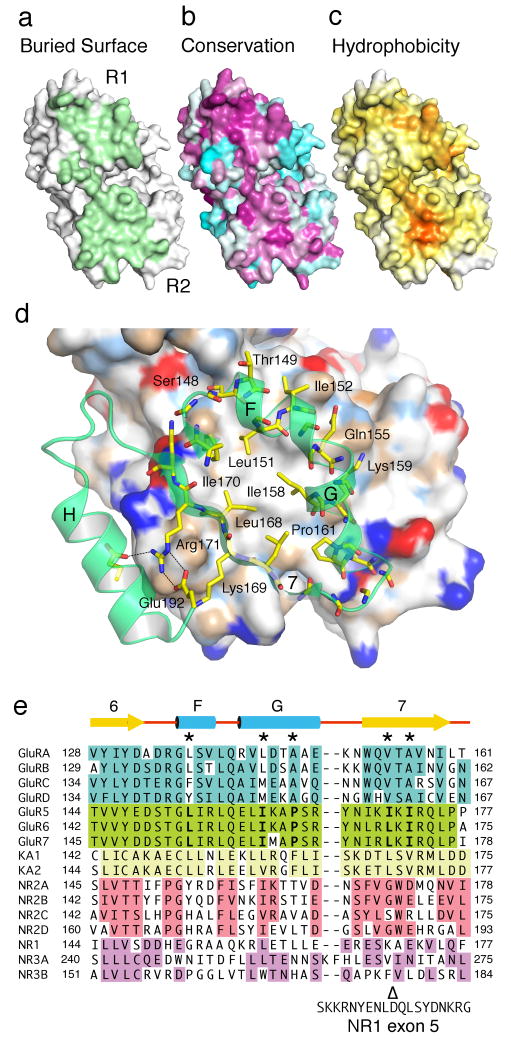
The dimer interface formed by domain R2 has a buried surface area of approximately 750 Å2 per monomer, approximately half of the dimer total (Fig. 6a). Notably, the buried surface area of domain R2 alone has nearly the same size as the total buried area in mGluR dimers7,20,22. Although the domain R1 and R2 dimer surfaces of non-NMDA receptors both show high sequence conservation scores (Fig. 6b) a residue by residue analysis of the free energy for transfer from water to octanol for surface exposed residues23 reveals the presence of a patch in domain R2 which is the most prominent hydrophobic surface feature present in a GluR6 ATD monomer (Fig. 6c). The central hydrophobic patch is formed by residues Leu168 and Ile170 in β strand 7, and by Leu151, Ile158 and Pro161 from helix F & G (Fig. 6d). Ile170 of one monomer stacks with Ile158 of the other; similarly, Leu151 of one chain stacks against Leu151 of the dimer partner. The hydrophobic patch is surrounded by peripheral charged residues forming intermolecular hydrogen bond interactions. On helix G the side chain amide of Gln155 interacts with the hydroxyl group of Tyr145 in the dimer partner, while the side chain amino group of Lys159 interacts with the side chain carbonyl oxygen of Gln172. β strand 7 is amphipathic with hydrophobic residues facing the dimer center, while the side facing the solvent exposed surface of helix H forms a series of intramolecular salt bridges and hydrogen bonds which stabilize the dimer surface of each subunit (Fig. 6d).
Figure 6.
The R2 domain has a hydrophobic patch absent in NMDA receptors. Molecular surface for a GluR6 ATD protomer viewed face on to the dimer interface and colored by (a) buried surface area, (b) sequence conservation scores a calculated from alignments of GluR1-4, GluR5-7 and the KA1 and KA2 subunits, and (c) hydrophobicity. (d) Expanded view of the R2 domain dimer interface, with the rear subunit colored by chemical property; a ring of hydrophobic residues on the interior faces of helices F, G and strand 7 packs against their counterparts in the opposite subunit. (e) Amino acid sequence alignments for the domain 2 dimer surface in five iGluR gene families, with sequence identities within each family indicated by colored backgrounds; five hydrophobic residues which form the dimer interface in GluR6 are conserved in the seven AMPA and kainate receptor genes (positions marked by *), while NMDA receptors contain charged or polar residues at four of the corresponding positions; Δ indicates the point of insertion between Lys170 and Ala171 of 21 amino acids encoded by exon 5 in the NR1b splice variant.
Amino acid sequence alignments reveal that, for the nine AMPA and kainate receptor genes, Leu151 in helix F is highly conserved, while Ile158 and Pro161 in helix G are replaced by either leucine, valine, methionine, alanine or phenylalanine (Fig. 6e and Supplementary Fig. 2). In β strand 7 Leu168 and Ile170 are replaced by either valine, leucine or alanine. In contrast, NMDA receptors have lowest homology with GluR6 on the domain R2 dimer surface, and for both the glutamate binding NR2 and glycine binding NR1 and NR3 subunits several of these hydrophobic residues are replaced by hydrophilic or charged residues, while in the NR1b splice variant β strand 7 is interrupted by the insertion of a highly charged segment of 21 amino acids which regulate sensitivity to polyamines and protons (Fig. 6e). The GluR5 kainate receptor also undergoes alternative splicing which results in insertion of 15 amino acids in the ATD of the GluR5-2 splice variant24; this insertion occurs at the solvent exposed lateral face of domain R2, in the middle of the penultimate β-strand of the GluR6 ATD structure, and in contrast to the NR1b splice variant is unlikely to play any role in dimer assembly.
Discussion
Our results provide insight into the structural organization of the amino terminal domain of iGluRs; reveal features not apparent from homology models based on LIVBP and mGluR1; and highlight the continued need for high resolution crystal structures of neurotransmitter receptors and other signaling molecules in order to understand molecular signaling mechanisms in the brain.
Dimer surfaces differ in iGluR subtypes and mGluRs
The iGluR dimer assembly revealed by our structures agrees well with the results of functional mapping experiments which used GluR3 and GluR6 chimeras (Supplementary Fig. 5). The buried surface area in the GluR6 ATD dimer is approximately 75% larger than that found in dimers of the mGluR1 glutamate complex in structures labeled as the closed-open active A form7, and 240% larger than for the open-open relaxed R form obtained with the antagonist S-MCPG22. This difference reflects the contribution of domain R2 to dimer formation by GluR6 ATDs. A slice through the two fold axis of molecular symmetry reveals smooth surfaces in both domains R1 and R2 in the dimer interface, while the solvent exposed faces of the protein are filled with pockets and protrusions, creating a rough molecular surface less likely to support extensive interactions between protomers (Supplementary movie). The molecular surfaces of both mGluRs and LIVBP in regions corresponding to the GluR6 ATD dimer interface are quite different. The dimer surface of mGluR1 is formed largely by domain 1, which has a hydrophobic buried surface resembling that in domain R2 of GluR6, while the mGluR1 surface of domain 2, facing the axis of symmetry, is studded with hydrophilic residues; for example Glu238 and Lys245 replace Leu151 and Ile158, which form key hydrophobic contacts in the GluR6 dimer interface. We note that in NMDA receptors, and especially the NR1b subunit with the exon 5 splice variant, the surface of domain R2 appears to be more similar to that found in mGluRs.
ATD structure in tetrameric iGluRs
Although iGluRs are tetrameric assemblies, negative stain electron microscopy images of intact AMPA receptors support a dimer of dimers assembly of the ATD, and reveal elongated bipartite densities above the ligand binding domains and ion channel25. In agreement with the electron microscopy analysis, there are no obvious surface features on the GluR6 ATD dimer crystal structure which would enable close apposition of four subunits in a tightly packed tetramer with four fold symmetry. Within a dimer of dimers assembly it seems probable the pairs of dimers assemble via limited contacts on the lateral edges of domain R2 to generate the V-shaped structures seen in electron microscopy images25. However, in other AMPA receptor single particle reconstructions the top surface is more closed indicating, closer contacts between the dimer pairs, but still with 2-fold rather than 4-fold symmetry, also consistent with a dimer of dimers26,27.
Multiple functional roles for the ATD in iGluRs
The ligand binding sites of mGluRs and G-protein coupled GABA receptors contain highly conserved residues most of which are present also in periplasmic proteins which bind polar and hydrophobic amino acids. An 8 amino acid sequence motif containing these elements has been used in bioinformatic studies to identify a family of 146 candidate amino acid binding family 3 G-protein coupled receptors28. Strikingly, none of the ATDs of vertebrate iGluRs contain this signature sequence, making it very unlikely that they bind amino acids, consistent with the result that [3H]glutamate does not bind to the GluR4 ATD14.
The intermediate extent of domain closure for the GluR6 ATD compared to the apo and ligand bound forms of mGluR1 and LIVBP is probably close to the native conformation, since it is also seen in GluR2 ATD crystal structures. Although the GluR6 ATD protomers in the MPD and tartrate crystal forms are slightly more open, by 7° and 11°, compared to the GluR2 ATD, this can be traced to structural differences in loop2; in GluR6 this loop acts as a clamp on domain R2 which prevents further closure (Supplementary Fig. 6), while in GluR2 the loop is shortened by 4 residues and projects laterally, forming contacts with domain R1. Loop 2 is absent in LIVBP, and based on sequence alignments also in NMDA receptors, while in mGluR1 it forms an elaborate substructure on the lateral edge of lobe 1.
Although it is possible that the ATDs of the kainate and AMPA receptor families of iGluRs could bind unidentified ligands, several lines of evidence suggests that they have evolved to act primarily as assembly signals, the most striking of which is the high affinity for dimer formation. In contrast, the Kd for dimer formation by the GluR6 S1S2 glutamate binding domain is in the mM range19. Ligand induced conformational changes involving movement of domain R2, like those found in periplasmic proteins and mGluRs, are compromised by the tight packing of the GluR6 and GluR2 dimer interfaces, and especially by the large hydrophobic patch on domain R2. In addition, the projection of loop 1 into the cleft between domains R1 and R2 of the dimer partner subunit, and interdomain contacts made by loop 2, which in GluR6 projects downwards to make contacts with helix I in domain R2 would further hinder domain opening and closing (Supplementary Fig. 6). In contrast, for mGluRs and other family 3 G-protein coupled receptors, domain 2 plays little role in dimer assembly and is free to move in response to the binding of glutamate or GABA. Loss of ligand binding activity for proteins with the periplasmic binding protein fold is not without precedent and for G-protein GABA receptors, which are obligate heterodimers, only the GB1 subunit binds GABA while the GB2 subunit has low sequence conservation of amino acid binding site residues29,30. In contrast, it is well established that several allosteric modulators of NMDA receptor activity bind to the ATDs, and it is likely that these trigger domain closure producing ligand induced conformational changes like those observed in mGluR dimers31.
Methods
Protein Preparation
The GluR6 R1R2 ATD (residues 1-389) with a C-terminal LELVPRGS-His8 affinity tag and thrombin cleavage site was generated by PCR. Following subcloning, we confirmed the sequence of amplified segments. GluR6 R1R2 was expressed as a secreted protein in transiently transfected wild type HEK293 cells or HEK293 cells lacking N-acetylglucosaminyltransferase I (GnTI) activity and hence unable to synthesize complex N-glycans13. Adherent monolayers cultured in triple layer flasks (Nunc) were grown to ∼ 90% confluency in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum and 2 mM L-glutamine and transiently transfected32 with the “PEI-MAX” form of polyethyleneimine (Polysciences). Media was harvested five days after transfection; Tris-HCl (pH 8.0) and NaCl were added to final concentrations of 50 and 200 mM. GluR6 ATD was eluted at 200 mM imidazole from a Ni2+ charged 1 ml HiTrap chelating HP column (Amersham). The eluate was digested at room temperature with thrombin at 1:400 ratio for 90 minutes and then with Endoglycosidase H at 1:10 ratio for 120 minutes, and further purified by SP Sepharose ion-exchange chromatography. SEC-MALS/RI/UV (Superdex 200 HR 10/30 column, Mini Dawn Treos, Optilab rEX, Wyatt Technology), matrix assisted laser desorption ionization (MALDI PerSeptive Biosystems Voyager-DE) and electrospray ionization (ESI) mass spectrometery (micromass Q-Tof micro with nanolock) were used to estimate MW. Purified protein was concentrated by shock elution from an SP Sepharose column, dialyzed against a storage buffer (20 mM sodium acetate, pH 5.0, 200 mM NaCl, 1 mM EDTA), flash frozen in liquid nitrogen at 2 mg ml-1 and stored at −80 °C.
Crystallization and structure determination
GluR6 ATD (2mg ml-1) in storage buffer was crystallized at 293 °K by vapor diffusion in hanging drops against 0.6 M sodium potassium tartrate, pH 6.0. An additional crystal form was obtained with 100 mM Bicine, pH 9.0, 10% MPD. Crystals were cryoprotected by soaking in mother liquor supplemented with either increasing concentrations of glycerol or MPD to a final values of 18% and 30% v/v.
X-ray diffraction data was collected at APS beamline ID22 at 100 °K using a MAR 300 CCD detector and a wavelength of 1 Å (Table 1). Data was indexed, scaled and merged using HKL200033. The GluR6 ATD tartrate form structure was solved by molecular replacement with Phaser34 using unpublished coordinates for a GluR2ATD monomer as a search probe16. Phaser identified two copies of the GluR6 ATD resulting in a Matthews coefficient of 5.2 with 76.4% solvent content, possibly explaining the observed diffraction resolution limit. The MPD crystal form also contained a dimer in the asymmetric unit, but the unit cell was shortened by 30 Å along the c axis, with a resulting decrease in the Matthews coefficient to 4.0, solvent content of 69.4%. The solutions were verified by inspection of crystal packing and electron density maps (Supplementary Fig. 1). Density modification, model building and refinement was performed with PHENIX35, initially using simulated annealing to overcome model bias. Non-crystallographic symmetry restraints were applied and three TLS groups per monomer identified using motion determination analysis36 were used in refinement together with riding hydrogens. The maps were of reasonable quality for the given resolution, and additional model building into σ A weighted Fo-Fc and 2Fo-Fc and composite omit maps was performed using COOT37, coupled with cycles of crystallographic refinement, resulting in values for Rwork and Rfree of 21.2% and 24.1%, and 21.1 and 25.4% for the tartrate and MPD structures respectively (Table 1). Calculations with MOLPROBITY38 revealed that 95.4% and 95.9% of residues were in the preferred regions of the Ramachandran plot for the tartrate and MPD structures38. Additional crystallographic calculations were performed using CCP439 and the USF suite40. Dimer contact surface areas were calculated using AREAIMOL with a point density value of 20. Figures were prepared using PyMol41 and MOLSCRIPT42. Sequence conservation plots were calculated using ConSurf43.
Analytical ultracentrifugation
Experiments were carried out in a ProteomeLab XL-I analytical ultracentrifuge (Beckman Coulter, Palo Alto, Ca) following standard protocols with a buffer containing 200 mM NaCl, 20 mM Na acetate, 1 mM EDTA, pH 5.0. SV experiments were conducted at 20 °C at a rotor speed of 50,000 rpm, using 0.11 ml samples with 3-fold dilution in 3 mm centerpieces and 0.4 ml samples with 10-fold and 30-fold dilution in 12 mm centerpieces, respectively44. Concentration gradients were measured at 1 min intervals using interference optics and fit with sedimentation coefficient distributions c(s) using maximum entropy regularization on a confidence level of P = 0.90 in SEDFIT45, yielding residuals with rmsd of ∼ 0.003 fringes. The peak locations and the apparent average molar mass values of the reaction boundaries were consistent with a rapid monomer-dimer equilibrium. The c(s) peaks were integrated to determine the weighted-average sedimentation coefficients sw, and the isotherm sw(c) was modeled in SEDPHAT using a monomer-dimer model46. Statistical parameter errors were determined by Monte-Carlo analysis. Theoretical s-values were estimated from the crystal structures of the dimer and monomer using HYDROPRO47. This resulted in values of 3.45 – 3.53 S for the monomer and 5.43 – 5.54 S for the dimer, respectively, dependent on the conformation of 6 vector encoded C-terminal residues not resolved in the crystal structure. For the WT glycosylated protein, the M2/3 scaling law predicts ∼ 7% higher s-values, but with some uncertainty due to the opposing additional effects of the carbohydrate contributions to density and translational friction.
Sedimentation equilibrium (SE) experiments were carried out using ‘aged’ cell assemblies equipped with sapphire windows, filled with 0.15 ml of samples at a 10-fold range of loading concentrations48. Equilibrium was attained sequentially at rotor speeds of 12,000 rpm, 17,000 rpm, and 23,000 rpm, at 10 °C. The radial signal profiles were acquired using interference and absorbance optics at 250 and 280 nm followed by water blank subtraction. The molar interference signal increment was determined from a global multi-signal SE analysis on the basis of the theoretical extinction coefficient predicted from the amino acid composition. Eight interference profiles at different loading concentrations and rotor speeds were loaded in SEDPHAT and modeled with a monomer-dimer or monomer-dimer-tetramer model, respectively, using mass conservation constraints49, fixing all species' buoyant molar masses to the expected values. Statistical parameter errors were estimated using the projection method and F-statistics.
Supplementary Material
Acknowledgments
We thank Dr. E. Gouaux for sharing GluR2 ATD coordinates; Carla Glasser and Andrea Balbo for technical assistance; Drs. D. Leahy, A. Plested and C. Chaudhry for advice and discussion; Drs. P. Seeburg and S. Heinemann for the gift of the wild type iGluR plasmids; Drs. S. Hansen and Philip Reeves (MIT) for the gift of GnTI- cells; Dr. Howard Jaffe, Protein/Peptide sequencing facility, NINDS, for performing mass spectral analysis and N-terminal sequencing; Drs. T. Kawate and E. Gouaux for the gift of Endo H and PNGase F clones; Dr. P. Paoletti for providing coordinates for NR2A and NR2B ATD models; Dr. Jose Garcia de la Torre for the program HYDROPRO. Nucleic acid sequencing was performed by the NINDS DNA sequencing facility. Synchrotron diffraction data was collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This work was supported by the intramural research program of NICHD, NIH, DHHS (MLM).
Footnotes
Accession codes. Protein Data Bank: Coordinates and structure factors have been deposited with accession codes 3H6G and 3H6H.
Author Contributions: Biochemistry and structural biology was performed by JK; AUC by PS; MLM assisted with data collection, structure solution and analysis; RJ solved the GluR2 structure used for molecular replacement; all authors contributed to data analysis and interpretation.
References
- 1.Watkins JC, Evans RH. Excitatory amino acid transmitters. Ann Rev Pharmacol Toxicol. 1981;21:165–204. doi: 10.1146/annurev.pa.21.040181.001121. [DOI] [PubMed] [Google Scholar]
- 2.Hollmann M. Structure of ionotropic glutamate receptors. In: Jonas P, Monyer H, editors. Ionotropic glutamate receptors in the CNS, Handbook of Experiment Pharmacology. Vol. 141. Springer-Verlag; Berlin: 1999. pp. 3–98. [Google Scholar]
- 3.Pin JP, Galvez T, Prezeau L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol Ther. 2003;98:325–54. doi: 10.1016/s0163-7258(03)00038-x. [DOI] [PubMed] [Google Scholar]
- 4.Nakanishi N, Shneider NA, Axel R. A family of glutamate receptor genes: evidence for the formation of heteromultimeric receptors with distinct channel properties. Neuron. 1990;5:569–81. doi: 10.1016/0896-6273(90)90212-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stern-Bach Y, et al. Agonist-selectivity of glutamate receptors is specified by two domains structurally related to bacterial amino acid binding proteins. Neuron. 1994;13:1345–1357. doi: 10.1016/0896-6273(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 6.O'Hara PJ, et al. The ligand-binding domain in metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron. 1993;11:41–52. doi: 10.1016/0896-6273(93)90269-w. [DOI] [PubMed] [Google Scholar]
- 7.Kunishima N, et al. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–7. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 8.Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 9.Masuko T, et al. A regulatory domain (R1-R2) in the amino terminus of the N-methyl-D-aspartate receptor: effects of spermine, protons, and ifenprodil, and structural similarity to bacterial leucine/isoleucine/valine binding protein. Mol Pharmacol. 1999;55:957–969. doi: 10.1124/mol.55.6.957. [DOI] [PubMed] [Google Scholar]
- 10.Paoletti P, et al. Molecular organization of a zinc binding n-terminal modulatory domain in a NMDA receptor subunit. Neuron. 2000;28:911–925. doi: 10.1016/s0896-6273(00)00163-x. [DOI] [PubMed] [Google Scholar]
- 11.Marinelli L, et al. Homology modeling of NR2B modulatory domain of NMDA receptor and analysis of ifenprodil binding. ChemMedChem. 2007;2:1498–510. doi: 10.1002/cmdc.200700091. [DOI] [PubMed] [Google Scholar]
- 12.Mony L, et al. Structural basis of NR2B-selective antagonist recognition by NMDA receptors. Mol Pharmacol. 2009;75:60–74. doi: 10.1124/mol.108.050971. [DOI] [PubMed] [Google Scholar]
- 13.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99:13419–24. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuusinen A, Abele R, Madden DR, Keinanen K. Oligomerization and ligand-binding properties of the ectodomain of the alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunit GluRD. J Biol Chem. 1999;274:28937–28943. doi: 10.1074/jbc.274.41.28937. [DOI] [PubMed] [Google Scholar]
- 15.Wells GB, Lin L, Jeanclos EM, Anand R. Assembly and ligand binding properties of the water-soluble extracellular domains of the glutamate receptor 1 subunit. J Biol Chem. 2001;276:3031–6. doi: 10.1074/jbc.M006668200. [DOI] [PubMed] [Google Scholar]
- 16.Jin R, et al. Crystal structure and association behavior of the GluR2 amino terminal domain. Embo J. 2009 doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kantardjieff KA, Rupp B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 2003;12:1865–71. doi: 10.1110/ps.0350503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer ML. Crystal Structures of the GluR5 and GluR6 Ligand Binding Cores: Molecular Mechanisms Underlying Kainate Receptor Selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 19.Weston MC, Schuck P, Ghosal A, Rosenmund C, Mayer ML. Conformational restriction blocks glutamate receptor desensitization. Nature Structural and Molecular Biology. 2006;13:1120–7. doi: 10.1038/nsmb1178. [DOI] [PubMed] [Google Scholar]
- 20.Muto T, Tsuchiya D, Morikawa K, Jingami H. Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc Natl Acad Sci U S A. 2007;104:3759–64. doi: 10.1073/pnas.0611577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papadakis M, Hawkins LM, Stephenson FA. Appropriate NR1-NR1 disulfide-linked homodimer formation is requisite for efficient expression of functional, cell surface N-methyl-D-aspartate NR1/NR2 receptors. J Biol Chem. 2004;279:14703–12. doi: 10.1074/jbc.M313446200. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchiya D, Kunishima N, Kamiya N, Jingami H, Morikawa K. Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+ Proceedings of the National Academy of Sciences of the United States of America. 2002;99:2660–2665. doi: 10.1073/pnas.052708599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fauchere JL, Pliska V. Hydrophobic paramete π of amino acid side chains from partitioning of N-acetyl-amino amides. Eur J Med Chem. 1983;18:369–375. [Google Scholar]
- 24.Bettler B, et al. Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron. 1990;5:583–595. doi: 10.1016/0896-6273(90)90213-y. [DOI] [PubMed] [Google Scholar]
- 25.Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–9. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- 26.Tichelaar W, Safferling M, Keinanen K, Stark H, Madden DR. The Three-dimensional Structure of an Ionotropic Glutamate Receptor Reveals a Dimer-of-dimers Assembly. J Mol Biol. 2004;344:435–42. doi: 10.1016/j.jmb.2004.09.048. [DOI] [PubMed] [Google Scholar]
- 27.Midgett CR, Madden DR. The quaternary structure of a calcium-permeable AMPA receptor: conservation of shape and symmetry across functionally distinct subunit assemblies. J Mol Biol. 2008;382:578–84. doi: 10.1016/j.jmb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 28.Acher FC, Bertrand HO. Amino acid recognition by Venus flytrap domains is encoded in an 8-residue motif. Biopolymers. 2005;80:357–66. doi: 10.1002/bip.20229. [DOI] [PubMed] [Google Scholar]
- 29.Kniazeff J, Galvez T, Labesse G, Pin JP. No ligand binding in the GB2 subunit of the GABA(B) receptor is required for activation and allosteric interaction between the subunits. J Neurosci. 2002;22:7352–61. doi: 10.1523/JNEUROSCI.22-17-07352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rondard P, et al. Functioning of the dimeric GABA(B) receptor extracellular domain revealed by glycan wedge scanning. Embo J. 2008;27:1321–32. doi: 10.1038/emboj.2008.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gielen M, et al. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–50. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;277:307–344. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, et al. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 36.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–50. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–9. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The CCP4 suite: programs for protein crystallography. Acta Crystallog sect D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 40.Kleywegt GJ, Zou JY, Kjeldgaard M, Jones TA, Around O. Crystallography of Biological Macromolecules. F. Kluwer Academic Publishers; Dordrecht: 2001. pp. 353–356. [Google Scholar]
- 41.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific Palo Alto; CA, USA: 2002. [Google Scholar]
- 42.Kraulis PJ. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J App Crystal. 1991;24:946–950. [Google Scholar]
- 43.Landau M, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown PH, Balbo A, Schuck P. Characterizing protein-protein interactions by sedimentation velocity analytical ultracentrifugation. Curr Protoc Immunol. 2008;Chapter 18, Unit 18 15 doi: 10.1002/0471142735.im1815s81. [DOI] [PubMed] [Google Scholar]
- 45.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–24. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 47.Garcia De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys J. 2000;78:719–30. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balbo A, Brown PH, Braswell EH, Schuck P. Measuring protein-protein interactions by equilibrium sedimentation. Curr Protoc Immunol. 2007;Chapter 18, Unit 18 8 doi: 10.1002/0471142735.im1808s79. [DOI] [PubMed] [Google Scholar]
- 49.Vistica J, et al. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326:234–56. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.