Abstract
Rotavirus is the most common cause of acute gastroenteritis among infants and young children throughout the world, but rotavirus cases in developing countries account for nearly all of the ∼600,000 annual deaths. We studied the epidemiology of rotavirus in 22 rural communities in northern coastal Ecuador over a five-year period. From 250 rotavirus positive stool specimens, the percentage that could not be RT-PCR genotyped for VP4 and VP7 was 77% and 63%, respectively. The possibility of sample degradation was considered but discounted after an experimental examination of rotavirus stability and EM visualization of rotavirus-like particles in several untypeable samples. Finally, alternate primers were used to amplify Ecu534, a sample that was untypeable using most published VP4 and VP7 primers. Characterization of the VP7, VP4, and VP6 full gene segments revealed novel genotypes and nucleotide mismatches with most published primer sequences. When considered with other findings, our results suggest that primer mismatch may be a widespread cause of genotyping failure, and might be particularly problematic in countries with greater rotavirus diversity. The novel sequences described in this study have been given GenBank accession numbers EU805775 (VP7), EU805773 (VP4), EU805774 (VP6) and the RCWG has assigned them novel genotypes G20P[28]I13, respectively.
Keywords: rotavirus, novel genotype, genotyping failure, primer design, RT-PCR, Ecuador
Introduction
Globally, group A rotavirus is responsible for about half of all hospitalizations due to diarrhea and causes more than 600,000 deaths annually, 80% of which are in South Asia and Sub-Saharan Africa (Parashar et al., 2006). Rehydration therapy is a simple and effective treatment, and in countries with well-developed health care infrastructure, mortality is low. In the United States, despite an estimated three million episodes of rotavirus requiring more than 60,000 hospitalizations, rotavirus accounts for only 20–40 deaths each year (Glass et al., 2006; Parashar et al., 2006).
Project EcoDeSS began in 2003 to better understand the epidemiological factors of diarrheal disease the rural, coastal province of Esmeraldas, Ecuador (Eisenberg et al., 2006). This population-based, case-control study collects fecal specimens from all symptomatic residents (regardless of etiologic agent) and from three healthy controls for each case. During the course of the study, Endara et al. (2007) reported rotavirus genotypes for samples collected during 2005 from the rural study location and metropolitan Quito. Genotype G9 was identified in 72% and 90% of these samples, respectively. This finding was in concert with the rapid emergence of G9 reported in many locations throughout the world, for example, Thailand (Khamrin et al., 2006), Brazil (Santos and Hoshino, 2005), and Portugal (Rodrigues et al., 2007).
In Ecuador, as elsewhere in the world, there is evidence that genotype G9 may be declining from its peak prevalence. A recent study in Ecuador has reported G9 at only 43% among samples collected in 10 clinics during 2006 (Naranjo et al., 2008) and our data show that in 2007, G9 prevalence fell to below 10% (unpublished data). A rapid decline was also recently reported in Thailand, where G9 rotavirus peaked at >90% prevalence in 2001–02, but accounted for less than a third of typed samples in the two following years (Khamrin et al., 2007).
Curiously, the decline of G9 rotavirus detected by the EcoDeSS field study was concomitant with a dramatic increase in the rate of untypeable samples. Here, untypeable is defined as a rotavirus positive sample (as ascertained by immunochromatographic assay) failing to yield detectable or interpretable PCR product from the secondary amplification of the primer-specific RT-PCR typing methods for the VP4 (Gentsch et al., 1992) and VP7 (Gouvea et al., 1990) genome segments.
During the 2006–2007 field seasons, approximately 89% and 65% of rotavirus positive samples were untypeable for the VP4 and VP7 genes, respectively. This high rate of typing failure was in contrast to samples collected during the 2005 field season, when Endara et al. (2007) reported untypeable VP4 and VP7 genes among only 26% and 21% of the rural samples. The failure rate from the urban isolates was even lower, 7% and 3%, respectively. One potential explanation for the increase of the genotyping failure rate is that this region of Ecuador harbors high rotavirus genotype diversity, which was temporarily replaced when the G9 strain swept through the region.
Group A rotaviruses may be classified into subgroup (SG) on the basis of their VP6 gene, which encodes the middle capsid protein. VP6 serotyping differentiates group A rotaviruses into four subgroups, based on reactivity with two antibodies: I, II, I+II, and “non-I, non-II” (Greenberg et al., 1983). Studies of VP6 amino acid sequence have observed two VP6 “genogroups”, where genogroup I corresponds to SG I, and genogroup II incorporates the other three SGs (Iturriza-Gómara et al., 2002; Kerin et al., 2007). More recently, 12 nucleotide sequence-based VP6 genotypes were recognized by the Rotavirus Classification Working Group (RCWG) (Matthijnssens et al., 2008b). The VP7 and VP4 genes, which encode the outer capsid G and P proteins, are today also routinely genotyped by RT-PCR and/or sequencing in lieu of serotyping. There are currently 19 VP7 genotypes and 27 VP4 genotypes recognized by the RCWG.
To better understand the cause of rotavirus typing failure in rural Ecuador, a concerted effort was made to characterize a rotavirus positive fecal sample, Ecu534, which was taken from a symptomatic patient but was untypeable using standard VP4 and VP7 primers. Upon finding primers capable of amplifying internal fragments of the VP4 and VP7 genes, a single primer amplification technique was used to obtain the full-length sequences, which were then compared to the sequences of the primers that had failed. In addition, a brief experiment was carried out to ascertain the stability of raw stool samples stored for several days at temperatures similar to the tropical conditions encountered in coastal Ecuador.
Results
Rotavirus stability in raw fecal specimens
The length of time that three rotavirus positive test stool specimens were stored at 30 °C had no effect on subsequent RT-PCR amplification of the VP4 and VP7 gene segments. All three time points (5, 10, and 15 days) for all three samples produced RT-PCR products of equal intensity, when visualized by gel electrophoresis (data not shown). All three samples and time points were also positive by immunochromatographic strip test. Additionally, serial dilutions of one fecal sample (1:10 and 1:100) also produced positive RT-PCR products, although the 1:100 dilution was negative by immunochromatographic strip test.
VP4 amplification and complete sequencing
Table 1 shows the nucleotide sequences of the six VP4 primer pairs commonly cited in the literature, plus a primer pair from the present study. The primer sequences are shown with the complementary sequence from the VP4 gene of isolate Ecu534. No amplification products were detected when primer pairs Con3/Con2, 1–17F/1099R2, 1004F/1937R, F4/C8, or VP4F/VP4R were used (Table 1). All five published primer pairs that failed to amplify Ecu534 have one nucleotide mismatch among the three bases at the 3’ end of one of the primer oligos. Although the sixth published primer pair, HC5/HC3, was not tested in the present study, it had no 3’ mismatches, suggesting it would have successfully amplified the segment. By chance, an amplicon was generated using primers VP4P8F/VP4P8R, despite the fact they were designed in our laboratory for P[8]-specific amplification. The reverse primer VP4P8R matched the Ecu534 template, but the forward primer VP4P8F matched at only eight of 20 nucleotides. Critically, seven of those matches were at the 3’ end of the primer. The 350 bp fragment, though much smaller than the expected size of 1.6kb, was gel isolated, cloned, and sequenced. The result revealed a rotavirus sequence with approximately 60–75% nucleotide identity with other group A VP4 genotypes. Additional primers were designed within the sequence fragment for the single primer amplification technique, producing a complete sequence.
Table 1.
Mismatch of the Ecu534 VP4 sequence with previously published primers. The top sequence is the primer and the bottom sequence is the intended primer binding site in the Ecu534 VP4 sequence. The vertical lines show matches, the asterisks show mismatches, and the colons show partial matches of degenerate bases. The 5’ most base position is shown for the Ecu534 template.
 |
Although not the intended binding site for the forward primer, the site was confirmed by analysis of amplicon sequencing data.
The complete VP4 genome segment of Ecu534 was 2362 bp long and was assigned GenBank accession number EU805773. Table 2 shows the percent identity of the VP4 coding nucleotide sequence (cds) and deduced amino acid sequence (aa), compared with 28 rotavirus VP4 gene sequences downloaded from GenBank that were selected to represent each of the recognized VP4 genotypes. The first 247 amino acids of the full-length VP4 gene, which constitute the VP8* subunit, is also compared. The maximum aa percent identity was 80% when compared with simian rotavirus isolate SA11 (genotype P[2]), but the percent identity with feline isolate FRV64 (genotype P[3]) was nearly the same. Nucleotide and peptide sequence BLAST searches of GenBank did not reveal any significantly closer matches than the best matches displayed in Table 2. The RCWG has assigned novels genotype P[28] to the Ecu534 VP4 sequence.
Table 2.
Percent identity of isolate Ecu534 VP4 coding nucleotide (cds) and amino acid (aa) sequence compared with 27 rotavirus VP4 genotypes from GenBank. The aa identity for the VP8* subunit of VP4 is also shown. The most similar two sequences are shown in bold.
| Genotype | hosta | Accession no. | isolate name | VP4 cds | VP4 aa | VP8* aa |
|---|---|---|---|---|---|---|
| P[1] | bo | Y00127 | C486 | 72% | 78% | 67% |
| P[2] | si | X14204 | SA11 | 73% | 80% | 69% |
| P[3] | fe | D14723 | FRV64 | 73% | 80% | 68% |
| P[3] | ca | D13400 | K9 | 72% | 79% | 67% |
| P[4] | hu | AB118025 | DS-1 | 68% | 70% | 58% |
| P[5] | bo | M63267 | UK-or-B641 | 67% | 70% | 57% |
| P[6] | hu | AF079356 | US1205 | 68% | 71% | 56% |
| P[7] | po | X13190 | OSU | 71% | 76% | 67% |
| P[8] | hu | L34161 | Wa | 67% | 69% | 54% |
| P[9] | hu | D90260 | K8 | 68% | 70% | 58% |
| P[10] | hu | M60600 | 69M | 72% | 78% | 68% |
| P[11] | hu | L07934 | 116E | 60% | 59% | 40% |
| P[12] | eq | D16342 | FI23 | 71% | 76% | 64% |
| P[13] | po | L07886 | MDR-13 | 70% | 74% | 61% |
| P[14] | hu | D14032 | Mc35 | 67% | 71% | 59% |
| P[15] | ov | L11599 | Lp14 | 73% | 77% | 69% |
| P[16] | mu | U08419 | EB | 67% | 74% | 59% |
| P[17] | av | AB009632 | PO-13 | 62% | 61% | 39% |
| P[18] | eq | D13399 | L338 | 72% | 74% | 64% |
| P[19] | hu | D38052 | Mc323 | 70% | 72% | 60% |
| P[20] | mu | U08424 | EHP | 69% | 75% | 66% |
| P[21] | bo | AF237665 | Hg18 | 70% | 72% | 56% |
| P[22]b | la | AF526373 | 308/01 | 64% | 61% | 60% |
| P[23]b | po | AY768809 | 34461-4 | 67% | 69% | 66% |
| P[24] | si | AY596189 | TUCH | 72% | 77% | 64% |
| P[25] | hu | AY773004 | Dhaka6 | 68% | 68% | 54% |
| P[26] | po | DQ061053 | 134/04-15 | 70% | 75% | 64% |
| P[27] | po | DQ534016 | CMP034 | 69% | 72% | 56% |
| P[28] | hu | EU805773 | Ecu534 | - | - | - |
Host abbreviations: av, avian; bo, bovine; ca, canine; ch, chicken; eq, equine; fe, feline; hu, human; la, lapine; mu, murine; ov, ovine; po, porcine; si, simian; ty, turkey.
Genotype represented by strain with incomplete sequence data.
Figure 1 shows a phylogenetic tree of isolate Ecu534 VP4 nucleotide sequence, compared with the other 27 existing genotypes. Several sequences for each genotype were selected to provide a representative sample of the sequence diversity that exists within and between the established genotypes.
Figure 1. Unrooted phylogenetic tree of VP4 coding sequence.
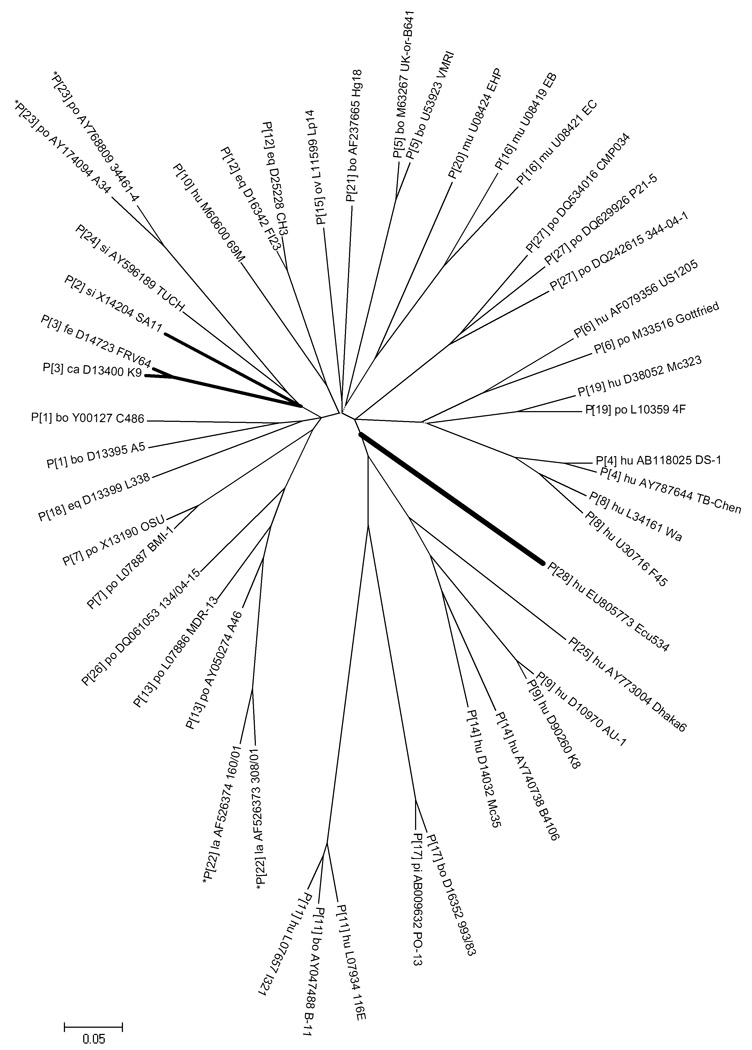
For each genotype, several representative sequences from GenBank are shown to provide perspective on the relative distance within and between genotypes. (The sequence alignment is available from the authors by request.) Strain Ecu534 (genotype P[28]) and three top BLAST hits are highlighted with bold branches. Evolutionary distances are in the units of base substitutions per site and were computed using the Maximum Composite Likelihood method (Tamura et al., 2004), as implemented by MEGA4 (Tamura et al., 2007). The optimal tree was constructed using the Neighbor-Joining method (Saitou and Nei, 1987). Nucleotide positions with missing data or gaps were eliminated only in pairwise sequence comparisons, which mainly affected the incomplete P[22] and P[23] genotype sequences (indicated by asterisks). All nodes have ≥90% bootstrap support, except for the three very short internal branches indicated by dotted lines.
VP7 amplification and complete sequencing
Table 3 shows the nucleotide sequences of six VP7 primer pairs cited in the literature. The primer sequences are shown with the complementary VP7 sequence of isolate Ecu534. No amplification products were detected when primer pairs Beg9/End9, LID1/G922, and sBeg9/RVG9 were used (Table 3). All three of these pairs had one nucleotide mismatch among the three bases at the 3’ end of one of the primer oligos. Although primer pairs VP7F/VP7R and 9con1L/VP7-RDg were not tested, VP7F/VP7R also shows nucleotide mismatches among the last three bases, and so would have been unlikely to produced an amplicon. Of the primer pairs tested, only 9con1/9con2 produced a detectable amplicon. Here, the reverse primer 9con2 matched perfectly with the Ecu534 template and forward primer 9con1 had only three mismatches, mostly in the 5’ half of the oligo. The resulting amplicon was directly sequenced, and from the sequence, additional primers were designed for the single primer amplification technique, producing a complete sequence.
Table 3.
Mismatch of the Ecu534 VP7 sequence with previously published primers. The top sequence is the primer and the bottom sequence is the intended primer binding site in the Ecu534 VP4 sequence. The vertical lines show matches, the asterisks show mismatches, and the colons show partial matches of degenerate bases. The 5’ most base position is shown for the Ecu534 template.
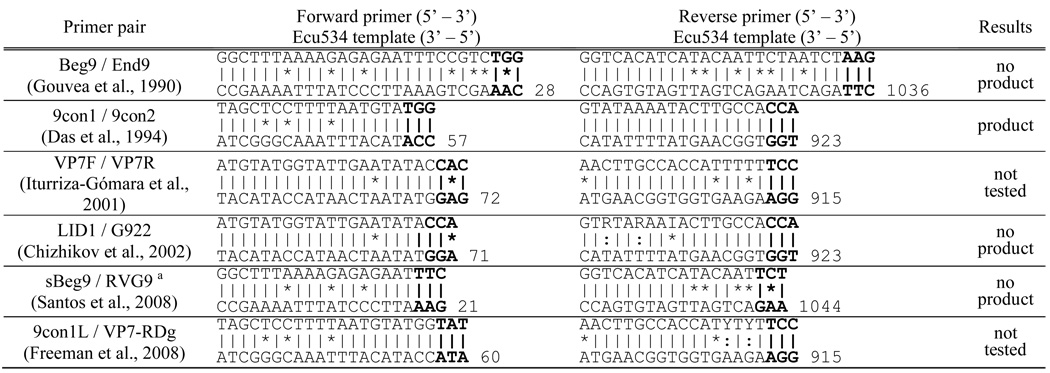 |
Santos et al. suggested using primers sBeg9 (Gouvea et al., 1994) and RVG9 (Gouvea et al., 1990) together.
The complete sequence of the VP7 genome segment of Ecu534 was 1062 bp long and was assigned GenBank accession number EU805775. Table 4 shows the percent identity of the VP7 coding nucleotide sequence (cds) and the deduced amino acid (aa) sequence, compared with 16 rotavirus VP7 genotypes, selected to represent the breadth of VP7 genotype diversity. The maximum percent aa identity was 87.7%, when compared with the G3 sequence from human isolate CMH222. GenBank contained several other G3 sequences with nearly the same percent aa identity, including simian isolate SA11 and canine isolate RV52/96. The RCWG has assigned novel genotype G20 to the Ecu534 VP7 sequence.
Table 4.
Percent identity of isolate Ecu534 VP7 coding nucleotide (cds) and amino acid (aa) sequence compared with 19 rotavirus VP7 genotypes from GenBank. The most similar three sequences are shown in bold.
| Genotype | Hosta | Accession no. | Isolate name | VP7 cds | VP7 aa |
|---|---|---|---|---|---|
| G1 | hu | K02033 | Wa | 72% | 78% |
| G2 | hu | D50124 | KUN | 70% | 71% |
| G3 | bo | DQ487203 | B31 | 74% | 84% |
| G3 | la | AF528204 | 30/96 | 76% | 86% |
| G3 | po | DQ256503 | CMP099 | 76% | 86% |
| G3 | si | V01190 | SA11-H96 | 77% | 87% |
| G3 | ca | AF271090 | RV52/96 | 78% | 86% |
| G3 | hu | AY707792 | CMH222 | 78% | 88% |
| G4 | po | X06759 | Gottfried | 72% | 74% |
| G5 | po | X04613 | OSU | 75% | 82% |
| G6 | bo | D12710 | KN-4 | 74% | 82% |
| G6 | bo | X00896 | UK | 74% | 82% |
| G7 | ch | X56784 | CH2 | 61% | 59% |
| G8 | hu | L20882 | HAL1166 | 74% | 80% |
| G9 | hu | AF060487 | US1205 | 74% | 84% |
| G10 | bo | X57852 | B223 | 73% | 81% |
| G10 | hu | D14033 | Mc35 | 75% | 82% |
| G11 | hu | AY773003 | Dhaka6 | 74% | 83% |
| G12 | po | DQ204743 | RU172 | 74% | 79% |
| G13 | po | D13549 | L338 | 73% | 75% |
| G14 | eq | D25229 | CH3 | 73% | 79% |
| G15 | bo | AF237666 | Hg18 | 70% | 74% |
| G16 | mu | AF039220 | EDIM | 74% | 83% |
| G17 | ty | S58166 | Ty-1 | 64% | 61% |
| G18 | po | D82979 | PO-13 | 64% | 59% |
| G19 | av | AB080738 | Ch-1 | 62% | 58% |
| G20 | hu | EU805775 | Ecu534 | - | - |
| n/ab | bo | AB259665 | Tak2 | 67% | 65% |
Host abbreviations: av, avian; bo, bovine; ca, canine; ch, chicken; eq, equine; hu, human; la, lapine; mu, murine; po, porcine; si, simian; ty, turkey.
Genotype not yet assigned to this isolated by the RCWG due to incomplete sequence. Comparison of this strain with Ecu534 was done with the available incomplete sequence.
Figure 2 shows a phylogenetic tree of isolate Ecu534 VP7 nucleotide sequence, compared with existing genotypes. Several sequences for each genotype were selected to provide a representative sample of the of sequence diversity that exists within and between the established genotypes.
Figure 2. Unrooted phylogenetic tree of VP7 coding sequence.
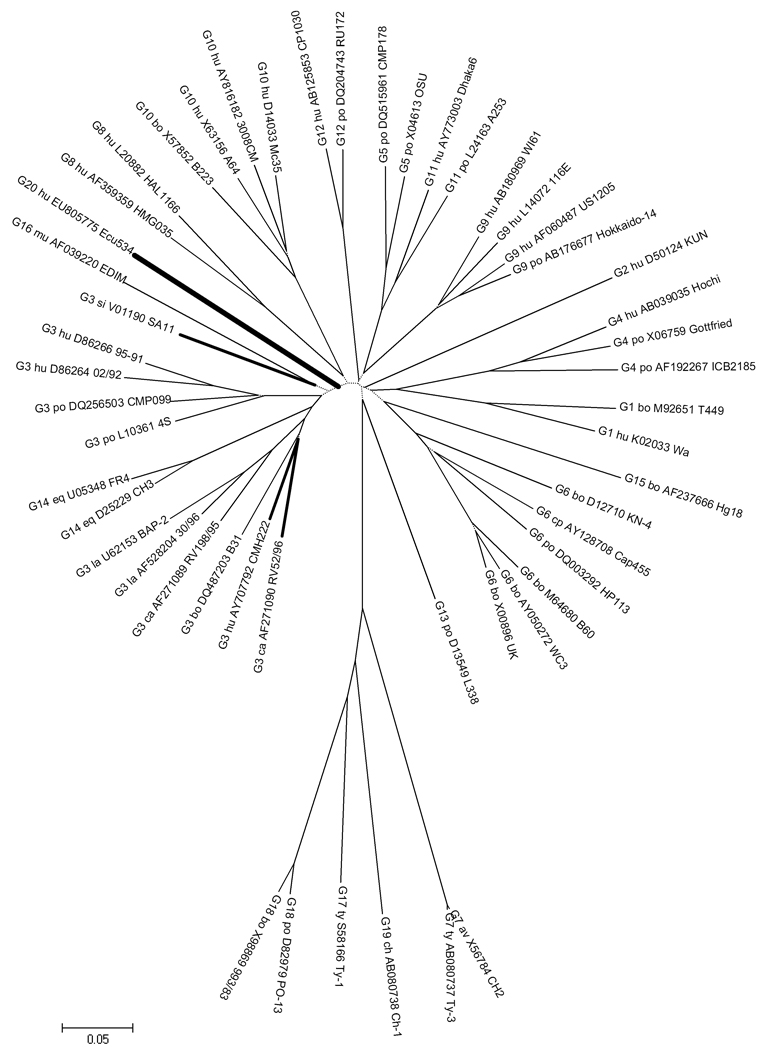
For each genotype, several representative sequences from GenBank are shown to provide perspective on the relative distance within and between genotypes. (The sequence alignment is available from the authors by request.) Strain Ecu534 (genotype G20) and most similar sequences are highlighted with bold branches. See Figure 1 caption for tree construction details. All nodes have ≥90% bootstrap support, except for the short internal branches indicated by dotted lines.
VP6 amplification and sequencing
The complete sequence of the VP6 genome segment of Ecu534 was 1357 bp long and was assigned GenBank accession number EU805774. Because most known complete VP6 sequences are 1356 bp long, the insertion of an additional adenine in the 3’ untranslated region was positively confirmed by resequencing. Table 5 shows the percent identity of the VP6 coding nucleotide sequence (cds) and the deduced amino acid sequence (aa), compared with four prototype VP6 sequences and the nearest BLAST hit, isolate 4S. The maximum percent identity by amino acid sequence was 85%, when compared with strain Hosokawa (SG II, genotype I1) and several porcine strains (genotype I5). The RCWG has assigned novel genotype I13 to the Ecu534 VP6 sequence.
Table 5.
Percent identity of isolate Ecu534 VP6 coding nucleotide (cds) and amino acid (aa) sequence compared with 12 rotavirus VP6 genotypes from GenBank. Four of the most similar sequences are shown in bold.
| Genotype | Hosta | Accession no. | Isolate name | VP6 cds | VP6 aa |
|---|---|---|---|---|---|
| I1 | hu | U36240 | E210 | 75% | 84% |
| I1 | hu | DQ870492 | Hosokawa | 75% | 85% |
| I1 | hu | EF426125 | US8970 | 75% | 84% |
| I1 | hu | K02086 | Wa | 74% | 84% |
| I1 | po | D00326 | Gottfried | 75% | 84% |
| I2 | hu | D00325 | 1076 | 75% | 83% |
| I2 | hu | AY787645 | TB-Chen | 75% | 84% |
| I2 | hu | AF079357 | US1205 | 75% | 82% |
| I3 | hu | DQ490538 | AU-1 | 75% | 84% |
| I4 | av | D16329 | PO-13 | 69% | 72% |
| I5 | po | L29184 | 4F | 77% | 84% |
| I5 | po | L29186 | 4S | 77% | 84% |
| I5 | po | AF317125 | A411 | 75% | 85% |
| I5 | po | DQ534018 | CMP034 | 76% | 84% |
| I6 | eq | D00323 | FI14 | 76% | 82% |
| I7 | mu | U36474 | EW | 72% | 83% |
| I8 | hu | DQ288659 | CMH222 | 76% | 84% |
| I9 | si | AY594670 | TUCH | 75% | 84% |
| I10 | ov | L11595 | Lp14 | 75% | 83% |
| I11 | ch | X98870 | Ch-1 | 67% | 70% |
| I12 | hu | N/A | KTM368 | 74% | 82% |
| I13 | hu | EU805774 | Ecu534 | - | - |
Host abbreviations: av, avian; ch, chicken; eq, equine; hu, human; mu, murine; ov, ovine.
Figure 3 shows a phylogenetic tree of isolate Ecu534 VP6 coding nucleotide sequence, compared with existing genotypes. Several sequences for each genotype were selected to provide a representative sample of the of sequence diversity that exists within and between the four established subgroups and 12 established genotypes.
Figure 3. Unrooted phylogenetic tree of VP6 coding sequence.
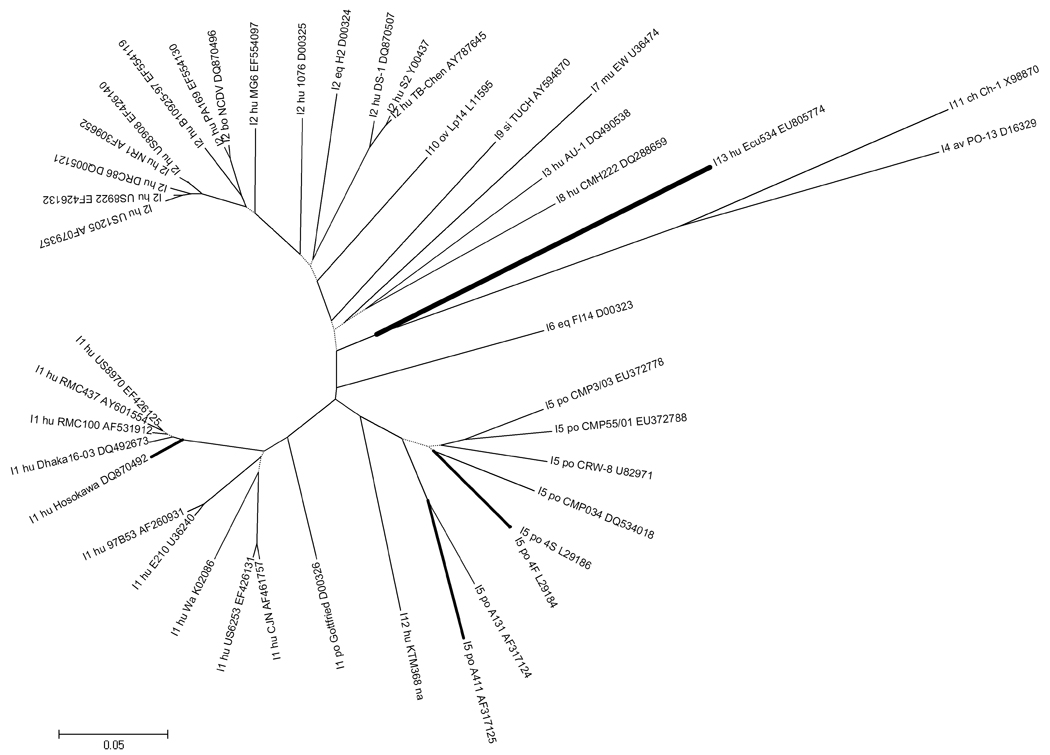
Representative sequences from GenBank of all 13 known VP6 genotypes are shown to provide perspective on the relative distance within and between genotypes. (The sequence alignment is available from the authors by request.) Genogroup I strains are in the top portion of the figure (DS-1-like, genotype I2) and genogroup II strains are found in the bottom portion of the figure (Wa-like, genotype I1). Strain Ecu534 (genotype I13) and most similar sequences are highlighted with bold branches. See Figure 1 caption for tree construction details.
Screening of other untypeable samples
Out of 27 previously untypeable samples that were re-screened with newly designed VP4 and VP7 primers specific for Ecu534, only one produced positive results. Seven previously typed samples were also re-screened with the new primers, all producing negative results. Ecu534, included as a positive control, produced positive results.
Discussion
The present study establishes that rotavirus isolate Ecu534, previously untypeable in our laboratory, is a novel strain and possesses VP4 and VP7 nucleotide sequences unlikely to be amplified by most (9 out of 12) published primer pairs. In the process of troubleshooting the genotyping failure, alternative explanations were explored. Our experiment on degradation suggests that the doublestranded, triple-encapsulated genome is relatively sturdy, maintaining integrity even in cases of sample mishandling (i.e., elevated temperatures for extended periods), a result in agreement with several more remarkable reports of rotavirus genome stability (Rahman et al., 2004; Whittier et al., 2004) and infectivity (Espinosa et al., 2008; Fischer et al., 2002; Ramos et al., 2000) after long periods of time.
The genetic distances between Ecu534 VP4, VP7, and VP6 nucleotide sequences and established genotypes suggest this isolate represents a novel strain. The maximum aa identity for VP4 (80%, Table 2), VP7 (88%, Table 4), and VP6 (85%, Table 5) are all below the conventional threshold (89%) to establish new genotypes (Gorziglia et al., 1990). Phylogenetic analysis also supports the novelty of Ecu534, as its branch does not root among its closest matches for the VP4 (Figure 1), VP7 (Figure 2), or VP6 (Figure 3) trees. Ecu534 VP7, VP4, and VP6 sequences have been assigned the new genotype G20P[28]I13 by the Rotavirus Classification Working Group (RCWG) (Matthijnssens et al., 2008b).
Serotype assignment of Ecu534 was not possible due to the lack of reference antibodies in our laboratory. Amino acid sequence analysis of antigenically important regions offers clues, but no definitive conclusions. For example, although the VP7 protein of Ecu534 shares 86–88% identity with other G3 isolates, it has numerous substitutions in three regions thought to be most critical in epitope determination (Figure 4), including positions 147 and 208 (Ciarlet et al., 1996; Trinh et al., 2007).
Figure 4. Alignment of VP7 amino acid sequences associated with epitope determination.
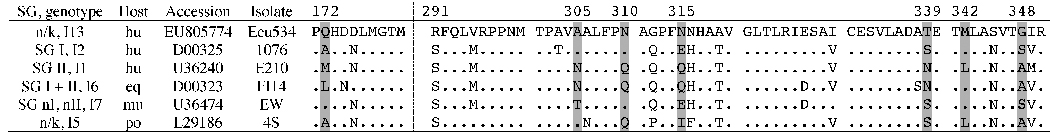
Alignment of the VP7 amino acid sequences of Ecu534 with G3 genotype representatives. Antigenic regions A, B, and C are marked.
The VP6 gene of Ecu534 does not group closely with isolates representing the four known SGs and 12 known genotypes (Table 5). The VP6 amino acid sequence of Ecu534 possesses the residues characteristic of SG I at positions 305 (Ala) and 310 (Asn) (Figure 5) (Iturriza-Gómara et al., 2002; Kerin et al., 2007; Tang et al., 1997). However, these positions are known only to be prerequisites, not determinants, for monoclonal antibody binding (Buragohain et al., 2008), so we do not conclude that Ecu534 would exhibit SG I reactivity. Moreover, its atypical residues at other antigenically important positions (172, 315, 339, and 348) and its relatively great genetic distance from other known strains (Figure 3 and Table 5) both discourage SG prediction based on a few amino acid positions.
Figure 5. Alignment of VP6 amino acid sequences associated with epitope determination.
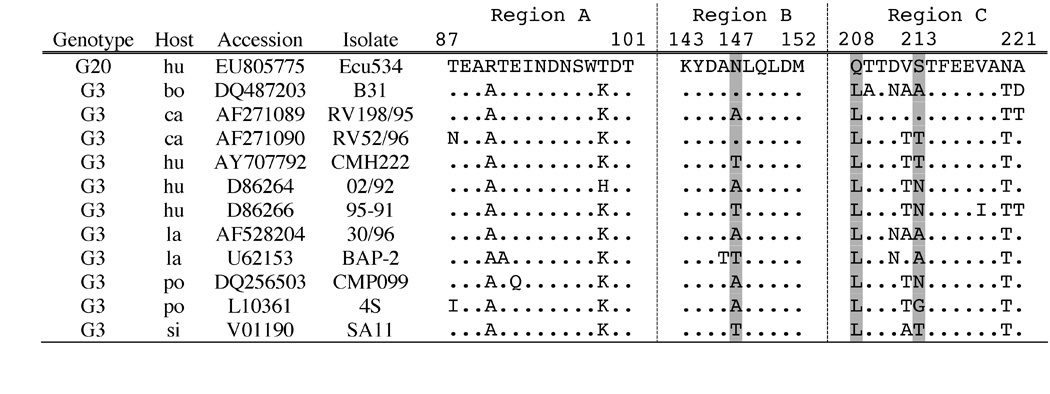
Alignment of the VP6 amino acid sequences of Ecu534 with five subgroup representatives. Positions 171–180 and 291–350 are shown. Positions associated with subgroup specificity are marked with asterisks. See text for details.
The high rate of primer-based genotyping failure is an unresolved problem in rotavirus research. Not surprisingly, it has often been attributed to single nucleotide polymorphisms at the primer binding sites (Adah et al., 1997; Espinola et al., 2008; Griffin et al., 2002; Rahman et al., 2005b). Attempts to cope with this challenge include primer redesign (Banyai et al., 2003; Cunliffe et al., 1999; Iturriza-Gómara et al., 2000; Iturriza-Gómara et al., 2004; Santos et al., 2003), microarray-based typing and detection (Chizhikov et al., 2002; Honma et al., 2007; Lovmar et al., 2003; Santos et al., 2008), as well as qPCR approaches for detection, using the more highly conserved VP2 (Gutierrez-Aguirre et al., 2008) and NSP3 (Freeman et al., 2008) gene segments. Robust sets of detection primers are welcome and needed tools for rotavirus research.
Recently, Simmonds et al. (2008) designed and tested new primers for the initial amplification of the VP4 gene. The new primers were designed by identifying consensus regions using more sequences (mainly from USA, UK, and India) than were available 16 years earlier when the Con3/Con2 primers were designed. Overall, the new primers successfully typed 64% of 366 previously untypeable isolates from geographically diverse locations. Interestingly, the improvement in the UK (92%), USA (72%, which includes some non-USA samples), and Australia (75%) was better than in India (49%), Namibia (23%), and Ghana (54%). Although this valuable study discusses several possible causes for this differential, such as different screening techniques and different sample preparation techniques, another explanation that should be considered is that the rotavirus samples from India and Africa are more diverse and thus less likely to be amplified with a given set of primers.
Two decades of molecular genotyping studies have greatly expanded our appreciation for role of genomic reassortment, mixed infections, and interspecies transmission in generating the tremendous diversity of rotavirus genotypes, and there is evidence that some of these processes are amplified in developing countries (for a comprehensive review, see Gentsch et al., 2005). Likewise, while RT-PCR genotyping failure is common in general (many studies report failure rates of 10–30%), the highest rates tend to be seen in developing countries (Fischer et al., 2000; summary table in Gentsch et al., 2005; Gladstone et al., 2008; Laird et al., 2003; Parra et al., 2005; Ramachandran et al., 1996). Animal rotavirus isolates are also difficult to type by RT-PCR using known primers; a failure rate of 73% was recently reported from a study of porcine rotavirus in Argentina (Parra et al., 2008). Many of the newest rotavirus genotypes were discovered in developing countries or from animal studies. To what extent do primer-based genotyping methods bias our understanding of rotavirus diversity?
From 250 rotavirus-positive stool samples collected in rural Ecuador, 153 were untypeable using conventional RT-PCR genotyping; isolate Ecu534 accounts for only one of these samples. We are intrigued by the following observations: samples we collected from an urban hospital showed a much lower rate of genotyping failure than those collected from the rural communities; the peak incidence of G9 rotavirus in 2005 corresponded with a low rate of genotyping failure; and sample degradation appeared to be an unlikely culprit. Because the novel strain was found in only one additional (out of 27) untypeable samples, further investigation is clearly necessary to know how many of the total remaining 151 untypeable samples exhibit other novel sequences, and to understand the relative diversity among this potential pool of novel strains. If the diversity is limited, it may be possible to design new primers for more reliable genotyping. However, if the genetic diversity of untypeable isolates is high, alternative approaches may be necessary; i.e., no primer design will successfully type many of the isolates. Even if an isolate is genotypically rare and found mainly in provincial settings or animal reservoirs, as a whole these novel genotypes are likely to be important players in the evolutionary epidemiology of a recombining RNA virus like rotavirus.
Material and methods
Initial amplification and sequencing of Ecu534
Ecu534 was a rotavirus-positive fecal sample collected from a person with diarrhea in the Esmeraldas province of Ecuador in July, 2006. It was one of ∼3100 samples, and one of 250 rotavirus-positive samples, collected as part of the EcoDeSS project. Samples were collected, tested, transported, stored, and RNA extracted as described previously (Endara et al., 2007). Attempts were made to RT-PCR amplify the RNA using published primers for the VP4, VP7, and VP6 genes. For VP4 and VP7 genes, various published primer pairs were used (see Table 1 and Table 3). For the VP6 gene, primers GEN-VP6F (5’ GGCTTTWAAACGAAGTCTTC 3’) and GEN-VP6R (5’ GGTCACATCCTCTCACT 3’) were used (Matthijnssens et al., 2008a). Changes were made to the PCR reaction conditions, such as lowering the annealing temperature to encourage primer-template hybridization, and adding DMSO to discourage formation of RNA secondary structure.
To prepare the template for the RT-PCR reaction, 1 µL of DMSO was mixed with 4 µL of the RNA sample. The mixture was heated to 99 °C for 1 min and then immediately quenched in ice water. The sample was then used in a 20 µL (final volume) reaction containing: 1X buffer, 0.4 mM each dNTP, 0.6 µM each primer, 4 units RNasin (Promega, Madison, WI), and 0.8 µL OneStep enzyme mix (QIAGEN, Valencia, California). The temperature cycling was 50 °C for 60 min, 95 °C for 15 min, 35 cycles of (94 °C for 30 sec, 50 °C for 30 sec, 72 °C for 1:45 min), followed by 72 ° for 7 min. The PCR amplicons were purified with the QIAquick PCR purification kit (QIAGEN) and were directly sequenced if possible, or were gel extracted with the QIAquick gel extraction kit (QIAGEN) prior to direct sequencing, if necessary. Sequencing was performed at the UC Berkeley DNA Sequencing Facility.
Linker ligation and full length cDNA synthesis
Primers were designed from the internal fragments of the VP4 and VP7 genes for use with the “single primer amplification technique” of Lambden et al. (1992). The approach exploits the double-stranded RNA genome by ligating a “linker” oligonucleotide to the 3’ end of each strand, and which has a 3’ amine modification to prevent auto-ligation. The linker oligo then provides a primer binding site that can be used in conjunction with a specific primer at a binding site of known sequence.
Attoui et al. (2000) presented a modification of the Lambden technique, to shorten the protocol and reduce the handling steps for the ligated RNA. A modification of this procedure was used in the present study. In brief, the TGP-linker oligo (Matthijnssens et al., 2006) was ligated onto the 3’ ends of the RNA in a ligation reaction mixture containing approximately 50 ng RNA, 1X ligation buffer, 1µM TGP linker, 20 units T4 RNA ligase 2 (NEB, Ipswich, MA), 20 units RNasin (Promega), and 30 µg/mL of BSA (Promega) in a final volume of 50 µL. The reaction mixture was incubated at 16 °C for 16 hours. The RNA was then processed with the RNeasy kit (QIAGEN) according to the manufacturer's instructions. DMSO was added to the RNA elutant to a concentration of 15% and mixed well. The mixture was then heated at 99 °C for 1 min, followed immediately by an ice water quench.
Synthesis of cDNA and amplification reaction was carried out in one tube using QIAGEN OneStep RT-PCR Kit, according to the manufacturer’s protocol. The reactions contained 1 µM of the primer TGP-out (Matthijnssens et al., 2006) and an equal concentration of a primer designed from the internal sequence of Ecu534. The resulting PCR amplicon was sequenced directly, by primer walking from the internal primers towards the 5’ and 3’ ends. Quality control for sequence data submitted to GenBank was provided by sequencing both strands (plus and minus) from two independent RT-PCR reactions, for an average of 4x coverage. Due to the single primer amplification technique, the 20–40 bp at the extreme 5’ and 3’ ends of the genome segments were sequenced at only 2x coverage, again from two independent RT-PCR reactions to help eliminate sequencing errors.
Screening of other untypeable samples
A set of detection primers was designed from the novel VP4 and VP7 sequences of isolate Ecu534. RT-PCR reactions were carried out as described above, using 27 of the 152 previously untypeable RNA samples. Seven of the 97 previously typeable samples, plus Ecu534 itself, were also used. RTPCR product was visualized on a 1.5% agarose gel. A positive result was counted if a sample showed a band of similar size as Ecu534.
Phylogenetic analysis
Individual sequencing reads were assembled by the phred-phrap-consed suite of programs (Ewing et al., 1998; Gordon et al., 1998). The resulting VP4, VP6, and VP7 sequences were aligned using MAFFT version 6 (Katoh et al., 2002) with sequences collected from GenBank to represent each of the currently known genotypes. BioEdit was used for visual inspection of the alignment and to calculate nucleotide and amino acid identity. MEGA4 was used to construct neighbor-joining phylogenetic trees (Tamura et al., 2007). The sequence alignment files, including the accession numbers of the sequences used for genotype reference purposes, are available from the authors by request.
Rotavirus stability in raw fecal specimens
Three fecal samples, positive for rotavirus by an immunochromatographic test (RIDA Quick Rotavirus, R-Biopharm AG, Darmstadt, Germany) were collected from a hospital in California, USA. The fecal samples were from children aged 12, 14, and 24 months, and each had been stored at 4 °C for 24–48 hours prior to collection. Each sample was then stored in 2 mL plastic tubes at 30 °C. After 5, 10, and 15 days, an aliquot of 200 mg was removed from each tube and frozen to −80 °C. After several weeks at −80 °C, all aliquots were thawed, retested with an immunochromatographic strip, and purified for RNA using a column-format kit. Briefly, a 20% suspension was made by adding 800 µL of 50 mM tris buffer to the 200 mg fecal aliquot. The suspension was vortexed vigorously for 1 min, centrifuged at 8000 g for 1 min, and 560 µL of supernatant was removed and processed with a QIAamp Viral RNA Mini Kit (QIAGEN, Valencia, California) according to the manufacturer’s protocol.
An additional aliquot of one of the three fecal samples was made on the day of collection. This aliquot was diluted 1:10 and 1:100, and both dilutions were frozen to −80 °C. Like the neat samples, the dilutions were thawed after several weeks, retested with an immunochromatographic strip, and processed as described above. (The dilutions were considered neat for the purposes of the RNA extraction, i.e., the same 10% suspension was made.)
Acknowledgments
We thank the Ecologia, Desarrollo, Salud, y Sociedad (EcoDeSS) project field team for their invaluable contribution collecting the field data. This work was supported by a National Institute of Allergy and Infectious Diseases Grant R01-AI050038.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Owen D. Solberg, Email: solberg@berkeley.edu.
Maria Eloisa Hasing, Email: meloisa@hasing.com.
Gabriel Trueba, Email: gabriel@usfq.edu.ec.
Joseph N. S. Eisenberg, Email: jnse@umich.edu.
Works Cited
- Adah MI, Rohwedder A, Olaleyle OD, Werchau H. Nigerian rotavirus serotype G8 could not be typed by PCR due to nucleotide mutation at the 3' end of the primer binding site. Arch Virol. 1997;142(9):1881–1887. doi: 10.1007/s007050050206. [DOI] [PubMed] [Google Scholar]
- Attoui H, Billoir F, Cantaloube JF, Biagini P, de Micco P, de Lamballerie X. Strategies for the sequence determination of viral dsRNA genomes. J Virol Methods. 2000;89(1–2):147–158. doi: 10.1016/s0166-0934(00)00212-3. [DOI] [PubMed] [Google Scholar]
- Banyai K, Gentsch JR, Griffin DD, Holmes JL, Glass RI, Szucs G. Genetic variability among serotype G6 human rotaviruses: identification of a novel lineage isolated in Hungary. J Med Virol. 2003;71(1):124–134. doi: 10.1002/jmv.10462. [DOI] [PubMed] [Google Scholar]
- Buragohain M, Cherian SS, Prabhakar G, Chitambar SD. VP6 capsid protein of chicken rotavirus strain CH2: Sequence, Phylogeny and In Silico antigenic analyses. Virus Res. 2008 doi: 10.1016/j.virusres.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Chizhikov V, Wagner M, Ivshina A, Hoshino Y, Kapikian AZ, Chumakov K. Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization. J Clin Microbiol. 2002;40(7):2398–2407. doi: 10.1128/JCM.40.7.2398-2407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarlet M, Hidalgo M, Liprandi F. Cross-reactive, serotype-and monotype-specific neutralization epitopes on VP7 of serotype G3 and G5 porcine rotavirus strains. Arch Virol. 1996;141(3–4):601–614. doi: 10.1007/BF01718320. [DOI] [PubMed] [Google Scholar]
- Cunliffe NA, Gondwe JS, Broadhead RL, Molyneux ME, Woods PA, Bresee JS, Glass RI, Gentsch JR, Hart CA. Rotavirus G and P types in children with acute diarrhea in Blantyre, Malawi, from 1997 to 1998: predominance of novel P[6]G8 strains. J Med Virol. 1999;57(3):308–312. [PubMed] [Google Scholar]
- Das BK, Gentsch JR, Cicirello HG, Woods PA, Gupta A, Ramachandran M, Kumar R, Bhan MK, Glass RI. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994;32(7):1820–1822. doi: 10.1128/jcm.32.7.1820-1822.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg JN, Cevallos W, Ponce K, Levy K, Bates SJ, Scott JC, Hubbard A, Vieira N, Endara P, Espinel M, Trueba G, Riley LW, Trostle J. Environmental change and infectious disease: how new roads affect the transmission of diarrheal pathogens in rural Ecuador. Proc Natl Acad Sci U S A. 2006;103(51):19460–19465. doi: 10.1073/pnas.0609431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endara P, Trueba G, Solberg OD, Bates SJ, Ponce K, Cevallos W, Matthijnssens J, Eisenberg JN. Symptomatic and subclinical infection with rotavirus P[8]G9, rural Ecuador. Emerg Infect Dis. 2007;13(4):574–580. doi: 10.3201/eid1304.061285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinola EE, Parra GI, Russomando G, Arbiza J. Genetic diversity of the VP4 and VP7 genes affects the genotyping of rotaviruses: analysis of Paraguayan strains. Infect Genet Evol. 2008;8(1):94–99. doi: 10.1016/j.meegid.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Espinosa AC, Mazari-Hiriart M, Espinosa R, Maruri-Avidal L, Méndez E, Arias CF. Infectivity and genome persistence of rotavirus and astrovirus in groundwater and surface water. Water Res. 2008;42(10–11):2618–2628. doi: 10.1016/j.watres.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8(3):175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fischer TK, Steinsland H, Molbak K, Ca R, Gentsch JR, Valentiner-Branth P, Aaby P, Sommerfelt H. Genotype profiles of rotavirus strains from children in a suburban community in Guinea-Bissau, Western Africa. J Clin Microbiol. 2000;38(1):264–267. doi: 10.1128/jcm.38.1.264-267.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer TK, Steinsland H, Valentiner-Branth P. Rotavirus particles can survive storage in ambient tropical temperatures for more than 2 months. J Clin Microbiol. 2002;40(12):4763–4764. doi: 10.1128/JCM.40.12.4763-4764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MM, Kerin T, Hull J, McCaustland K, Gentsch J. Enhancement of detection and quantification of rotavirus in stool using a modified real-time RT-PCR assay. J Med Virol. 2008;80(8):1489–1496. doi: 10.1002/jmv.21228. [DOI] [PubMed] [Google Scholar]
- Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30(6):1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, Jain V, Cunliffe NA, Nakagomi O, Kirkwood CD, Fischer TK, Parashar UD, Bresee JS, Jiang B, Glass RI. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J Infect Dis. 2005;192 Suppl 1:S146–S159. doi: 10.1086/431499. [DOI] [PubMed] [Google Scholar]
- Gladstone BP, Iturriza-Gomara M, Ramani S, Monica B, Banerjee I, Brown DW, Gray JJ, Muliyil J, Kang G. Polymerase chain reaction in the detection of an 'outbreak' of asymptomatic viral infections in a community birth cohort in south India. Epidemiol Infect. 2008;136(3):399–405. doi: 10.1017/S0950268807008709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass RI, Parashar UD, Bresee JS, Turcios R, Fischer TK, Widdowson MA, Jiang B, Gentsch JR. Rotavirus vaccines: current prospects and future challenges. Lancet. 2006;368(9532):323–332. doi: 10.1016/S0140-6736(06)68815-6. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8(3):195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Gorziglia M, Larralde G, Kapikian AZ, Chanock RM. Antigenic relationships among human rotaviruses as determined by outer capsid protein VP4. Proc Natl Acad Sci U S A. 1990;87(18):7155–7159. doi: 10.1073/pnas.87.18.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28(2):276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvea V, Santos N, Timenetsky Mdo C. Identification of bovine and porcine rotavirus G types by PCR. J Clin Microbiol. 1994;32(5):1338–1340. doi: 10.1128/jcm.32.5.1338-1340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg HB, Valdesuso J, van Wyke K, Midthun K, Walsh M, McAuliffe V, Wyatt RG, Kalica AR, Flores J, Hoshino Y. Production and preliminary characterization of monoclonal antibodies directed at two surface proteins of rhesus rotavirus. J Virol. 1983;47(2):267–275. doi: 10.1128/jvi.47.2.267-275.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DD, Nakagomi T, Hoshino Y, Nakagomi O, Kirkwood CD, Parashar UD, Glass RI, Gentsch JR. Characterization of nontypeable rotavirus strains from the United States: identification of a new rotavirus reassortant (P2A[6],G12) and rare P3[9] strains related to bovine rotaviruses. Virology. 2002;294(2):256–269. doi: 10.1006/viro.2001.1333. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Aguirre I, Steyer A, Boben J, Gruden K, Poljsak-Prijatelj M, Ravnikar M. Sensitive detection of multiple rotavirus genotypes with a single reverse transcription-real-time quantitative PCR assay. J Clin Microbiol. 2008;46(8):2547–2554. doi: 10.1128/JCM.02428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma S, Chizhikov V, Santos N, Tatsumi M, Timenetsky Mdo C, Linhares AC, Mascarenhas JD, Ushijima H, Armah GE, Gentsch JR, Hoshino Y. Development and validation of DNA microarray for genotyping group A rotavirus VP4 (P[4], P[6], P[8], P[9], and P[14]) and VP7 (G1 to G6, G8 to G10, and G12) genes. J Clin Microbiol. 2007;45(8):2641–2648. doi: 10.1128/JCM.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriza-Gómara M, Cubitt D, Desselberger U, Gray J. Amino acid substitution within the VP7 protein of G2 rotavirus strains associated with failure to serotype. J Clin Microbiol. 2001;39(10):3796–3798. doi: 10.1128/JCM.39.10.3796-3798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriza-Gómara M, Green J, Brown DWG, Desselberger U, Gray JJ. Diversity within the VP4 gene of rotavirus P[8] strains: Implications for reverse transcription-PCR genotyping. J Clin Microbiol. 2000;38(2):898–901. doi: 10.1128/jcm.38.2.898-901.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriza-Gómara M, Kang G, Gray J. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J Clin Virol. 2004;31(4):259–265. doi: 10.1016/j.jcv.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Iturriza-Gómara M, Wong C, Blome S, Desselberger U, Gray J. Molecular characterization of VP6 genes of human rotavirus isolates: correlation of genogroups with subgroups and evidence of independent segregation. J Virol. 2002;76(13):6596–6601. doi: 10.1128/JVI.76.13.6596-6601.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerin TK, Kane EM, Glass RI, Gentsch JR. Characterization of VP6 genes from rotavirus strains collected in the United States from 1996–2002. Virus Genes. 2007;35(3):489–495. doi: 10.1007/s11262-007-0119-7. [DOI] [PubMed] [Google Scholar]
- Khamrin P, Peerakome S, Tonusin S, Malasao R, Okitsu S, Mizuguchi M, Ushijima H, Maneekarn N. Changing pattern of rotavirus G genotype distribution in Chiang Mai, Thailand from 2002 to 2004: decline of G9 and reemergence of G1 and G2. J Med Virol. 2007;79(11):1775–1782. doi: 10.1002/jmv.20960. [DOI] [PubMed] [Google Scholar]
- Khamrin P, Peerakome S, Wongsawasdi L, Tonusin S, Sornchai P, Maneerat V, Khamwan C, Yagyu F, Okitsu S, Ushijima H, Maneekarn N. Emergence of human G9 rotavirus with an exceptionally high frequency in children admitted to hospital with diarrhea in Chiang Mai, Thailand. J Med Virol. 2006;78(2):273–280. doi: 10.1002/jmv.20536. [DOI] [PubMed] [Google Scholar]
- Laird AR, Ibarra V, Ruiz-Palacios G, Guerrero ML, Glass RI, Gentsch JR. Unexpected detection of animal VP7 genes among common rotavirus strains isolated from children in Mexico. J Clin Microbiol. 2003;41(9):4400–4403. doi: 10.1128/JCM.41.9.4400-4403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambden PR, Cooke SJ, Caul EO, Clarke IN. Cloning of noncultivatable human rotavirus by single primer amplification. Journal of Virology. 1992;66(3) doi: 10.1128/jvi.66.3.1817-1822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovmar L, Fock C, Espinoza F, Bucardo F, Syvanen AC, Bondeson K. Microarrays for genotyping human group a rotavirus by multiplex capture and type-specific primer extension. J Clin Microbiol. 2003;41(11):5153–5158. doi: 10.1128/JCM.41.11.5153-5158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gomara M, Maes P, Patton JT, Rahman M, Van Ranst M. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol. 2008a;82(7):3204–3219. doi: 10.1128/JVI.02257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Banyai K, Estes MK, Gentsch JR, Iturriza-Gomara M, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Patton JT, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Desselberger U, Van Ranst M. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol. 2008b;153(8):1621–1629. doi: 10.1007/s00705-008-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Rahman M, Martella V, Xuelei Y, De Vos S, De Leener K, Ciarlet M, Buonavoglia C, Van Ranst M. Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission. J Virol. 2006;80(8):3801–3810. doi: 10.1128/JVI.80.8.3801-3810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo A, Cedeno C, Teran E, Castello A. Prevalence of VP4 and VP7 genotypes of human rotavirus in Ecuadorian children with acute diarrhea. J Med Virol. 2008;80(6):1106–1111. doi: 10.1002/jmv.21181. [DOI] [PubMed] [Google Scholar]
- Parashar UD, Gibson CJ, Bresse JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg Infect Dis. 2006;12(2):304–306. doi: 10.3201/eid1202.050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra GI, Bok K, Martinez V, Russomando G, Gomez J. Molecular characterization and genetic variation of the VP7 gene of human rotaviruses isolated in Paraguay. J Med Virol. 2005;77(4):579–586. doi: 10.1002/jmv.20495. [DOI] [PubMed] [Google Scholar]
- Parra GI, Vidales G, Gomez JA, Fernandez FM, Parreno V, Bok K. Phylogenetic analysis of porcine rotavirus in Argentina: increasing diversity of G4 strains and evidence of interspecies transmission. Vet Microbiol. 2008;126(1–3):243–250. doi: 10.1016/j.vetmic.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Rahman M, Goegebuer T, De Leener K, Maes P, Matthijnssens J, Podder G, Azim T, Van Ranst M. Chromatography paper strip method for collection, transportation, and storage of rotavirus RNA in stool samples. Journal of Clinical Microbiology. 2004;42(4):1605–1608. doi: 10.1128/JCM.42.4.1605-1608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Matthijnssens J, Nahar S, Podder G, Sack DA, Azim T, Van Ranst M. Characterization of a novel P[25],G11 human group a rotavirus. J Clin Microbiol. 2005a;43(7):3208–3212. doi: 10.1128/JCM.43.7.3208-3212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Sultana R, Podder G, Faruque AS, Matthijnssens J, Zaman K, Breiman RF, Sack DA, Van Ranst M, Azim T. Typing of human rotaviruses: nucleotide mismatches between the VP7 gene and primer are associated with genotyping failure. Virol J. 2005b;2:24. doi: 10.1186/1743-422X-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran M, Das BK, Vij A, Kumar R, Bhambal SS, Kesari N, Rawat H, Bahl L, Thakur S, Woods PA, Glass RI, Bhan MK, Gentsch JR. Unusual diversity of human rotavirus G and P genotypes in India. J Clin Microbiol. 1996;34(2):436–439. doi: 10.1128/jcm.34.2.436-439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos AP, Stefanelli CC, Linhares RE, de Brito BG, Santos N, Gouvea V, de Cassia Lima R, Nozawa C. The stability of porcine rotavirus in feces. Veterinary Microbiology. 2000;71(1–2):1–8. doi: 10.1016/s0378-1135(99)00140-6. [DOI] [PubMed] [Google Scholar]
- Rodrigues F, Iturriza M, Gray J, Januario L, Lemos L. Epidemiology of rotavirus in Portugal: G9 as a major cause of diarrhoea in non-hospitalised children. J Clin Virol. 2007;40(3):214–217. doi: 10.1016/j.jcv.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Santos N, Honma S, Timenetsky Mdo C, Linhares AC, Ushijima H, Armah GE, Gentsch JR, Hoshino Y. Development of a microtiter plate hybridization-based PCR-enzyme-linked immunosorbent assay for identification of clinically relevant human group A rotavirus G and P genotypes. J Clin Microbiol. 2008;46(2):462–469. doi: 10.1128/JCM.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005;15(1):29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- Santos N, Volotao EM, Soares CC, Albuquerque MC, da Silva FM, Chizhikov V, Hoshino Y. VP7 gene polymorphism of serotype G9 rotavirus strains and its impact on G genotype determination by PCR. Virus Res. 2003;93(1):127–138. doi: 10.1016/s0168-1702(02)00318-0. [DOI] [PubMed] [Google Scholar]
- Simmonds MK, Armah G, Asmah R, Banerjee I, Damanka S, Esona M, Gentsch JR, Gray JJ, Kirkwood C, Page N, Iturriza-Gomara M. New oligonucleotide primers for P-typing of rotavirus strains: Strategies for typing previously untypeable strains. J Clin Virol. 2008;42(4):368–373. doi: 10.1016/j.jcv.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101(30):11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, Gilbert JM, Matsui SM, Greenberg HB. Comparison of the rotavirus gene 6 from different species by sequence analysis and localization of subgroup-specific epitopes using site-directed mutagenesis. Virology. 1997;237(1):89–96. doi: 10.1006/viro.1997.8762. [DOI] [PubMed] [Google Scholar]
- Trinh QD, Pham NT, Nguyen TA, Phan TG, Khamrin P, Yan H, Hoang PL, Maneekarn N, Li Y, Kozlov V, Kozlov A, Okitsu S, Ushijima H. Amino acid substitutions in the VP7 protein of human rotavirus G3 isolated in China, Russia, Thailand, and Vietnam during 2001–2004. J Med Virol. 2007;79(10):1611–1616. doi: 10.1002/jmv.20931. [DOI] [PubMed] [Google Scholar]
- Whittier CA, Horne W, Slenning B, Loomis M, Stoskopf MK. Comparison of storage methods for reverse-transcriptase PCR amplification of rotavirus RNA from gorilla (Gorilla g. gorilla) fecal samples. J Virol Methods. 2004;116(1):11–17. doi: 10.1016/j.jviromet.2003.10.003. [DOI] [PubMed] [Google Scholar]


