Altered glucose metabolism in cancer cells is termed the Warburg effect, which describes the propensity for most cancer cells to take up glucose avidly and convert it primarily to lactate, despite available oxygen 1,2. Notwithstanding the renewed interest in the Warburg effect, cancer cells also depend on continued mitochondrial function for metabolism, specifically glutaminolysis that catabolizes glutamine to generate ATP and lactate3. Glutamine, which is highly transported into proliferating cells4,5, is a major source for energy and nitrogen for biosynthesis, and a carbon substrate for anabolic processes in cancer cells, but the regulation of glutamine metabolism is not well understood1,6. Here, we report that the c-Myc (or Myc) oncogenic transcription factor, which is known to regulate miRNAs7,8 and stimulate cell proliferation9, transcriptionally represses miR-23a and miR-23b, resulting in greater expression of their target protein, mitochondrial glutaminase (GLS). This leads to up-regulation of glutamine catabolism10. GLS converts glutamine to glutamate that is further catabolized through the TCA cycle for the production of ATP or serves as substrate for glutathione synthesis11. The unique means by which Myc regulates GLS reported herein uncovers a previously unsuspected link between Myc regulation of miRNAs, glutamine metabolism, and energy and reactive oxygen species (ROS) homeostasis.
Oncogenes and tumor suppressors have now been linked to the regulation of glucose metabolism, thereby connecting genetic alterations in cancers to their glucose metabolic phenotype1,2. In particular, the MYC oncogene produces Myc protein that directly regulates glucose metabolic enzymes as well as genes involved in mitochondrial biogenesis9,12. In this regard, we sought to determine the role of the MYC in altering the mitochondrial proteome to further understand regulation of tumor metabolism. We studied the human P-493 B cells that bear a tetracycline-repressible MYC construct, such that tetracycline withdrawal results in rapid induction of Myc and mitochondrial biogenesis, followed by cell proliferation12,13. By comparing the mitochondrial proteome from tetracycline-treated and untreated cells with high Myc expression, we found 8 mitochondrial proteins that are distinctly differentially expressed in response to Myc (Figures 1a, 1b and Tables S1). We note that mitochondrial glutaminase or GLS (MW∼58kDa) was increased ∼10-fold in response to Myc. As such, we determined the response of glutaminase to Myc induction in a time-course study using anti-GLS antibody10 (Figure 1c) and found that GLS levels diminish with decreased Myc expression and recover upon Myc re-induction. However, the level of the mitochondrial protein TFAM remained virtually unaltered. GLS levels also correlate with Myc levels in another human B cell line (CB33) and one (CB33-Myc) with constitutive Myc expression14. Since human prostate cancer is linked to Myc expression15, we sought to determine whether reduction of Myc expression by siRNA in the human PC3 prostate cancer cell line was also associated with reduction of GLS expression (Figure 1d). Similar to the human lymphoid cells, the PC3 cells also displayed a correlation between Myc and GLS levels.
Figure 1.
1a. The expanded insets of 2-dimensional gels reveal the induction of glutaminase (GLS; highlighted by white circles) by Myc in P493-6 B cells. For each condition, 350μg of mitochondrial protein lysate was resolved on 18 cm immobilized pH gradient (IPG) strips as the first dimension followed by 10% Bis-Tris SDS-PAGE as the second dimension, which is marked by molecular mass markers. Protein spots were visualized by silver staining. Six independent biological experiments were performed for each condition. Table S1 summarizes the identity of the spots with the same numbering system as depicted in the figure.
1b. Immunoblot with anti-GLS antibody of a 1-dimensional SDS-PAGE gel of mitochondrial proteins (20 μg/lane) validates the induction of GLS by Myc discovered in Figure 1a. TFAM represents a control mitochondrial protein.
1c. P493-6 cells were treated with tetracycline (Tet) for different lengths of time to inhibit Myc expression or were treated first with tetracycline for 48h and then washed (Wash) to remove tetracycline with the times after wash out indicated. Cells were then harvested for immunoblot assay for GLS or c-Myc. Anti-tubulin antibody and anti-TFAM were used for loading controls.
1d. Human CB33 lymphoblastoid cells, CB33-Myc cells, and PC3 cells transfected with siRNA against c-Myc (siMYC) or control siRNA (siCont) were used for immunoblot assays. Experiments were replicated with similar results.
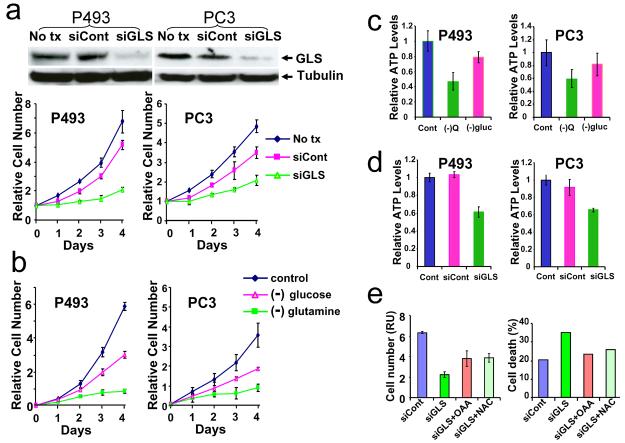
We then sought to determine whether the dramatic alteration of GLS levels in response to Myc is functionally linked to Myc-induced cell proliferation. While there are two major known tissue-specific GLS isoforms, GLS1 and GLS216,17, our data showed that only GLS1 is predominantly expressed in P493-6 or PC3 cells (Figure S1). We first determined whether gain of GLS1 function through overexpression in PC3 cells would rescue the diminished growth rate associated with siRNA-mediated reduction of Myc (Figure S2) and found that ectopic GLS1 expression alone is insufficient to stimulate growth. In light of the observation that no single gene could substitute for Myc 18,19 and that Myc is a pleiotropic transcription factor9, this outcome was not particularly surprising. As such, we reduced the expression of GLS1 (herein referred to as GLS) by RNA interference (siGLS) and found that P-493-6 cell proliferation is markedly attenuated by siGLS but not by control siRNA (Figure 2a). Likewise, proliferation of the human PC3 prostate cancer cell line was diminished by siGLS (Figure 2a), indicating that GLS is necessary for cell proliferation.
Figure 2.
2a. Top panel, immunoblots document that GLS protein level was diminished by transfecting the cells with siRNA for GLS1 (siGLS) as compared to non transfection (No tx) or control siRNA (siCont); Lower panel, Growth inhibition of P493 and PC3 cells by siGLS. The results shown are mean ± SD, n=3.
2b. Growth inhibition of P493 and PC3 cells cultured under control, glucose- or glutamine- deprived conditions. The results shown are mean ± SD, n=3.
2c. Cells were cultured with normal medium or medium without glucose ((−)gluc)or glutamine ((−)Q) for 48h and harvested for ATP assay as described in Methods section. The results shown (mean ± SD, n=2) were relative ATP levels per microgram total protein normalized to the control (Cont) normal medium group.
2d. ATP levels in control cells (Cont) or cells transfected with siRNA for GLS (siGLS) or control siRNA (siCont). 72h after transfection, cells were harvested for ATP assay. The results shown (mean ± SD, n=2) were relative ATP levels per microgram total protein normalized to the non transfected control group (Cont).
2e. Cells were transfected with siRNA for GLS (siGLS) or control siRNA (siCont) and cultured with 10mM N-acetylcysteine (NAC), or 5mM oxaloacetate (OAA), or no addition control. Left panel shows cell counts (mean ± SD, n=5) of different groups at 72h after transfection; complete cell growth curve is available in Figure S5. Right panel shows percentage cell death at 72h after transfection. Percentage Cell death indicates Annexin positive plus Annexin V and 7-AAD positive cells. Primary data are shown in Figure S4.
All experiments in Figure 2 were repeated at least twice. All experiments with P493 cells were in the absence of tetracycline. (*) denotes mean (± SD) that is significantly different (P < 0.05 by t test).
Because glutamine is converted by GLS to glutamate for further catabolism by the TCA cycle and previous studies indicate that overexpression of Myc sensitizes human cells to glutamine withdrawal induced apoptosis11, we determined the metabolic responses of P493-6 or PC3 cells to glutamine deprivation (Figure 2b). The growth of both cell lines was diminished significantly by glutamine withdrawal and moderately with glucose withdrawal. Glutamine withdrawal also resulted in a decrease in ATP levels (Figure 2c) associated with a diminished cellular oxygen consumption rate (Figures S3a and S3b). Reduction of GLS by RNAi also reduced ATP levels (Figure 2d). Because glutamine is a precursor for glutathione20, glutathione levels were measured by flow cytometry and were found diminished with glutamine withdrawal or RNAi mediated reduction of GLS (Figure S4 and Table S2) that is also associated with an increase in ROS levels (Figure S3c) and cell death in the P493-6 cells (Figures 2e and S5). It is notable shortly after the MYC proto-oncogene was discovered, that the MC29 retrovirus which bears the v-myc oncogene was found to enhance glutamine catabolism and mitochondrial respiration in transplantable avian liver tumor cells21. Thus, our findings functionally link historical observations with Myc, glutaminase and glutamine metabolism.
Since GLS catabolizes glutamine for ATP and glutathione synthesis, its reduction affects proliferation and cell death presumably through depletion of ATP and augmentation of ROS, respectively. Hence, we sought to rescue the P493-6 cells with the TCA cycle metabolite oxaloacetate (OAA) and the oxygen radical scavenger N-acetylcysteine (NAC)11. Both OAA and NAC partially rescued the decreased proliferation and death of P493-6 cells deprived of GLS (Figures 2e and S6). Likewise, OAA and NAC both partially rescued glutamine-deprived P493-6 cells (Figures S5 and S6). These findings support the notion that glutamine catabolism through GLS is critical for cell proliferation induced by Myc and protection against ROS generated by enhanced mitochondrial function in response to Myc11,20.
Given that GLS is critical for cell proliferation and is induced by Myc, we determined the mechanism by which Myc regulates GLS. Because Myc is a transcription factor9, we hypothesized that Myc transactivates GLS directly as a target gene. Despite the presence of a canonical Myc binding site (5′-CACGTG-3′) in the GLS gene intron 1, GLS mRNA levels do not respond to alterations in Myc levels in the P493-6 cells, suggesting that GLS is regulated at the post-transcriptional level (Figure 3a). As such, we hypothesized that GLS could be regulated by miRNAs that are in turn directly regulated by Myc. The TargetScan algorithm predicts that miR-23a and miR-23b could target the GLS 3′UTR seed sequence. Intriguingly, our earlier studies uncovered that both miR-23a and miR-23b are suppressed by Myc in P493-6 cells7, and both miR-23s are decreased in human prostate cancers22, which are associated with elevated Myc expression15.
Figure 3.
3a. P493-6 cells were treated with tetracycline (Tet) for different lengths of time to inhibit Myc expression or were treated first with tetracycline for 48h and then washed (Wash) to remove tetracycline with the times after wash out indicated. RNA was then harvested for real-time PCR for GLS1 as described in Methods section. Data are shown as mean ± SD, n = 3 PCR reactions.
3b. Northern analysis of miR-23a and miR-23b expression in P493 cells treated with or without tetracycline for 24h and then transfected with anti-sense miR-23a and miR-23b LNAs or Scrambled control LNA. 48h after transfection, cells were harvested and northern blot assays were performed as described in the Methods section. U6 snRNA probe was used as the loading control.
3c. Chromatin immunoprecipitation (ChIP) assay with P493 cells documents Myc binding to the promoter region of C9orf3, whose transcript is processed to miR-23b. The positions of the amplicons are depicted in the cartoon of the C9orf3 gene below the bar graphs (mean ± SD, n=3) demonstrating the binding of Myc in the amplicon 2 region in a tetracycline dependent manner. Anti-HGF serves as a non-specific antibody control.
3d. Inhibition of GLS-3′-UTR luciferase reporter by miR-23. Upper panel. Glutaminase reporter (wild-type GLS-3′UTR or mutant Mut-GLS-3′-UTR) or control (PGL3) luciferase constructs were transfected into MCF-7 cells with the following oligonucleotides: scrambled control LNA (cont-LNA) nucleotide or antisense (miR-23-LNA). The ratio of normalized reporter to control luciferase activity is shown. Cells were co-transfected either with 100ng reporter vectors and 4ng pSV-Renilla, and further co-transfected with 10nM LNA antisense for miR-23 or control LNA. After 24h, luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega). Data shown are luciferase activity (RLU = relative light unit) normalized to control group (mean ± SD, n=4). (*) denotes mean (± SD) that is significantly different (P < 0.05 by t test). Lower panel illustrates miR-23a, miR-23b, GLS-3′UTR and MutGLS-3′UTR sequences.
3e. Analysis of GLS protein levels in P493 and PC3 cells treated with control (Cont-LNA) or antisense miR-23 LNAs (miR-23-LNA). Left panel, P493 cells were treated with or without tetracycline for 24h and then transfected with antisense miR-23a and miR-23b LNAs or Scramble control probe. After 72h, cells were harvested for immunoblot assay with anti-Myc and anti-GLS antibodies. Tubulin serves as a loading control. Right panel, PC3 cells were transfected with siRNA for MYC (siMyc) or control siRNA (siCont). After 24h, cells were transfected with LNA knockdown probes for miR-23a and miR-23b or Scramble control probe. Cells were cultured for 72h and then were harvested for immunoblot assay. Each experiment was repeated twice with a representative experiment shown.
To verify that miR-23a and miR-23b (herein referred to as miR-23) are suppressed by Myc and could be diminished by antisense miR-23 locked nucleic acid (LNA) oligomers, northern analysis was performed and miR-23 was suppressed by Myc and profoundly diminished by antisense miR-23 LNAs (Figure 3b). Using quantitative real-time PCR, we document (Figure S7) that miR-23 levels increased with diminished Myc expression and then decreased upon Myc re-induction in a manner that is compatible with the GLS protein levels seen in Figure 1c. We also found an inverse relationship between Myc and the levels of miR-23a and miR-23b in the CB33 human lymphoid cells and PC3 prostate cancer cell line (Figure S8). Furthermore, Myc directly bound the transcriptional unit, C9orf3, encompassing miR-23b, as demonstrated for other Myc miRNA targets7 by chromatin immunoprecipitation (Figure 3c). Because the transcriptional unit involving miR-23a has not been mapped, we did not study miR-23a in this context. These observations document that Myc represses miR-23a and miR23b, which appear to be directly regulated by Myc.
We determined next whether miR-23 targets and inhibits the expression of GLS through the 3′UTR. In this regard, we cloned the 3′UTR sequence of GLS including the predicted binding site for miR-23 to the pGL3 luciferase reporter vector and transfected MCF-7 cells, which is known to express miR-2323. The GLS 3′UTR inhibited luciferase activity in a fashion that was blocked by co-transfection with the antisense miR-23 LNAs, but not with control LNAs (Figure 3d). We next mutated the predicted binding site by a site-directed mutagenesis strategy8 and observed that mutant 3′UTR did not inhibit luciferase activity as the wild-type sequence did. Using these reporters in PC3 cells, we observed that siRNA-mediated diminished Myc expression resulted in decreased luciferase activity with wild-type but not with the mutant 3′UTR reporter (Figure S9). Most importantly, diminished GLS protein level, which follows decreased Myc expression (Figure 1c), was rescued by antisense miR-23 LNAs (Figure 3e). The antisense miR-23 LNAs also partially rescued the diminished GLS level associated with RNAi-mediated reduction of Myc expression in the PC3 cells (Figure 3e).
We also examined events upstream and downstream of GLS24 and found that the glutamine transporter SLC7A5 is induced by Myc in P493-6 cells at the transcriptional level (5-fold by nuclear run-on, unpublished data) with a >7-fold induction of its mRNA level. The glutamine transporter ASCT2 is induced by Myc at the mRNA level by 2-fold, whereas glutamate dehydrogenase mRNA levels appear unaltered (unpublished data). Furthermore, we found that elevated levels of Myc protein in human prostate cancer correspond to levels of GLS, which was not increased in the accompanying normal tissue from the same patients (Figure S10). Intriguingly, miR-23a and miR-23b are significantly decreased in human prostate cancer as compared with normal prostate tissue22. It is notable that loss of GLS function by antisense suppression significantly inhibited the tumorigenesis of Ehrlich ascites tumor cells in vivo25. Our findings here uncover a pathway by which Myc suppression of miR-23, which targets GLS, enhances glutamine catabolism through increased mitochondrial glutaminase expression. Taken together, these observations provide a regulatory mechanism involving Myc and miRNAs for elevated expression of glutaminase and glutamine metabolism in human cancers.
Methods Summary
Human cell lines were cultured under standard conditions. Isolation of mitochondria, enrichment for mitochondrial proteins, and proteomic analysis were performed as described26-29. RNA interference experiments and luciferase reporter analysis of miRNA activity were as reported8,30. Flow cytometric analyses of reactive oxygen species, cell death and glutathione level were performed as described11,30. Human samples were acquired with the approval of the Johns Hopkins University School of Medicine Institutional Review Board.
Supplementary Material
Acknowledgements
The authors want to thank Lee Blosser and Ada Tam for their generous help in flowcytometry analysis and Huiyuan Zhang for her expertise in statistical analysis. This work was partially supported by NIH Awards NHLBI NO1-HV-28180, NCI R01CA051497, NCI R01CA57341, NCI R01CA120185, Rita Allen Foundation, Leukemia and Lymphoma Society, and Sol Goldman Center for Pancreatic Cancer Research.
Footnotes
The authors have no competing financial interest.
References
- 1.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13:472–82. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 3.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–50. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254:2669–76. [PubMed] [Google Scholar]
- 5.Gallagher FA, Kettunen MI, Day SE, Lerche M, Brindle KM. 1C MR spectroscopy measurements of glutaminase activity in human hepatocellular carcinoma cells using hyperpolarized 13C-labeled glutamine. Magn Reson Med. 2008;60:253–7. doi: 10.1002/mrm.21650. [DOI] [PubMed] [Google Scholar]
- 6.Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu Rev Nutr. 1995;15:133–59. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- 7.Chang TC, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 9.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–66. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kita K, Suzuki T, Ochi T. Down-regulation of glutaminase C in human hepatocarcinoma cell by diphenylarsinic acid, a degradation product of chemical warfare agents. Toxicol Appl Pharmacol. 2007;220:262–70. doi: 10.1016/j.taap.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–34. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuhmacher M, et al. Control of cell growth by c-Myc in the absence of cell division. Curr Biol. 1999;9:1255–8. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- 14.Lombardi L, Newcomb EW, Dalla-Favera R. Pathogenesis of Burkitt lymphoma: expression of an activated c-myc oncogene causes the tumorigenic conversion of EBV-infected human B lymphoblasts. Cell. 1987;49:161–70. doi: 10.1016/0092-8674(87)90556-3. [DOI] [PubMed] [Google Scholar]
- 15.Gurel B, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156–67. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Gomez C, et al. Co-expression of glutaminase K and L isoenzymes in human tumour cells. Biochem J. 2005;386:535–42. doi: 10.1042/BJ20040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner A, McGivan JD. Glutaminase isoform expression in cell lines derived from human colorectal adenomas and carcinomas. Biochem J. 2003;370:403–8. doi: 10.1042/BJ20021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berns K, Hijmans EM, Koh E, Daley GQ, Bernards R. A genetic screen to identify genes that rescue the slow growth phenotype of c-myc null fibroblasts. Oncogene. 2000;19:3330–4. doi: 10.1038/sj.onc.1203639. [DOI] [PubMed] [Google Scholar]
- 19.Nikiforov MA, et al. Complementation of Myc-dependent cell proliferation by cDNA expression library screening. Oncogene. 2000;19:4828–31. doi: 10.1038/sj.onc.1203880. [DOI] [PubMed] [Google Scholar]
- 20.Lora J, et al. Antisense glutaminase inhibition decreases glutathione antioxidant capacity and increases apoptosis in Ehrlich ascitic tumour cells. Eur J Biochem. 2004;271:4298–306. doi: 10.1111/j.1432-1033.2004.04370.x. [DOI] [PubMed] [Google Scholar]
- 21.Matsuno T, Satoh T, Suzuki H. Prominent glutamine oxidation activity in mitochondria of avian transplantable hepatoma induced by MC-29 virus. J Cell Physiol. 1986;128:397–401. doi: 10.1002/jcp.1041280308. [DOI] [PubMed] [Google Scholar]
- 22.Porkka KP, et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–5. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 23.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bode BP. Recent molecular advances in mammalian glutamine transport. J Nutr. 2001;131:2475S–85S. doi: 10.1093/jn/131.9.2475S. discussion 2486S-7S. [DOI] [PubMed] [Google Scholar]
- 25.Lobo C, et al. Inhibition of glutaminase expression by antisense mRNA decreases growth and tumourigenicity of tumour cells. Biochem J. 2000;348(Pt 2):257–61. [PMC free article] [PubMed] [Google Scholar]
- 26.Rabilloud T, et al. The mitochondrial antioxidant defence system and its response to oxidative stress. Proteomics. 2001;1:1105–10. doi: 10.1002/1615-9861(200109)1:9<1105::AID-PROT1105>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 27.Anderson TJ, et al. Discovering robust protein biomarkers for disease from relative expression reversals in 2-D DIGE data. Proteomics. 2007;7:1197–207. doi: 10.1002/pmic.200600374. [DOI] [PubMed] [Google Scholar]
- 28.Kersey PJ, et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–8. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 29.Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–36. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 30.Gao P, et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell. 2007;12:230–8. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.