Abstract
An analysis of the effect of an oil spill on mangrove sediments was carried out by contamination of mesocosms derived from two different mangroves, one with a history of contamination and one pristine. The association between N2 fixers and hydrocarbon degradation was assessed using quantitative PCR (qPCR) for the genes rrs and nifH, nifH clone library sequencing and total petroleum hydrocarbon (TPH) quantification using gas chromatography. TPH showed that the microbial communities of both mangroves were able to degrade the hydrocarbons added; however, whereas the majority of oil added to the mesocosm derived from the polluted mangrove was degraded in the 75 days of the experiment, there was only partially degradation in the mesocosm derived from the pristine mangrove. qPCR showed that the addition of oil led to an increase in rrs gene copy numbers in both mesocosms, having almost no effect on the nifH copy numbers in the pristine mangrove. Sequencing of nifH clones indicated that the changes promoted by the oil in the polluted mangrove were greater than those observed in the pristine mesocosm. The main effect observed in the polluted mesocosm was the selection of a single phylotype which is probably adapted to the presence of petroleum. These results, together with previous reports, give hints about the relationship between N2 fixation and hydrocarbon degradation in natural ecosystems.
Keywords: Diazotrophs, Mangrove sediment, Microbial community, Oil degradation
Introduction
Mangroves are highly productive coastal ecosystems (Jennerjahn and Ittekkot 2002); they comprise 60–75% of the world’s tropical and subtropical coastlines (Holguin et al. 2001). As a consequence of the large productivity and net nutrient export, they participate in functions of adjacent environments (Wieder and Lang 1982). For these reasons, mangrove habitats have been pinpointed as critical environments needing urgent attention and conservation (Al-Sayed et al. 2005; Duke et al. 2007).
In regard to oil pollution, mangroves are especially affected by the oil hydrocarbons (Lewis 1983). A large part of the oil spilled in the sea may converge to the shore. When the oil reaches a mangrove forest, it may persist there due to the limited wash by tidal action resulting from the vegetation density and the slow microbial decomposition that take place in anaerobic sediments (Lewis 1983).
Mangrove forests can carry out N and P cycling in an efficient and tightly regulated manner (Holguin et al. 2001). The export of organic matter and the fast consumption of N compounds in the sediment make N a limited element in this environment (Holguin et al. 1992; Holguin et al. 1999; Holguin et al. 2001). Moreover, it has been observed that nitrogen fixation is responsible for the support of these ecosystems (Holguin et al. 2001).
A major limitation to the natural degradation of hydrocarbons in most ecosystems is the unbalanced C:N:P ratio caused by the high C content of the pollutant. This leads to the fast consumption of the nitrogen and phosphorus available to the hydrocarbonoclastic populations present in the ecosystem causing the degradation to become slower or even cease. However, with the break in the C:N:P ratio after an oil spill (Musat et al. 2006), it has been postulated that there would be association between hydrocarbonoclastic and diazotrophic populations (Head et al. 2006) to symbiotically degrade hydrocarbons.
We aimed to shed light on the possible interaction between hydrocarbon-degrading organisms and the input of nitrogen from diazotrophs in mangroves. Thus, two different mangroves (one pristine and another one that had suffered a recent oil spill) were sampled and mesocosms were constructed to simulate the effect of oil on the diazotrophic communities and to evaluate the hydrocarbon degradation by the microbial communities.
Materials and methods
Sampling
Sediments for mesocosm construction were collected from two different mangroves located in the ‘Todos os Santos’ bay at the northeast coast of Brazil. The sediments were obtained from locations outside the mangrove forest to avoid effects of rhizospheric bacterial populations in the mesocosm. From these mangrove areas, 20 kg of the water-saturated top sediment layer (0–10 cm) was collected and homogenised. All samples were kept cold (4°C) until arrival in the laboratory. The pristine sample was collected at the following coordinates S12°43′46.7′′, W38°30′09.1′′, the polluted one was collected at S12°24′38.1′′, W38°33′49.9′′.
Mesocosm design
Four mesocosms were set up in order to allow the application of two different treatments. Each mesocosm consisted of a 20-L aquarium containing 10 L of wet sediment (1.2 Kg/L dry wt.). In order to mimic tidal action, two pumps were connected to the mesocosms, one of which was used to pump artificial seawater (Natural Sea Salt Mix, Oceanic Systems, Texas, USA) into the aquarium (roughly 5 L each time) and another one was used to remove the water after 6 h. This produced an artificial tide that kept the sediment under 5 cm of water for 6 h each time. This procedure was repeated every 6 h. The artificial seawater (20 L) was stored in a 20-L bottle, and 10 L of that water was refreshed every 5 days. A light: dark regime of 12 h of light and 12 h of darkness was used to mimic the light period of the ‘Todos os Santos’ bay through the use of timers and three 100 W tungsten light sources hung approx 30 cm above the mesocosms. After setting up each mesocosm, these were left functioning for 3D in order to stabilise and mimic the stratification found in this environment before the onset of the experiment.
One mesocosm of each mangrove sediment received 2% (v/v) heavy oil and the other one was left without the addition of petroleum (control). Samples were collected before the addition of oil (T-0), as well as on days one (T-1), seven (T-7), 15 (T-15), 27 (T-27), 45 (T-45), 65 (T-65) and 75 (T-75) after contamination. At each sampling, 10 g of wet sediment was taken from three points at the mesocosm surface (0–5 cm) during the low tide and these samples were homogenised prior to DNA extraction.
Total petroleum hydrocarbons (TPH) quantification
TPH quantifications were carried out on 100 g sediment samples collected from surface sediments (0–5 cm depth) and the TPHs were extracted from this samples using an Accelerated Solvent Extractor model 200 (Dionex, USA), as described previously (dos Santos 2007). The extract was then concentrated on a Turbovap Model II to a volume of 1 mL (Seabra 2005). This concentrated material was then run on an HP5890 Gas chromatograph (equipped with a HP-5 column), as described in the USEPA 8015B manual (USEPA-8015B, 1996). In order to avoid differences in the concentration of hydrocarbons caused by the uneven spread of the oil on the surface of the sediment, we chose to compare the relative amounts of hydrocarbons in the sample in spite of the absolute quantities.
DNA extraction
Three replicate samples from each mangrove mesocosm at each time point were collected for DNA extraction. The isolation of total DNA from sediment samples was carried out using the FastDNA® Spin Kit for Soil (BIO101, Califórnia, USA) following the manufacturer’s protocol. DNA samples were purified using the Wizard® Genomic DNA Purification System (Promega, The Netherlands). The quality and quantity of DNA were checked using electrophoresis on agarose gels. DNA was, on average, 20 Kb in size and yields ranged from 25 to 50 μg/g of sediment material (dry wt).
Q-PCR
To determine the number of copies of the gene that encodes the rRNA 16S (rrs) present in each sample, a Real-time PCR system was applied. We used primers P1 (5′-CCT ACG GGA GGC AGC AG-3′) and P2 (5′-ATT ACC GCG GCT GCT GG-3′;) (Muyzer et al. 1993) in a PCR reaction of 10 μL containing 5 μL of 2× PCR buffer (QuantiTect SYBR Green PCR Kit, Qiagen, The Netherlands), 0.2 μM of each primer and 1 μL of sample DNA (100× dilution). PCR was carried out on an Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems, USA) apparatus. Thermal cycling was as follows: 95°C for 15 min; 30 cycles of 94°C for 1 min, 55°C for 1 s, 72°C for 1 min, with a dissociation curve after the final cycle to evaluate the formation of primer dimer or other byproducts. Data collection took place during the annealing step of the reaction.
A standard curve was constructed based on a clone containing an rrs insert. For the absolute quantification of each PCR reaction, dilutions of the standard curve (107, 108, 1010 and 1011 copies/μL) were added in triplicates.
To quantify the number of copies of nifH genes in the total community DNA, a Real-time PCR using primers FGPH19 (Simonet et al. 1991) and PolR (Poly et al. 2001) was established. The PCR reaction mix (10 μL) contained 5 μL of 2× PCR buffer (QuantiTect SYBR Green PCR Kit Qiagen, The Netherlands), 0.5 μM of each primer and 1 μL of total community DNA. PCR cycling, data collection and dissociation curve were carried out as described above. In all reactions, triplicates of the standard curve (105, 106, 109 and 1010 copies/μL) were added. To test whether differences were significant between treatments, one-way ANOVA was applied using GraphPad Prism version 4.00 (GraphPad Software, San Diego California USA).
Cloning, sequencing and sequence analysis
Four samples were selected for nifH based clone library analysis. These were: Polluted + oil, T-0 (Poll T-0), Polluted + oil, T-75 (Poll T-75), Pristine + oil, T-0 (Prist T-0) and Pristine + oil, T-75 (Prist T-75). The nifH genes from total community DNA were amplified using primers FGPH19 (Simonet et al. 1991) and PolR (Poly et al. 2001) as previously described (Diallo et al. 2004). The PCR reaction mix (25 μL) contained 10.2 mM Tris, 2.5 mM MgCl2, 50 mM KCl, 0.2 mM of each dNTP, 0.5 μM of each primer and 2.5 U of Taq DNA polymerase. PCR was carried out on a Perkin–Elmer GeneAmp PCR System 9700 (Perkin–Elmer Applied Biosystems, Nieuwerkerk a/d IJsel, the Netherlands) and thermal cycling was as follows: 95°C for 5 min; 30 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 1 min with a final elongation step of 72°C for 7 min. These 320 bp PCR products were ligated into the pGEM-T easy vector (Promega, Madison, USA) according to the manufacturer’s protocol. The ligation products were introduced into Escherichia coli MM294 competent cells (Sylphium Life Sciences, Groningen, The Netherlands) by transformation according to the supplier’s protocol, after which cells were plated onto selective media (Sambrook and Russell, 2001). Following growth at 37°C, white colonies were randomly picked, and plasmids with inserts of the correct size were isolated using the Cetyl Trimethyl Ammonium Bromide (CTAB) isolation method (Sambrook and Russell 2001). Sequencing reactions were performed on the plasmid material according to the Perkin–Elmer ABI Prism protocol (Applied Biosystems, Foster City, USA) using primer SP6. Sequence runs were done on an ABI377 DNA sequencer (Applied Biosystems, Foster City, USA).
All chromatograms were analysed for sequence quality using Bioedit (Hall 1999). Sequences containing ambiguities were not further analysed. Sequences were searched for vector contamination in NCBI’s VecScreen (http://www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html) and contaminant sequences were omitted. The sequences were then checked for the presence of chimeras using the Greengenes chimera-check tool (http://greengenes.lbl.gov/cgi-bin/nph-bel3_interface.cgi, [DeSantis et al. 2006]). The non-chimeric non-vector sequences were then classified based on the most similar sequence from a cultivated organism using the BLAST-n tool from NCBI (Altschul et al. 1990). When no cultivated organism was found between the 50 best matches the best match was then used for classification. The sequences obtained were aligned using ClustalX (Thompson et al. 1997) and the output file was used in Phylip 3.66 to construct a distance matrix using the Jukes and Cantor distance model (Jukes and Cantor, 1969). These were then used in the software DOTUR (Schloss and Handelsman 2005) to identify operational taxonomic units (OTUs) and generate estimations of richness and diversity, as well as rarefaction curves. A Neighbor-Joining tree of 86 nifH sequences was constructed using MEGA 4.0. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. The sequences reported in this study were deposited on GenBank under the accession numbers FJ669424-FJ669486.
Results
Oil degradation assessed by total petroleum hydrocarbons (TPH) quantification
TPH was measured to evaluate oil degradation during the course of the mesocosm experiment (Fig. 1). The absolute quantities observed were not entirely concordant due to the uneven spread of the oil at the surface of the sediment. Hence, a relative quantification approach was taken to analyse the data. The TPH profiles observed at T-0 were similar in both mesocosms (Fig. 1a, b). These can be attributed to artificial contamination at the onset of the mesocosm experiment. No clear effect of the previous contamination could be observed in the Polluted-T0 sample (Fig. 1b) when compared to the Pristine-T0 one (Fig. 1b).
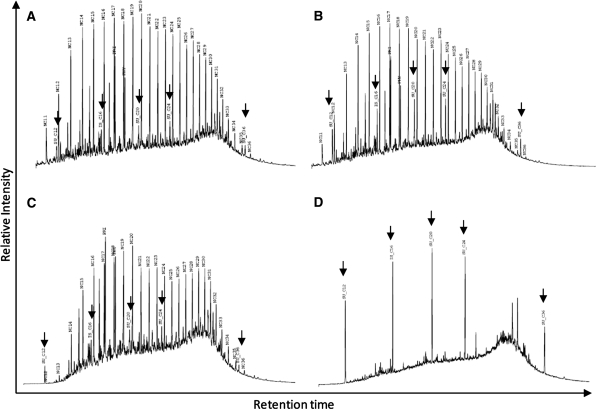
Fig. 1.
Total petroleum hydrocarbons quantification for samples: a Pristine T0; b polluted T0; c Pristine T75; d polluted T75. Peaks indicated by arrows represent the injected standards
After 75 days, the TPH profiles for both mesocosms differed greatly (Fig. 1c, d). Comparing the initial profile for the pristine mangrove with the T75 one, we could observe the preferential degradation of some PHs (Fig. 1a, c, respectively). At first glance, all PHs seemed to be degraded to some extent. However, hydrocarbons between nC20 and nC28 revealed a larger decrease when compared to other hydrocarbons such as pristane (Pri) and phytane (Phy). Pri and Phy were more resilient to degradation in this environment than the other resolved hydrocarbons. Additionally, longer PHs (>nC28) became more abundant at T75 relatively to the other PHs, hence they were less degraded then the remainder of the PHs.
On the other hand, the hydrocarbon degradation observed in the polluted mangrove mesocosm was almost complete (Fig. 1d). A so-called unresolved complex mixture (UCM) was the only measurable PH fraction. This indicates a near total degradation of the oil contaminating the mesocosm. This result was also visually observed by the nearly complete disappearance of the oil layer on the sediment.
Quantitative effect of oil addition on the microbial community
In order to quantify the copy numbers of particular genes occurring in the total community DNA we applied two Real-time PCR protocols based on well-established primer sets (Muyzer et al. 1993; Diallo et al. 2004). The first one aimed to detect the number of copies of the 16S rRNA gene (rrs) in each of the samples (Fig. 2a, b) while the second one was designed to quantify the nifH gene.
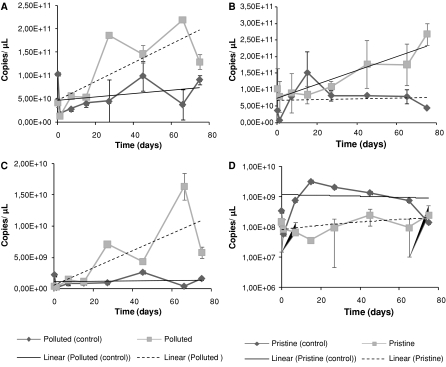
Fig. 2.
Q-PCR quantifications of rrs and nifH from total community amplifications; arrs quantification from the polluted mangrove mesocosms; brrs quantification from the pristine mangrove mesocosms; cnifH quantification from the polluted mangrove mesocosms; dnifH quantification from the pristine mangrove mesocosms. Trend line for control mesocosms are shown in black. The dotted line represents the trend line for the mesocosms with the addition of oil
Figure 2a shows the gene copy numbers for both mesocosms based on the polluted mangrove (control and contaminated). Although there was some variation in these numbers throughout the experiment, the general trend observed in both mesocosms was different. The trend lines for both mesocosms showed that the mesocosm that had received oil underwent a greater increase in gene copy numbers than the control. During the first 21 days of the experiment, both mesocosms showed similar amounts of rrs genes (≈5.00 × 1011 copies/μL). Both mesocosms revealed maxima of around 1011 copies/μL, being that the control mesocosm had a maximum of 1.03 × 1011 copies/μL at the onset of the experiment while the one that received oil had a maximum of 2.20 × 1011 at 66 days.
These patterns were not observed in the mesocosms that had been based on the pristine mangrove sediment (Fig. 2b). There was a small increase in the number of copies of the rrs gene during the first 21 days of the experiment, and after this period the numbers increased to near that observed for the polluted mangrove systems (Fig. 2a). Moreover, these values were higher than those observed for the control mesocosm (Fig. 2b), which were stable during the course of the experiment. The pristine mesocosm that received oil had, at 75 days, the highest number of rrs copies (2.67 × 1011 copies/μL) across all samples. According to one-way ANOVA, all treatments were significantly different (P = 0.0051).
The patterns observed for the nifH gene were quite different (Fig. 2c, d). Although the nifH gene counts observed for the mesocosms based on the polluted mangrove (Fig. 2c) resembled those observed for the rrs genes (Fig. 2a), the patterns observed for the pristine mesocosms differed greatly between the nifH and rrs genes (Fig. 2d, b, respectively).
Figure 2c shows that, as observed for the rrs gene (Fig. 2a), an increase in nifH gene copy numbers occurred in the mesocosm that had received oil. In addition, the control mesocosm showed similar rrs and nifH abundances. The maximum nifH gene copy number observed for the mesocosm with oil addition was more than ten times higher than that of the control (1.65 × 1010 copies/μL). The average nifH/rrs ratio for the control mesocosm shows that 2.00% of the total population carried nifH genes, while for the mesocosm that had received oil this average was 3.33%, indicating an increase in the proportion of the community carrying nifH genes after oil addition.
In contrast, the mesocosms based on the pristine sediments displayed a possible negative impact of oil on the populations carrying nifH genes. Even though the rrs gene numbers increased over time (Fig. 2b) the amount of nifH observed remained stable throughout the 75 days of experiment. This effect was corroborated by comparing the average nifH/rrs ratio observed in the mesocosm that had received oil with that of the control. The average percentages found were 0.20 and 1.20% respectively. Absolute nifH quantifications were also subjected to one-way ANOVA. This test showed significant differences between the treatments (P = 0.0237).
Selection of a specific nifH cluster by oil
In order to assess the relationship between oil addition and the populations capable of fixing nitrogen, four rrs clone libraries were constructed. Both mesocosms that received oil addition were sampled at the beginning of the experiment and at its termination.
A total of 46 clones were sequenced; the number of OTUs per treatment (similarity level >90%) ranged from 15 to 2 (Poll T0 and Poll T75, respectively) (Table 1). The libraries based on the polluted mesocosm (Poll T0 and Poll T75) showed great declines in the number of OTUs observed during the course of the mesocosm (15 to 2 OTUs). Moreover, an even greater decline was observed when considering the estimators of richness Chao1 and ACE: these estimators indicated a richness ranging from 54 ± 28.911 to 73.270 ± 45.398 (Chao1 and ACE, respectively) at T-0 which dropped to 2.0 after 75 days.
Table 1.
Diversity and richness estimation* for nifH clone libraries
| Clones sampled | Observed OTUs | Simpson | Shannon (H′) | Chao1 | ACE | |
|---|---|---|---|---|---|---|
| Poll T0 | 18 | 15 | 0.026 | 2.630 ± 0.358 | 54 ± 28.911 | 73.270 ± 45.398 |
| Poll T75 | 20 | 2 | 0.731 | 0.422 ± 0.288 | 2.00 ± 0.00 | 2.00 ± 0.00 |
| Prist T0 | 15 | 7 | 1.580 | 0.228 ± 0.228 | 17.00 ± 8.067 | 21.00 ± 11.626 |
| Prist T75 | 10 | 4 | 0.466 | 0.940 ± 0.618 | 7.00 ± 2.626 | 12.857 ± 7.866 |
Sequences were clustered on OTUs with >90% similarity
A similar selection of nifH groups was observed in the pristine mesocosm (Table 1). From the seven OTUs observed at T0, the richness dropped to four at T75. The number of OTUs thus did not decrease drastically, but through the analysis of Simpson′s Evenness Index we found that evenness decreased over time. Hence, there is a dominance of specific OTUs in the T75 community.
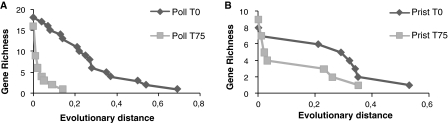
The selection of specific OTUs was stronger in the polluted mangrove than in the pristine one. Figure 3 illustrate how this selection acted on the basis of sequences clustered using several evolutionary distances. The Poll T75 library (Fig. 1a) showed a sudden drop in richness at distances between 0.0 and 0.1, while Poll T0 presented a smoother decline to 0.5. On the other hand, both libraries prepared from the pristine mangrove showed similar profiles on their curves (Fig. 3b). However, after an abrupt fall at distances between 0.0 and 0.1, the gene richness was lower for the Prist T75 library throughout all evolutionary distances.
Fig. 3.
Richness of nifH gene types at different evolutionary distances. a Observed richness for the Polluted mesocosm; b observed richness for the Pristine mesocosm
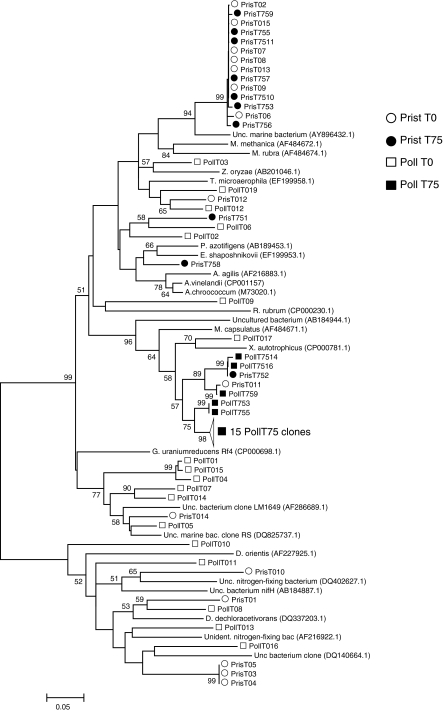
Blast-N searches (data not shown) indicate that the majority of the sequences belonged to the nitrogenase clusters I and III (Zehr et al. 2003). The majority of the sequences (≈60%) obtained from the Poll T0 library belonged to cluster III, similar to nifH genes from Geobacter uraniumreducens and Desulfovibrio dechloroacetivorans (deltaproteobacteria) (Fig. 4). A similar result was obtained from the Prist T0 library: 55% of the clones belonged to cluster III, however they were more similar to Desulfosporosinus orientis, a member of the Firmicutes (Fig. 4).
Fig. 4.
Neighbor-joining tree of 86 nifH sequences. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. Branche lengths are in the units of number of base substitutions per site
As shown above, the libraries constructed for samples collected at the end of the experiment (Poll T75 and Prist T75) displayed a major decrease of nifH gene diversity. Based on Blast-N results, this is caused by a shift in the types of sequences observed. In none of the two libraries, sequences related to cluster III were retrieved (Fig. 4), corroborating the previous results that indicate a greater effect of the addition of hydrocarbons on populations carrying the nifH gene in the polluted sample than in the pristine one.
Discussion
Hydrocarbon degradation is a natural process in all ecosystems, however factors such as the type of pollutant, its availability and the presence of hydrocarbonoclastic microorganisms are among the most important variables associated with the outcome of the process (Alexander 1994). We observed that in both mesocosms efficient biodegradation took probably place. However, the polluted mangrove mesocosm revealed a faster degradation of oil when compared with the pristine mesocosm. The more efficient degradation of pollutants in environments with previous contact with the pollutant is often related to a higher number of hydrocarbonoclastic bacteria in this environments (Olivera et al. 1997; Johnsen and Karlson 2005; Polymenakou et al. 2006). This is caused by a previous selection of populations capable of utilizing these compounds efficiently (Margesin et al. 2003; Labbe et al. 2007).
The effect of the oil on the bacterial community was evaluated by assessing their 16S rRNA gene (rrs) copy numbers. This showed that the bacterial community responded positively to the presence of a high amount of complex carbons in both mescosm systems (Pristine and Polluted). However, although it has been observed that counts of heterotrophic (Ramsay et al. 2000; Maruyama et al. 2003) and hydrocarbon-degrading bacteria (Ramsay et al. 2000; Maruyama et al. 2003; Vinas et al. 2005; Uyttebroek et al. 2007) increase with the addition of oil, the total amount of bacteria is usually not affected (Ka et al. 2001; Maruyama et al. 2003). As has been observed previously (Ka et al. 2001), the rDNA copy numbers remained stable after the addition of oil while the rRNA/rDNA ratio increased, indicating that the hydrocarbon amounts were not correlated with an increase in the number of individuals in the community but with an increase in the activity of these organisms. However, the results presented in this study indicate that the hydrocarbons might be used by some bacterial populations to increase abundance.
On the other hand, the degradation of the oil compounds should readily consume most of the nitrogen available in the sediment. Hence, since mangrove forests are often deficient in nitrogen (Sengupta and Chaudhuri 1991; Holguin et al. 1992; Alongi et al. 1993; Vazquez et al. 2000), the expected degradation should be lower than the observed one. However, in previous observations the rate of nitrogen fixation in the mangrove sediments was high due to the amount of organic matter present (Potts 1984). For this reason, as proposed earlier (Head et al. 2006), the input of carbon was expected to affect the population of nitrogen-fixing bacteria.
The nifH gene quantification through qPCR indicated that the bacterial populations capable of fixing N2 increased slightly in the polluted mangrove mesocosm after the addition of oil, following the same pattern as observed for the 16S rRNA gene (rrs). Therefore, no selection was observed in regard to the nifH gene in this system. On the other hand, the pristine mangrove showed a increase in rrs gene copy numbers and almost no change in nifH gene numbers, Indicating that in this mesocosm system nitrogen fixation may have suffered some inhibition by the presence of the hydrocarbons. In a previous study, Musat et al. (2006) observed that nitrogen fixation was not stimulated by the addition of oil to a sediment, much like our observations on the behavior of the pristine sample. On the other hand, the polluted mesocosm showed a different pattern, as in this mangrove there was an apparent stimulus of these organisms. In addition, it has been observed that ammonia oxidation was dramatically reduced when hydrocarbons were added to a pristine soil while in a soil with a history of oil pollution, this process was stimulated (Deni and Penninckx 1999).
The majority of the sequences presented here were related to the Alpha- and Gammaproteobacteria, that are known to dominate in aerobic marine environments as well as in mangrove sediments (Bostrom et al. 2007; Flores-Mirales et al. 2007). However, no sequences related to the Cyanobacteria were observed in any of the libraries, in spite of the fact that this group is often associated with oil-impacted environments (Sorkhoh et al. 1992; Hoffmann 1996; Höpner et al. 1996; Perez-Vargas et al. 2000; Sanchez et al. 2005; Musat et al. 2006) and has been appointed as responsible for the bulk of the N2 fixation in such environments (Musat et al. 2006).
Although the addition of oil incited a significant change in the numbers of both rrs and nifH genes (as shown by ANOVA), it did not induce a significant change in the community structure of the pristine mangrove mesocosm. This finding, in association with the less effective hydrocarbon degradation observed in this system, gave hints on the association of these two processes.
Nitrogen fixation is rarely observed in petroleum-polluted environments despite the frequent limitation of nitrogen (Musat et al. 2006). Moreover, where it was observed, it is not possible to correlate nitrogen fixation with hydrocarbon degradation (Laguerre et al. 1987; Toccalino et al. 1993; Piehler et al. 1999; Eckford et al. 2002; Prantera et al. 2002). However, as pointed out by Musat et al. (2006), the occurrence of N2 fixation in environments impacted by oil spills leads us to believe that this process would (still) be the source of N, and this may be extended to any polluted environment.
Acknowledgments
RT acknowledges the financial support by PDEE-CAPES, FAPERJ, CNPq and the Soil Biotechnology Foundation (Groningen, NL). The authors would like to thank Dr. C. D da Cunha for the assistance in the mesocosm design, N. O. Franco for help in the mesocosm design and maintenance and E.A. Amazonas for help in the sampling.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Alexander M (1994) Biodegradation and bioremediation. Academic Press, San Diego
- Alongi DM, Christoffersen P, Tirendi F (1993) The influence of forest type on microbial-nutrient relationships in tropical mangrove sediments. J Exp Mar Biol Ecol 171:201–223. doi:10.1016/0022-0981(93)90004-8 [DOI]
- Al-Sayed HA, Ghanem EH, Saleh KM (2005) Bacterial community and some physico-chemical characteristics in a subtropical mangrove environment in Bahrain. Mar Pollut Bull 50:147–155. doi:10.1016/j.marpolbul.2004.10.002 [DOI] [PubMed]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed]
- Bostrom KH, Riemann L, Kuhl M, Hagstrom A (2007) Isolation and gene quantification of heterotrophic N2-fixing bacterioplankton in the Baltic sea. Environ Microbiol 9:152–164. doi:10.1111/j.1462-2920.2006.01124.x [DOI] [PubMed]
- Deni J, Penninckx MJ (1999) Nitrification and autotrophic nitrifying bacteria in a hydrocarbon-polluted soil. Appl Environ Microbiol 65:4008–4013 [DOI] [PMC free article] [PubMed]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16 s rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi:10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed]
- Diallo MD, Willems A, Vloemans N, Cousin S, Vandekerckhove TT, de Lajudie P, Neyra M, Vyverman W, Gillis M, Van der Gucht K (2004) Polymerase chain reaction denaturing gradient gel electrophoresis analysis of the N2-fixing bacterial diversity in soil under Acacia tortilis ssp raddiana and Balanites aegyptiaca in the dryland part of senegal. Environ Microbiol 6:400–415. doi:10.1111/j.1462-2920.2004.00577.x [DOI] [PubMed]
- dos Santos RM (2007) Avaliação da adição do pó da casca de coco verde, como material estruturante, na biorremediação de solo contaminado com petróleo. Universidade Federal do Rio de Janeiro, Rio de Janeiro
- Duke NC, Meynecke JO, Dittmann S, Ellison AM, Anger K, Berger U, Cannicci S, Diele K, Ewel KC, Field CD, Koedam N, Lee SY, Marchand C, Nordhaus I, Dahdouh-Guebas F (2007) A world without mangroves? Science 317:41–42. doi:10.1126/science.317.5834.41b [DOI] [PubMed]
- Eckford R, Cook FD, Saul D, Aislabie J, Foght J (2002) Free-living heterotrophic nitrogen-fixing bacteria isolated from fuel-contaminated Antarctic soils. Appl Environ Microbiol 68:5181–5185. doi:10.1128/AEM.68.10.5181-5185.2002 [DOI] [PMC free article] [PubMed]
- Flores-Mirales AL, Winans SC, Holguin G (2007) Molecular characterization of diazotrophic and denitrifying bacteria associated with mangrove roots. Appl Environ Microbiol 73:7308–7321. doi:10.1128/AEM.01892-06 [DOI] [PMC free article] [PubMed]
- Hall TA (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nucleic Acid Symp Ser 41:95–98
- Head IM, Jones DM, Roling WFM (2006) Marine microorganisms make a meal of oil. Nat Rev Microbiol 4:173–182. doi:10.1038/nrmicro1348 [DOI] [PubMed]
- Hoffmann L (1996) Recolonisation of the intertidal flats by microbial mats after the gulf war oil spill. In: Krupp F, Abuzinada AH, Nader IA (eds) A marine wildlife sanctuary for the Arabian gulf, NCWCD. Senckenbergische Nat.forschende Gesellschaft, Riyadh, Frankfurt
- Holguin G, Guzman MA, Bashan Y (1992) 2 new nitrogen-fixing bacteria from the rhizosphere of mangrove trees—their isolation, identification and in vitro interaction with rhizosphere Staphylococcus sp. FEMS Microbiol Ecol 101:207–216 [DOI]
- Holguin G, Bashan Y, Mendoza-Salgado R, Amador E, Toledo G, Vázquez P, Amador A (1999) La microbiología de los manglares Bosques en la frontera entre el mar y la tierra. Cienc Desarro 25:26–35
- Holguin G, Vazquez P, Bashan Y (2001) The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: an overview. Biol Fertil Soils 33:265–278. doi:10.1007/s003740000319 [DOI]
- Höpner T, Yousef M, Berthe-Corti L, Felzmann H, Struck H, Al-Thukair A (1996) Cyanobacterial mats on oil-polluted sediments-start of a promising self-remediation process? In: Krupp F, Abuzinada HA, Nader IA (eds) A marine sanctuary for the Arabian gulf, NCWCD. Senckenbergische Nat.forschende Gesellschaft, Riyadh, Frankfurt, pp 85–95
- Jennerjahn TC, Ittekkot V (2002) Relevance of mangroves for the production and deposition of organic matter along tropical continental margins. Naturwissenschaften 89:23–30. doi:10.1007/s00114-001-0283-x [DOI] [PubMed]
- Johnsen AR, Karlson U (2005) PAH degradation capacity of soil microbial communities—does it depend on PAH exposure? Microb Ecol 50:488–495. doi:10.1007/s00248-005-0022-5 [DOI] [PubMed]
- Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Hn M (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
- Ka JO, Yu Z, Mohn WW (2001) Monitoring the size and metabolic activity of the bacterial community during biostimulation of fuel-contaminated soil using competitive PCR and RT-PCR. Microb Ecol 42:267–273. doi:10.1007/s00248-001-0003-2 [DOI] [PubMed]
- Labbe D, Margesin R, Schinner F, Whyte LG, Greer CW (2007) Comparative phylogenetic analysis of microbial communities in pristine and hydrocarbon-contaminated alpine soils. FEMS Microbiol Ecol 59:466–475. doi:10.1111/j.1574-6941.2006.00250.x [DOI] [PubMed]
- Laguerre G, Bossand B, Bardin R (1987) Free-living dinitrogen-fixing bacteria isolated from petroleum refinery oily sludge. Appl Environ Microbiol 53:1674–1678 [DOI] [PMC free article] [PubMed]
- Lewis RRI (1983) Impact of oil spills on mangrove forests. In: Teas HJ (ed) Tasks for vegetation science vol. 8 (biology and ecology of mangroves), vol 8. Dr. W. Junk, The Hague, pp 171–183
- Margesin R, Labbe D, Schinner F, Greer CW, Whyte LG (2003) Characterization of hydrocarbon-degrading microbial populations in contaminated and pristine alpine soils. Appl Environ Microbiol 69:3085–3092. doi:10.1128/AEM.69.6.3085-3092.2003 [DOI] [PMC free article] [PubMed]
- Maruyama A, Ishiwata H, Kitamura K, Sunamura M, Fujita T, Matsuo M, Higashihara T (2003) Dynamics of microbial populations and strong selection for Cycloclasticus pugetii following the Nakhodka oil spill. Microb Ecol 46:442–453. doi:10.1007/s00248-002-3010-z [DOI] [PubMed]
- Musat F, Harder J, Widdel F (2006) Study of nitrogen fixation in microbial communities of oil-contaminated marine sediment microcosms. Environ Microbiol 8:1834–1843. doi:10.1111/j.1462-2920.2006.01069.x [DOI] [PubMed]
- Muyzer G, Dewaal EC, Uitterlinden AG (1993) Profiling of complex microbial-populations by denaturing gradient gel-electrophoresis analysis of polymerase chain reaction-amplified genes-coding for 16 s ribosomal-RNA. Appl Environ Microbiol 59:695–700 [DOI] [PMC free article] [PubMed]
- Olivera NL, Esteves JL, Commendatore MG (1997) Alkane biodegradation by a microbial community from contaminated sediments in Patagonia, Argentina. Int Biodeterior Biodegradation 40:75–79. doi:10.1016/S0964-8305(97)00065-6 [DOI]
- Perez-Vargas J, Poggi-Varaldo HM, Calva-Calva G, Rios-Leal E, Rodriguez-Varquez R, Ferrera-Cerrato R, Esparza-Garcia F (2000) Nitrogen-fixing bacteria capable of utilising kerosene hydrocarbons as a sole carbon source. Water Sci Technol 42:407–410
- Piehler MF, Swistak JG, Pinckney JL, Paerl HW (1999) Stimulation of diesel fuel biodegradation by indigenous nitrogen fixing bacterial consortia. Microb Ecol 38:69–78. doi:10.1007/s002489900157 [DOI] [PubMed]
- Poly F, Monrozier LJ, Bally R (2001) Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol 152:95–103. doi:10.1016/S0923-2508(00)01172-4 [DOI] [PubMed]
- Polymenakou PN, Tselepides A, Stephanou EG, Bertilsson S (2006) Carbon speciation and composition of natural microbial communities in polluted and pristine sediments of the eastern mediterranean sea. Mar Pollut Bull 52:1396–1405. doi:10.1016/j.marpolbul.2006.03.021 [DOI] [PubMed]
- Potts M (1984) Nitrogen fixation in mangrove forests. In: Por FD, Dor I (eds) Hydrobiology of the mangal. The ecosystem of the mangrove forest. Junk, The Hague, pp 155–162
- Prantera MT, Drozdowicz A, Leite SG, Rosado AS (2002) Degradation of gasoline aromatic hydrocarbons by two N2-fixing soil bacteria. Biotechnol Lett 24:85–89. doi:10.1023/A:1013875431825 [DOI]
- Ramsay MA, Swannell RPJ, Shipton WA, Duke NC, Hill RT (2000) Effect of bioremediation on the microbial community in oiled mangrove sediments. Mar Pollut Bull 41:413–419. doi:10.1016/S0025-326X(00)00137-5 [DOI]
- Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
- Sanchez O, Diestra E, Esteve I, Mas J (2005) Molecular characterization of an oil-degrading cyanobacterial consortium. Microb Ecol 50:580–588. doi:10.1007/s00248-005-5061-4 [DOI] [PubMed]
- Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506. doi:10.1128/AEM.71.3.1501-1506.2005 [DOI] [PMC free article] [PubMed]
- Seabra PNC (2005) Aplicação de biopilha na biorremediação de solos contaminados com petróleo. Universidade Federal do Rio de Janeiro, Rio de Janeiro
- Sengupta A, Chaudhuri S (1991) Ecology of heterotrophic dinitrogen fixation in the rhizosphere of mangrove plant community at the Ganges river estuary in India. Oecologia 87:560–564. doi:10.1007/BF00320420 [DOI] [PubMed]
- Simonet P, Grojean MC, Misra AK, Nazaret S, Cournoyer B, Normand P (1991) Frankia genus-specific characterization by polymerase chain reaction. Appl Environ Microbiol 57:3278–3286 [DOI] [PMC free article] [PubMed]
- Sorkhoh N, Alhasan R, Radwan S, Hopner T (1992) Self-cleaning of the gulf. Nature 359:109. doi:10.1038/359109a0 [DOI]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The clustal-x windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi:10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed]
- Toccalino PL, Johnson RL, Boone DR (1993) Nitrogen limitation and nitrogen-fixation during alkane biodegradation in a sandy soil. Appl Environ Microbiol 59:2977–2983 [DOI] [PMC free article] [PubMed]
- Uyttebroek M, Spoden A, Ortega-Calvo JJ, Wouters K, Wattiau P, Bastiaens L, Springael D (2007) Differential responses of Eubacterial, Mycobacterium, and Sphingomonas communities in polycyclic aromatic hydrocarbon (PAH)-contaminated soil to artificially induced changes in PAH profile. J Environ Qual 36:1403–1411. doi:10.2134/jeq2006.0471 [DOI] [PubMed]
- Vazquez P, Holguin G, Puente ME, Lopez-Cortes A, Bashan Y (2000) Phosphate-solubilizing microorganisms associated with the rhizosphere of mangroves in a semiarid coastal lagoon. Biol Fertil Soils 30:460–468. doi:10.1007/s003740050024 [DOI]
- Vinas M, Sabate J, Espuny MJ, Solanas AM (2005) Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl Environ Microbiol 71:7008–7018. doi:10.1128/AEM.71.11.7008-7018.2005 [DOI] [PMC free article] [PubMed]
- Wieder RK, Lang GE (1982) A critique of the analytical methods used examining decomposition data obtained from litter bags. Ecology 63:1636–1642. doi:10.2307/1940104 [DOI]
- Zehr JP, Jenkins BD, Short SM, Steward GF (2003) Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol 5:539–554. doi:10.1046/j.1462-2920.2003.00451.x [DOI] [PubMed]