Abstract
Spontaneous preterm birth (PTB) before 37 completed weeks of gestation resulting from preterm labor (PTL) is a leading contributor of perinatal morbidity and mortality. Early identification of at-risk women by reliable screening tests could alleviate this health issue; however, conventional methods such as obstetric history and clinical risk factors, uterine activity monitoring, biochemical markers, and cervical sonography for screening women at risk for PTB have proven unsuccessful in lowering the rate of PTB. Cervicovaginal fluid (CVF) might prove to be a useful, readily available biological fluid for identifying diagnostic PTB biomarkers. Human columnar epithelial endocervical-1 (End1) and vaginal (Vk2) cell secretomes were employed to generate a stable isotope labeled proteome (SILAP) standard to facilitate characterization and relative quantification of proteins present in CVF. The SILAP standard was prepared using stable isotope labeling by amino acids in cell culture (SILAC) of End1 and Vk2 through seven passages. The labeled secreted proteins from both cell lines were combined and characterized by liquid-chromatography-tandem mass spectrometry (LC-MS/MS). 1211 proteins were identified in the End1-Vk2 SILAP standard, with 236 proteins being consistently identified in each of the replicates analyzed. Individual proteins were found to contain < 0.5 % of the endogenous unlabeled forms. Identified proteins were screened to provide a set of fifteen candidates that have either previously been identified as potential PTB biomarkers or could be linked mechanistically to PTB. Stable isotope dilution LC-multiple reaction monitoring (MRM/MS) assays were then developed for conducting relative quantification of the fifteen candidate biomarkers in human CVF samples from term and PTB cases. Three proteins were significantly elevated in PTB cases (desmoplakin isoform 1, stratifin, and thrombospondin 1 precursor), providing a foundation for further validation in larger patient cohorts.
Keywords: Cervicovaginal fluid, preterm birth, SILAC, biomarkers, mass spectrometry, multiple reaction monitoring, proteomics, LC-MS/MS, LC-MRM/MS
Introduction
PTL is characterized by the presence of regular uterine contractions causing cervical dilation before term gestation (between 20 and 37 weeks).1 Spontaneous PTB before 37 completed weeks of gestation results from PTL and is a leading contributor of perinatal morbidity and mortality.1,2 According to a report from the National Institute of Medicine on PTB, approximately half a million babies are born preterm annually in the US alone.3 Infants born preterm have a significantly higher risk of contracting infections as well as suffering from neuro-developmental disorders, and this serious medical issue carries an economic burden of approximately $26 billion/year in the US.3 Early identification of at-risk women by reliable screening tests could alleviate this health issue; however, conventional methods such as obstetric history and clinical risk factors, uterine activity monitoring, biochemical markers, and cervical sonography for screening women at risk for PTB have proven unsuccessful in lowering the rate of PTB.2
Recent advances in the application of various proteomics-based platforms have facilitated the process of discovery of novel protein biomarkers of PTB.4-7 Mass spectrometry-based shotgun proteomics methodology for identifying tryptic peptides has become a standard method for characterizing proteomes of biological fluids as well as tissues.8-10 Further advancements in multidimensional protein separation techniques have made it possible to dig even deeper to identify low abundance proteins and thereby increase the dynamic range of detection in complex biological samples such as plasma and serum.11,12 In addition, the application of SILAC technology allows for the generation of entire labeled proteomes that can be used as internal standards to facilitate accurate quantification of peptides, and proteins thereof, in biological samples.13,14 Finally, LC-MRM/MS methodology provides speed, sensitivity, selectivity and the ability to quantitate multiple peptides simultaneously.15,16 The present study was designed to probe human CVF samples for candidate protein biomarkers by combining the quantification capabilities of SILAC to generate a Stable Isotope Labeled Proteome (SILAP) standard and LC-MRM/MS methodology with enhanced search capabilities of multidimensional LC/MS/MS.
During pregnancy, the fetus is present in the amniotic sac, and is connected to the placenta by the umbilical cord.17 The placenta is an ephemeral organ present only during pregnancy, and is implanted in the wall of the uterus, the major female reproductive organ.17 The uterus is connected on one end to the cervix, which opens into the vagina, and is attached to the fallopian tubes at the other end.17 Although the biology of PTB is not well understood, it is clear that the complex interactions of signaling pathways in these reproductive organs are ultimately responsible for controlling PTL and PTB, thereby making these organs potential sites for biomarker discovery.2,18-21 CVF is a complex mixture of secretions from the vagina, endocervix, endometrial decidua and amniochorion; and therefore serves as an important diagnostic site to monitor maternal and fetal health in pregnant women.22 Collection of CVF is minimally invasive and relatively safe, as compared to collection of amniotic fluid by amniocentesis or tissue excisions from the placenta or uterus, and therefore is expected to be more readily available and potentially useful as a source of biomarkers in pregnant women.22 End1 and Vk2 cells were grown in SILAC conditions to generate a SILAP standard containing the secreted proteins.13,14,23 The rationale behind using End1 and Vk2 cells was that they are “normal” transformed cells of human origin, have a secretory phenotype, and therefore their secreted proteins would model the proteins present in human CVF.23 The labeled End1-Vk2 secretome was characterized by three dimensional (3D)-LC-MS/MS, fifteen potential candidate protein biomarkers of PTB were chosen, and stable isotope dilution LC-MRM/MS assays were designed to accurately conduct relative quantification of these candidate biomarkers in human PTB and term CVF samples using the SILAP internal standard.
Materials and Methods
Cell Lines and SILAC Secretome
Human immortalized End1 and VK2 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). Cells were grown in stable isotope-labeled serum-free DMEM/F12 media containing [13C6 15N2]-lysine and [13C6 15N1]-leucine (Cambridge Isotopes, Cambridge, MA). The SILAC media was supplemented with 50 kU/L penicillin, 50 mg/L streptomycin, 10 mL/L Stemline® keratinocyte growth supplement (Sigma-Aldrich, St. Louis, MO) containing bovine pituitary extract, epidermal growth factor, insulin, transferrin, hydrocortisone, epinephrine, and calcium chloride. Cells were passaged seven times and then supernatant was collected every alternate day, filtered through a 0.22 μm filter, concentrated through a 5 kD MW cut-off spin-filter (Millipore, Billerica, MA), pooled and stored at -80°C until analyzed. The isotopic purity (at least 99.5 %) of the End1 and Vk2 SILAC secretomes was confirmed as described previously.14
Multi-dimensional Liquid Chromatography
SILAC supernatants from the End1 and VK2 cells were each double depleted of the six high abundance plasma proteins (albumin, transferrin, haptoglobin, anti-trypsin, IgG, IgA) using a multiple affinity removal system (MARS Hu6) affinity LC column (4.6 × 100 mm) from Agilent Technologies (Palo Alto, CA). Depletion was performed according to the manufacturer’s instructions. Depleted supernatants were then concentrated as described above, protein concentrations were estimated by Coomassie Protein Assay (Pierce Scientific, Milwaukee, WI), and stored at -80°C. Equal amounts of protein (100 μg/cell line) from the immuno-depleted End1 and Vk2 SILAC-labeled supernatants were mixed together in a 1:1 ratio, henceforth referred to as the End1-Vk2 SILAC secretome. Proteins were precipitated using a standard methanol/chloroform protocol. For every 100 μL of sample, 400 μL methanol, 100 μL chloroform and 300 μL water were added stepwise, with thorough mixing at each step. Samples were centrifuged for 10 min at 12,000 × g, and the upper phase was removed. The protein precipitate was washed with 300 μL of methanol and then centrifuged for 10 min at 12,000 × g. Methanol was decanted and the protein pellets were allowed to air dry. Proteins were resolubilized in a 6 M urea/2 M thiourea solution, reduced for 30 min with 20 mM dithiothreitol (DTT) at room temperature, and then alkylated for 20 min with 50 mM iodoacetamide (IAA) at room temperature in the dark. The protein mixture was then diluted tenfold with 50 mM ammonium bicarbonate buffer, to reduce the final concentration of urea below 1 M and to adjust the pH to approximately 8.5, and digested with sequencing grade modified trypsin (Promega, Madison, WI) at 37°C for 16 hrs. Trypsin was added at a 1:20 w/w ratio of enzyme to protein sample being digested. The tryptic digest was lyophilized in a vacuum concentrator (Jouan, Winchester, VA); pH was lowered to 3 with 10 % formic acid, and diluted in strong cation exchange (SCX) mobile phase A (25 mM ammonium formate, 25 % acetonitrile, pH 3). SCX chromatography was performed on a PolySulfoethyl A column (100 mm × 2.1 mm, 5 μm 300 Å, PolyLC, The Nest Group, Southborough, MA) attached to an HP 1100 HPLC system (Agilent Technologies, Santa Clara, CA). A linear gradient was performed over 65 min from 100 % mobile phase A to 100 % mobile phase B (800 mM ammonium formate, 25 % acetonitrile, pH 5.8). For each sample, thirty-three 2-min fractions were collected and pooled into 9 fractions as follows: Fractions 1 through 6 were discarded; fractions 7 through 12 were used as individual fractions; fractions 13 through 18 were pooled into a single fraction; fractions 19 through 25 were pooled into a single fraction; and fractions 25 through 32 were pooled into a single fraction.. These 9 fractions were lyophilized and stored at -80°C until further analysis.
Mass Spectrometry
Individual SCX fractions (9 fractions/replicate; three independent replicates) from trypsin-digested End1-Vk2 SILAC secretome were resuspended in 0.1 % formic acid, 5 % acetonitrile, and further separated by online reversed-phase microflow LC and analyzed by tandem mass spectrometry in positive electrospray ionization MS/MS mode. Separation was performed at a flow rate of 16 μL/min, on a monomeric C18 Higgins column (100 mm × 0.5 mm, 5 μm, 200 Å, The Nest Group, Southborough, MA) with an Eksigent LC system equipped with an autosampler (Dublin, CA), on-line to an LTQ mass spectrometer (Thermo Scientific, Waltham, MA). Mobile phases used for separation were 0.1 % formic acid in water (A) and 0.1 % formic acid in acetonitrile (B). A linear gradient (0-50 % mobile phase B) was applied for 80 min, followed by a 10 min wash at 100 % mobile phase B and then equilibration for 10 min with 0 % mobile phase B. The LTQ was operated in full scan (m/z 300-2000) data-dependent mode set to automatically switch between MS and MS/MS acquisition. The five most intense precursor ions were dynamically isolated for fragmentation in the ion trap using collision induced dissociation (CID) with normalized collision energy of 35 %.
Peptide and Protein Identification
The MS/MS spectra were searched against an indexed human RefSeq database (version updated 11/2007, 33,439 entries) with TurboSEQUEST™ (Thermo Scientific, Waltham, MA, version 27.12) and Mascot™ (Matrix Science, Boston, MA, version 2.2.03). Strict trypsin cleavage rules with maximum of two missed cleavages, mass accuracy of 1 Da for the precursor and fragment ion, and variable modifications of methionine oxidation, carboxyamidomethlyation on cysteine, [13C6 15N1]-leucine and [13C6 15N2]-lysine were applied in the search criteria. The SEQUEST™ and Mascot™ output files were integrated into Scaffold version 2.01 (Proteome Software, Portland, OR) for validating MS/MS based peptide and protein identifications. Assignment of peptide sequences was performed using the PeptideProphet™ algorithm.24 PeptideProphet™ accounts for the distribution of scores over an entire dataset to calculate the probability of a correct assignment for every peptide. PeptideProphet™ calculates false-positive error rates at specific probability score cutoff values for each dataset.25 A minimum PeptideProphet™ probability score of ≥ 0.5 was used to remove low probability peptides. At this cutoff, the estimated false-positive error rate was 10.8 %. Protein identifications were accepted with a minimum ProteinProphet™ probability score of ≥ 0.8 and at least two identified unique peptides.26 For this dataset, a ProteinProphet™ probability score of > 0.8 corresponded to a false-positive error rate of 4 %. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. The entire work-up of the End1-Vk2 SILAC secretome has been outlined in Figure 1.
Figure 1.
General workflow depicting the generation and characterization of the End1-Vk2 SILAP standard. End1 and Vk2 cells were grown in SILAC conditions and their supernatants were individually depleted of the six most abundant serum proteins to generate the End1-Vk2 SILAC secretome. The SILAC secretome was digested with trypsin, and proteins were identified by reversed phase LC-MS/MS analysis, using the Sequest and Mascot search algorithms. The data were integrated into Scaffold, and the identified proteins were scrutinized using the selection criteria displayed in the scheme to characterize the End1-Vk2 SILAP standard.
Bioinformatics
The identified list of proteins that met the filtering criteria were analyzed for biological pathway association using the Kyoto Encyclopedia of Genes and Genomes (KEGG, release 47.1, updated 09/2008) database.27 KEGG is a database of manually curated biological pathways assigning proteins with known biological function to the different pathways, and is updated constantly. Since the raw MS/MS spectra were searched against the RefSeq database, the identified proteins were reported in terms of their gene, with their corresponding Genbank Geninfo Identifier (GI) accession numbers. The corresponding protein IDs were extracted from the GI accession numbers, and converted to the respective KEGG IDs using the Uniprot database.28 The KEGG IDs were then queried against each species-specific (Homo sapiens) biological pathway contained in the KEGG database to establish the biological pathway association of the identified proteins. The GI accession numbers that were not implicated to be associated with any known biological pathway in the KEGG database were searched against the Gene Ontology (GO) classification using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Functional Annotation Tool (http://david.abcc.ncifcrf.gov) and were assigned the most relevant biological process/molecular function.
LC-MRM/MS Assay
The End1-Vk2 SILAC secretome was characterized by full scan LC-MS/MS so that it could be used as a SILAP standard to quantitate candidate protein biomarkers in human CVF samples by LC-MRM/MS assay. Since the End1-Vk2 SILAP standard was stably labeled with [13C6 15N2]-lysine and [13C6 15N1]-leucine, it generated proteins and thereby peptides that were greater in mass than their endogenous counterparts due to a mass shift of 7 amu for each leucine and 8 amu for each lysine residue present in them. Therefore, the proteins and hence the peptides of the End1-Vk2 SILAP standard will be referred to as the “heavy” (isotopically labeled) forms as compared to the “light” (unlabeled) forms found naturally in the human CVF samples. Identified heavy proteins that met all the selection criteria described above were screened to create a set of fifteen candidates for designing LC-MRM/MS assays. Each of the proteins has been implicated previously as a potential biomarker of PTB (Table 1). MS/MS spectra of all the unique heavy peptides identified in these fifteen candidates were manually inspected, one best unique heavy peptide per protein was chosen, and the transition from the precursor ion to the most intense fragment ion for each of these fifteen heavy peptides was identified, thus generating a set of fifteen heavy MRM transitions. Peptides were chosen with a probability score of > 0.9, corresponding to a false-positive error rate of 2.1 %. A corresponding set of fifteen light MRM transitions for these selected peptides was generated using ProteinProspector® software (version 4.27, http://prospector.ucsf.edu). The LC-MRM/MS method was designed to analyze 10 transitions (light and heavy sets of 5 peptides) simultaneously in a single LC-MS run under the conditions described above, therefore each sample was injected three times to complete the 15-protein analysis in three sets. The order of sample injection was randomized in each MRM set to eliminate sample bias. Blank injections were made in each MRM set to check for any carry-over effects. The LC method used for the MRM assays was the same as that used for the LC-MS/MS analysis of End1-Vk2 SILAC secretome, as described above.
Table 1.
Fifteen-protein panel of candidate biomarkers selected for quantification in human CVF samples by LC-MRM/MS assay.
| GI Accession |
Protein name | KEGG Pathway or Biological Process/Molecular Function (GO) |
Ref |
|---|---|---|---|
| 16933542 | fibronectin 1 isoform 3 preproprotein | Cell Communication, Signal Transduction, Signaling Molecules & Interaction | 5,29,30 |
| 40317626 | thrombospondin 1 precursor | Cell Communication, Signal Transduction, Signaling Molecules& Interaction | 21 |
| 38045910 | laminin alpha 3 subunit isoform 1 | Cell Communication, Signal Transduction | 32 |
| 10835159 | plasminogen activator inhibitor-1 | Signal Transduction, Cell Growth and Death | 21,29 |
| 113422396 | fatty acid binding protein 5 (psoriasis-associated) | Signal Transduction | 5,22,29 |
| 4758236 | extracellular matrix protein 1 isoform 1 precursor | Signal Transduction | 5,22,29,30 |
| 5031875 | lamin A/C isoform 2 | Cell Communication | 5,29 |
| 5454052 | stratifin | Cell Growth and Death | 5 |
| 4507509 | tissue inhibitor of metalloproteinase 1 precursor | Cell Growth and Death | 21 |
| 4505863 | urokinase plasminogen activator preproprotein | Immune System | 21 |
| 57242755 | calsyntenin 1 isoform 2 | Cell Communication | - |
| 4503107 | cystatin C precursor | *Cysteine protease inhibitor | 30 |
| 4507065 | secretory leukocyte peptidase inhibitor precursor | *Endo-peptidase inhibitor | 21 |
| 23110960 | cathepsin L2 preproprotein | *Cathespsin L activity, Protein binding | 31 |
| 58530840 | desmoplakin isoform I | *Epidermis development, Keratinocyte differentiation, Peptide cross linking | 5,22,29 |
Collection and Processing of Clinical Samples
CVF samples were selected at random from among 2,100 subjects enrolled in a randomized controlled trial at the Hospital of the University of Pennsylvania. The study was approved by the HUP institutional review board and registered with www.clinicaltrials.gov (NCT00116974). Subjects who presented for prenatal care before 20 weeks’ gestation (confirmed by ultrasound) were enrolled in the study, and CVF samples were self-collected by subjects during routine prenatal visits between 24-28 weeks’ gestation. Samples were collected using Dacron-tipped swabs, placed in 500 μL phosphate-buffered saline, immediately flash frozen in liquid nitrogen and stored at -80°C. A nested case-control study was performed in which CVF samples were selected at random from five subjects who delivered preterm between 28-32 weeks’ gestation following spontaneous PTL and five subjects who delivered after 37 weeks’ gestation (controls). Proteins were extracted from these samples by the methanol/chloroform method as described above in the preparation of the End1-Vk2 SILAP standard, and stored at -80°C. For initial screening of fifteen candidate biomarkers by LC-MRM/MS assay, equal amounts of protein (200 μg/patient) from five control and five PTB subjects were spiked with an equal amount of undigested End1-Vk2 SILAP standard (200 μg/patient), reduced for 30 min with 20 mM DTT at room temperature, then alkylated for 20 min with 50 mM IAA at room temperature in the dark. Samples were then digested with trypsin (1:20 w/w ratio of enzyme to sample protein) at 37°C for 16 hrs, and desalted with Vydac-C18® macrospin cartridges (The Nest Group, Southborough, MA). Differences in the level of these 15 candidate biomarkers between the control and PTB subjects were compared statistically by non-parametric Mann-Whitney rank-sum test, with p-value < 0.05 considered as significant.
Results
Identification and Characterization of End1-Vk2 SILAP Standard by LC-MS/MS
The flow chart depicting the process of generation and characterization of End1-Vk2 SILAP standard has been outlined in Figure 1. LC-MS/MS analysis (full scan data dependent acquisition) identified a combined total of 1211 proteins in the three replicates. A Venn-diagram describing the distribution of proteins identified in the individual replicates is shown in Figure 2. Individually, 692 proteins were identified in replicate 1, 420 in replicate 2, and 779 proteins in replicate 3. Of the 1,211 proteins identified, 236 were identified in all three replicates (supplementary data).
Figure 2.
Venn diagram of identified proteins that met selection criteria in three independent replicates of End1-Vk2 SILAP standard by reversed phase LC-MS/MS analyses.
Biological Pathway Analysis of the Identified Proteins using the KEGG Database
For the 236 proteins identified in all three replicates, biological association to known functional pathways was established using the KEGG database. Since multiple GI accession numbers can correspond to a single protein entry, and vice versa, the dataset of 236 proteins actually corresponded to a total of 307 GI accession numbers (data not shown). Of the 307 GI accession numbers, 129 were found to be associated with at least one biological pathway, and several with multiple pathways. Table 2 lists the total number of GI accessions that were found associated with at least one biological pathway contained in the KEGG database.
Table 2.
KEGG pathway association of the 236-protein dataset identified by LC/MS/MS analysis in all of the three replicates of End1-Vk2 SILAP standard.
| KEGG Pathway | # of GI Accessions |
|---|---|
| 1. Metabolism | |
| Carbohydrate Metabolism | 17 |
| Energy Metabolism | 14 |
| Amino Acid Metabolism | 11 |
| Nucleotide Metabolism | 6 |
| Xenobiotics Biodegradation and Metabolism | 5 |
| Metabolism of Other Amino Acids | 3 |
| Metabolism of Cofactors and Vitamins | 1 |
| Biosynthesis of Secondary Metabolites | 1 |
| 2. Genetic information Processing | |
| Folding, Sorting and Degradation | 10 |
| Translation | 4 |
| 3. Environmental Information Processing | |
| Signaling Molecules and Interaction | 11 |
| Signal Transduction | 7 |
| 4. Cellular Processes | |
| Cell Communication | 28 |
| Immune System | 19 |
| Cell Motility | 16 |
| Cell Growth and Death | 13 |
| Endocrine System | 6 |
| Development | 3 |
Generation of the Fifteen Protein Panel of Candidate Biomarkers for PTB
A panel of fifteen candidate protein biomarkers was created for the development of LC-MRM/MS assays from the 236-protein dataset. The 236 identified proteins were searched against previously published studies related to either proteins or genes identified in human CVF, amniotic fluid, placental or uterine tissue samples with regards to their relevance to PTB or other pregnancy-related conditions. These comparisons were utilized to select a panel of fifteen relevant proteins for developing a quantitative LC-MRM/MS assay to measure these proteins in human CVF samples (Table 1).
Development of LC-MRM/MS Assay for the Fifteen Candidate Biomarker Panel
Table 3 displays the MS/MS fragmentation information of the fifteen proteins (one unique peptide/protein) required for developing the LC-MRM/MS assay. The MS/MS spectral data of each of the unique “heavy” peptides identified in the fifteen protein panel from the LC/MS/MS experiment of the End1-Vk2 SILAP standard was manually inspected to select the best one unique “heavy” peptide per protein. For each unique “heavy” peptide selected, the charge state and the mass to charge (m/z) values for the precursor ion as well as for the most intense fragment ion generated were obtained from the MS/MS data. These details were utilized to set up the “heavy” transitions for the LC/MRM/MS assays, as shown in Table 3. The corresponding “light” transitions for monitoring the fragmentation of the endogenous counterpart of these fifteen peptides in human CVF samples were computed using the ProteinProspector® software, and are represented in Table 3.
Table 3.
LC-MRM/MS assay set up for the fifteen-protein candidate biomarker panel. Fragmentation details of the peptides selected for the quantification by LC-MRM/MS assay of the fifteen-protein panel (one set of MRM transitions/one unique peptide/protein) were extracted from the LC-MS/MS data of the End1-Vk2 SILAP standard. C* refers to the carbamido-methylated form of cysteine amino acid residue obtained by treatment of protein samples with iodoacetamide. (K) and (L) refer to the stable isotope labeled [13C6 15N2]-lysine and [13C6 15N1]-leucine residues, respectively.
| Protein | Peptide | Precursor Charge |
Fragment Ion |
Light Transition | Heavy Transition | ||
|---|---|---|---|---|---|---|---|
| Precursor m/z |
Fragment m/z |
Precursor m/z |
Fragment m/z |
||||
| calsyntenin 1 isoform 2 | IPDGVVSVSP(K) | +2 | y10++ | 549.31 | 492.77 | 553.32 | 496.78 |
| cathepsin L2 preprotein | (L)YGANEEGWR | +2 | y8+ | 597.78 | 918.41 | 601.29 | 918.41 |
| cystatin C precursor | (L)VGGPMDASVEEEGVR | +2 | y14++ | 822.9 | 716.82 | 826.41 | 716.82 |
| desmoplakin isoform 1 | AITGFDDPFSG(K) | +2 | y10++ | 627.8 | 535.74 | 631.81 | 539.75 |
| extracellular matrix protein 1 isoform 1 precursor | NVA(L)VSGDTENA(K) | +2 | y8+ | 659.34 | 821.36 | 666.85 | 829.38 |
| fatty acid binding protein 5 (psoriasis associated) | TTQFSC*T(L)GE(K) | +2 | y8+ | 636.3 | 941.44 | 643.82 | 956.47 |
| fibronectin 1 isoform 3 preprotein | D(L)QFVEVTDV(K) | +2 | y8+ | 646.84 | 936.5 | 654.36 | 944.52 |
| lamin A/C isoform 2 | EGD(L)IAAQAR | +2 | y5+ | 522.28 | 516.29 | 525.79 | 516.29 |
| laminin alpha 3 subunit isoform 1 | VWQDAC*SP(L)P(K) | +2 | y9+ | 650.82 | 1015.49 | 658.34 | 1030.52 |
| plasminogen activator inhibitor-1 | FIINDWV(K) | +2 | y6+ | 517.79 | 774.41 | 521.79 | 782.43 |
| secretory leukocyte peptidase inhibitor precursor | C*(L)DPVDTPNPTR | +2 | y9+ | 692.83 | 996.51 | 696.34 | 996.51 |
| stratifin | Y(L)AEVATGDD(K) | +2 | y9+ | 591.29 | 905.42 | 598.8 | 913.44 |
| thrombospondin 1 precursor | TIVTT(L)QDSIR | +2 | y9+ | 623.85 | 1032.57 | 627.36 | 1039.59 |
| tissue inhibitor of metalloproteinase 1 precursor | GFQA(L)GDAADIR | +2 | y7+ | 617.31 | 717.35 | 620.82 | 717.35 |
| urokinase plasminogen activator preprotein | (L)NSNTQGEM(K) | +2 | y8+ | 561.27 | 894.4 | 568.78 | 902.41 |
LC-MRM/MS Assay for Fifteen Candidate Biomarkers in CVF Samples
LC-MRM/MS assays were performed to quantitate the peptide (of the fifteen protein panel) levels in the CVF samples obtained from five control subjects that delivered at term and five PTB subjects that delivered pre-term (cases). Since each sample was spiked with the same quantity of End1-Vk2 SILAP standard, two MRM transitions were monitored for each of the fifteen peptides in the CVF samples, one corresponding to the “light” form (L, endogenous peptide present in the samples), and the other corresponding to the “heavy” form (H, labeled peptide from the spiked End1-Vk2 SILAP standard). The area ratio of the “light” to the “heavy” peptide (L/H) for each of the fifteen proteins was calculated in each sample in the two groups (control and PTB), and the mean L/H ratios for the respective groups were computed accordingly. The spiking of SILAP standard in each sample allowed for normalizing and thereby quantitating the levels of these fifteen proteins in terms of their respective peptides. The relative fold-change in expression levels of the fifteen peptides between the two patient groups were determined using the mean L/H values obtained for the PTB and control groups respectively, and the corresponding p-value for each peptide was calculated by applying the Mann-Whitney rank-sum statistical test. A p-value of < 0.05 was considered as statistically significant.
Table 4 summarizes the LC-MRM/MS assay data for the fifteen proteins quantified in terms of their fifteen peptides (one unique peptide/protein). The fold-change data from Table 4 shows that the levels of desmoplakin isoform 1, stratifin, thrombospondin 1 precursor, fatty acid binding protein 5 (psoriasis associated), extracellular matrix protein 1 isoform 1 precursor, secretory leukocyte peptidase inhibitor precursor, and plasminogen activator inhibitor-1 peptides were observed to be higher in the PTB subjects as compared to the control subjects. In contrast, fibronectin 1 isoform 3 preprotein, urokinase plasminogen activator preprotein, cathepsin L2 preprotein, tissue inhibitor of metalloproteinase 1 precursor, and calsyntenin 1 isoform 2 peptides levels were lower in the PTB group as compared to the control group. Cystatin C precursor peptide levels were observed to be the same between the two groups. Laminin alpha 3 subunit isoform 1 and lamin A/C isoform 2 peptide levels were below the limits of detection in both patient groups, and therefore their fold change could not be computed.
Table 4.
Summary of the fifteen proteins analyzed by LC-MRM/MS assay in human CVF samples. The peak area ratios (L/H) were calculated for each peptide in each sample and used to compute the corresponding mean L/H ratios for the PTB and CONTROL groups, along with their standard deviation values (SD). The fold change in the PTB L/H ratio as compared to the CONTROL L/H ratio for the fifteen peptides was calculated. Mann-Whitney’s rank-sum t-test was applied to determine whether the mean ratios of the two groups were statistically different and to compute the p-values.
| PROTEIN | CONTROL | PTB | Fold Change |
p-value | ||
|---|---|---|---|---|---|---|
| Mean L/H | SD | Mean L/H | SD | |||
| desmoplakin isoform 1 | 0.004 | 0.006 | 0.262 | 0.309 | 70.7 | 0.01 |
| stratifin | 0.009 | 0.009 | 0.369 | 0.289 | 42.4 | 0.01 |
| thrombospondin 1 precursor | 0.009 | 0.008 | 0.048 | 0.024 | 5.1 | 0.01 |
| fatty acid binding protein 5 (psoriasis associated) | 0.240 | 0.316 | 1.722 | 3.473 | 7.2 | 0.84 |
| extracellular matrix protein 1 isoform 1 precursor | 0.040 | 0.009 | 0.148 | 0.200 | 3.7 | 0.69 |
| secretory leukocyte peptidase inhibitor precursor | 0.022 | 0.012 | 0.074 | 0.108 | 3.4 | 0.69 |
| plasminogen activator inhibitor-1 | 0.005 | 0.004 | 0.008 | 0.009 | 1.5 | 0.42 |
| fibronectin 1 isoform 3 preprotein | 0.300 | 0.314 | 0.098 | 0.184 | 0.3 | 0.10 |
| urokinase plasminogen activator preprotein | 0.033 | 0.056 | 0.011 | 0.010 | 0.3 | 1.00 |
| cathepsin L2 preprotein | 0.055 | 0.079 | 0.025 | 0.056 | 0.5 | 0.69 |
| tissue inhibitor of metalloproteinase 1 precursor | 0.080 | 0.088 | 0.039 | 0.037 | 0.5 | 0.42 |
| calsyntenin 1 isoform 2 | 0.177 | 0.049 | 0.131 | 0.085 | 0.7 | 0.42 |
| cystatin C precursor | 0.024 | 0.009 | 0.023 | 0.007 | 1.0 | 0.84 |
| laminin alpha 3 subunit isoform 1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.0 | NC |
| lamin A/C isoform 2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.0 | NC |
The p-value data from Table 4 showed that the calculated levels of three out of the fifteen proteins (desmoplakin isoform 1, stratifin, and thrombospondin 1 precursor) in the panel were found to be significantly different (p-value < 0.05) between the control and PTB groups. Desmoplakin isoform 1 peptide was quantified to be 70.7 fold higher in the PTB samples as compared to the control samples, and similarly, stratifin peptide (42.4 fold) and thrombospondin 1 precursor peptide (5.1 fold) levels were significantly greater in the PTB patient samples. Figure 3 displays the scatter-plot graph of the individual L/H area ratios calculated for desmoplakin isoform 1, stratifin, and thrombospondin 1 precursor peptides in the CVF samples obtained from the ten subjects (five controls and five PTB). Collectively, the data suggest that these three proteins could serve as potential markers for PTB, and should be investigated further in a larger cohort of patient samples.
Figure 3.
Scatter-plot graph for the peak area ratios (L/H) computed for desmoplakin isoform 1, stratifin, and thrombospondin 1 precursor peptides by LC-MRM/MS assay of human CVF samples. Equal amount of protein (200 μg/patient) obtained from CVF samples of ten human subjects (five CONTROL and PTB subjects each) were spiked with an equal amount of End1-Vk2 SILAP standard (200 μg/patient) and processed for LC-MRM/MS assay as described in the Materials and Methods section. Peak areas were computed for the “light” and “heavy” forms of the three peptides and used to determine the L/H ratio for these peptides in each patient sample in the two groups (CONTROL and PTB). Mann-Whitney’s rank-sum test was applied to determine if the peak area ratios were statistically different (p-value < 0.05) between the two groups for the three proteins. The horizontal bar in each group represents the Median value of the peak area ratios in that group.
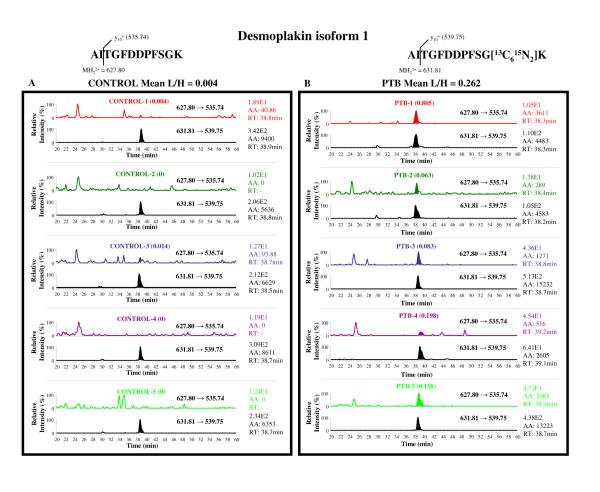
LC-MRM/MS chromatograms for desmoplakin isoform 1 and cystatin C precursor peptides from ten CVF samples are shown in Figures 4 and 5. Chromatograms of the CVF samples obtained from the five control subjects are shown in panel A and those from the five PTB subjects are shown in panel B. Two MRM transitions are displayed in each patient sample chromatogram; obtained by monitoring the “light” (upper channel) and the “heavy” (lower channel) transitions resulting from the fragmentation of the “light” and “heavy” peptides respectively. The peak area (AA) computed for the “light” transition represents the quantity of the endogenous peptide, and thereby the corresponding protein, present in that CVF sample and this value is normalized using the peak area calculated for the corresponding “heavy” transition which arises from the labeled peptide present in the spiked End1-Vk2 SILAP standard. This normalization allows for correcting any kind of sample losses that may have occurred due to sample processing, since each patient sample was spiked with an equal quantity of the End1-Vk2 SILAP standard right at the beginning of the assay procedure. Figure 4A clearly demonstrates that the desmoplakin isoform 1 peptide was below detection limits (regarded as a zero value) in three out of five control patient samples. The remaining two control patient samples contained very low levels of this peptide. In contrast, the desmoplakin isoform 1 peptide levels were significantly elevated in all five of the PTB patient samples (Figure 4B). This differential expression suggests that desmoplakin isoform 1 protein might become elevated before PTL and PTB (Table 4). Levels of cystatin C precursor peptide did not differ when comparing control (Figure 5A, Table 4) to PTB (Figure 5, Table 4) subjects.
Figure 4.
Chromatograms from the LC-MRM/MS assay of desmoplakin isoform 1 peptide (AITGFDDPFSGK) in human CVF samples. Protein (200 μg/patient) from CVF samples was spiked with End1-Vk2 SILAP standard (200 μg/patient) and processed for LC-MRM/MS assay as described in the Materials and Methods section. Panel A displays the chromatograms from five CONTROL subjects, and the panel B represents the five PTB patient chromatograms. The MRM transitions of the precursor ion (charge state of +2) to the y10+ fragment ion were monitored for the light (L, m/z 627.80 → 535.74) and the heavy (H, m/z 631.81 → 539.75) forms of the AITGFDDPFSGK peptide in each sample, thereby generating a pair of two chromatograms for each sample. For each pair, the upper chromatogram reflects the light peptide (present in human sample), and the lower chromatogram represents the labeled peptide from the End1-Vk2 SILAP standard. The retention times of the peptide in each sample are displayed along with the peak area (AA) and the total ion current generated in that transition. The peaks below the limit of detection were assigned a value of zero. Peak area ratios (L/H) were calculated for each patient sample, and the mean L/H values were computed for the CONTROL and PTB groups.
Figure 5.
Chromatograms from the LC-MRM/MS assay of cystatin C precursor peptide (LVGGPMDASVEEEGVR) in human CVF samples. Protein (200 μg/patient) from CVF samples was spiked with End1-Vk2 SILAP standard (200 μg/patient) and processed for LC-MRM/MS assay as described in the Materials and Methods section. Panel A displays the chromatograms from five CONTROL subjects, and the panel B represents the five PTB patient chromatograms. The MRM transitions of the precursor ion (charge state of +2) to the y14++ fragment ion were monitored for the light (L, m/z 822.90 → 716.82) and the heavy (H, m/z 826.41 → 716.82) forms of the LVGGPMDASVEEEGVR peptide in each sample, thereby generating a pair of two chromatograms for each sample. For each pair, the upper chromatogram reflects the light peptide (present in human sample), and the lower chromatogram represents the labeled peptide from the End1-Vk2 SILAP standard. The retention times of the peptide in each sample are displayed along with the peak area (AA) and the total ion current generated in that transition. Peak area ratios (L/H) were calculated for each patient sample, and the mean L/H values were computed for the CONTROL and PTB groups.
Discussion
A multi-dimensional LC-MS/MS platform for protein discovery along with in vitro SILAC models for peptide quantification have been employed to identify and quantify novel protein biomarkers of PTB in human biological samples (CVF). Several biological fluids, including CVF, plasma and amniotic fluid, and tissues including placenta, myometrium, cervix and uterus have all been investigated in the quest for PTB biomarkers.4-7,10,21,22,29,30 These previous studies have applied proteomics technologies to identify numerous peptides and their corresponding proteins in these biological samples by employing data dependent acquisition of the MS/MS spectra with the hope of discovering novel biomarkers of PTB.4-6,22,29,30 Operating the mass spectrometer in data dependent acquisition mode in tandem with separation of peptides by LC allows for estimating the total number of times a peptide is identified by an MS/MS spectra and thereby calculating its spectral count.33 The spectral count value can provide an estimate of the relative abundance of the protein in the different samples, and this has been the basis for biomarker discovery in most of the previous efforts.33 In other words, these previous efforts have relied on qualitative analyses to establish differential expression patterns of proteins between the term and PTB samples for biomarker discovery. Not surprisingly, although these prior studies have identified and tested several potential biomarkers (such as fetal fibronectin, corticotrophin-releasing hormone, interleukin-6 (IL-6), C-reactive protein, CD163, thrombin-antithrombin complex, IL-8, SERPINH1, tumor necrosis factor-alpha, monocytic chemotactic protein-1, matrix metalloproteinase-8, ferritin, placental alkaline phosphatases, and relaxin), with the exception of fetal fibronectin, currently there are no reliable biomarkers for PTB in clinical use.2,34-50
In order to make biomarker discovery a more successful and efficient endeavor, it is imperative to study readily available biological samples in individuals before the onset of clinical symptoms, and to be able to quantitate the protein concentrations in the biological samples in question. A unique feature of the present study is that self-collected CVF samples were used from asymptomatic women at 24-28 weeks’ gestation, five of whom ultimately delivered preterm (before 32 weeks) and five of whom delivered without complications at term (after 37 weeks). In general, quantitative protein expression profiles in biological samples are based upon generating peptide standards to be spiked in the biological samples to accurately calculate each corresponding protein concentration in the samples.15,16 However, biological samples are composed of numerous proteins. For example, comprehensive proteomic analyses of human CVF identified from 150 to 685 unique proteins in three separate studies.5,22,29 These findings not only highlight the complex composition of biological samples, but also demonstrate the high variability in protein identification amongst different studies. Therefore, generating peptide standards for quantitating such a large number of proteins in complex biological samples is not only an ambitious but also an economically challenging task.
The current study has employed SILAC methodology to establish a cell model that can be employed to generate a relevant SILAP standard for precise and accurate relative quantification of proteins in complex biological samples. The biological sample investigated in this study was human CVF collected from pregnant women who later delivered at term or preterm. Since there are already several studies that have reported the protein composition of human CVF, rather than focusing efforts on protein identification in the CVF samples, the current study was designed to devise stable isotope LC-MRM/MS based methodology to quantitate multiple proteins simultaneously in the CVF samples.5,7,22,29 Two cell lines grown in SILAC conditions were used to generate a stable labeled secretome. This secretome was characterized by multi-dimensional liquid chromatography tandem mass spectrometry and employed as a SILAP standard. End1 and Vk2 cells are transformed “normal” human cell lines of endocervical and vaginal origin respectively with a secretory phenotype, and therefore were chosen as an in vitro model of proteins present in human CVF.23 The SILAP standard proved to be a rich source of potential biomarkers, with 1211 total proteins identified among all replicates analyzed. 236 of these proteins were consistently identified in all replicates (Figure 2 and Supplementary data).
In order to quantify these 236 proteins by LC-MRM/MS, it would be necessary to monitor at least 236 pairs of transitions (one pair of transition/peptide/protein; each pair for the light and heavy form of the peptide). This presents a significant challenge not only from an instrumentation perspective, but also from a data handling and data processing perspective. During the LC-MRM/MS method development on the LTQ, it was observed that ten MRM transitions could be monitored successfully in a single segment in a run without any significant loss of signal or sensitivity (data not shown). Therefore, the LC-MRM/MS assay was designed to monitor five pairs of transitions (corresponding to the light and heavy form of the peptide) per single run, and thus it would require at least 48 runs to complete the 236-protein quantification. It was also observed that, with the current methodology, at least 100 μg of protein starting material was required per run to allow for quantification. Therefore, 48 runs would require almost 5 mg of protein per human CVF sample, an almost impossible task considering the low protein yield in CVF samples. Moreover, these details are for performing only one replicate analysis. Therefore, it seemed prudent to test our methodology on a small subset of proteins and to establish proof of principle. Since this was a pilot study, it was decided to monitor one transition per peptide, and to quantify one peptide per protein, for a set of fifteen proteins to test our methodology. No doubt, further validation studies would be required to confirm this methodology, by monitoring multiple transitions/peptide and tracking multiple peptides/protein. However, this raised another issue of which fifteen proteins to select from the 236-protein dataset. For this purpose, the 236-protein dataset was subjected to KEGG pathway database analysis to determine their biological relevance. Table 2 summarizes the data obtained from the KEGG analysis, and shows that almost 40 % of the proteins in this dataset were found to be associated with at least one known biological pathway. This indicates that the 236-protein dataset of the End1-Vk2 SILAP standard is a biologically relevant model. Furthermore, these proteins were compared against the known literature pertaining to female reproductive physiology with regards to PTB and other pregnancy related conditions, to narrow down the fourteen protein list that could be considered most relevant to our study (Table 1). In addition, calsyntenin 1 was monitored (Table 1) because it is an abundant protein in both cell types that is involved in cell-cell communication (Table 2). LC-MRM/MS assays (Table 4) were designed for quantifying this initial set of fifteen proteins by adding the End1-Vk2 SILAP standard to the human CVF samples (Table 3). Seven out of the fifteen proteins were elevated in PTB subjects and five proteins were observed to be lower in PTB samples (Table 4). Two of the proteins were below limits of detection, and therefore considered as absent in both the groups, and one protein, cystatin C precursor was unaltered amongst the two groups. This clearly highlights the utility and the robustness of the current methodology to screen multiple proteins simultaneously in complex biological samples in a quantitative fashion. However, it is critical to point out the preliminary nature of this study and that the primary purpose of this study was to test our methodology of utilizing a SILAP standard as a tool for identifying and quantifying potential biomarkers simultaneously by LC-MRM/MS assays in complex biological samples, such as CVF. Future studies will be designed to validate the results obtained in this pilot study. Validation will be performed by monitoring multiple transitions per peptide, tracking multiple peptides/protein, and evaluating a larger cohort of patients. As the linear ion trap instruments are limited by the number of reactions that can be monitored simultaneously, the current methodology will be transferred and validated on a triple quadrupole-based MS, which will allow for monitoring of a significantly higher number of reactions simultaneously. Absolute quantification will then be performed for biomarker proteins that are validated by this approach. This future study would involve synthesizing multiple tryptic peptides for the protein, generating the standard curves for their concentrations, spiking them into human CVF samples as internal standards, and thereby calculating the absolute level of these peptides in CVF samples by LC-MRM/MS assays.
Three proteins (desmoplakin isoform 1, stratifin, thrombospondin 1 precursor) were demonstrated to be significantly (p-value < 0.05) elevated in the PTB subjects as compared to the controls (Figure 3). Although desmoplakin isoform 1 has not yet been associated with any of the known biological pathways in the KEGG database (Table 1), it is well established that desmoplakin-1 is an obligate component of functional desmosomes.51 Desmosomes are intercellular junctions that tightly link adjacent cells, and are involved in regulating cell-cell communication, cell motility and cell adhesion.51 Desmoplakin-1 has been previously identified in human CVF and has also been found to be localized in the human fetal and amniotic membranes.52 Although it is unclear as to how desmoplakin-1 may be involved in PTB, one study had reported desmoplakin, based on its spectral counts, to be significantly down-regulated in PTL.5 Interestingly, we have demonstrated that desmoplakin isoform 1 was significantly elevated, by almost 70-fold, in PTB subjects. Stratifin, also referred to as the 14-3-3σ protein, was found to be associated with cell growth and death pathways in the KEGG database (Table 1). Members of the 14-3-3 protein family, including stratifin, have been shown to be negative regulators of cell proliferation, and some of their effects are by virtue of their ability to inhibit protein kinase C and binding to various members of the growth factor signaling pathways and cell cycle regulators.53 Although the mechanistic link between stratifin and PTB is not established, its levels have been documented to be elevated in PTL.5 From the KEGG analysis, it was observed that thrombospondin-1 precursor is known to be associated with biological pathways under the cell communication as well as signal transduction and signaling molecules and interaction categories (Table 1). It is an adhesive glycoprotein that mediates cell-to-cell and cell-to-matrix interactions and has the ability to bind to several extracellular proteins.54 It is noteworthy to point out that thrombospondin-1 can bind to fibronectin, one of the fifteen proteins in our study that was observed to be lower in the PTB subjects as compared to the controls (Table 4). This observation suggests that there might be a possibility of interaction between these two proteins, and whether this interaction has any role in PTB remains to be investigated. Furthermore, in previous reports that studied the role of myometrial tissue in PTL in humans as well as mice models, elevated levels of thrombospondin-1 were associated with labor as well as PTL.55,56
In summary, we have shown that a SILAP standard generated from End1 and Vk2 human cell lines can be employed to quantify multiple proteins simultaneously by LC-MRM/MS in human CVF. In theory, the SILAP standard can be used quantify 236 proteins in human CVF samples. The pilot analysis of a set of fifteen proteins in a small set of term and PTB subjects revealed that three proteins (desmoplakin isoform 1, stratifin, thrombospondin 1 precursor) were significantly elevated in subjects who later experienced PTB (Table 4). Based on the known literature, this LC-MRM/MS data is suggestive of the potential of these three proteins as biomarkers of PTB. In contrast, cystatin C precursor protein levels are not an indicator of increased risk of PTB (Table 4), and therefore can serve as a negative control as well as a quality control biomarker to show that CVF samples were collected and stored correctly. Further studies are underway to validate the utility of desmoplakin isoform 1, stratifin, and thrombospondin 1 precursor as PTB markers in a larger dataset of subjects.
Supplementary Material
Acknowledgements
Supported by the Pennsylvania Department of Health SAP# 4100020720 and by NIH grants U01HD050088, P30ES013508, and UL1RR024134. We acknowledge the receipt of an Institute for Translational Medicine and Therapeutics Research Fellowship (K.H.Y.).
Abbreviations
- 3D
three-dimensional
- ATCC
American Type Culture Collection
- CID
collision induced dissociation
- CVF
cervicovaginal
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DTT
dithithreitol
- ELISA
enzyme-linked immunosorbent assay
- End1
columnar epithelial endocervical-1
- GI
GenBank Geninfo Identifier
- GO
Gene Ontology
- IAA
iodoacetamide
- IL
interleukin
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LC
liquid chromatography
- MRM
multiple reaction monitoring
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- PTB
preterm birth
- PTL
preterm labor
- SCX
strong cation exchange
- SILAC
stable isotope labeling by amino acids in cell culture
- SILAP
stable isotope labeled proteome
- Vk2
vaginal mucosal-2
References
- (1).Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371(9606):75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Menon R. Spontaneous preterm birth, a clinical dilemma: etiologic, pathophysiologic and genetic heterogeneities and racial disparity. Acta. Obstet. Gynecol. Scand. 2008;87(6):590–600. doi: 10.1080/00016340802005126. [DOI] [PubMed] [Google Scholar]
- (3).Preterm birth: causes, consequences and prevention. Institute of Medicine. 2006 [Google Scholar]
- (4).Klein LL, Jonscher KR, Heerwagen MJ, Gibbs RS, McManaman JL. Shotgun proteomic analysis of vaginal fluid from women in late pregnancy. Reprod. Sci. 2008;15(3):263–273. doi: 10.1177/1933719107311189. [DOI] [PubMed] [Google Scholar]
- (5).Pereira L, Reddy AP, Jacob T, Thomas A, Schneider KA, Dasari S, Lapidus JA, Lu X, Rodland M, Roberts CT, Jr., Gravett MG, Nagalla SR. Identification of novel protein biomarkers of preterm birth in human cervical-vaginal fluid. J. Proteome. Res. 2007;6(4):1269–1276. doi: 10.1021/pr0605421. [DOI] [PubMed] [Google Scholar]
- (6).Butt RH, Lee MW, Pirshahid SA, Backlund PS, Wood S, Coorssen JR. An initial proteomic analysis of human preterm labor: placental membranes. J. Proteome. Res. 2006;5(11):3161–3172. doi: 10.1021/pr060282n. [DOI] [PubMed] [Google Scholar]
- (7).Rüetschi U, Rosén A, Karlsson G, Zetterberg H, Rymo L, Hagberg H, Jacobsson B. Proteomic analysis using protein chips to detect biomarkers in cervical and amniotic fluid in women with intra-amniotic inflammation. J. Proteome. Res. 2005;4(6):2236–2242. doi: 10.1021/pr050139e. [DOI] [PubMed] [Google Scholar]
- (8).Chen G, Pramanik BN. LC-MS for protein characterization: current capabilities and future trends. Expert. Rev. Proteomics. 2008;5(3):435–444. doi: 10.1586/14789450.5.3.435. [DOI] [PubMed] [Google Scholar]
- (9).Yang S, Huang CM. Recent advances in protein profiling of tissues and tissue fluids. Expert. Rev. Proteomics. 2007;4(4):515–529. doi: 10.1586/14789450.4.4.515. [DOI] [PubMed] [Google Scholar]
- (10).Kolialexi A, Mavrou A, Spyrou G, Tsangaris GT. Mass spectrometry-based proteomics in reproductive medicine. Mass. Spectrom. Rev. 2008 doi: 10.1002/mas.20181. Advance Access published online on July 10, 2008. [DOI] [PubMed] [Google Scholar]
- (11).Fang X, Zhang WW. Affinity separation and enrichment methods in proteomic analysis. J. Proteomics. 2008;71(3):284–303. doi: 10.1016/j.jprot.2008.06.011. [DOI] [PubMed] [Google Scholar]
- (12).Whiteaker JR, Zhang H, Eng JK, Fang R, Piening BD, Feng LC, Lorentzen TD, Schoenherr RM, Keane JF, Holzman T, Fitzgibbon M, Lin C, Zhang H, Cooke K, Liu T, Camp DG, 2nd, Anderson L, Watts J, Smith RD, McIntosh MW, Paulovich AG. Head-to-head comparison of serum fractionation techniques. J. Proteome. Res. 2007;6(2):828–836. doi: 10.1021/pr0604920. [DOI] [PubMed] [Google Scholar]
- (13).Ong SE, Mann M. Stable isotope labeling by amino acids in cell culture for quantitative proteomics. Methods. Mol. Biol. 2007;359:37–52. doi: 10.1007/978-1-59745-255-7_3. [DOI] [PubMed] [Google Scholar]
- (14).Yan Y, Weaver VM, Blair IA. Analysis of protein expression during oxidative stress in breast epithelial cells using a stable isotope labeled proteome internal standard. J. Proteome. Res. 2005;4(6):2007–2014. doi: 10.1021/pr050175d. [DOI] [PubMed] [Google Scholar]
- (15).Han B, Higgs RE. Proteomics: from hypothesis to quantitative assay on a single platform. Guidelines for developing MRM assays using ion trap mass spectrometers. Brief. Funct. Genomic. Proteomic. 2008 doi: 10.1093/bfgp/eln032. Advance Access published online on June 25, 2008. [DOI] [PubMed] [Google Scholar]
- (16).Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics. 2007;6:2212–2219. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Fox SI. Human Physiology. 9 ed McGraw-Hill Science/Engineering/Math; 2006. Reproduction. [Google Scholar]
- (18).Challis JRG, Matthews SG, Gibb W, Lye SJ. Endocrine and paracrine regulation of birth at term and preterm. Endocr. Rev. 2000;21(5):514–550. doi: 10.1210/edrv.21.5.0407. [DOI] [PubMed] [Google Scholar]
- (19).Slattery MM, Morrison JJ. Preterm delivery. Lancet. 2002;360(9344):1489–1497. doi: 10.1016/S0140-6736(02)11476-0. [DOI] [PubMed] [Google Scholar]
- (20).Lockwood CJ. The initiation of parturition at term. Obstet. Gynecol. Clin. North. Am. 2004;31(4):935–947. doi: 10.1016/j.ogc.2004.08.012. [DOI] [PubMed] [Google Scholar]
- (21).Hassan SS, Romero R, Tarca AL, Draghici S, Pineles B, Bugrim A, Khalek N, Camacho N, Mittal P, Yoon BH, Espinoza J, Kim CJ, Sorokin Y, Malone J., Jr. Signature pathways identified from gene expression profiles in the human uterine cervix before and after spontaneous term parturition. Am. J. Obstet. Gynecol. 2007;197(3):250.e1–250.e7. doi: 10.1016/j.ajog.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Dasari S, Pereira L, Reddy AP, Michaels JE, Lu X, Jacob T, Thomas A, Rodland M, Roberts CT, Jr., Gravett MG, Nagalla SR. Comprehensive proteomic analysis of human cervical-vaginal fluid. J. Proteome. Res. 2007;6(4):1258–1268. doi: 10.1021/pr0605419. [DOI] [PubMed] [Google Scholar]
- (23).Fichorova RN, Rheinwald JG, Anderson DJ. Generation of papillomavirus-immortalized cell lines from normal human ectocervical, endocervical, and vaginal epithelium that maintain expression of tissue-specific differentiation proteins. Biol. Reprod. 1997;57(4):847–855. doi: 10.1095/biolreprod57.4.847. [DOI] [PubMed] [Google Scholar]
- (24).Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- (25).Von Haller PD, Yi E, Donohoe S, Vaughn K, Keller A, Nesvizhskii AI, Eng J, Li XJ, Goodlett DR, Aebersold R, Watts JD. The application of new software tools to quantitative protein profiling via isotope-coded affinity tag (ICAT) and tandem mass spectrometry: I. Statistically annotated datasets for peptide sequences and proteins identified via the application of ICAT and tandem mass spectrometry to proteins copurifying with T cell lipid rafts. Mol. Cell. Proteomics. 2003;2(7):426–427. doi: 10.1074/mcp.D300002-MCP200. [DOI] [PubMed] [Google Scholar]
- (26).Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- (27).Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic. Acids. Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).UniProt Consortium The universal protein resource (UniProt) Nucleic. Acids Res. 2008;36(Database issue):D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Shaw JL, Smith CR, Diamandis EP. Proteomic analysis of human cervico-vaginal fluid. J. Proteome. Res. 2007;6(7):2859–2865. doi: 10.1021/pr0701658. [DOI] [PubMed] [Google Scholar]
- (30).Michaels JE, Dasari S, Pereira L, Reddy AP, Lapidus JA, Lu X, Jacob T, Thomas A, Rodland M, Roberts CT, Jr., Gravett MG, Nagalla SR. Comprehensive proteomic analysis of the human amniotic fluid proteome: gestational age-dependent changes. J. Proteome. Res. 2007;6(4):1277–1285. doi: 10.1021/pr060543t. [DOI] [PubMed] [Google Scholar]
- (31).Vazquez-Ortiz G, Pina-Sanchez P, Vazquez K, Duenas A, Taja L, Mendoza P, Garcia JA, Salcedo M. Overexpression of cathepsin F, matrix metalloproteinases 11 and 12 in cervical cancer. BMC. Cancer. 2005;5(1):68–78. doi: 10.1186/1471-2407-5-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wyatt SM, Kraus FT, Roh CR, Elchalal U, Nelson DM, Sadovsky Y. The correlation between sampling site and gene expression in the term human placenta. Placenta. 2005;26(5):372–379. doi: 10.1016/j.placenta.2004.07.003. [DOI] [PubMed] [Google Scholar]
- (33).Liu H, Sadygov RG, Yates JR. A Model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76(14):4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- (34).Vogel I, Thorsen P, Curry A, Sandager P, Uldbjerg N. Biomarkers for the prediction of preterm delivery. Acta. Obstet. Gynecol. Scand. 2005;84(6):516–525. doi: 10.1111/j.0001-6349.2005.00771.x. [DOI] [PubMed] [Google Scholar]
- (35).Leitich H, Egarter C, Kaider A, Hohlagschwandtner M, Berghammer P, Husslein P. Cervicovaginal fetal fibronectin as a marker for preterm delivery: a meta-analysis. Am. J. Obstet. Gynecol. 1999;180(5):1169–1176. doi: 10.1016/s0002-9378(99)70612-5. [DOI] [PubMed] [Google Scholar]
- (36).Lockwood CJ, Senyei AE, Dische MR, Casal D, Shah KD, Thung SN, Jones L, Deligdisch L, Garite TJ. Fetal fibronectin in cervical and vaginal secretions as a predictor of preterm delivery. N. Engl. J. Med. 1991;325(10):669–674. doi: 10.1056/NEJM199109053251001. [DOI] [PubMed] [Google Scholar]
- (37).Holzman C, Jetton J, Siler-Khodr T, Fisher R, Rip T. Second trimester corticotropin-releasing hormone levels in relation to preterm delivery and ethnicity. Obstet. Gynecol. 2001;97(5):657–663. doi: 10.1016/s0029-7844(00)01209-6. [DOI] [PubMed] [Google Scholar]
- (38).Romero R, Avila C, Santhanam U, Sehgal PB. Amniotic fluid interleukin 6 in preterm labor. Association with infection. J. Clin. Invest. 1990;85(5):1392–1400. doi: 10.1172/JCI114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Jacobsson B, Mattsby-Baltzer I, Hagberg H. Interleukin-6 and interleukin-8 in cervical and amniotic fluid: relationship to microbial invasion of the chorioamniotic membranes. BJOG. 2005;112(6):719–724. doi: 10.1111/j.1471-0528.2005.00536.x. [DOI] [PubMed] [Google Scholar]
- (40).Ozer KT, Kavak ZN, Gökaslan H, Elter K, Pekin T. Predictive power of maternal serum and amniotic fluid CRP and PAPP-A concentrations at the time of genetic amniocentesis for the preterm delivery. Eur. J. Obstet. Gynecol. Reprod. Biol. 2005;122(2):187–190. doi: 10.1016/j.ejogrb.2005.02.021. [DOI] [PubMed] [Google Scholar]
- (41).Vogel I, Grove J, Thorsen P, Moestrup SK, Uldbjerg N, Møller HJ. Preterm delivery predicted by soluble CD163 and CRP in women with symptoms of preterm delivery. BJOG. 2005;112(6):737–742. doi: 10.1111/j.1471-0528.2005.00557.x. [DOI] [PubMed] [Google Scholar]
- (42).Rosen T, Kuczynski E, O’Neill LM, Funai EF, Lockwood CJ. Plasma levels of thrombin-antithrombin complexes predict preterm premature rupture of the fetal membranes. J. Matern. Fetal. Med. 2001;10(5):297–300. doi: 10.1080/714904361. [DOI] [PubMed] [Google Scholar]
- (43).Elovitz MA, Baron J, Phillippe M. The role of thrombin in preterm parturition. Am. J. Obstet. Gynecol. 2001;185(5):1059–1063. doi: 10.1067/mob.2001.117638. [DOI] [PubMed] [Google Scholar]
- (44).Macones GA, Parry S, Elkousy M, Clothier B, Ural SH, Strauss JF., 3rd A polymorphism in the promoter region of TNF and bacterial vaginosis: preliminary evidence of gene-environment interaction in the etiology of spontaneous preterm birth. Am. J. Obstet. Gynecol. 2004;190(6):1504–1508. doi: 10.1016/j.ajog.2004.01.001. [DOI] [PubMed] [Google Scholar]
- (45).Wang H, Parry S, Macones G, Sammel MD, Kuivaniemi H, Tromp G, Argyropoulos G, Halder I, Shriver MD, Romero R, Strauss JF., 3rd A functional SNP in the promoter of the SERPINH1 gene increases risk of preterm premature rupture of membranes in African Americans. Proc. Natl. Acad. Sci. U S A. 2006;103(36):13463–13467. doi: 10.1073/pnas.0603676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Wang H, Parry S, Macones G, Sammel MD, Ferrand PE, Kuivaniemi H, Tromp G, Halder I, Shriver MD, Romero R, Strauss JF., 3rd Functionally significant SNP MMP8 promoter haplotypes and preterm premature rupture of membranes (PPROM) Hum. Mol. Genet. 2004;13(21):2659–2669. doi: 10.1093/hmg/ddh287. [DOI] [PubMed] [Google Scholar]
- (47).Yoon BH, Oh SY, Romero R, Shim SS, Han SY, Park JS, Jun JK. An elevated amniotic fluid matrix metalloproteinase-8 level at the time of mid-trimester genetic amniocentesis is a risk factor for spontaneous preterm delivery. Am. J. Obstet. Gynecol. 2001;185(5):1162–1167. doi: 10.1067/mob.2001.117678. [DOI] [PubMed] [Google Scholar]
- (48).Tamura T, Goldenberg RL, Johnston KE, Cliver SP, Hickey CA. Serum ferritin: a predictor of early spontaneous preterm delivery. Obstet. Gynecol. 1996;87(3):360–365. doi: 10.1016/0029-7844(95)00437-8. [DOI] [PubMed] [Google Scholar]
- (49).Meyer RE, Thompson SJ, Addy CL, Garrison CZ, Best RG. Maternal serum placental alkaline phosphatase level and risk for preterm delivery. Am. J. Obstet. Gynecol. 1995;173(1):181–186. doi: 10.1016/0002-9378(95)90187-6. [DOI] [PubMed] [Google Scholar]
- (50).Vogel I, Thorsen P, Hundborg HH, Uldbjerg N. Prediction of preterm delivery using changes in serum relaxin in low risk pregnancies. Eur. J. Obstet. Gynecol. Reprod. Biol. 2006;128(12):113–118. doi: 10.1016/j.ejogrb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- (51).Green KJ, Parry DA, Steinert PM, Virata ML, Wagner RM, Angst BD, Nilles LA. Structure of the human desmoplakins. Implications for function in the desmosomal plaque. J. Biol. Chem. 1990;265(19):11406–11407. [PubMed] [Google Scholar]
- (52).Ockleford C, Malak T, Hubbard A, Bracken K, Burton SA, Bright N, Blakey G, Goodliffe J, Garrod D, d’Lacey C. Confocal and conventional immunofluorescence and ultrastructural localisation of intracellular strength-giving components of human amniochorion. J. Anat. 1993;183(Pt 3):483–505. [PMC free article] [PubMed] [Google Scholar]
- (53).van Heusden GP. 14-3-3 proteins: regulators of numerous eukaryotic proteins. IUBMB. Life. 2005;57(9):623–629. doi: 10.1080/15216540500252666. [DOI] [PubMed] [Google Scholar]
- (54).Kazerounian S, Yee KO, Lawler J. Thrombospondins in cancer. Cell. Mol. Life. Sci. 2008;65(5):700–712. doi: 10.1007/s00018-007-7486-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Haddad R, Romero R, Gould BR, Tromp G, Gotsch F, Edwin SS, Zingg HH. Angiogenesis gene expression in mouse uterus during the common pathway of parturition. Am. J. Obstet. Gynecol. 2008;198(5):539, e1–8. doi: 10.1016/j.ajog.2007.11.021. [DOI] [PubMed] [Google Scholar]
- (56).Morimoto T, Head JR, MacDonald PC, Casey ML. Thrombospondin-1 expression in human myometrium before and during pregnancy, before and during labor, and in human myometrial cells in culture. Biol. Reprod. 1998;59(4):862–870. doi: 10.1095/biolreprod59.4.862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.