Infection following total joint arthroplasty is difficult to diagnose and treat; a nascent body of evidence from studies of prosthetic joint infections suggests that biofilm bacteria are the underlying cause1-3. We describe the case of a patient who had chronic recurring symptoms of infection that persisted for years following total elbow arthroplasty despite numerous medical and surgical interventions. Confocal microscopy performed on fluid, tissue, and cement at the final surgical revision demonstrated viable bacteria in biofilm aggregates. Reverse transcriptase-polymerase chain reaction analysis confirmed the presence of metabolically active Staphylococcus aureus. These observations comprise compelling evidence that viable biofilm bacteria play an important role in refractory infection following joint arthroplasty.
Chronic infection following joint replacement is increasingly thought to result from the presence of bacterial biofilm communities attached to the implant1-3. Biofilm bacteria are vastly more resistant to conventional antibiotic therapy than are their single planktonic counterparts (unattached solitary bacteria living freely) and are typically difficult to culture by conventional microbiological methods4,5. The biofilm paradigm can explain contradictory signs and symptoms that suggest infection but are often associated with negative cultures6,7. Moreover, biofilm infections are difficult to detect by simple Gram stain and culture techniques but can persist as a nidus of infection from which recurrent acute exacerbations may arise through episodic planktonic “showering”8.
Interestingly, although many articles discuss biofilms as being an important factor in infections affecting orthopaedic implants9,10, the evidence remains largely anecdotal, deriving from paradoxical clinical signs and symptoms, from the detection of bacterial aggregates in sonicate from ex vivo implants11,12, or from in vitro experiments with use of medical materials13. Only a few studies have provided direct evidence of attached biofilms associated with ex vivo implants, and those studies have all dealt with hip prostheses3,14,15.
We have previously used a combination of confocal microscopy, fluorescent in situ hybridization, and polymerase chain reaction to provide convincing evidence for a biofilm etiology in chronic otitis media4. In the present study, we applied similar techniques to directly demonstrate viable Staphylococcus aureus biofilms on operative specimens in a case of total elbow arthroplasty wherein a methicillin-resistant Staphylococcus aureus infection had eluded detection despite ten separate attempts at culture, including intraoperative cultures on four different occasions. Ultimately, operative specimens that were examined with use of confocal microscopy and reverse transcriptase-polymerase chain reaction-based analysis demonstrated the presence of viable Staphylococcus aureus biofilm bacteria associated with the retained cement in the humerus. The patient was informed that data concerning the case would be submitted for publication, and he consented. This study received institutional review board approval.
Case Report
Clinical History
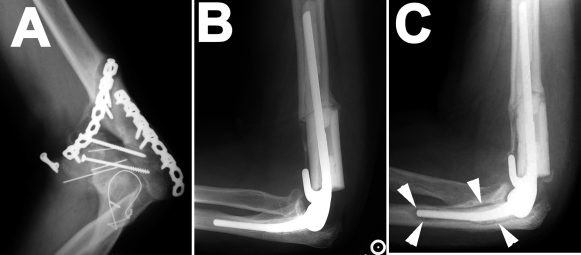
A forty-seven-year-old man presented with left elbow pain five years after undergoing open reduction and internal fixation of a comminuted left distal humeral fracture. The patient was a laborer and had continued working with the injured arm despite discomfort. Radiographs demonstrated nonunion of the distal part of the humerus along with hardware failure (Fig. 1, A). The hardware was surgically removed; an intraoperative Gram stain was negative, with fewer than five white blood cells per high-powered field. Given the extent of the tissue inflammation that was observed, it was believed that the safest course would be to remove all hardware, reactive tissue, and devascularized bone. The void left by removal of the bone was filled with a cement spacer impregnated with vancomycin (3.5 g). Final bacterial cultures showed no growth.
Fig. 1.
Radiographs documenting the total elbow arthroplasty before and after revision. A: Radiograph demonstrating nonunion of the distal part of the humerus and loose hardware. There is no evidence of periosteal reaction, which would suggest osteomyelitis. B: Lateral radiograph of elbow following total elbow arthroplasty with placement of an intercalary tibial allograft. C: Lateral radiograph of elbow thirteen months after the total elbow arthroplasty. An area of lucency (arrowheads) surrounds the ulnar component.
One month later, the patient underwent a total elbow arthroplasty with allograft (Fig. 1, B). The immediate postoperative course was uneventful. Eight months later, the patient complained of recurrent left elbow pain. Routine aerobic and anaerobic cultures of an aspirate from the elbow demonstrated no bacterial growth. Due to persistent symptoms, the patient underwent irrigation and débridement and the radial head was resected. Aerobic and anaerobic bacterial cultures again were negative both for the aspirate and tissue samples (no growth).
Thirteen months after the total elbow arthroplasty, the elbow pain recurred. Radiographs showed evidence of loosening about the ulnar component. The humeral component appeared to be well-fixed (Fig. 1, C). An irrigation and débridement was again performed. The implants and cement were removed; a cement spacer that was impregnated with tobramycin (prepared according to the manufacturer's directions) was placed. Final bacterial cultures of samples taken from within the humeral shaft, from the medullary canal of the ulna, and from tissue about the elbow joint again showed no growth. The patient was taken back to the operating room three and one-half months later for removal of the cement spacer. Final bacterial cultures at this point were also negative (no growth).
Five months later, the patient returned with a one-week history of fever and malaise. He had diffuse pain and swelling in the region of the midbrachium. Cloudy fluid was draining from a small sinus track on the anterolateral aspect of the arm. The patient was returned to the operating room for débridement of the wound. Retained cement was removed from the humerus. Intraoperative cultures from aspirated joint fluid at this time did grow methicillin-resistant Staphylococcus aureus. This isolate was found to be susceptible to vancomycin, gentamicin, clindamycin, and sulfamethoxazole-trimethoprim and resistant to nafcillin and erythromycin. Fungal cultures and mycobacterial cultures were all negative (no growth). An eight-week course of intravenous vancomycin and oral rifampin was completed (the patient had previously received multiple other courses of antibiotics empirically as well, usually consisting of intravenous cefazolin followed by oral cephalexin, although ceftriaxone and gatifloxacin had also been briefly administered).
Two years later (at the time of the most recent follow-up), the patient had no evidence of infection. He has a flail elbow resulting in an arm that is only useful when the elbow is supported in a hinged brace.
Confocal Laser Scanning Microscopy and Viability Staining
Tissue and cement recovered during the final revision surgery were collected in sterile tubes and placed on ice. Prior to staining, samples were rinsed by immersion in Ringer solution (one-quarter strength), blotted to remove excess fluid, and then transferred to a 100-mm Petri dish. A dab of Lubriseal grease (Thomas Scientific, Swedesboro, New Jersey) was smeared on the bottom of each Petri dish; the specimen was gently pressed onto the grease, avoiding contact with the center, to immobilize the specimen for microscopic examination. Tissue and cement samples were stained with use of the LIVE/DEAD BacLight kit (Molecular Probes, Eugene, Oregon) by drop-pipetting the manufacturer's recommended concentration directly onto the specimens. Specimens were incubated for fifteen minutes in the dark at room temperature. Excess stain was rinsed away by flooding the plate with phosphate-buffered saline solution and then aspirating. The specimens were submerged in phosphate-buffered saline solution before they were histologically examined with use of a Leica DM RXE upright microscope attached to a TCS SP2 AOBS confocal system (Leica Microsystem, Exton, Pennsylvania). The 488-nm line of the Kr/AG-laser was used as the excitation wavelength and the detector wavelength windows were set such that the “live” stain (SYTO 9; Molecular Probes) appeared green and the “dead” stain (propidium iodide) appeared red. The surface of the bone cement was imaged in reflected confocal mode and made to appear blue in the images. Thus, fresh specimens were examined in their fully hydrated state with minimal preparation.
Nucleic Acid Isolation and Reverse Transcriptase-Polymerase Chain Reaction
Nucleic acid isolations from bacterial reference strains (Staphylococcus aureus American Type Culture Collection [ATCC] 25923 and Staphylococcus epidermidis ATCC 35984) were performed as described7. For the clinical specimens, 200 μL of aspirate fluid obtained from the operative site prior to open surgery was placed in 1 mL of RNAlater (Ambion, Austin, Texas) and stored at −70°C. Cells were pelleted, and 480 μL of hot phenol buffer7 was added. This resuspension then was subjected to phenol-chloroform extraction as previously described7. Recovered nucleic acids, both wound-derived and from the reference bacterial strains noted above, were treated with deoxyribonuclease (DNase) (as per the specifications of the TURBO DNase kit; Ambion) and evaluated for integrity with use of an Agilent bioanalyzer (Model 2100; Agilent, Palo Alto, California), which confirmed little to no degradation. Reverse transcription and subsequent polymerase chain reaction on the recovered ribonucleic acids were then carried out as described7. A set of negative controls to test for contaminating DNA (sterile water was added in place of reverse transcriptase) was also carried out simultaneously. Following reverse transcriptase-polymerase chain reaction, the amplimers were subjected to 1% agarose-gel electrophoresis and visualized with ethidium bromide. The specific primer sequences for Staphylococcus aureus and Staphylococcus epidermidis have been previously described7. In addition, we used a generic staphylococcal primer set (GF-1/GR-2) directed against the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene16 and a 23S rRNA Staphylococcus aureus-specific primer set (532720-GGACGACATTAGACGAATCA-532739 and 534038-CGGGCACCTATTTTCTATCT-534019), which were developed and validated in the present study under the same polymerase-chain-reaction-cycling conditions as previously described7.
Results
Confocal Microscopy
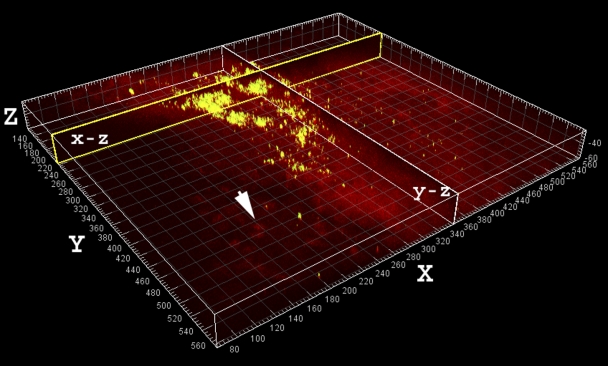
At the time of the final operation, confocal laser scanning microscopy of fluid aspirated from the site to be surgically resected revealed the presence of cocci ranging from single cells to large aggregates of grape-like clusters (Fig. 2). The largest clumps were approximately 100 μm in diameter and were estimated to contain many hundreds to thousands of cocci. The bacteria in these clusters stained with SYTO 9 (green) but not with propidium iodide (red), signifying that they were viable.
Fig. 2.
Large aggregate of viable (greenish-yellow) cocci in the aspirate stained with use of Molecular Probes LIVE/DEAD viability kit. The largest detected clumps were up to 100 μm in diameter and had a heterogeneous morphology consistent with that of “in vitro” grown Staphylococcus aureus biofilms. The aggregates may represent clumps of bacteria that had shed naturally from the biofilm, possibly contributing to systemic symptoms (e.g., fever). Sagittal sections through the clumps along the x and y axes are shown in the vertical planes, labeled “x-z” and “y-z.” The human nuclei (arrow) and associated tissue stained red. Scale: major divisions = 10 μm.
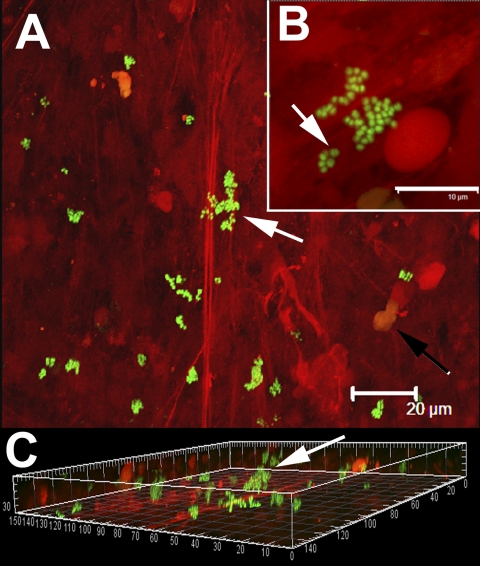
Examination of tissue associated with the implant revealed similar clusters of viable bacteria (stained green) that were adherent to the tissue (Fig. 3). Host nuclei and fibrous material stained red with propidium iodide. Bacterial clusters protruded from the surface of the host tissue and were similar in appearance to biofilm clusters grown in vitro8,17. Some of the cocci were in the process of division, which was consistent with the results of viability staining. The distribution of biofilm was patchy, with some sites containing large clusters and other regions showing no evidence of infection.
Fig. 3.
Single cells and clumps of viable cocci (stained green with use of Molecular Probes LIVE/DEAD viability kit) attached to tissue displaying characteristic staphylococcal morphology. Human tissue stained red with propidium iodide. A: Low-power image showing bacterial cells (arrow) attached to fibrous material (red striation). The nuclei of human cells were also visible (black arrow). Scale bar = 20 μm. B: Higher-power magnification of a group of cocci attached to fibrous material. Some cells were in the process of division (arrow), indicating that they were viable, which was consistent with the results of viability staining and culturing. Scale bar = 10 μm. C: Three-dimensional orthogonal projection of panel “A” showing that the biofilm clumps (arrow) were attached and protruding from the fibrous material. Scale: major divisions = 10 μm.
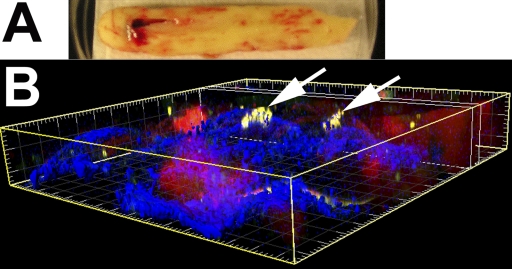
Examination of the residual bone cement revealed both single bacterial coccal cells and clusters of biofilm bacteria attached to the irregular and porous surface (Fig. 4). The clusters were as much as approximately 10 μm in diameter and contained between ten and fifty cells. The distribution of biofilm was patchy, similar to that seen in the tissue samples. Consistent with what was seen in the tissue and aspirate samples, the cocci stained green, indicating viability. The clumps had three-dimensional structure, evidencing an extracellular matrix, and protruded from the surface of the cement. Host cells, visualized by red-stained nuclei, were also seen in close proximity to the cement surface.
Fig. 4.
Viable Staphylococcus aureus biofilm cocci attached to a piece of bone cement that was removed during the surgical revision. A: Macroscopic view of the cement immersed in buffer in a 10-cm-diameter Petri plate. The specimen was oriented for subsequent confocal microscopic observation with use of a water-immersion objective. B: Microscopic three-dimensional orthogonal view showing the clumps of biofilm (greenish-yellow and indicated by arrows) attached to the surface of the bone cement (blue), as constructed from a confocal microscopy stack. Scale: major divisions = 10 μm.
Reverse Transcriptase-Polymerase Chain Reaction and Polymerase Chain Reaction
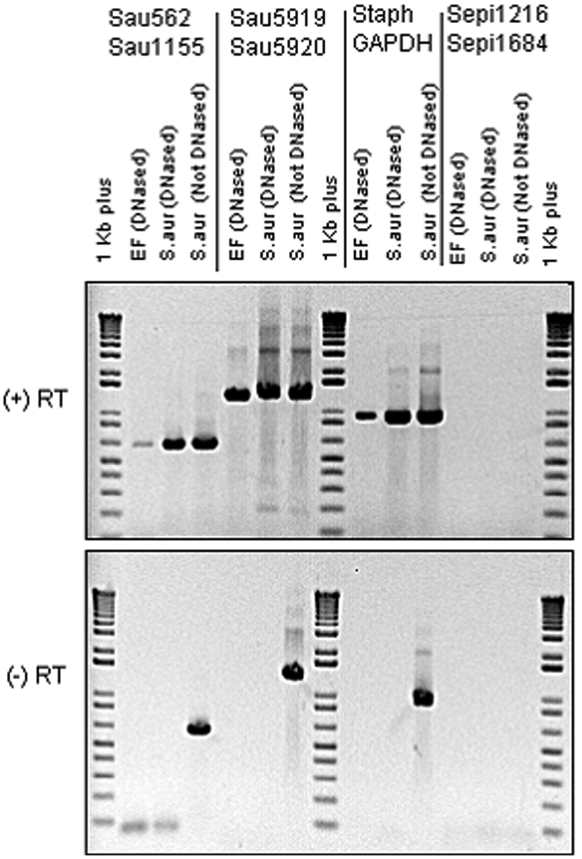
The polymerase chain reaction (reverse transcriptase-polymerase chain reaction) assay for Staphylococcus aureus and Staphylococcus epidermidis in tissue from the elbow showed polymerase-chain-reaction amplimers of the expected molecular weight when a Staphylococcus aureus primer or nonspecific Staphylococcus primers were used on wound material from the patient (as well as on a reference Staphylococcus aureus control strain) (Fig. 5). In contrast, no amplimer was seen when Staphylococcus epidermidis-specific primers were used, signifying that no Staphylococcus epidermidis was present. When no reverse transcriptase enzyme was added, the only reactions that produced amplimer were the non-DNase-treated control reactions. The absence of amplimers from the DNase-treated clinical specimens when reverse transcriptase was omitted, together with the positive reverse transcriptase-polymerase chain reaction results from the DNase-treated clinical specimens, proved that bacterial messenger ribonucleic acid (mRNA) was present.
Fig. 5.
Agarose gel electrophoresis of amplimers following polymerase chain reaction and reverse transcriptase-polymerase chain reaction. The upper panel shows the results for reverse transcriptase-polymerase chain reaction; the lower panel, for polymerase chain reaction with no reverse transcriptase. The primer sets are noted at the top. The nucleic acid templates, and whether they have been treated with DNase, are noted above each sample lane. “EF” signifies the clinical sample obtained from elbow-fluid aspirate. In the reverse transcriptase (+RT) lanes, all three Staphylococcus aureus primer sets (first three columns) yielded amplimer in both the aspirate and positive controls. In the lanes with no reverse transcriptase (-RT), only the non-DNase-treated specimens yielded amplimer, as expected. No amplimer was obtained with the Staphylococcus epidermidis primers (last column) in either the clinical specimen or the Staphylococcus aureus negative control, demonstrating that the Staphylococcus epidermidis primers used were species-specific and that no Staphylococcus epidermidis was detected in our clinical sample. Lanes 1, 8, and 15 were “1 kilobase (kb) plus” molecular weight standards. RT = reverse transcriptase; Sau = Staphylococcus aureus primer set; Staph GAPDH = Staphylococcal primer set (GF-1/GR-2) directed against glyceraldehyde-3-phosphate dehydrogenase gene; and Sepi = Staphylococcus epidermidis primer set.
Discussion
We have identified viable staphylococcal biofilms with use of confocal laser scanning microscopy-based in situ visualization techniques in explanted tissues and cement from a patient who underwent the removal of total elbow arthroplasty material following a long and complicated clinical course. The micrographic findings were confirmed by both culture and reverse transcriptase-polymerase chain reaction-based diagnostics. To our knowledge, this is the first reported demonstration of a bacterial infection associated with an elbow arthroplasty that fulfills all of the established criteria for a biofilm infection4,6, including direct examination of an infected tissue, revealing pathogenic bacteria in clusters attached to a surface; infection localized to a particular anatomic site; and persistent infection despite exposure to antibiotic agents to which the causative organism was sensitive in planktonic form.
The patient's clinical course was marked by recurrent episodes of symptoms that were consistent with infection despite multiple courses of antibiotics and multiple operative interventions; however, bacterial cultures continued to be negative on testing throughout. This clinical progression comports entirely with the behavior expected when there is a chronic underlying biofilm infection that is resistant to standard antibiotic therapy and recalcitrant to standard culture and that periodically erupts in acute exacerbations. Interestingly, the bacteria were able to survive on the cement, even though the cement was impregnated with tobramycin. Antibiotic resistance in biofilm bacteria of up to 1000 times that of planktonic bacteria has been extensively documented in experiments in vitro5,18,19.
The demonstration of live cocci on both fibrous tissue in the wound and on the foreign body suggests that a well-developed biofilm can adhere both to the implant and to the soft-tissue sheathing that typically envelops it. In orthopaedic infections, biofilms are usually discussed in the context of growth directly on the foreign body1,20. Our data suggest that the tissue that envelops the foreign body may also act as a reservoir for pathogenic biofilms.
The present study also describes the use of reverse transcriptase-polymerase chain reaction to document the presence of viable Staphylococcus aureus bacteria (but not Staphylococcus epidermidis) in the clinical samples. The finding of bacterial mRNA is prima facie evidence that viable bacteria were present in the joint at the time of explant since bacterial mRNA has a half-life that is typically measured in seconds and/or minutes and would therefore not be detectable if the bacteria were not viable21. While we believe that the combination of confocal microscopy and reverse transcriptase-polymerase chain reaction is a powerful tool for the direct detection of biofilms, the approach is not without limitations. Confocal microscopy is a somewhat specialized technique and, although now commonly used in research, this equipment is not generally available in hospital diagnostic laboratories. Secondly, a thorough examination of specimens is often time-consuming due to the patchy nature and microscopic scale of biofilms. Even with a highly trained operator, examination of a single specimen might take hours. While some techniques for specimen handling, staining, and examination with use of confocal microscopy are directly transferable from conventional histologic techniques, to move this technique from a research tool to a routine clinical diagnostic modality will require the development of standardized methods and specialized training.
Finally, although our results were obtained with use of intraoperative samples, similar methodologies could be applied to joint-fluid aspirates obtained in the clinic, and these methodologies may, with further study and validation, provide a useful adjunct to the decision-making process of the clinician confronted with this difficult problem. 
Acknowledgments
Note: The authors thank Dr. Luanne Hall-Stoodley and Dr. J. Christopher Post for their assistance with this report.
Disclosure: In support of their research for or preparation of this work, one or more of the authors received, in any one year, outside funding or grants in excess of $10,000 from the NIH: NIDCD and NIDCR (K08 DE014780, SK), Allegheny-Singer Research Institute, HRSA C76HF06163 (GDE), DC 04173 (GDE), RO1 DC005659 (JCP). In addition, one or more of the authors or a member of his or her immediate family received, in any one year, payments or other benefits of less than $10,000 or a commitment or agreement to provide such benefits from commercial entities (Stryker and Medtronics). No commercial entity paid or directed, or agreed to pay or direct, any benefits to any research fund, foundation, division, center, clinical practice, or other charitable or nonprofit organization with which the authors, or a member of their immediate families, are affiliated or associated.
Investigation performed at the Center for Genomic Sciences, Allegheny-Singer Research Institute, Allegheny General Hospital, Pittsburgh, Pennsylvania
References
- 1.Costerton JW. Biofilm theory can guide the treatment of device-related orthopaedic infections. Clin Orthop Relat Res. 2005;437:7-11. [DOI] [PubMed] [Google Scholar]
- 2.Ehrlich GD, Hu FZ, Shen K, Stoodley P, Post JC. Bacterial plurality as a general mechanism driving persistence in chronic infections. Clin Orthop Relat Res. 2005;437:20-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gristina AG, Costerton JW. Bacterial adherence to biomaterials and tissue. The significance of its role in clinical sepsis. J Bone Joint Surg Am. 1985;67:264-73. [PubMed] [Google Scholar]
- 4.Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, Forbes M, Greenberg DP, Dice B, Burrows A, Wackym PA, Stoodley P, Post JC, Ehrlich GD, Kerschner JE. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA. 2006;296:202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135-8. [DOI] [PubMed] [Google Scholar]
- 6.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677-701. [DOI] [PubMed] [Google Scholar]
- 7.Stoodley P, Kathju S, Hu FZ, Erdos G, Levenson JE, Mehta N, Dice B, Johnson S, Hall-Stoodley L, Nistico L, Sotereanos N, Sewecke J, Post JC, Ehrlich GD. Molecular and imaging techniques for bacterial biofilms in joint arthroplasty infections. Clin Orthop Relat Res. 2005;437:31-40. [DOI] [PubMed] [Google Scholar]
- 8.Fux CA, Wilson S, Stoodley P. Detachment characteristics and oxacillin resistance of Staphylococcus aureus biofilm emboli in an in vitro catheter infection model. J Bacteriol. 2004;186:4486-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mack D, Rohde H, Harris LG, Davies AP, Horstkotte MA, Knobloch JK. Biofilm formation in medical device-related infection. Int J Artif Organs. 2006;29:343-59. [DOI] [PubMed] [Google Scholar]
- 10.Zimmerli W, Trampuz A, Ochsner PE. Prosthetic-joint infections. N Engl J Med. 2004;351:1645-54. [DOI] [PubMed] [Google Scholar]
- 11.Tunney MM, Patrick S, Curran MD, Ramage G, Hanna D, Nixon JR, Gorman SP, Davis RI, Anderson N. Detection of prosthetic hip infection at revision arthroplasty by immunofluorescence microscopy and PCR amplification of the bacterial 16S rRNA gene. J Clin Microbiol. 1999;37:3281-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trampuz A, Piper KE, Hanssen AD, Osman DR, Cockerill FR, Steckelberg JM, Patel R. Sonication of explanted prosthetic components in bags for diagnosis of prosthetic joint infection is associated with risk of contamination. J Clin Microbiol. 2006;44:628-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neut D, Hendriks JG, van Horn JR, van der Mei HC, Busscher HJ. Pseudomonas aeruginosa biofilm formation and slime excretion on antibiotic-loaded bone cement. Acta Orthop. 2005;76:109-14. [DOI] [PubMed] [Google Scholar]
- 14.Neut D, van Horn JR, van Kooten TG, van der Mei HC, Busscher HJ. Detection of biomaterial-associated infections in orthopaedic joint implants. Clin Orthop Relat Res. 2003;413:261-8. [DOI] [PubMed] [Google Scholar]
- 15.Tunney MM, Patrick S, Gorman SP, Nixon JR, Anderson N, Davis RI, Hanna D, Ramage G. Improved detection of infection in hip replacements. A currently underestimated problem. J Bone Joint Surg Br. 1998;80:568-72. [DOI] [PubMed] [Google Scholar]
- 16.Yugueros J, Temprano A, Sánchez M, Luengo JM, Naharro G. Identification of Staphylococcus spp. by PCR-restriction fragment length polymorphism of gap gene. J Clin Microbiol. 2001;39:3693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rupp CJ, Fux CA, Stoodley P. Viscoelasticity of Staphylococcus aureus biofilms in response to fluid shear allows resistance to detachment and facilitates rolling migration. Appl Environ Microbiol. 2005;71:2175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones SM, Morgan M, Humphrey TJ, Lappin-Scott H. Effect of vancomycin and rifampicin on methicillin-resistant Staphylococcus aureus biofilms. Lancet. 2001;357:40-1. [DOI] [PubMed] [Google Scholar]
- 19.Saginur R, Stdenis M, Ferris W, Aaron SD, Chan F, Lee C, Ramotar K. Multiple combination bactericidal testing of staphylococcal biofilms from implant-associated infections. Antimicrob Agents Chemother. 2006;50:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehrlich GD, Hu FZ, Post JC. Role for biofilms in infectious disease. In: Ghannoum M, O'Toole GA, editors. Microbial biofilms. Washington, DC: ASM Press; 2004. p 332-58.
- 21.Post JC, Aul JJ, White GJ, Wadowsky RM, Zavoral T, Tabari R, Kerber B, Doyle WJ, Ehrlich GD. PCR-based detection of bacterial DNA after antimicrobial treatment is indicative of persistent, viable bacteria in the chinchilla model of otitis media. Am J Otolaryngol. 1996;17:106-11. [DOI] [PubMed] [Google Scholar]