Abstract
Parathyroid hormone (PTH) regulates bone remodeling and calcium and phosphate homeostasis. PTH actions are mediated by type I PTH/PTH-related peptide receptor (PTH1R). There has been no commercially available, specific antibody to detect human PTH1R expression so far. Flag-tagged human PTH1R construct, converting the sequence DKEAPTGS (residues 94–101) in the exon E2 region of PTH1R to DYKDDDDK of Flag epitope, was generated by using PCR overlap extension or ligase enzyme for two-fragment assembly. We found that Flag-tagged PTH1R assembled by ligase was easy to be manipulated, but its efficiency was lower than that of PCR overlap extension. The PTH1R plasmids generated by both techniques were expressed successfully in vitro and in vivo and possessed the same physiological function as wild-type PTH1R. The Flag-tagged PTH1R construct will provide invaluable tools for study of PTH1R signaling and trafficking.
Keywords: polymerase chain reaction, protein expression, ligation, type I PTH/PTH-related peptide receptor
INTRODUCTION
Type I parathyroid hormone (PTH)/PTH-related peptide receptor (PTH1R) belongs to class B of G protein-coupled receptors.1 PTH1R is present primarily in the bone and kidney. Interaction with its cognate ligands leads to activation of Gs and Gq, with consequent stimulation of adenylate cyclase and phospholipase C. A cascade of cell-specific events ensues that regulate bone remodeling and extracellular calcium and phosphate homeostasis.2
PTH1R activation, desensitization, internalization, and recycling proceed in a cyclical manner. PTH1R internalization and recycling are studied conventionally by 125I-labeled PTH binding to the receptor.3 The receptor trafficking can be studied by ELISA or flow cytometry using epitope antibodies.4,5 An epitope-tagged receptor can also be used for the study of protein-protein interaction and receptor dimerization.6 Generation of epitope-tagged PTH1R will play an important role for the study of PTH1R signaling and trafficking. Deletion of residues 61–105, which are encoded by exon E2 in the PTH1R gene, did not affect the receptor binding and its function.7 To our knowledge, there is no Flag-tagged PTH1R construct, where Flag epitope locates in the exon E2 region.8 Here, we generated Flag-tagged PTH1R by incorporating Flag epitope (DYKDDDDK) into the exon E2 region. Two-fragment ligation to assemble Flag-tagged PTH1R was compared by using PCR overlap extension or ligase. The constructs of Flag-tagged PTH1R assembled by two different approaches were expressed successfully in vitro and in vivo. There was no difference for PTH-stimulated adenylyl cyclase and the activation of ERK1/2 in human embryonic kidney (HEK)293 cells transfected with wild-type PTH1R or Flag-tagged PTH1R assembled by two different approaches.
MATERIALS AND METHODS
Materials
T4 ligase, EcoRI, HindIII, and Kpn1 were purchased from New England Biolabs, Inc. (Beverly, MA). The transcription and translation (TNT) reticulocyte lysate kit was provided from Promega (Madison, WI). pcDNA3.1(+) vector was purchased from Invitrogen (Carlsbad, CA). Anti-p44/p42 MAPK (ERK1/2) and phospho-p44/42 MAPK (pERK1/2; Thr202/Tyr204) polyclonal antibodies were purchased from Cell Signaling Technology (Beverly, MA). HRP-conjugated sheep anti-mouse antibody was from Amersham Biosciences (Piscataway, NJ). FuGENE6 was purchased from Roche Applied Science (Indianapolis, IN). Protease inhibitor cocktail set I was from EMD Biosciences (San Diego, CA). Shrimp alkaline phosphatase was purchased from Roche Applied Science (Germany). Human [Nor8,18,Tyr34] PTH (1–34) was provided from Bachem (Torrance, CA). The QIAprep miniprep kit and QIAquick gel extraction kit were purchased from Qiagen Sciences (Germantown, MD). All other reagents were from Sigma-Aldrich (St. Louis, MO).
PCR Condition
After an initial denaturation of 2 min at 95°C, the amplifications were carried out for 30 cycles using PFU turbo polymerase (Stratagene, La Jolla, CA) as follows: denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 2.5 min. The final step was followed by an additional extension at 72°C for 10 min.
In Vitro Translation
In vitro translation reactions were performed according to the manufacturer's instructions. Briefly, the samples of the translation mixture containing 25 μl rabbit reticulocyte lysate, 1 μl T7 RNA polymerase, 1 μl 1 mM amino acids mixture (minus methionine), 2 μl [35S]methionine (10.5 mCi/ml, MP Biomedicals, Inc., Invine, CA), and 2 μl 1 μg plasmid DNA were incubated for 90 min at 30°C. Following incubation, 10 μl each translation reaction was mixed with 10 μl 2× SDS-loading buffer, and the samples were subjected to electrophoresis in a 10% SDS-polyacrylamide gel. The gels were dried, exposed on a PhosphorImager cassette, and scanned with a Storm 860 system.9
Cell Culture, Transfection, and Immunoblotting
HEK293 cells were cultured in DMEM medium with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified atmosphere consisting of 5% CO2 at 37°C. Transfection of HEK293 cells was performed as described previously.2 Briefly, the cells were transiently transfected with 1.0 μg DNA/well in six-well plates or 0.25 μg DNA/well in 24-well plates with empty vector [pcDNA3.1(+)], plasmids of wild-type PTH1R, Flag-tagged PTH1R, or Flag-tagged β-arrestin 1 by using FuGENE 6. After 48 h transfection, the cells were lysed with Nonidet P-40 (NP-40) lysis buffer (50 mM Tris, 150 mM NaCI, 5 mM EDTA, 0.5% NP-40), supplemented with protease inhibitor cocktail I, and incubated for 15 min on ice. Total lysates were analyzed by SDS-PAGE gels and transferred to Immobilon-P membranes (Millipore, Bedford, MA) using the semi-dry method (Bio-Rad, Hercules, CA). Membranes were blocked overnight at 4°C with 5% nonfat dried milk in Tris-buffered saline plus Tween-20 and incubated with different antibodies (polyclonal anti-ERK1/2 antibody at 1:1000, anti-pERK1/2 antibody at 1:500, or anti-Flag mAb at 1:500) for 2 h at room temperature. The membranes were then washed and incubated with 1:5000 dilution of goat anti-rabbit IgG or sheep anti-mouse IgG conjugated to HRP for 1 h at room temperature. Protein bands were visualized with a luminol-based ECL substrate.
Adenylyl Cyclase Activity
Adenylyl cyclase activity was determined by assay of cAMP accumulation as described previously.3 Briefly, HEK293 cells in 24-well plates were labeled with 0.5 μCi [3H]-adenine for 2 h. The cells were then treated with vehicle or 100 nM PTH (1–34) in the presence of phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine (1 mM) for 15 min. The reaction was terminated by addition of 1 M TCA. cAMP was isolated by the two-column method.
RESULTS AND DISCUSSION
PCR to Amplify Three Different Sizes of Fragments
Lee et al.7 reported that deletion of the internal sequence (residues 61–105) of PTH1R did not affect receptor binding and function. There are several different strategies to generate a Flag epitope in the internal position of PTH1R. Point mutation is impractical for the mutation of 8 aa needed for the Flag epitope. Two-fragment fusion by ligase requires the same restriction site in both fragments.10,11 PCR overlap extension may also be used to assemble two fragments.12,13 Comparison of the two approaches for generating constructs from two fragments has not been reported. We substituted the Flag epitope at aa position 94–101 of wild-type of PTH1R (Fig. 1). There is a unique KpnI restriction enzyme site within position 102–104, so we do not need to introduce a new restriction site in each fragment. Four oligonucleotide primers were designed (Table 1). We used wide-type pcDNA1-human PTH1R without epitope (provided by Dr. Thomas J. Gardella, Harvard Medical School, Boston, MA) as the template to amplify three fragments by PCR (Fig. 2A). The first fragment (F1) of 0.3 kb size, starting from the beginning of PTH1R to aa position 106 and containing Flag epitope and KpnI restriction site, was generated using primers of F1F and F1R. The second fragment (F2) is a 1.5-kb PCR product from aa position 100 until the end of PTH1R, containing the last two amino acids of the Flag epitope and KpnI restriction site, generated using primers of F2F and F2R. The last fragment (F3) is 1.8 kb full-length wild-type PTH1R without epitope, generated using primers of F1F and F2R.
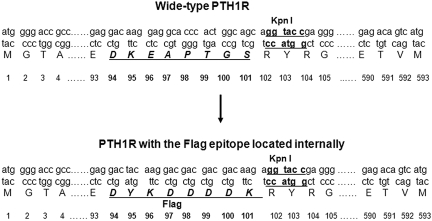
FIGURE 1.
Schematic diagram of the Flag epitope in the aa position of 94–101 of PTH1R. Wild-type human PTH1R has 593 aa. The sequence DKEAPTGS (residues 94–101) in the exon E2 region of PTH1R was substituted for DYKDDDDK of the Flag epitope.
Table 1.
Primers for Flag-tagged or wide-type PTH1R
| Name | Forward or Reverse | Sequence |
|---|---|---|
| F1F | Forward | 5′-gcg gcg aag ctt gcg atg ggg acc gcc cgg atc-3′ |
| F1R | Reverse | 5′-gcg ccc tcg gta cct ctt gtc gtc gtc gtc ctt-3′ |
| F2F | Forward | 5′-gac aag agg tac cga ggg cgc ccc tgt ctg ccg-3′ |
| F2R | Reverse | 5′-gcg gcg gaa ttc tca cat gac tgt ctc cca ctc-3′ |
The oligonucleotide primers containing HindIII, KpnI, and EcoRI restriction enzyme sites are underlined. The nucleotide sequence encoding the Flag epitope is shown in bold.
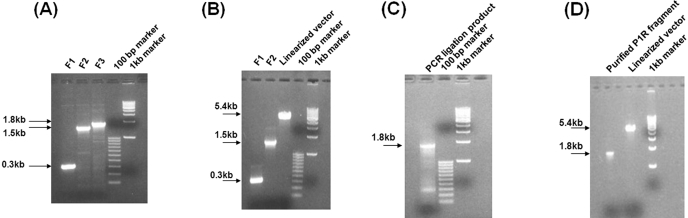
FIGURE 2.
Flag-tagged PTH1R was generated by PCR overlap extension or ligase for two-fragment assembly. (A) Three fragments of F1 (0.3 kb), F2 (1.5 kb), and F3 (1.8 kb) were amplified by PCR and run by agarose gel electrophoresis. (B) Purified products of F1, F2, and linearized vector (5.4 kb) were run by gel electrophoresis. (C) A second PCR step was performed using template of purified F1 and F2 and the primers of F1F and F2R. The PCR product was run by gel electrophoresis. (D) Purified, full-length Flag-PTH1R fragment (1.8 kb) by PCR overlap extension and linearized vector was run by gel electrophoresis
Two Fragments of F1 and F2 and Linearized Vector Were Assembled Sequentially by Ligase to Generate Flag-PTH1R Construct
Purified PCR products of F1 and F2 were digested with HindIII and KpnI or KpnI and EcoRI, respectively, for 1 h at 37°C. pcDNA3.1(+) vector was digested with EcoRI and HindIII under the same conditions. The digested products were purified and run by gel electrophoresis (Fig. 2B). The F1 and F2 were first fused by ligase at room temperature for 2 h. The linearized vector, which was treated by shrimp alkaline phosphatase, was then added to the mixture (molar ratio for F1, F2, and linearized vector is 5:5:1) and ligated at 16°C overnight. The ligation mixture was then transformed to competent cells (Invitrogen) and screened by ampicillin. There were 18 single colonies grown in a 10-cm agar plate. Six single colonies were picked. Plasmid DNA was prepared and digested with HindIII and EcoRI. All single colonies released 1.8 kb bands (data not shown), suggesting that the Flag-tagged PTH1R fragment was inserted into the pcDNA3.1(+) vector. The accuracy of the plasmid was confirmed further by sequencing (ABI PRISM 377, Applied Biosystems, Foster City, CA) and subsequent sequence alignment (National Center for Biotechnology Information BLAST) with human PTH1R (GenBank Accession No. L04308) to assure the fidelity of the construct.
Two Fragments of F1 and F2 Were Fused by PCR Overlap Extension
As PCR products of F1 and F2 had 18 bases overlapping, the second PCR amplification was done by using a template of purified F1 and F2 and the primers of F1F and F2R. The band of 1.8 kb full-length PTH1R and another unspecific band were found (Fig. 2C). The 1.8-kb fragment was prepared and digested with HindIII and EcoRI and then subcloned into the linearized vector pcDNA3.1(+) (Fig. 2D) with a molar ratio of 3:1. Sixty-eight single colonies were screened in a 10-cm agar plate, suggesting that the Flag-PTH1R fragment was easier to insert by PCR overlap extension than by ligase assembly. Six single colonies were picked up, and prepared plasmids were digested by HindIII and EcoRI. Bands (1.8 kb) were released from all six colonies (data not shown). The accuracy of the plasmids was confirmed by sequencing.
The fragment of F3 without epitope was subcloned into pcDNA3.1(+) to engineer wild-type PTH1R plasmid, which was used as a function control of Flag-tagged PTH1R.
Expression of PTH1R Constructs Was Detected In Vitro and In Vivo
As detection of protein expression in vitro does not require antibody, wild-type PTH1R without epitope (WT-P1R) and Flag-PTH1R assembled by ligase (Flag-P1R-L) or by PCR overlap extension (Flag-P1R-P) were first translated in vitro by using the TNT reticulocyte lysate kit. Flag-P1R-L, Flag-P1R-P, and WT-P1R expressed the same size protein (∼70 kDa) (Fig. 3, upper), suggesting that Flag-tagged PTH1R was translated successfully in vitro.
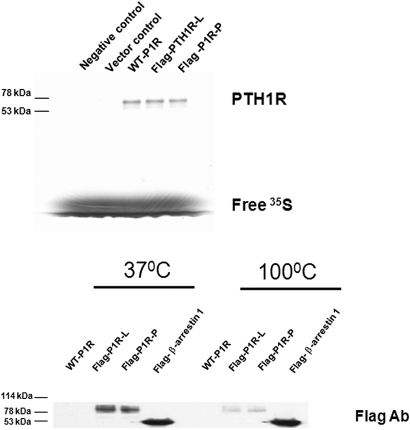
FIGURE 3.
Detection of the expression of WT-PTH1R and Flag-tagged PTH1R in vitro and in vivo. (Upper) The negative control, vector control, WT-P1R, and Flag-P1R-L (Flag-PTH1R-L) or Flag-P1R-P were translated in vitro by using the TNT reticulocyte lysate kit, as described in Materials and Methods. The samples were subjected to electrophoresis in a 10% SDS-polyacrylamide gel. (Lower) HEK293 cells were transiently transfected with WT-P1R, Flag-P1R-L, Flag-P1R-P, and Flag-β-arrestin 1. After 48 h transfection, the cell lysates were analyzed by SDS-PAGE. A representative immunoblot using Flag antibody is shown. Molecular mass standard sizes in kilodaltons (kDa) are shown on the left.
We next detected in vivo expression of Flag-tagged PTH1R constructs by using immunoblot analysis. Flag-β-arrestin 1 (positive control), WT-P1R, Flag-P1R-L, and Flag-P1R-P construct were transiently transfected to HEK293 cells. After 48 h transfection, the cell lysates were analyzed by SDS-PAGE gels. The results showed that an anti-Flag mAb (Sigma-Aldrich Catalog No. F1804) could detect Flag-β-arrestin 1 and Flag-P1R-L and Flag-P1R-P (about 90 kDa) (Fig. 3, lower). The size of Flag-P1R expressed in vivo is larger than that in vitro, because of receptor post-translation modification. We also found that the expression of Flag-P1R-L and Flag-P1R-P was decreased significantly after lysates were boiled for 3 min compared with the treatment of 37°C for 30 min, whereas Flag-β-arrestin 1 was not affected by the same condition. These findings suggest the stability of membrane receptor PTH1R is more sensitive to heat treatment than that of cytoplasmic protein β-arrestin 1. The mechanism of difference to heat treatment needs to be elucidated.
The Function of PTH1R Constructs Was Detected in HEK293 Cells
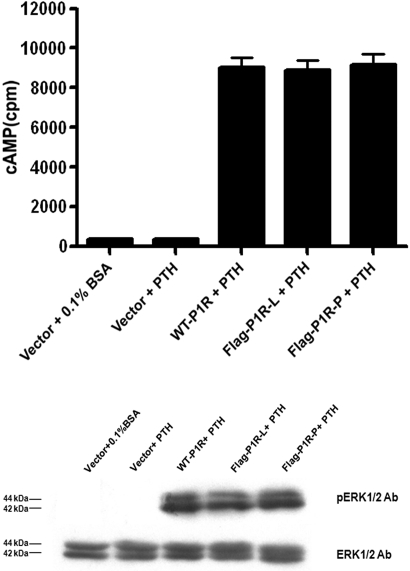
PTH1R interaction with its cognate ligand PTH leads to activation of G protein, which subsequently regulates intracellular effectors such as adenylate cyclase and phospholipase C. The activity of adenylate cyclase was detected by stimulation of PTH (1–34), a biologically active peptide fragment, in HEK293 cells transfected with vector, WT-P1R, Flag-P1R-L, and Flag-P1R-P. PTH (100 nM, 15 min) could not induce cAMP production in HEK293 cells transfected with control vector (Fig. 4, upper), as the cells do not express an endogenous PTH receptor. PTH induced the same amount of cAMP production in HEK293 cells transfcted with Flag-P1R-L and Flag-P1R-P compared with WT-P1R.
FIGURE 4.
Detection of PTH-stimulated adenylyl cyclase and ERK1/2 phosphorylation. HEK293 cells were transiently transfected with WT-P1R, Flag-P1R-L, and Flag-P1R-P. After 48 h transfection, the cells were incubated with vehicle (0.1% BSA) or PTH (100 nM). (Upper) cAMP accumulation was measured as described in Materials and Methods. Data were summarized as the mean ± se of three independent experiments. (Lower) A representative immunoblot showed that PTH stimulated ERK1/2 phosphorylation (Lower, upper panel). The amount of ERK1/2 expression in each sample was used as an internal loading control (Lower, lower panel).
The effects of PTH depend in part on the activation of ERK1/2.14 Similar results were found for PTH-stimulated ERK1/2 phosphorylation in HEK293 cells expressing different PTH1R plasmids (Fig. 4, lower). Together, these findings confirmed that the Flag epitope located internally did not affect the receptor function.7
In summary, the procedure by PCR overlap extension is complicated, but efficiency is higher than that of ligase assembly (Table 2). Flag-tagged PTH1R constructs generated by both strategies were not only expressed successfully in vitro and in vivo but also possessed the same biological function as the wild-type PTH receptor.
Table 2.
Comparison of two-fragment assembly by using PCR overlap extension or ligase
| Assembly by PCR overlapping | Assembly by ligase |
|---|---|
| Two fragments must have several amino acids overlapping in their terminus. | Two fragments must have one same restriction enzyme site in their terminus. |
| Two fragments are amplified by PCR. The purified two fragments are assembled by a second PCR step and then subcloned into the vector. | Two fragments are amplified by PCR. The purified two fragments and vector digested by restriction enzymes are assembled sequentially by ligase. |
| The procedure is complicated, but efficiency is higher. | The procedure is simple, but efficiency is lower. |
ACKNOWLEDGMENT
We thank Dr. Peter A. Friedman for advice and help in the completion of this work.
REFERENCES
- 1.Friedman PA.PTH revisited. Kidney Int Suppl 2004;October:S13–S19 [DOI] [PubMed] [Google Scholar]
- 2.Wang B, Yang Y, Friedman PA.Na/H exchange regulatory factor 1, a novel AKT-associating protein, regulates extracellular signal-regulated kinase signaling through a B-Raf-mediated pathway. Mol Biol Cell 2008; 19: 1637– 1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang B, Bisello A, Yang Y, Romero GG, Friedman PA.NHERF1 regulates parathyroid hormone receptor membrane retention without affecting recycling. J Biol Chem 2007;282:36214–36222 [DOI] [PubMed] [Google Scholar]
- 4.Daunt DA, Hurt C, Hein L, Kallio J, Feng F, Kobilka BK.Subtype-specific intracellular trafficking of α2-adrenergic receptors. Mol Pharmacol 1997; 51: 711– 720 [DOI] [PubMed] [Google Scholar]
- 5.Seck T, Baron R, Horne WC.Binding of filamin to the C-terminal tail of the calcitonin receptor controls recycling. J Biol Chem 2003;278:10408–10416 [DOI] [PubMed] [Google Scholar]
- 6.Brock C, Oueslati N, Soler S, Boudier L, Rondard P, Pin JP.Activation of a dimeric metabotropic glutamate receptor by intersubunit rearrangement. J Biol Chem 2007; 282: 33000– 33008 [DOI] [PubMed] [Google Scholar]
- 7.Lee C, Gardella TJ, Abou-Samra AB, et al. Role of the extracellular regions of the parathyroid hormone (PTH)/PTH-related peptide receptor in hormone binding. Endocrinology 1994;135:1488–1495 [DOI] [PubMed] [Google Scholar]
- 8.Tazawa H, Takahashi S, Zilliacus J.Interaction of the parathyroid hormone receptor with the 14–3-3 protein. Biochim Biophys Acta 2003; 1620: 32– 38 [DOI] [PubMed] [Google Scholar]
- 9.Wang B, Ma L, Tao X, Lipsky PE.Triptolide, an active component of the Chinese herbal remedy Tripterygium wilfordii Hook F, inhibits production of nitric oxide by decreasing inducible nitric oxide synthase gene transcription. Arthritis Rheum 2004;50:2995–2303 [DOI] [PubMed] [Google Scholar]
- 10.Schneider H, Harbottle RP, Yokosaki Y, Kunde J, Sheppard D, Coutelle C.A novel peptide, PLAEIDGIELTY, for the targeting of α9β1-integrins. FEBS Lett 1998; 429: 269– 273 [DOI] [PubMed] [Google Scholar]
- 11.Sorensen HP, Mortensen KK.Advanced genetic strategies for recombinant protein expression in Escherichia coli. J Biotechnol 2005;115:113–128 [DOI] [PubMed] [Google Scholar]
- 12.Liang Q, Chen L, Fulco AJ.An efficient and optimized PCR method with high fidelity for site-directed mutagenesis. PCR Methods Appl 1995; 4: 269– 274 [DOI] [PubMed] [Google Scholar]
- 13.Jensen PH, Weilguny D.Combination primer polymerase chain reaction for multi-site mutagenesis of close proximity sites. J Biomol Tech 2005;16:336–340 [PMC free article] [PubMed] [Google Scholar]
- 14.Sneddon WB, Liu F, Gesek FA, Friedman PA.Obligate mitogen-activated protein kinase activation in parathyroid hormone stimulation of calcium transport but not calcium signaling. Endocrinology 2000; 141: 4185– 4193 [DOI] [PubMed] [Google Scholar]