Summary
Muscle contraction involves the interaction of the myosin heads of the thick filaments with actin subunits of the thin filaments. Relaxation occurs when this interaction is blocked by molecular switches on these filaments. In many muscles, myosin-linked regulation involves phosphorylation of the myosin regulatory light chains (RLC). Electron microscopy of vertebrate smooth muscle myosin molecules (regulated by phosphorylation) has provided insight into the relaxed structure, revealing that myosin is switched off by intramolecular interactions between its two heads, the free-head and the blocked head. Three-dimensional reconstruction of frozen-hydrated specimens reveals that this asymmetric head interaction is also present in native thick filaments of tarantula striated muscle. Our goal here has been to elucidate the structural features of the tarantula filament involved in phosphorylation-based regulation. A new reconstruction reveals intra- and intermolecular myosin interactions in addition to those seen previously. To help interpret the interactions, we sequenced the tarantula RLC, and fitted to the reconstruction an atomic model of the myosin head that included the predicted RLC atomic structure and an S2 crystal structure. The fitting suggests an intramolecular interaction between the cardiomyopathy loop of the free-head and its own S2 and two intermolecular interactions—between the cardio-loop of the free head and the ELC of the blocked head, and between the Leu-305 - Gln-327 “interaction loop” (loop I) of the free-head and the N-terminal fragment of the RLC of the blocked-head. These interactions, added to those previously described, would help to switch off the thick filament. Molecular dynamics simulations suggest how phosphorylation could increase the helical content of the RLC N-terminus, weakening these interactions, thus releasing both heads and activating the thick filament.
Keywords: Myosin regulation, thick filament, cryo-EM, myosin regulatory light chain, phosphorylation
Introduction
Striated muscle is formed by two sets of overlapping filaments, the thick myosin-containing and the thin actin-containing filaments. During contraction the two sets actively slide past each other, shortening the sarcomere. The myosin heads are helically ordered on the backbone of relaxed thick filaments. When filaments are activated the heads detach from the backbone and become disordered. Sliding force is produced when these heads cyclically attach to and pull on the thin filaments1.
Contraction of most muscles is regulated via Ca2+-controlled molecular switches located on either or both sets of filaments2. While the mechanism of actin-linked regulation by troponin and tropomyosin is well understood3, 4, our knowledge of the mechanism of myosin-linked regulation is less complete5, 6. It can occur by direct Ca2+ binding to the essential light chains (ELC – see below)5 (scallop striated muscle); or by regulatory light chain (RLC) phosphorylation (invertebrate striated muscle7,8 and vertebrate smooth muscle9). While RLC phosphorylation occurs in many muscles, its functional importance varies. In the case of vertebrate striated muscle, for example, phosphorylation appears to modulate contraction, but is not essential for activity6.
Myosin contains two heavy chains, and two pairs of light chains (the ELC and the RLC)10,11. One RLC and one ELC are non-covalently bound to the heavy chain α-helix in the lever arm of each myosin head10, stabilizing it and forming the regulatory domain11,12. The RLC has two domains9 connected by a “linker helix”, and an N-terminal fragment (NTF) or extension; all are important for regulation. The NTF varies in length, depending on species, and includes the phosphorylation site(s). While the NTF is absent from S1 crystal structures10, its structure has emerged from EPR of smooth muscle myosin13; revealing that it acts as a distinct phosphorylation domain, changing from solvent-inaccessible and weakly helical when unphosphorylated, to solvent accessible with helical order and increased rotational mobility when Ser-19 is phosphorylated. Electron microscopy (EM) of chicken smooth muscle myosin 14–17 has revealed that in the switched off (dephosphorylated) state the two heads establish an asymmetric, ‘interacting-head’ structure in which actin-binding activity of one head (‘blocked’) is sterically blocked by binding of its actin-binding interface to the converter domain of the other head. Actin-binding activity of this head is not blocked (it is therefore called the ‘free’ head), while its ATPase activity is inhibited, by prevention of converter movements needed for phosphate release. In addition to interaction between the two motor domains, there is also an interaction between the blocked head motor domain and the free head ELC. This interacting-head structure, deduced from 2D-crystals14–16 and single molecule17 studies, has also been shown to be present in native thick filaments of tarantula striated muscle18. It is therefore not an artifact of myosin isolation. In the filament, three additional interactions are seen18: one intramolecular, between the blocked head and its S2 (subsequently also seen in single molecule studies17), and two intermolecular, between the blocked head SH3 domain and the S2 from its axially adjacent neighbor, and between the blocked head ELC and the axially neighboring free head motor domain. The interacting-head motif has now been observed in other isolated myosin molecules (scallop19, tarantula, Limulus and mouse skeletal and cardiac20), and in thick filaments from mouse cardiac muscle21, Limulus and scallop striated muscle22, and scorpion striated muscles (Sanchez et al. in preparation), supporting the concept18,23, 24 that this motif is highly conserved and underlies the relaxed state of thick filaments in both smooth and striated muscles over a wide range of species.
When regulated thick filaments are activated by phosphorylation of their RLCs, the interaction between the heads is broken14,16 and the heads are released from the filament surface, becoming disordered8,25, 26. Breakage of the interaction could occur through a disorder-to-order transition of the phosphorylation domain13, apparently by establishing an Arg-16/Ser-19 salt-bridge27, a sequence known to be essential for regulation by Ser-19 phosphorylation28.
Our goal here has been to elucidate the mechanism by which phosphorylation-induced changes in the RLC activate the thick filament by releasing the heads from each other and from the filament surface so that they can interact with the thin filament. To achieve this, we have calculated a new 3D-map of tarantula filaments that better defines the RLC region, revealing two new interactions with functional significance. We propose a mechanism to explain how RLC phosphorylation weakens these inhibitory head interactions, releasing them so they can interact with the thin filament.
Results
Three-dimensional reconstruction of frozen-hydrated tarantula thick filaments
We carried out a new 3D reconstruction of tarantula filaments by the IHRSR technique, using an initial reference viewed at 0° or tilted up to ± 12°, to account for possible filament tilt in the images (see Methods)29–31. The reconstruction was more detailed, clearly showing two new interactions in addition to those seen previously18 (see Introduction)—an intramolecular density (a, fig. 1) between the motor domain of the free head and its own S2, and an intermolecular density (b, fig. 1) between the RLC of the blocked head and the motor domain of the free head of the axially adjacent molecule.
Fig. 1.

3D-reconstruction of the frozen-hydrated tarantula thick filament, filtered to 2 nm resolution (see suppl. fig. 3 and suppl. methods), showing four helices of interacting-head motifs (one in yellow). The 3D-map segment shows four 14.5 crowns of interacting-heads. Bar 14.5 nm.
We carried out atomic fitting to aid in the interpretation of these interactions. Using the atomic model determined previously18, part of the RLC volume is not filled. (b, fig. 1). A simple explanation is that the atomic model used18 had a smaller RLC than tarantula. The molecular mass of the tarantula RLC has been reported to be 26 kDa8 whereas the chicken smooth muscle RLC32 used is only 20 kDa. To determine whether this difference could account for the extra volume, we determined the sequence of the tarantula striated muscle RLC, and performed an improved atomic fitting of the 3D-map.
Sequence of tarantula RLC
We cloned the RLC gene of the tarantula Avicularia avicularia from RNA isolated from the Flexor Metatarsus Longus striated muscle of the legs. The sequence consists of 196 amino acids, and has a molecular mass of 21,679.2 Da, including a 52 aa NTF (fig. 2). This is about 4 kDa smaller than the 26 kDa estimated by SDS PAGE8, and about 2 kDa bigger than the 20 kDa of the smooth muscle RLC32. The NTF molecular mass of 5,671.4 Da, about a quarter of the RLC molecule, is absent from the RLC atomic structure used for the previous fitting18. A histogram of the aa length of the NTFs from the reported 110 RLC amino acid sequences (suppl. fig. 1) shows that these fragments can be either short (8 – 27 aa, 100 species) or long (43 – 61 aa, 10 species, fig. 3a) as in tarantula. In almost all reported RLC sequences with short NTFs the putative phosphorylatable serine is located in the same homologous position with a closely similar MLCK consensus sequence33 KKRXXSXBB (where X: any amino acid, B: any hydrophobic amino acid A, V, F, I or L). Tarantula myosin has two phosphorylation sites34 and mass spectrometry studies show that the first site phosphorylated is Ser-35 and the second Ser-45, and that some basal (mono-) phosphorylation occurs (at Ser-35) even in relaxed filaments (Brito et al. in preparation). The agreement with the MLCK consensus region (fig. 2) is better for Ser-45 than for Ser 35. In common with tarantula, all the RLC sequences with long NTFs (fig. 3a) have one serine homologous to Ser-45 which is located inside an MLCK consensus regions33, and all but one (Riftia), have an additional putative phosphorylatable serine located within ± 1 aa of the homologous Ser-35 tarantula position.
Fig. 2.
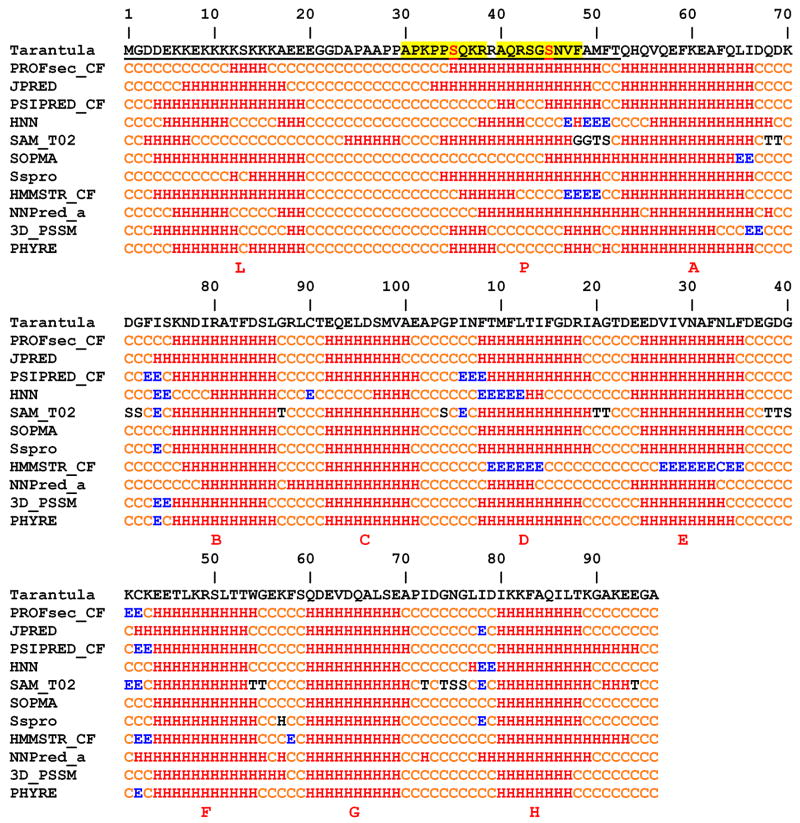
First line: Amino acid sequence of tarantula RLC. The 52 aa NTF is underlined. Lines 2–12: Prediction of secondary structure using the programs PROFsec35, JPRED69, PSIPRED70, HNN71, SAM72, SOPMA73, Sspro74, HMMSTR75, NNPred76, 3D_PSSM77 and PHYRE78. All programs predict ten helices, A–H corresponding to four EF hands motifs10 and L and P located in the NTF. Helix L, rich in positively charged Lys residues, is connected through a Pro-Ala repeat coil linker to the phosphorylation helix P, containing the two phosphorylatable serines (in red, Ser-35, Ser-45) each located in a MLCK consensus sequence33 (highlighted in yellow). H = helix, E = strand, C = coil, G = 3/10 helix, T = H-bonded turn and S = bend.
Fig. 3.
(a) Sequence alignment of RLCs for the 10 reported species with long NTFs. The two tarantula phosphorylatable serines (Ser-35 and Ser-45) are marked with arrows, and their MLCK consensus sequences33 with red boxes. UniProtKB ID code sequences (striated): Bombyx mori (Q1HPS0), Gryllotalpa orientalis (Q49M29), Riftia pachyptila (Q9GUA2), Clonorchis sinensis (Q2YHG1), Schistosoma japonicum (Q5DB55), Aedes aegypti (Q17HX1), Culex pipiens (Q45FA2), Anophelex gambae (Q7PUV3) and Drosophila melanogaster (P18432). (b) Sequence alignment of S2 from human cardiac36 and chicken smooth32 muscles. The S2 kink positions are indicated with red arrows: Met-877 (human36) and His-888 (chicken32). The black boxes show the two negatively charged ring36 1 (Glu-905 – Asp-917) and ring 236 (Glu-932 – Gln-946) of S2.
Predicted atomic structure of the tarantula RLC
As the crystallographic structure of the tarantula RLC has not been solved, we have attempted to predict its tertiary structure using homology modelling methods on the basis of known template structures35, using the sequence we have determined (see Methods). The predicted structure consists of three domains (fig. 4). Domains 1 and 2 each comprise two EF-hand helix-loop-helix motifs, so that there are four helices A–D in domain 1 and four helices E–H in domain 2, connected by a linker helix cf. 9. The known divalent cation (Ca2+ or Mg2+) binding site in the chicken skeletal RLC is located in the first EF motif (helices A–B)10 (although sequence analysis suggests that this is nonfunctional as a cation binding site in tarantula (Zhu et al, in preparation). Domain 3 or the “phosphorylation domain”13 comprises the NTF, whose structure can not be predicted using such homology methods, as the NTF has not been reported in any myosin crystallographic studies. Therefore we used programs to predict its secondary structure (fig. 2). These algorithms predicted ten helical regions for the complete RLC sequence, eight of them confirming the helices A-H revealed in the X-ray structure10, and two (helix L and helix P, fig. 2 and 4), located in the NTF domain, in agreement with previous predictions27. Helix L is rich in positively charged Lys residues, and is connected through a Pro-Ala repeat linker to the phosphorylation helix P, which contains the two phosphorylatable serines Ser-35 and Ser-45. This NTF secondary structure (fig. 2) was used to predict the structure of domain 3 using the ab initio tertiary structure predictor PredictProtein server35. As shown in fig. 4, the helix L, linker and helix P of the final predicted domain 3 comprised Asp-3–Gly-21, Gly-22–Pro-33 and Pro-34–Gln-41, respectively.
Fig. 4.
Predicted atomic structure for the tarantula RLC obtained using the PredictProtein server35. The structure has three domains: domain 1 (helices A–D), domain 2 (helices E–H) and domain 3 (helices L and P). The predicted secondary structure for the 52 aa NTF (domain 3 or “phosphorylation domain”13) is formed by a positively charged helix L and helix P (with phosphorylatable Ser-35 and Ser-45), connected by a Pro-Ala coil linker. The 8.5 nm IQ motif helix of the myosin heavy chain (Glu-812 – Phe-855) is shown in light gray.
Interacting-heads atomic model
The Wendt et al.15 HMM atomic model included an S2 domain, but this was arbitrarily positioned as its density was not directly visible in the reconstruction. In contrast, in our previous 3D-reconstruction18, a clear rod-like volume of density, running from the junction of the light chain domains towards the filament backbone, was identified as the first portion of the myosin S2 tail. For the present work, we used an initial model for atomic fitting comprising the refined chicken smooth muscle structure as determined by cryo-EM16(fig 5a) and the human cardiac S2 crystal structure36 (fig. 5c); see Methods). The intervening aa’s connecting them were energy minimized using molecular dynamics (MD, fig. 5a, see Methods). We replaced the RLC with the tarantula RLC predicted atomic structure including the NTF (fig. 4). The complete initial hybrid model used for atomic fitting is shown in fig. 5a. The sequence numbering used in this work is that corresponding to chicken16 for both HMM heavy chains, including in this numbering the S2 sequence (which is from human36), and the tarantula (fig. 2) for the RLC.
Fig. 5.
(a) Starting HMM atomic model used for flexible fitting built using the predicted structure of the tarantula RLC (fig. 4), the structure of human S236 and the chicken smooth muscle HMM atomic model (without the S2)16. The kink (red spheres) in the S2 was located at Met-888, in agreement with Blankenfeldt et al. 200636 (see fig. 3b and 5c). The connected skeleton of 31 positional markers used for flexible fitting, located inside the density subset of the interacting-heads, is shown with gray spheres and rods. (b) Final HMM atomic model after the flexible fitting. The RLC of the blocked and free heads are shown in yellow and red, with their corresponding NTFs in tan (blocked) and pink (free). The ELC of the blocked and the free heads are shown in orange and purple. The heavy chain of the blocked and free head are shown in green and blue. The 3D-map is shown as a pale gray surface in (a) and (b). (c) A comparison of the crystal structure of the human S236 before (red α-helices) and after (blue and green α-helices) the flexible fitting, which only changed the position of the coil-coiled α-helices. The arrow head indicates that the position of the kink (Met-888) does not changed before and after the flexible fitting. C-terminus are Lys-974 and Leu-972 (top). N-terminus is Pro-849 (red spheres, bottom). The connection of the two α-helices of the S2 with the two α-helices of the interacting-heads regulatory domains was done between Ser-853 and Gln-852. The connection of the S2 N-terminal with the C-terminal structure of the heads required some unbending and torsion in the 6 (blocked head) or 8 (free head) intervening aa’s (see Results). (d) The nine interactions of the interacting-head motif in a relaxed thick filament, as viewed (towards) the filament surface (cf. 5a and b from above the filament surface). The five intramolecular interactions (a, d, e, f, g) are between: (a) the CM loop of the free head motor domain (blue) and the S2 of the interacting-head motif (blue); (d) the blocked head motor domain (green) and the free head motor domain16 (blue), (e) the blocked head motor domain (green) and the free head ELC16 (purple), (f) the blocked head motor domain (green) and the S2 of the interacting-head motif36 (blue and green), and (g) the blocked head ELC (orange) and the S2 of the interacting-head motif (blue). Four intermolecular interactions (b/b′, c/c′, h) are between : (b/b′) the blocked head RLC NTF (b, tan; b′, gold) and the I loop of the motor domain of the neighbor free head (b, gold, b′, blue); (c/c′) the blocked head ELC18 (c, orange, c′, gold) and the C loop of the neighboring free head (c, gold, c′, blue); (h) the SH3 of the blocked head motor domain18 (green) with the S2 of the neighbor interacting-head (pale blue backbone surface). An interaction between the blocked head ELC (orange) and the backbone is not shown. The 3D-map is shown as a pale blue (backbone S2) or gold surface (up and low interacting-head pairs).
Atomic fitting of the interacting-heads atomic model to the 3D-reconstruction
The atomic model described above was flexibly fitted to the volume comprising the interacting-heads (fig. 1) of the 3d-map using Situs37 (see Methods). The best fitting achieved using 31 positional markers (fig. 5a) is shown in fig. 5b. The fitting reveals details of the two new interactions described above (in addition to those described previously18), an intramolecular interaction between the free head motor domain and its S2 (a, fig. 5b, 6ab), and an intermolecular interaction between the RLC of the blocked head and the motor domain of the axially adjacent free head (b, fig. 5b, 6ab). The spatial arrangement of these interactions is shown in the stereo views of fig. 6ab, along with the interaction previously described between the ELC of the blocked head and the motor domain of the neighboring free head (c, fig 5b, 6ab). After the flexible fitting (fig. 5b) both the blocked and the free head structures remained similar to the myosin “closed” conformation seen in the smooth muscle model16 and in the original tarantula fitting18 (fig. 6c); however the part of the free head heavy chain α-helix associated with the regulatory domain had an orientation midway between the closed and transition state pre-power stroke structures 1BR1 and 1DFL (fig. 6c). A comparison of this new model (fig. 5b) with the refined smooth muscle model16 (derived from PDB 1I84, without S2) and the atomic model published previously18 (with S2) is shown in supp. fig. 2.
Fig. 6.
(a) Stereo-pair of flexible atomic fitting of two adjacent interacting-heads on one helical track of the tarantula filament. Three interactions are labeled, an intramolecular interaction ‘a’, and intermolecular interactions ‘b’ and ‘c’18. The 3D-map is shown as a pale gray surface. (b) Stereo-pair of intramolecular interaction ‘a’ between the CM or cardiomyopathy loop (yellow ribbon) of free head (Ile-407 – Val-425 highlighted using balls and sticks, Arg-411 residue is in yellow) with its own S2 (blue a-helix), intermolecular interaction ‘b’ between the NTF (tan) of the RLC (yellow) of the blocked head with the neighbour motor domain free head I loop (Leu-305 – Gln-327 shown in balls and sticks, green ribbon), and intermolecular interaction ‘c’ between the ELC of the blocked head (orange) and the C loop (Gln-361 – Asp-380 shown in balls and sticks, red ribbon) neighbour motor domain free head (blue)18. Glu-932 – Glu-946 of the negatively charged ring 236 of S2 (fig. 3b) are shown as balls and sticks. Ser-35 and Ser-45 are shown as pale blue and pale magenta spheres, respectively. (c) Stereo-pair of a comparison of the blocked (green) and free (yellow) heads (fig. 5b) from tarantula myosin, with the published crystal structures known as the pre-power stroke closed (1BR1)63 (red) and transition structures (1DFL)66 (pale yellow), and the near(1DFK)66/post(2MYS)10/like(2OY6)88 rigor (pink/light blue/purple) or detached internally uncoupled (1B7T)79 (dark blue) open structures. The structures of the blocked and free heads used for the flexible fitting were assumed initially to be in the closed conformation15 (fig. 5a). For clarity the ELC and RLC have been removed. Note that the regulatory domain a-helices for the 1BR1, 1DFL, and the blocked and the free head are in the same plane. The difference between the blocked and the free head regulatory domain a-helices is mostly a change in angular orientation.
The flexible fitting modifies the orientation of the cardiomyopathy (or CM) loop (Lys-407 – Val-415) and the cardio (or C) loop (Gln-361 – Asp-380) of the free head by displacing them so that both loops fit better into the intramolecular tubes of density (a,c fig. 6b). Thus the motor domain of the free head interacts with the negatively charged ring 236 (Glu-932 – Gln-946 see fig. 3b) of its own S2 (interaction “a”). The flexible fitting also modifies the RLC NTF from its initial predicted structure (fig. 4, 5a), to a final structure that better fits the intermolecular tube of density (b, fig 5b, 6b). An intermolecular interaction occurs between the Leu-305 – Gln-327 Interaction loop (or I loop) of the free head and Met-1 – Asp-3 of the NTF helix L and Pro-25 – Ser-35 of the NTF helix P of the adjacent blocked head RLC (b, fig. 6b). The new fitting also suggests that the intermolecular interaction previously described18 is between the ELC of the blocked head and the C loop of the adjacent free head (c, fig. 5b, 6ab).
Atomic model of tarantula thick filament
Fig. 6a shows two levels of interacting-heads of a relaxed tarantula filament. Four helices38 of interacting-heads occur on the filament surface. The interacting-heads appear to be kept in the helically ordered arrangement by the establishment of the nine interactions shown in fig. 5d. Five interactions are intramolecular, one between the CM loop of the free head and its own S2 (a, fig. 5d); one between the motor domains of the free and blocked heads15–18, 30(d, fig. 5d), another between the motor domain of the blocked head and the ELC of the free head16, 18(e, fig. 5d), one between the motor domain of the blocked head and its own S236 (f, fig. 5d), and one between the ELC of the blocked head and its own S2 (g, fig. 5d). Four interactions are intermolecular, an interaction between the RLC NTF of the blocked head and the I loop of the adjacent free head (b and b′, fig. 5d), an interaction between the ELC of the blocked head and the C loop of the adjacent free head18 (c and c′, fig. 5d), one between the SH3 domain of the blocked head and the S2 of the adjacent myosin molecule18 (h, fig. 5d), and one (not shown) between the ELC of the blocked head and the backbone. The five intramolecular interactions presumably stabilize each interacting-head motif as a rigid unit, while the helical organization of motifs is established by formation of the four intermolecular interactions. The intramolecular interaction “a” and the two intermolecular interactions (h, and the one -not shown- between the blocked head ELC and the backbone, fig. 5d) would help to anchor each interacting-head motif to the backbone. Taken together, these interactions appear to keep the myosin heads close to the thick filament surface, and to minimize actin interaction and ATPase activity in the relaxed state.
Discussion
The 3D-reconstruction
The 3D-map together with flexible atomic fitting reveals a new intramolecular interaction, between the motor domain of the blocked head and its S2 (a, fig. 1), and a new intermolecular interaction, between the blocked head RLC and the free head motor domain (b, fig. 1), in addition to the interactions previously described16–18,36 (fig. 1). The map shows specific density protrusions that match well with the fitted atomic structure (fig. 5b). The interaction between the blocked head RLC and the free head motor domain (b, fig. 1, 5bd, 6ab) may be a key element in the phosphorylation-mediated release of the myosin heads from the filament surface that occurs on activation8.
The density in the RLC region of the reconstruction (b, fig. 1) is explained in part by the greater mass of the tarantula (21,679.6 Da) compared with the smooth muscle RLC. This difference is included in the 5,671.4 Da NTF, representing 26% of the RLC mass, which is absent from crystal structures of the myosin head. The tarantula RLC (196 aa) is one of the longest RLCs sequenced so far (111 species, range: 153–222 aa), its 52 aa NTF belonging to the class of long NTFs (43–61 aa, fig. 3a, suppl. fig. 1). The functional significance of the diversity in NTF structures and lengths (especially between the long and short classes) and its relation to regulation is unknown.
The structure of the interacting-heads motif
In the absence of a crystallographic structure for tarantula HMM, we built a hybrid HMM model (fig. 5a) for atomic fitting by including the crystallographic structure of human cardiac S236, the refined chicken smooth muscle interacting-heads structure16, and the tarantula RLC predicted atomic structure (fig. 4). Flexible fitting of this model produced a final pseudo-atomic model (fig. 5b) which matched well with the 3D-map. The new model (fig. 5b) agreed well with the general shape of the previous interacting-head atomic models16,18 (suppl. fig. 2). The main differences were in the precise S2 position, the orientation of the CM loop, and in the RLC regions of both regulatory domains. Because we do not have a complete tarantula crystallographic structure available yet, the precise amino acids involved in the interactions shown in fig. 5d can not be deduced from our current atomic fitting, although the loops involved can in principle be identified when their densities protrude in a characteristic way, as discussed below. We are sequencing the tarantula ELC and myosin heavy chain to improve the initial atomic model and the flexible atomic fitting (Zhu et al., in preparation, Patiño et al. in progress).
Both α-helices of the S2 atomic model were connected to the respective heavy chain a-helices of the regulatory domains of the interacting-heads (fig. 5ac). The connection of the S2 N-terminal Ser-853 (fig. 3b, fig. 5c) with the Gln-852 C-terminal of the head structure shown in fig. 5a required an unbending and torsion in the 6 and 8 intervening aa’s of the free and blocked head S2 N-terminals respectively (fig. 5c), as shown in the structure of the head-tail junction (fig. 5b). The flexing changed the relative position of the coil-coiled α-helices but not the kink position (Met-888, fig. 3b, fig. 5abc). The final structure showed an excellent fit of the coiled-coil α-helix into the tube of density (fig. 5ab). This unbending and torsion is in agreement with the results from crystallographic studies of the N-terminal 10–14 residues of the regulated scallop myosin S2 peptide construct39 which showed they were disordered. In contrast, the N-terminal 126 residues of the unregulated human cardiac myosin S2 peptide were found by crystallographic studies to be a straight parallel dimeric coiled coil36. Recent crystallographic and thermal studies of scallop myosin S2 peptide constructs40 suggest that the N-terminal region of the S2 in regulated myosins has less stability than unregulated myosin S2, indicating that the uncoiling of the N-terminal region of the S2 may be key to its function in regulated myosins40.
It has been shown previously that helical ordering of the myosin heads in thick filaments requires the “closed” conformation of the switch 2 element of the nucleotide pocket, preventing phosphate release41,42. It has also been shown that blebbistatin stabilizes helical ordering by promoting the switch 2 closed state43–45. The model shown in fig. 5b offers further insights into the specific conformation of the blocked and free heads in the relaxed filament. After flexible fitting (fig. 5b) both head structures remain similar to the closed conformation of the original HMM atomic model used, in agreement with these earlier results (fig. 6c) and with our previous fitting18 showing that the blocked head was in the closed conformation (i.e. MgADP-AlF4, PDB 1BR1)16 (fig. 6c). However, the free head, also in the closed conformation, shows a difference in orientation of its heavy chain regulatory domain, which is midway between the closed 1BR1 and transition 1DFL structures (fig. 6c). Fitting of the CM loop of the free head to the 3D-map required some rearrangement of the 50K domain of the head, including some closure of the cleft between the upper and lower parts of the 50K domain. These structural differences between the free and blocked head structures, derived from the 3D-map may have functional implications. One possibility is that the apparent cleft closure and the specific CM loop orientation is required to guide the free head, detached after the power stroke, to establish the precise electrostatic docking interaction “a” (fig. 1, 5b, 6ab) onto ring 236 of S2, a requisite for reforming the helices of interacting-heads. These structural differences could also preset the order in which the two heads are released on phosphorylation. The free head would be the one conformationally and properly located to be released first to interact with actin, followed by the blocked head (if required).
The structure of the blocked and free head regulatory domains
The predicted NTF tertiary structure was positioned in an arbitrary initial configuration (fig. 4, 5a) for the two heads and became rearranged during the flexible fitting and the associated MD energy minimization (fig. 5b, 6ab and 7abc). The linker length for both heads increased after the flexing while the length of helices L and P decreased (Table 1). The accuracy of the secondary structure prediction for the NTF is only 80%; therefore the beginnings and endings of the two α-helices may vary by ± 3 amino acids. As a consequence of the flexing, the tertiary structure of the NTF of the blocked head RLC appears to be more compact (a, fig. 7) than the free head NTF (c, fig. 7). The final atomic fitting (fig. 5b) shows that the positioning of both RLCs on the free and blocked heads is not symmetric, in agreement with the asymmetry of the two-headed motif as a whole, including the asymmetric positioning of Ser-35 and Ser-45 on the two RLCs (fig. 6b).
Fig. 7.
The structure of the tarantula RLC in the atomic model following flexible fitting. The top row a–c shows the flexed RLCs (in ribbons) from the blocked (a, yellow/orange), blocked and free (b) and free (c, red/pink) heads. The NTF structures are asymmetric: the blocked head NTF (a, orange) exhibits a more compact conformation than the free head (c, pink). The second, third and fourth rows show the RLC electrostatic surface charges (blue positive, red negative) for the whole RLCs (d–f), the RLC without their NTFs (g–i) and the NTFs alone (j–l). The bottom row (m,n) shows the elongated NTF (m, ribbon; n, surface charge; arrows: Ser-35, Ser-45). In relaxed muscle the NTF of the dephosphorylated blocked head RLC (a,d) exhibits a compact conformation (j) that enables the interactions shown in fig. 5d, while the NTF of the basally Ser-35 phosphorylated free head RLC (c,f) is partially elongated (l). Both positively charged helices L are packed –due to complementary charges (see suppl. movie 1)- against the blocked head domain 1 (cf54, see discussion). If phosphorylation in Ser-45 of the free head RLC produces elongation of its NTF (m,n), this could release the free head, exposing the blocked head NTF, which in turn could be phosphorylated at Ser-45, elongated (m,n) and the blocked head released. This mechanism could explain how RLC phosphorylation weakens the head-head interactions, releasing them so they can interact with the thin filament.
Table 1.
Extension of the NTF helices L, P and linker before and after the flexible fitting
| Helix L | Linker | HelixP | |
|---|---|---|---|
| Before flexing (both heads) | Asp-3–Gly-21 18 aa | Gly-22–Pro-33 11 aa | Pro-34–Gln-41 7 aa |
| After flexing (blocked head) | Glu-5–Lys-11 6 aa | Lys-12–Pro-33 21 aa | Pro-34–Ala-40 6 aa |
| After flexing (free head) | Asp-4-Lys-15 11 aa | Lys-16-Ser-35 19 aa | Glu-36-Arg-39 3 aa |
Table shows starting and ending aa’s as well as the aa length for helix L, P and linker.
Both phosphorylatable serines of the free head RLC seem to be freely accessible to the MLCK as they are located in an exposed surface of helix P (fig. 6b). The corresponding serines on the blocked head are located in the more hidden region of the intermolecular interaction with the neighboring motor domain I-loop (fig. 6b), and a close free head NTF facing it (fig. 7b), which could interfere with the MLCK access (fig. 6b). Differential MLCK access to the free and blocked serines may determine the phosphorylation sequence of the two heads. Two MLCKs have been reported in mouse skeletal muscle46 : a skMLCK present in adult skeletal muscle and a smMLCK present in smooth, non-muscle, cardiac and skeletal muscle. Mass spectrometry results46 suggest that skMLCK and smMLCK differentially phosphorylate the Ser-16 or Ser-15 in mouse skeletal muscle, favoring the possibility that two MLCKs may also exist in tarantula striated muscle (Zhu et al. personal communication); one phosphorylating the homologous Ser-35 (possibly presetting the basal phosphorylation level), and another phosphorylating the homologous Ser-45 (enabling the release of the heads when required after repetitive stimulation)
The new interactions
Flexible fitting of the atomic structure suggests the nature of the two new interactions, an intramolecular interaction between the CM loop of the free head and its own S2 (a, fig 6b), and an intermolecular interaction between the I loop of the free head and part of helix L and helix P of the NTF of the adjacent blocked head (b, fig. 6). It also reveals that the interaction between the blocked head ELC and the adjacent free head18 occurs specifically between the C loop of the free head and the adjacent ELC (c, fig. 6).
The tube of density between the motor domain of the free head and its own S2 (a, fig. 5d) is well fitted with the CM loop. The positively charged CM loop is located in the tube of density such that it faces ring 2 of the closest α-helix of the coiled coil located ~ 0.5 nm away. Ring 2 is one of the three negatively charged rings described in the human S2 crystal36. This intramolecular interaction could help to electrostatically tack the free head to the closest position in the backbone of the thick filament, by docking onto its own tail. This intramolecular interaction has not been seen in isolated myosin molecules17,19,20 suggesting that its formation requires the presence of a positioning backbone framework. R403 mutants were the first mutations to be shown to cause Familial Hypertrophic Cardiomyopathy (FHC) by linkage analysis47. This finding led to the adoption of the nomenclature “Cardiomyopathy Loop” to designate the loop on which the residue occurs. The close proximity and possible interaction of four S2 mutations (E924K, E927K, E930K, and E935K) to this CM loop” (fig 8) may be helpful in understanding how mutations in the rod region of human cardiac myosin lead to disease. One intramolecular interaction (f, fig. 5d) has been reported36 between ring 1 (Glu-905 – Asp-917, fig. 3b) of the S2 and the loop 2 Trp-625 - Ser-647 of the actin binding interface of the blocked head motor domain, suggesting that electrostatic interactions play an important role in stabilizing the OFF state. The intermolecular interactions between the motor domain of the free head and the ELC (c, c′, fig. 5d) and RLC (b, b′, fig. 5d) of the neighboring blocked head are well fitted with the C loop and the I loop of the free head (fig. 5d). A third intermolecular interaction (h, fig. 5d)18, probably helps to anchor the SH3 domain of the blocked head onto the neighboring S2 as it descends towards the thick filament backbone.
Figure 8.
Five of the reported mutations in the myosin head heavy chain and S2 associated with FHC. Myosin heavy chain: R403Q, R403L or R403W (orange sphere) in the CM loop (yellow); and E924K (grey spheres), E927K (pink spheres), E930K (cyan spheres) and E935K (yellow spheres) in the negatively charged ring 2 of S236, facing the CM loop. The side chain orientations at the interface of the loop may be unreliable as this is not a structure determination down to atomic detail, but a prediction at amino acid level of detail. See legend of fig. 6b.
Ordering of the heads in relaxed thick filaments
Myosin heads protrude from their helical origins on the filament backbone every 14.5 nm axially36,48–50, 50. Once interacting-head motifs have formed at these origins during relaxation, how do these flexibly attached motifs form into the ordered helices of the relaxed state? This presumably involves the intermolecular interactions we have described. Each motif appears to bind to an adjacent S2 via the SH3 motif of its blocked head18 (h, fig. 5d), while axially adjacent motifs interact via the interactions between the ELC of the blocked head and the C loop of the free head18 (c, c′ fig. 5d), and the NTF of the RLC of the blocked head and the I loop of the adjacent free head (b, b′ fig. 5d). These three intermolecular interactions may serve to tack down the flexibly attached heads in regular positions reflecting their origins in the filament backbone49, at the same time keeping the heads away from the actin filaments and inhibiting their ATPase activity. This stable conformation of the relaxed thick filament would help maintain the shutdown state.
The structural mechanism of thick filament activation
The structural basis of myosin activation via RLC phosphorylation, leading to release of myosin heads from their helices in the thick filament, remains unknown51. The effect of RLC phosphorylation in striated muscle has been explained as a loosening of the myosin heads from the thick filament backbone8,25, 26,52–54 causing disordering. It has been suggested that the restricted mobility of the relaxed state may be due to electrostatic interactions between positively charged amino acids near the phosphorylatable serine of the RLC and an adjacent structure54, and that reduction of the net charge by phosphorylation would be the simplest mechanism to weaken this effect54. The positively charged amino acids in the NTF have been shown to be important for catalytic activity of MLCK55.
The 52 aa long tarantula NTF belongs to the long N-terminal class (fig. 3a, suppl. fig. 1), which all contain many Lys and Arg near the N-terminus (fig. 3a). There are 14 positively charged aa in the NTF of tarantula, which, together with the 8 negatively charged aa, gives a total net positive charge of 6. Phosphorylation of Ser-35 could reduce this to 4 positive charges and additional phosphorylation at Ser-45 to only 2 positive charges. Phosphorylation could induce the formation of two Arg/phospho-Ser salt bridges in helix P similar to the Arg-16/phospho-Ser-19 for the smooth muscle RLC27, one possibly between Arg-42 and phospho-Ser-45, and another –less strong- between Lys-32 and phospho-Ser-35. MD simulations of the de- and phosphorylated tarantula NTF are in progress to test this point. The structural model proposed by Nelson et al.13 suggests that phosphorylation of the smooth muscle NTF increases its helical order, internal dynamics and solvent accessibility causing a decrease of head-head interactions and activating myosin. MD simulations27 have helped to refine this model. It was found that phosphorylation causes a disorder-to-order transition in residues Lys-11 to Ala-17, converting a dynamically unstructured region to a stable α-helix, with formation of a salt bridge between phosphoserine-19 and Arg-16. Atomic fitting of the tarantula reconstruction shows that the dephosphorylated blocked head NTF fits well into the density corresponding to the new intermolecular interaction (b, fig. 6b) in the tarantula filament switched off state. In this case, phosphorylation of the RLC NTF at the homologous Ser-45 could also induce a disorder-to-order transition, weakening the intermolecular interaction, as with smooth muscle myosin.
The atomic structure shown in fig. 5b suggests that the 52 aa blocked head NTF (27 aa longer than the short 25 aa long smooth muscle NTF), compacts itself (fig. 7a) in the small intermolecular interaction density “b” shown in fig. 6b. In contrast, the free head NTF, which is not engaged in any intermolecular interaction, adopts the structure shown in fig 7c, which is less compact than that in the blocked head (fig. 7a), but not the fully elongated structure expected when it is Ser-45 phosphorylated (fig. 7m,n). Under relaxing conditions, the heads are ~35–50% mono-phosphorylated34, and LC-MS/MS shows that this is on Ser-35 (Brito et al., in preparation). It seems likely that this would represent mainly phosphorylation of the free head, based on the greater accessibility of its phosphorylation sites (fig. 6b). If so, this would imply that 70–100% of the free heads are basally phosphorylated. This structural information (fig. 6b, 7abc) together with EPR13 and MD evidence27 suggests that in tarantula striated muscle, the compact NTF (fig. 7a) of the unphosphorylated blocked head and the less compact NTF (fig. 7c) of the Ser-35 mono-phosphorylated free head could elongate when they are Ser-45 phosphorylated (fig. 7m,n), weakening the interactions and releasing the heads.
Putting together biochemical8,34, in vitro motility (Guerrero et al, in preparation) and LC-MS/MS results (Brito et al., in preparation), we suggest that in the relaxed state, tarantula striated muscles exhibit a degree of basal Ser-35 phosphorylation in their free heads (presetting the basal level of force production), which are able to interact rapidly with actin when the troponin-tropomyosin switch on the thin filaments is activated by Ca2+. Phosphorylation of Ser-45 of the free and blocked RLCs on longer term activation could recruit additional heads, increasing force.
Material and Methods
Sequencing
To clone the RLC gene (rlcAa), total RNA was isolated from the Flexor Metatarsus Longus striated muscle of the legs of the tarantula Avicularia avicularia in the presence of Trizol® (Invitrogen). An initial cDNA was obtained using oligo d(T) and an internal degenerate primer, designed against the uniquely conserved region of published RLCs. Nucleotide sequence analysis of the 315-bp fragment amplified allowed the design of 2 gene specific primers over the Carboxy-terminus (GSP2: 5′-GGTGCAAAAGAAGAGGGAGCT-3′). The cDNA for the entire rlcAa transcript was obtained using these gene specific reverse primers and the 5′RACE kit (Invitrogen). The amplified 591-bp product was sequenced and verified with available RLC sequences. The RLC sequences from tarantula striated muscle and other species with long NTFs were aligned using ClustalX56 (fig. 3a).
Image processing and 3d reconstruction
Low-dose electron micrographs of frozen-hydrated thick filaments18 (1008 filaments halves) were digitized at 0.248 nm/pixel using a Nikon Super Coolscan 8000 ED scanner. Filaments were aligned with the bare zone at the top, to ensure correct polarity in subsequent steps. A total of 15,504 segments, each 62 nm long, with an overlap of 55.8 nm, and containing ~40,000 unique pairs of interacting-heads went into the reconstruction. Three-dimensional single particle reconstruction was carried out by a modification of the IHRSR method29–31,57, 58, using SPIDER59. As an initial reference model we used the tarantula negatively stained 3D-map38,60–62, which was axially rotated, axially shifted and also out of plane tilted30,31 up to ±12° for projection matching, giving a total of 45×7×13=4,095 projections (13 tilted projections ±12° every 2°, 45 reference rotated projections (0–90 degrees, 2° rotation angle), and 7 image axial shifts of 2.2 nm. The new 3D-map shown in fig. 1 combines about 10,700 out of 15,504 filament segments, a yield of 69% of included segments. The 3D-map was Fourier filtered to 2 nm resolution (see suppl. fig. 3a and supplementary methods).
Atomic model for 3D fitting
We created a hybrid atomic model of HMM using the refined chicken smooth muscle HMM atomic model16 (derived from the PDB 1I8415, atomic coordinates kindly provided by Dr Kenneth Taylor) in which we replaced the human cardiac myosin S2 crystallographic structure36 and both RLC structures by the predicted RLC structure from tarantula striated muscle, including a predicted NTF secondary structure (fig. 4). In this model the two heads consist of the high resolution X-ray structure of the smooth muscle motor domain plus ELC (called MDE) with bound MgADP.AlF4− (PDB 1BR1)63; together with the skeletal muscle RLC and its associated heavy chain (PDB 2MYS)10. The motor domain conformation therefore corresponds to the structure of the myosin head in which the -phosphate pocket is closed, and the lever arm is “up” approximately perpendicular to the thin filament axis, as reported for the ADP.AlF4−, ADP.Vi and ADP.BeFx states64–67. The HMM model consists of two of these S1s with their COOH termini moved closer together to achieve a better fit, accomplished by a ~20° rotation of the light chain binding domain15,16,18. S2 atomic model: Wendt et al (2001)15 were uncertain of the position of S2 in their atomic model, while in the thick filament reconstruction18, a rod-like volume of density about 30 nm long was observed running from the junction of the light chain domains towards the backbone and therefore identified as the first portion of S2. The human cardiac S2 crystallographic structure (human cardiac β-myosin II S2- Δ, 126 aa, P838–K963)36 has been reported, and we have used it (atomic coordinates kindly provided by Drs Ilme Schlichting and Wulf Blankenfeldt) to match this rod-like density. This density in the 3D-map is not straight (fig. 1), but has a bend of ~23° about ~5.7 nm from the head-tail junction. We made a preliminary visual adjustment of the S2 atomic structure with a kink at Met-88836, to match this bend and the head-tail junction in the 3D-map. Then we connected the two α-helices of the S2 atomic model to the two α-helices of the interacting-heads as shown in fig. 5a, and perform energy minimization of the intervening aa’s by MD using the Chimera MD module68. RLC Secondary Structure Prediction: We predicted the secondary structure of the tarantula myosin RLC from its aa sequence (fig. 2) using eleven different program packages: PROFsec35, JPRED69, PSIPRED70, HNN71, SAM72, SOPMA73, Sspro74, HMMSTR75, NNPred76, 3D_PSSM77 and PHYRE78. The predicted structure was similar in all cases. For tarantula, the myosin RLC included ten helices: eight (A–H, fig. 2) correspond to the two-helices of each of four EF hand motifs as determined by X-ray crystallography10, and two - that we have called L and P- are located in the NTF (fig. 2). The prediction of these new helices L and P is in agreement with other secondary structure predictions27 and MD simulations27. RLC Predicted Atomic Model: We used the tarantula RLC sequence to obtain the predicted atomic model (fig. 4) from Tyr-53 to Ala-196, using the PredictProtein server35, choosing the scallop RLC (PDB 1B7T)79 x-ray crystallographic structure. The predicted secondary structure of the 52 aa NTF was used to predict the tertiary structure of this region using de novo and template modeling methods35. The complete 196 amino acid tarantula RLC atomic model was assembled bonding both the homology RLC predicted atomic structure and the de novo predicted structure model of the NTF as shown in fig. 4. The NTF model was positioned in an arbitrary initial configuration, to allow a further rearrangement during the flexing fitting procedure (fig. 5a).
Flexible atomic fitting
We first attempted a rigid-body fitting of the atomic model shown in fig. 5a into the 3D map using the “colores” tool of Situs80. This allowed us to create a mask for a single HMM unit by low-pass filtering of the docked atomic structure (yellow in fig. 1). The mask was used with Situs tools to segment and subtract densities from neighboring symmetry-related subunits38 to obtain the density of a single HMM from the helical 3D map. This single HMM map can then be compared to the atomic structure. Rigid body docking of the atomic model to the single HMM map was not satisfactory with respect to the position and direction of the S2 subunit, the RLC conformations of both heads, and the conformation of the 50K domain of the free head, even when performed independently for each structural subunit. Therefore we subjected the predicted atomic model (fig. 5b) to flexible docking to characterize the observed changes. The flexible docking procedure is based on a connected “motion capture” network of identified features within the atomic model81. The atomic model is allowed to move according to displacements tracked by 31 control points defined by the network, in order to find the best match to the cryo-EM map (fig. 5b). The number of control points was judged to be sufficient for capturing the shape details of the single HMM map which occupies a volume of 620 nm3 at the isocontour level shown (fig. 5b). The number of independent pieces of information contained in the 3D-map is then 620/(2)3 = 77.5. This number comprises an upper bound for the number of recognizable features in this particular volume. Our conservative choice of 31 points (corresponding to a spatial resolution of 2.7 nm in our reduced network) was slightly below this upper bound to avoid an over fitting of the data.
The longitudinal distance constraints in the motion capture network were assigned manually82,83 by following the connectivity of the polypeptide chain to ensure robustness of the control points during the shape change. We found by trial and error that robustness was best achieved by affording more flexibility to the RLC regions and to the 50K domain in the free head by eliminating some constraints on the motion in these regions. The final network used for the automated flexing is shown in fig. 5b. We performed the flexing by adding a constraint energy function to the Hamiltonian of a MD simulation that penalizes global shape differences between the data sets81. In the MD run we added 116 buried water molecules predicted by DOWSER84 to the system, resulting in a total system size of 25,715 atoms. We expect that at 2 nm resolution (see suppl. fig. 4a) the flexing faithfully reproduces conformational differences with a precision of 0.3 nm if atomic structures are locally conserved81. Side chains are rearranged automatically to accommodate global conformational changes. Otherwise, the algorithm leaves the initial structure intact on the local level. Whether this assumption holds depends on the extent of the conformational rearrangement, which is not known a priori. However, it has been shown that only about 7% of protein domain rearrangements documented in the PDB are irregular motions where the tertiary structure is significantly perturbed85. Therefore, it is plausible, at least for predominantly hinge- and shear-type domain motions exhibited by HMM, that the low-resolution flexible fitting is performed with a precision of single amino acid residues. Inspection of the computer generated flexed model (not shown) revealed slight discrepancies between a low-pass filtered atomic model and the full helical 3D-map due to the omission of symmetry-related contacts. The flexing is limited to single molecules37,81,82 and required the above masking and editing of subunits. Also, the modeled N-terminal RLC region of the blocked head exhibited an unsatisfactory overlap with the corresponding EM density before flexing, due to structure prediction (see above) that did not consider shape information. Since the motion capture assumes that the shape of features remains similar during flexing (a requirement for finding a robust representation for the deformation), the flexing initially overcompensated the shape discrepancy with an unrealistic compression of the N-terminal region of the blocked RLC. We have therefore slightly modified the original approach for the present case by manually accounting for the initial discrepancies due to symmetry and modeling. Discrepancies were first computed and visualized based on difference maps between low-pass filtered atomic structures and the 3D-map. Subsequently, the network distance constraints were relaxed and control points in the most flexible RLC regions, in the free head, and in the S2 region were moved manually to minimize observed discrepancies. This manual “touch-up” of the computer generated model was judged independently by three authors (W.W., L.A., and R.P.). The manual, difference map-assisted movements of control points were on the order of the docking precision (rms deviation 0.38 nm) and small compared to the overall motion. The final atomic model is shown in fig. 5b and the final flexing-induced rms deviation in this atomic model was 0.74 nm. The atomic models were displayed using Chimera68 and Visual Molecular Dynamics86. The surface charge was calculated using the program APBS: Adaptative Poisson-Boltzmann Solver87 (a VMD plugin).
Supplementary Material
Supplementary Data
Supplementary data associated with this article can be found, in the online version. The tarantula myosin regulatory light chain DNA sequence has been deposited at the GenBank (accession no. EU090070). The frozen-hydrated tarantula thick filament 3D-map has been deposited in the EM Data (EMDB) (ID. Code EMD-1535). The atomic coordinates of the tarantula myosin interacting-heads have been deposited in the RCSB/PDB (ID. Code 3DTP).
Acknowledgments
We thank Dr. Neal Epstein, Dr. Ulf Lunberg, Dr. Jose Reinaldo Guerrero, M. Sc. Reicy Brito and M. Sc. Sol Patiño for discussions, Dr. Kenneth Taylor for providing the unpublished atomic coordinates of the refined smooth muscle heavy meromyosin16, and Drs. Ilme Schlichting and Wulf Blankenfeldt for providing the atomic coordinates of the human cardiac S2 structure36, Drs. D. Thomas and M. Espinoza-Fonseca for their help on MD simulations of the tarantula RLC NTF and Dr. Ulrich Meißner for providing the solid model of the tilted-corrected tarantula thick filament 3D-map. We thank Dr. Michael Shatz (Image Science Software GmbH, Berlin, Germany) for his kind help on the implementation of the IMAGIC program. Electron microscopy was carried out in the Core Electron Microscopy Facility of the University of Massachusetts Medical School, supported in part by Diabetes Endocrinology Research Center grant DK32520. This work was supported in part by NIH grant R01GM62968, Alfred P. Sloan Foundation (BR-4297) and Human Frontier Science Program RGP0026/2003 (to W.W.); NIH grant AR34711 (to R.C.); FONACIT, Venezuela (to R.P.) and Howard Hughes Medical Institute (HHMI), U.S.A. (to R.P.).
Abbreviations
- aa
amino acid
- ATP
adenosine triphosphate
- C loop
cardio loop
- CM loop
cardiomyopathy loop
- ELC
essential light chain
- EM
electron microscopy
- EPR
electron paramagnetic resonance
- FSC
Fourier shell correlation
- FHC
familial hypertrophic cardiomyopathy
- HMM
heavy meromyosin
- I loop
interacting loop
- IHRSR
iterative helical real space reconstruction
- IVMA
in vitro motility assay
- LC-MS/MS
liquid chromatography mass spectrometry
- MD
molecular dynamics
- MLCK
myosin light chain kinase
- MLCP
myosin light chain phosphatase
- NTF
N-terminal fragment
- RLC
regulatory light chain
- S1
subfragment 1
- S2
subfragment 2
Footnotes
Accession numbers
The tarantula myosin regulatory light chain DNA sequence has been deposited at the GenBank (accession no. EU090070). The frozen-hydrated tarantula thick filament 3D-map has been deposited in the EM Data (EMDB) (ID. Code EMD-1535). The atomic coordinates of the tarantula myosin II interacting-heads have been deposited in the RCSB/PDB (ID. Code 3DTP).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Craig R, Padrón R. Molecular Structure of the Sarcomere. In: Engel AG, Franzini-Armstrong C, editors. Myology. McGraw-Hill, Inc; New York: 2004. pp. 129–166. [Google Scholar]
- 2.Lehman W, Szent-Gyorgyi AG. Regulation of muscular contraction. Distribution of actin control and myosin control in the animal kingdom. J Gen Physiol. 1975;66:1–30. doi: 10.1085/jgp.66.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vibert P, Craig R, Lehman W. Steric-model for activation of muscle thin filaments. J Mol Biol. 1997;266:8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 4.Poole KJ, Lorenz M, Evans G, Rosenbaum G, Pirani A, Craig R, Tobacman LS, Lehman W, Holmes KC. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J Struct Biol. 2006;155:273–284. doi: 10.1016/j.jsb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 5.Szent-Gyorgyi AG. Regulation by myosin: how calcium regulates some myosins, past and present. Adv Exp Med Biol. 2007;592:253–264. doi: 10.1007/978-4-431-38453-3_21. [DOI] [PubMed] [Google Scholar]
- 6.Sweeney HL, Bowman BF, Stull JT. Myosin light chain phosphorylation in vertebrate striated muscle: regulation and function. Am J Physiol. 1993;264:C1085–C1095. doi: 10.1152/ajpcell.1993.264.5.C1085. [DOI] [PubMed] [Google Scholar]
- 7.Sellers JR. Phosphorylation-dependent regulation of Limulus myosin. J Biol Chem. 1981;256:9274–9278. [PubMed] [Google Scholar]
- 8.Craig R, Padron R, Kendrick-Jones J. Structural changes accompanying phosphorylation of tarantula muscle myosin filaments. J Cell Biol. 1987;105:1319–1327. doi: 10.1083/jcb.105.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chacko S, Conti MA, Adelstein RS. Effect of phosphorylation of smooth muscle myosin on actin activation and Ca2+ regulation. Proc Natl Acad Sci U S A. 1977;74:129–133. doi: 10.1073/pnas.74.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 11.Sweeney HL, Houdusse A. Mammalian muscle myosin. In: Engel AG, Franzini-Armstrong C, editors. Myology. McGraw-Hill; New York: 2004. pp. 167–186. [Google Scholar]
- 12.Kwon H, Goodwin EB, Nyitray L, Berliner E, O’Neall-Hennessey E, Melandri FD, Szent-Gyorgyi AG. Isolation of the regulatory domain of scallop myosin: role of the essential light chain in calcium binding. Proc Natl Acad Sci U S A. 1990;87:4771–4775. doi: 10.1073/pnas.87.12.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson WD, Blakely SE, Nesmelov YE, Thomas DD. Site-directed spin labeling reveals a conformational switch in the phosphorylation domain of smooth muscle myosin. Proc Natl Acad Sci U S A. 2005;102:4000–4005. doi: 10.1073/pnas.0401664102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wendt T, Taylor D, Messier T, Trybus KM, Taylor KA. Visualization of head-head interactions in the inhibited state of smooth muscle myosin. J Cell Biol. 1999;147:1385–1390. doi: 10.1083/jcb.147.7.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci U S A. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Wendt T, Taylor D, Taylor K. Refined model of the 10S conformation of smooth muscle myosin by cryo-electron microscopy 3D image reconstruction. J Mol Biol. 2003;329:963–972. doi: 10.1016/s0022-2836(03)00516-3. [DOI] [PubMed] [Google Scholar]
- 17.Burgess SA, Yu S, Walker ML, Hawkins RJ, Chalovich JM, Knight PJ. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol. 2007;372:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 18.Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padron R. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 19.Jung HS, Burgess SA, Billington N, Colegrave M, Patel H, Chalovich JM, Chantler PD, Knight PJ. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc Natl Acad Sci U S A. 2008;105:6022–6026. doi: 10.1073/pnas.0707846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung HS, Komatsu S, Ikebe M, Craig R. Head-Head and Head-Tail Interaction: A General Mechanism for Switching Off Myosin II Activity in Cells. Mol Biol Cell. 2008;19:3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci U S A. 2008;105:2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao FQ, Woodhead JL, Craig R. Head-head interaction characterizes the relaxed state of scallop and Limulus muscle myosin filaments. Biophys J Supplement Abstract. 2008:630-Pos. doi: 10.1016/j.jmb.2008.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craig R, Woodhead JL. Structure and function of myosin filaments. Curr Opin Struct Biol. 2006;16:204–212. doi: 10.1016/j.sbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Sellers JR, Knight PJ. Folding and regulation in myosins II and V. J Musc Res Cell Motil. 2008;28:363–370. doi: 10.1007/s10974-008-9134-0. [DOI] [PubMed] [Google Scholar]
- 25.Levine RJ, Chantler PD, Kensler RW, Woodhead JL. Effects of phosphorylation by myosin light chain kinase on the structure of Limulus thick filaments. J Cell Biol. 1991;113:563–572. doi: 10.1083/jcb.113.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine RJ, Kensler RW, Yang Z, Stull JT, Sweeney HL. Myosin light chain phosphorylation affects the structure of rabbit skeletal muscle thick filaments. Biophys J. 1996;71:898–907. doi: 10.1016/S0006-3495(96)79293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espinoza-Fonseca LM, Kast D, Thomas DD. Molecular dynamics simulations reveal a disorder-to-order transition on phosphorylation of smooth muscle myosin. Biophys J. 2007;93:2083–2090. doi: 10.1529/biophysj.106.095802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikebe M, Morita J. Identification of the sequence of the regulatory light chain required for the phosphorylation-dependent regulation of actomyosin. J Biol Chem. 1991;266:21339–21342. [PubMed] [Google Scholar]
- 29.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 30.Burgess S, Walker M, Knight PJ, Sparrow J, Schmitz S, Offer G, Bullard B, Leonard K, Holt J, Trinick J. Structural studies of arthrin: monoubiquitinated actin. J Mol Biol. 2004;341:1161–1173. doi: 10.1016/j.jmb.2004.06.077. [DOI] [PubMed] [Google Scholar]
- 31.Pomfret AJ, Rice WJ, Stokes DL. Application of the iterative helical real-space reconstruction method to large membranous tubular crystals of P-type ATPases. J Struct Biol. 2007;157:106–116. doi: 10.1016/j.jsb.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maita T, Chen JI, Matsuda G. Amino-acid sequence of the 20 000-molecular-weight light chain of chicken gizzard-muscle myosin. Eur J Biochem. 1981;117:417–424. doi: 10.1111/j.1432-1033.1981.tb06354.x. [DOI] [PubMed] [Google Scholar]
- 33.Kemp BE, Pearson RB. Protein kinase recognition sequence motifs. Trends Biochem Sci. 1990;15:342–346. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- 34.Hidalgo C, Craig R, Ikebe M, Padron R. Mechanism of phosphorylation of the regulatory light chain of myosin from tarantula striated muscle. J Muscle Res Cell Motil. 2001;22:51–59. doi: 10.1023/a:1010388103354. [DOI] [PubMed] [Google Scholar]
- 35.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blankenfeldt W, Thoma NH, Wray JS, Gautel M, Schlichting I. Crystal structures of human cardiac beta-myosin II S2-Delta provide insight into the functional role of the S2 subfragment. Proc Natl Acad Sci U S A. 2006;103:17713–17717. doi: 10.1073/pnas.0606741103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wriggers W, Chacon P. Modeling tricks and fitting techniques for multiresolution structures. Structure. 2001;9:779–788. doi: 10.1016/s0969-2126(01)00648-7. [DOI] [PubMed] [Google Scholar]
- 38.Crowther RA, Padron R, Craig R. Arrangement of the heads of myosin in relaxed thick filaments from tarantula muscle. J Mol Biol. 1985;184:429–439. doi: 10.1016/0022-2836(85)90292-x. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Brown JH, Reshetnikova L, Blazsek A, Farkas L, Nyitray L, Cohen C. Visualization of an unstable coiled coil from the scallop myosin rod. Nature. 2003;424:341–345. doi: 10.1038/nature01801. [DOI] [PubMed] [Google Scholar]
- 40.Brown JH, Yang Y, Reshetnikova L, Gourinath S, Suveges D, Kardos J, Hobor F, Reutzel R, Nyitray L, Cohen C. An unstable head-rod junction may promote folding into the compact off-state conformation of regulated myosins. J Mol Biol. 2008;375:1434–1443. doi: 10.1016/j.jmb.2007.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zoghbi ME, Woodhead JL, Craig R, Padron R. Helical order in tarantula thick filaments requires the “closed” conformation of the myosin head. J Mol Biol. 2004;342:1223–1236. doi: 10.1016/j.jmb.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 42.Xu S, Offer G, Gu J, White HD, Yu LC. Temperature and ligand dependence of conformation and helical order in myosin filaments. Biochemistry. 2003;42:390–401. doi: 10.1021/bi026085t. [DOI] [PubMed] [Google Scholar]
- 43.Xu S, Gu J, White H, Offer G, Yu L. Structural effects on myosin of three ATPase inhibitors. Biophys J Supplement Abstract. 2005:610-P. [Google Scholar]
- 44.Xu S, White HD, Offer GW, Yu LC. Stabilisation of the helical order of myosin filaments by blebbistatin. Biophys J Supplement, Abstract. 2008:629-Pos. [Google Scholar]
- 45.Zhao FQ, Padron R, Craig R. Blebbistatin Stabilizes the Helical Order of Myosin Filaments by Promoting the Switch 2 Closed State. Biophys J. 2008;95:3322–3329. doi: 10.1529/biophysj.108.137067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhi G, Ryder JW, Huang J, Ding P, Chen Y, Zhao Y, Kamm KE, Stull JT. Myosin light chain kinase and myosin phosphorylation effect frequency-dependent potentiation of skeletal muscle contraction. Proc Natl Acad Sci U S A. 2005;102:17519–17524. doi: 10.1073/pnas.0506846102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, Seidman JG. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 48.Huxley HE, Brown W. The low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J Mol Biol. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- 49.McLachlan AD, Karn J. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature. 1982;299:226–231. doi: 10.1038/299226a0. [DOI] [PubMed] [Google Scholar]
- 50.Decker B, Kellermayer MS. Periodically arranged interactions within the myosin filament backbone revealed by mechanical unzipping. J Mol Biol. 2008;377:307–310. doi: 10.1016/j.jmb.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 51.Ikebe M. Regulation of the function of mammalian myosin and its conformational change. Biochem Biophys Res Commun. 2008;369:157–164. doi: 10.1016/j.bbrc.2008.01.057. [DOI] [PubMed] [Google Scholar]
- 52.Metzger JM, Greaser ML, Moss RL. Variations in cross-bridge attachment rate and tension with phosphorylation of myosin in mammalian skinned skeletal muscle fibers. Implications for twitch potentiation in intact muscle. J Gen Physiol. 1989;93:855–883. doi: 10.1085/jgp.93.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barany K, Barany M, Gillis JM, Kushmerick MJ. Phosphorylation-dephosphorylation of the 18,000-dalton light chain of myosin during the contraction-relaxation cycle of frog muscle. J Biol Chem. 1979;254:3617–3623. [PubMed] [Google Scholar]
- 54.Sweeney HL, Yang Z, Zhi G, Stull JT, Trybus KM. Charge replacement near the phosphorylatable serine of the myosin regulatory light chain mimics aspects of phosphorylation. Proc Natl Acad Sci U S A. 1994;91:1490–1494. doi: 10.1073/pnas.91.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kemp BE, Stull JT. Myosin light chain kinases. In: Kemp BE, editor. Peptides and Protein Phosphorylation. CRC; Boca Raton, FL: 1990. pp. 115–133. [Google Scholar]
- 56.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Egelman EH. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J Struct Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 58.Egelman EH. Single-particle reconstruction from EM images of helical filaments. Curr Opin Struct Biol. 2007;17:556–561. doi: 10.1016/j.sbi.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 60.Padron R, Alamo L, Guerrero JR, Granados M, Uman P, Craig R. Three-dimensional reconstruction of thick filaments from rapidly frozen, freeze-substituted tarantula muscle. J Struct Biol. 1995;115:250–257. doi: 10.1006/jsbi.1995.1049. [DOI] [PubMed] [Google Scholar]
- 61.Padron R, Alamo L, Murgich J, Craig R. Towards an atomic model of the thick filaments of muscle. J Mol Biol. 1998;275:35–41. doi: 10.1006/jmbi.1997.1448. [DOI] [PubMed] [Google Scholar]
- 62.Offer G, Knight PJ, Burgess SA, Alamo L, Padron R. A new model for the surface arrangement of myosin molecules in tarantula thick filaments. J Mol Biol. 2000;298:239–260. doi: 10.1006/jmbi.2000.3664. [DOI] [PubMed] [Google Scholar]
- 63.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 64.Gulick AM, Bauer CB, Thoden JB, Rayment I. X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain. Biochemistry. 1997;36:11619–11628. doi: 10.1021/bi9712596. [DOI] [PubMed] [Google Scholar]
- 65.Fisher AJ, Smith CA, Thoden JB, Smith R, Sutoh K, Holden HM, Rayment I. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP. BeFx and MgADP. AlF4-Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 66.Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bauer CB, Holden HM, Thoden JB, Smith R, Rayment I. X-ray structures of the apo and MgATP-bound states of Dictyostelium discoideum myosin motor domain. J Biol Chem. 2000;275:38494–38499. doi: 10.1074/jbc.M005585200. [DOI] [PubMed] [Google Scholar]
- 68.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 69.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 70.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 71.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 72.Karplus K, Karchin R, Barrett C, Tu S, Cline M, Diekhans M, Grate L, Casper J, Hughey R. What is the value added by human intervention in protein structure prediction? Proteins Suppl. 2001;5:86–91. doi: 10.1002/prot.10021. [DOI] [PubMed] [Google Scholar]
- 73.Geourjon C, Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 74.Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33:W72–W76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bystroff C, Thorsson V, Baker D. HMMSTR: a hidden Markov model for local sequence-structure correlations in proteins. J Mol Biol. 2000;301:173–190. doi: 10.1006/jmbi.2000.3837. [DOI] [PubMed] [Google Scholar]
- 76.Kneller DG, Cohen FE, Langridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J Mol Biol. 1990;214:171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- 77.Kelley LA, MacCallum RM, Sternberg MJ. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 78.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 79.Houdusse A, Kalabokis VN, Himmel D, Szent-Gyorgyi AG, Cohen C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell. 1999;97:459–470. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- 80.Chacon P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J Mol Biol. 2002;317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- 81.Wriggers W, Chacón P, Kovacs J, Tama T, Birmanns S. Topology representing Neural Networks Reconcile Biomolecular Shape, Structure, and Dynamics. Neurocomputing. 2004;56:365–379. [Google Scholar]
- 82.Darst SA, Opalka N, Chacon P, Polyakov A, Richter C, Zhang G, Wriggers W. Conformational flexibility of bacterial RNA polymerase. Proc Natl Acad Sci U S A. 2002;99:4296–4301. doi: 10.1073/pnas.052054099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- 84.Zhang L, Hermans J. Hydrophilicity of cavities in proteins. Proteins. 1996;24:433–438. doi: 10.1002/(SICI)1097-0134(199604)24:4<433::AID-PROT3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 85.Gerstein M, Krebs W. A database of macromolecular motions. Nucleic Acids Res. 1998;26:4280–4290. doi: 10.1093/nar/26.18.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 87.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang Y, Gourinath S, Kovacs M, Nyitray L, Reutzel R, Himmel DM, O’Neall-Hennessey E, Reshetnikova L, Szent-Gyorgyi AG, Brown JH, Cohen C. Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor. Structure. 2007;15:553–564. doi: 10.1016/j.str.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 89.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 90.de Jong AF, van Dyck D. Ultimate resolution and information in electron microscopy. II: The information limit of transmission electron microscopes. Ultramicroscopy. 1993;49:66–80. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary data associated with this article can be found, in the online version. The tarantula myosin regulatory light chain DNA sequence has been deposited at the GenBank (accession no. EU090070). The frozen-hydrated tarantula thick filament 3D-map has been deposited in the EM Data (EMDB) (ID. Code EMD-1535). The atomic coordinates of the tarantula myosin interacting-heads have been deposited in the RCSB/PDB (ID. Code 3DTP).