Abstract
Vibrios are natural inhabitants of aquatic environments and form symbiotic or pathogenic relationships with eukaryotic hosts. Recent studies reveal that the ability of vibrios to form biofilms – i.e. matrix-enclosed, surface-associated communities– depends upon specific structural genes (flagella, pili, and exopolysaccharide biosynthesis) and regulatory processes (two-component regulators, quorum sensing, and c-di-GMP signaling). In this review, we compare and contrast mechanisms and regulation of biofilm formation by Vibrio species, with a focus on Vibrio cholerae, Vibrio parahaemolyticus, Vibrio vulnificus, and Vibrio fischeri. While many aspects are the same, others differ dramatically. Critical questions that remain to be answered regarding the molecular underpinnings of Vibrio biofilm formation will also be discussed.
Vibrios and biofilms
Vibrio species are ubiquitous in aquatic ecosystems. While many Vibrio species are free living, a small group can form pathogenic or symbiotic interactions with eukaryotic hosts. Indeed, these Vibrio species alternate between growth within their hosts and prolonged survival in aquatic habitats. V. cholerae, for example, causes periodic occurrences of the severe diarrheal disease cholera. These epidemics typically result from consumption of drinking water contaminated with the pathogen. In between epidemics, V. cholerae survives within brackish water [1].
Like V. cholerae, the pathogens Vibrio parahaemolyticus and Vibrio vulnificus are most often delivered to human hosts through the consumption of contaminated water or food, particularly raw seafood. V. parahaemolyticus is responsible for the most common Vibrio-associated, seafood-borne gastroenteritis [2]. V. vulnificus causes gastroenteritis, severe wound infections and septicemia in susceptible hosts [3]. The symbiont Vibrio fischeri similarly alternates between free-living and host-associated lifestyles [4].
Adaptation of Vibrio species to changing parameters of the aquatic ecosystem as well as to those of their respective hosts is critical to their survival and colonization success. One key factor for environmental survival and transmission is the ability to form biofilms – i.e. matrix-enclosed, surface-associated communities. The biofilm mode of growth is the preferred lifestyle in the microbial world as it enhances growth and survival by providing access to nutrients and protection from predators and antimicrobial compounds (reviewed in Ref. [5]).
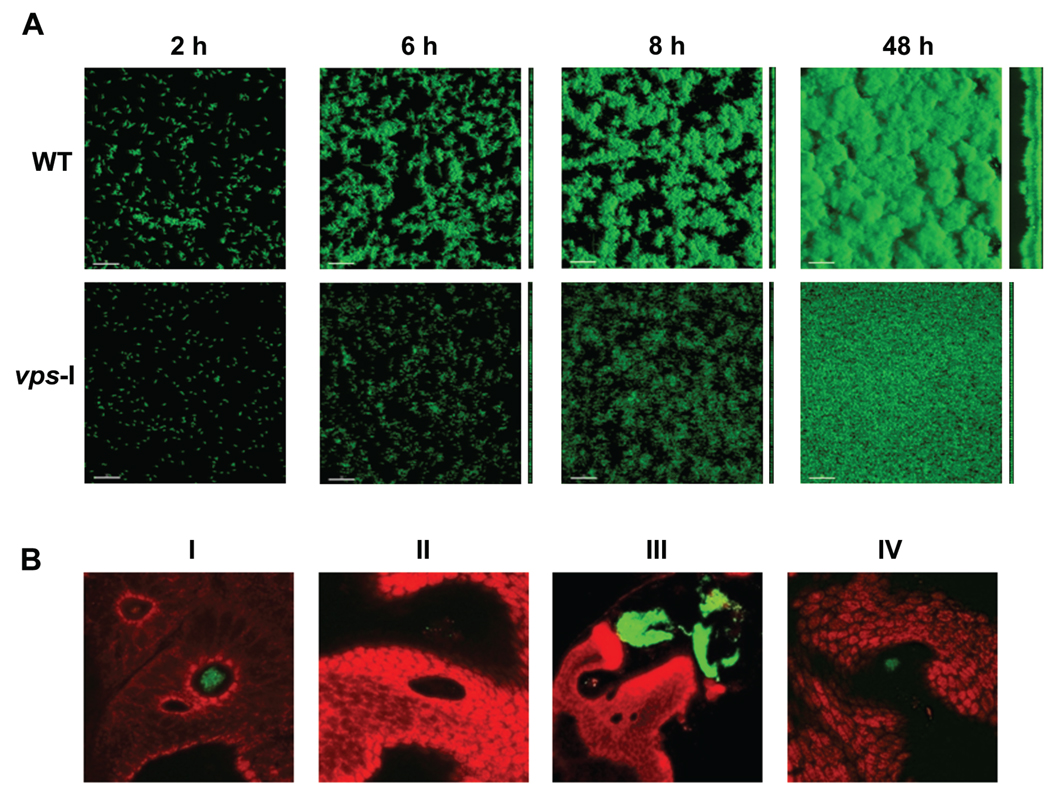
The biofilm forming capacity of V. cholerae is well documented, both in natural habitats and under laboratory conditions (Figure 1a) [1, 6, 7]. Stool samples from cholera patients, for example, contain not only planktonic V. cholerae, but also biofilm-like aggregates that are significantly more infectious [8]. Furthermore, removal from water of particles >20 µm in diameter can reduce cholera incidence by 48% [9]. Taken together, these studies highlight the importance of the biofilm growth mode in both the intestinal and aquatic phases of V. cholerae’s life cycle. Biofilm formation also plays a key role in host colonization by V. fischeri (Figure 1b) [10, 11]. It is likely to also be important for the ecology, transmission and/or virulence of V. vulnificus and V. parahaemolyticus, which are found on surfaces of plankton and colonize shellfish [2, 3], however, this area of research remains underdeveloped.
Figure 1. Biofilms of V. cholerae and V. fisheri.
(a)Three-dimensional biofilm structures of green fluorescent protein (GFP) tagged wild type V. cholerae and a vps-I cluster mutant (unable to produce VPS) formed after 2, 6, 8 and 48 h post-inoculation in once-through flow cell. Images were acquired with confocal laser scanning microscopy (CSLM) and top-down and side views of biofilms are shown. Scale bar indicates 30 µm. Data (but not images) are from Yildiz et al., 2008 [66].
(b) Biofilm-like aggregate formation on the light organ of squid. Newly hatched squid were inoculated with GFP tagged wild-type cells (I) or sypN polysaccharide mutant carrying vector control (II), and wild-type cells (III) or sypN mutant overexpressing the histidine kinase RscS (IV). Between 2–6 hour post inoculation, squids were stained with Cell Tracker Orange (red color) and aggregate formation by V. fisheri strains was analyzed by CSLM. Data (but not images) are from Yip et al., 2006 [11].
In recent years, numerous studies have investigated biofilm formation in Vibrio species. Many of these studies rely on colony morphology as an indicator of biofilm formation, including translucent (TR), opaque (OP) and rugose or wrinkled colonies; these and other methods for investigating Vibrio biofilms are described in Box 1. These studies have identified many key proteins, including those involved in the biosynthesis of flagella, pili, and polysaccharides, and in the regulators that control their expression, predominantly two-component and quorum sensing regulators and the small signaling molecule c-di-GMP. In this review, we will compare and contrast mechanisms and regulation of biofilm formation in Vibrio species, focusing on V. cholerae, V. parahaemolyticus, V. vulnificus, and V. fischeri as they are the species most intensively studied for biofilm formation.
Box 1. Experimental analysis of biofilm formation
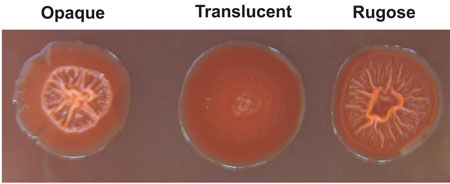
There is a correlation between biofilm matrix production and colony morphology. Cells able to produce exopolymers have corrugated (also termed wrinkled or rugose), or in some cases mucoid, colony morphologies. Thus, changes in colony morphology have been extensively utilized to identify biofilm matrix components. For example, V. parahaemolyticus undergoes a reversible phase variation between opaque (OP) and translucent (TR) colony types. OP colonies tend to result from increased polysaccharide production, and loss of the involved polysaccharide locus results in colonies that are TR [37, 51]. Besides OP and TR morphotypes, V. parahaemolyticus forms rugose colonies that exhibit increased CPS production compared to parental TR or OP strains [69]. V. vulnificus produces OP, TR and rugose colony morphotypes [33, 61, 72], while V. cholerae undergoes phase variation to switch between the smooth and rugose colony morphotypes [7].
Biofilms formed at solid-liquid interfaces have been analyzed under static or flow conditions [66]. For static culture conditions, microorganisms are grown in microtiter dishes or test tubes and the extent of biofilm formation is followed by staining of the surface-associated biofilm with crystal violet. Pellicle formation has also been used to analyze biofilms formed at air-liquid interfaces under static conditions. To evaluate biofilm architecture, biofilms that form in static cultures or in the ‘once-through’ flow cell reactor can be analyzed using confocal laser scanning microscopy (CSLM), usually utilizing strains that express fluorescent proteins.
Figure legend, Box 1
Representative colony phenotypes of V. parahaemolyticus. Colony phenotypes of opaque, translucent and rugose strains of V. parahaemolyticus grown on Congo red plates are shown.
Flagella are involved in initial stages of biofilm formation by Vibrio spp
Biofilm formation begins when a bacterium reaches and attaches to a surface. After the initial attachment, subsequent formation of microcolonies and three-dimensional (3-D) structures is mediated by movement and growth of attached bacteria. In many bacteria, flagella-mediated motility promotes the initial stages of biofilm formation, generally by enhancing movement towards and along the surface [12]. In vibrios, the impact of motility appears to extend beyond attachment.
In V. cholerae, loss of flagellar genes generally results in decreased attachment, although the contribution of the flagellum varies between strains [6, 13]. In V. cholerae O139, time lapse video microscopy [13] revealed that initial interactions between wild-type cells and a surface occurred rapidly: bacteria near the surface swam in circles, then exhibited more restricted movement. Some cells, tethered to the surface, alternated between short periods of jerky movement and no movement. The numbers of surface associated and immobilized bacteria increased over time, with subsequent formation of microcolonies and classical 3-D structures. A V. cholerae O139 flaA mutant (lacking the major flagellin subunit), however, did not undergo this pattern of biofilm formation. Instead, the mutant aggregated in liquid culture, then subsequently settled onto the surface as immobilized clusters of cells [13]. After this settling, the biofilms developed relatively normally, suggesting that the flaA mutant could form biofilms if it was allowed sufficient time.
The planktonic aggregation of flaA mutants was subsequently attributed to increased production of an exopolysaccharide termed VPS. Consistent with this result, these mutants formed rugose colonies. Surprisingly, any of several paralyzed (mot) mutants, which produce a flagellum but cannot rotate it, formed smooth colonies and poor biofilms [13, 14]. A flaA motX double mutant also produced smooth colonies, indicating that disruption of mot can eliminate the VPS-inducing signal caused by loss of the flagellum [14]. Thus, V. cholerae could use the flagellar motor not only to promote motility but also to transmit a signal indicating interaction with a surface [14].
Such a possibility would not be unprecedented: V. parahaemolyticus uses its motor to decide when to switch from a swimming cell (polarly flagellated) to a swarmer cell (with lateral flagella) capable of moving on surfaces or in highly viscous media [15]. V. parahaemolyticus also uses its polar flagellum to promote biofilm formation [16]. flgE and flgD (hook) mutants are defective in attachment, pellicle formation (forming ‘speckled’ pellicles), and biofilm formation [16]. The severity of the defect depended on the strain background. For example, whereas translucent (TR) fla mutants in submerged cultures could not adhere, opaque (OP) fla mutants could adhere, but could not form complex 3-D structures, even with prolonged incubation. These data suggest a role for the polar flagellum beyond biofilm initiation and/or in controlling other factors [16].
In V. vulnificus and V. fischeri, flagellum-mediated motility also promotes biofilm formation. For example, a V. vulnificus flgE (hook) mutant is defective for attachment both to polystyrene and to glass wool [17]. In V. fischeri, the flrC regulator is also required for biofilm formation [18]; prolonged incubation could overcome some, but not all of the defect.
Thus, motility plays a key role in biofilm formation in the vibrios, as has been seen for other biofilm models. Interestingly, the role of motility appears to extend beyond simply allowing the cell to reach the right location. Understanding “other” role(s) for motility and/or the motility apparatus during biofilm development remains an important area of investigation.
Pili promote cell-surface and/or cell-cell interactions
In V. cholerae, at least three types of pili contribute to biofilm formation: mannose-sensitive haemagglutinin type IV pili (MSHA), toxin co-regulated pili (TCP), and chitin-regulated pili (ChiRP) (formerly termed PilA) [19–22]. The relative importance of these pili varies under different conditions, and from strain to strain. MSHA, for example, is critical for early attachment to abiotic surfaces in V. cholerae O1 El Tor strains, yet initial studies revealed no role in strain O139 [13]. Subsequent work revealed a role for the O139 MSHA pilus structural gene mshA in monolayer formation, and demonstrated that the mshA mutant could bypass this stage by aggregating as planktonic cells and subsequently settling and forming 3-D biofilms [23]. Similarly, conflicting results for the importance of MSHA pili have been obtained for biofilm formation on various chitin substrates [19, 20, 22]. Thus, the contribution of MSHA to biofilm formation is impacted by both environmental and genetic factors.
The classic, virulence-associated pilus of V. cholerae, TCP, is involved in microcolony formation on an environmentally relevant substrate, chitin. A TCP pili mutant of El Tor N16961 formed monolayers, but not microcolonies, on a chitin substrate [21]. Recent work revealed that MSHA and TCP pili are inversely controlled at multiple levels [24], suggesting the possibility that the two pilus types sequentially promote monolayer and 3-D biofilm formation.
A role for ChiRP is less clear. ChiRP was required for competitive attachment to a chitin surface, crab shell, but largely unnecessary for individual attachment to chitin particles [20]. It is speculated that ChiRP might play a role other than adherence, such as orienting the cell optimally for chitin degradation [20].
V. parahaemolyticus also employs MSHA and ChiRP for biofilm formation. MSHA mutants form substantially reduced biofilms — a defect that could be overcome by prolonged incubation — and defective, speckled pellicles [16]. Like V. cholerae, strain background influenced the severity of the defect. ChiRP mutants fail to progress past monolayer formation [25]. In addition, both MSHA and ChiRP contribute to attachment to chitin particles [25].
In V. vulnificus, the type IV pilus structural protein PilA and, to a greater degree, the pre-pilin peptidase PilD, contribute to binding both to abiotic surfaces and to human epithelial cells [26, 27]. The difference in relative importance of the two genes could be attributed to the retention by the pilA mutant of other types of pili, and/or to the role of PilD in processing other secreted proteins [26, 27]. PilA and PilD are also necessary for prolonged attachment to oysters [28].
A role for pili in biofilm formation in V. fischeri has not been determined but is expected, given that the genome encodes eight putative pili loci, two of which contribute to efficient symbiotic colonization [29, 30].
In general, while similar pili are used by vibrios for attachment, it appears that the genetic context of the cell and the type of surface it encounters (and/or other clues from the environment) can substantially influence the relative importance and thus usage of a particular type of pili for attachment.
Polysaccharides are a major component of Vibrio biofilms
Production of mature biofilms requires extracellular matrix components that hold the cells together and keep the biofilm attached to the surface. The capsular polysaccharide (CPS) or exopolysaccharide (EPS, or VPS in V. cholerae) loci involved in biofilm formation have been identified from numerous Vibrio spp. Expression of these loci is frequently correlated with biofilm-associated changes in colony morphology, in particular OP, rugose, or wrinkled colonies (Box 1).
vps and vps-like loci
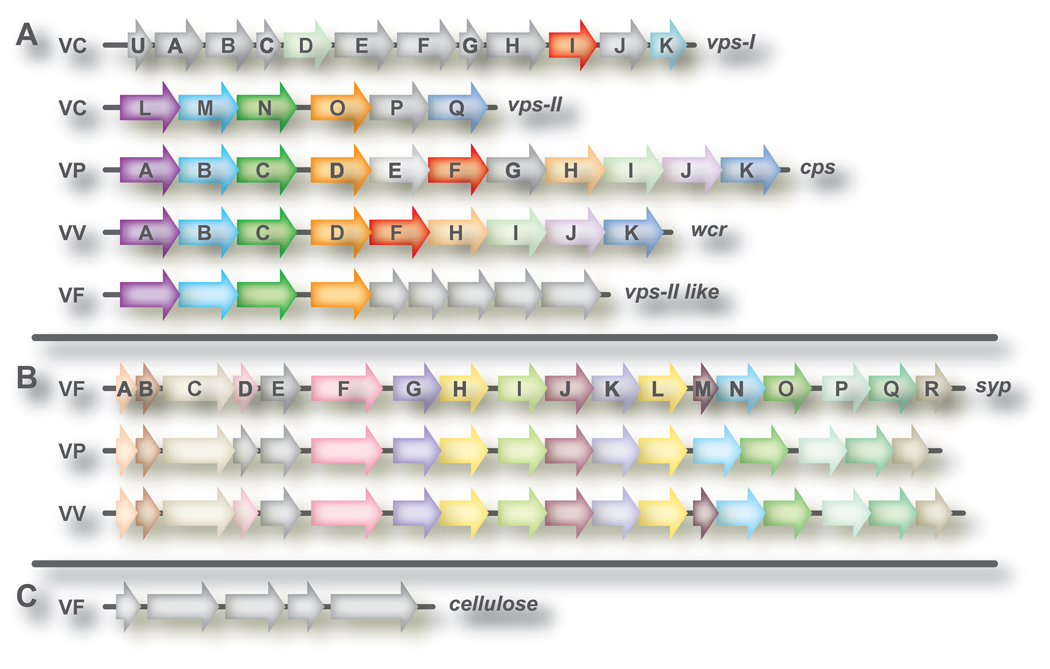
In V. cholerae O1 El Tor A1552, biofilm formation depends upon two linked loci, vps-I and vps-II (collectively termed vps), which encode structural proteins responsible for EPS production [7] (Figure 2a). vps-I and vps-II are separated by six genes (rbmA-F) that also are involved in biofilm formation but not polysaccharide production, with one exception: disruption of rbmB (a predicted VPS lyase) leads to an accumulation of polysaccharide [31]. VPS from V. cholerae O1 El Tor A1552 primarily contains glucose and galactose [7], with N-acetyl glucosamine, mannose and xylose representing minor constituents. This VPS is associated with rugose colony formation, matrix production, pellicles, 3-D biofilms, and resistance to chlorine [7, 32]. Loss of vps eliminates these phenotypes.
Figure 2. Polysaccharide loci in Vibrio spp.
The 3 major loci with established roles in biofilm formation in vibrio spp. are shown: (a) vps and vps-like, (b) syp and syp-like, and (c) cellulose. In V. cholerae, the large vps locus encompasses 2 sub-loci, vps-I and vps-II; the polysaccharide loci of other vibrios is more similar to vps-II than to vps-I and thus for clarity these sub-loci are separated. (a) V. cholerae (VC) vps locus (vps-I (VC0916-VC0927) and vps-II (VC0934-VC0938), V. parahaemolyticus (VP) cps locus, V. vulnificus (VV) wcr locus (VVA0395-VVA0387; VV21582-VV21574), and a vps-II-like locus in V. fischeri (VF) (VF0352-VF0344). (b) V. fischeri syp locus (VFA1020-VFA1037), and similar loci in V. parahaemolyticus (VP1476-1458) and V. vulnificus (VV12658-VV12674). (c). V. fischeri cellulose locus (VFA0885-VFA0881). Genes (not drawn to scale) are represented by arrows. Gray arrows represent genes that are dissimilar to others in the same panel, while those with the same color exhibit sequence similarity. Genes are named as labeled.
The vps locus is conserved, in part, in other Vibrios [29, 33, 34] (Figure 2a). No role in biofilm formation for the vps-like locus of V. fischeri has been observed to date [35]. Similarly, little is known about the V. vulnificus locus, wcr, other than that it is associated with the formation of both OP and rugose colonies [33, 36]. wcr most resembles the V. parahaemolyticus locus, cps, which produces a CPS (CPSA), rather than an EPS like V. cholerae [37]. cps is required for OP colony morphology and pellicle formation, as its loss disrupts these phenotypes [34]. OP colonies have increased cps gene expression and produce more CPSA [34, 37]. CPSA contains approximately equal amounts of fucose, galactose, glucose, and N-acetylglucosamine, making it distinct from V. cholerae EPS [16, 37]. Its production appears to require activated sulphur, as cysteine mutants fail to synthesize CPSA [16].
syp and syp-like loci
Biofilm formation in V. fischeri depends upon an 18 gene cluster of polysaccharide biosynthetic genes (syp) and associated regulators [11, 38]. This locus is lacking in V. cholerae, but is conserved in V. parahaemolyticus and V. vulnificus (Figure 2b). Induction of syp expression results in wrinkled colonies, pellicle formation and matrix production; loss of specific syp genes largely restores the wild-type phenotype [11, 18]. The Syp polysaccharide has not been purified, but appears to contain glucose or α-linked mannose [11]
Other polysaccharide loci
Biofilm formation by V. fischeri also depends upon a cellulose gene cluster similar to that found in Salmonella but absent in other vibrios [35] (Figure 2c). Numerous other polysaccharides and polysaccharide loci have been uncovered in Vibrio spp. (e.g. [39, 40]), although few have been investigated for roles in biofilm formation. Where investigated, a surprising number appear to have negative roles. For example, the group 1 CPS in V. vulnificus is associated with the production of OP colonies, but the loss of a representative gene increases attachment [41, 42]. V. cholerae O139 contains a locus with genes for CPS and lipopolysaccharide (LPS) O antigen biosynthesis that also plays a negative role in biofilm formation [43]. A third example is a putative O antigen CPS locus of V. parahaemolyticus, VP0214-VP0237, whose loss increases attachment [16]. A deeper understanding of the roles of these possible polysaccharide loci awaits further investigation.
The vibrio species studied to date produce distinct exopolysaccharides, and some if not all have the potential to produce multiple types of polysaccharides. Because biofilms gain their structural integrity largely from the exopolysaccharide matrix component, these studies indicate a diversity of biofilms are produced. It is possible that these biofilms thus may play an important role in niche selection.
Transcriptional control of exopolysaccharide production genes
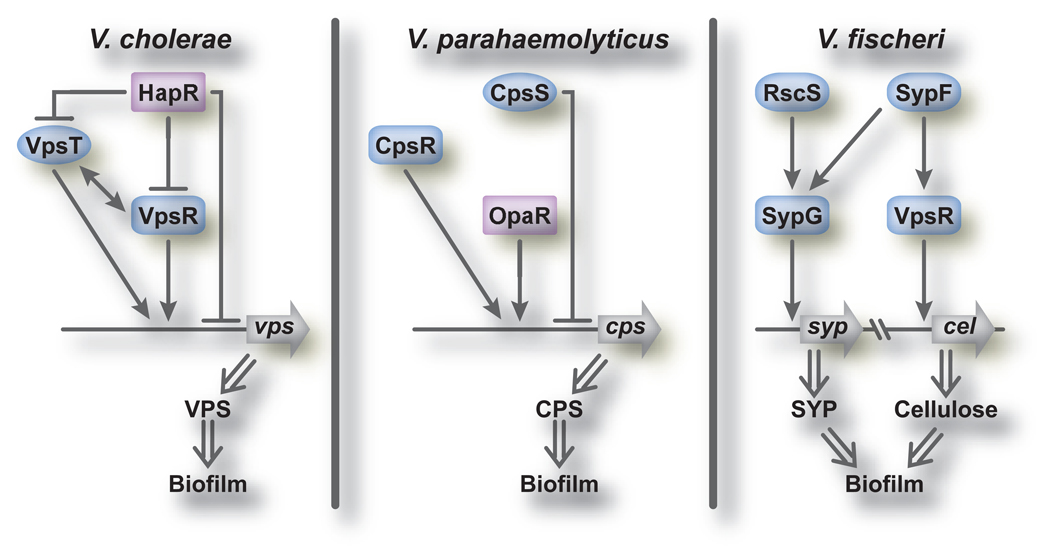
In Vibrio species, regulation of exopolysaccharide production and biofilm formation is complex, and involves numerous transcriptional regulators, particularly two-component signal transduction and quorum sensing regulators (Figure 3). In a typical two-component system, a stimulus detected by a sensor histidine kinase (HK) is transformed into a cellular signal by a phosphorelay event that involves autophosphorylation of the HK at a conserved histidine residue. The phosphoryl group is then passed to a response regulator (RR) at a conserved aspartate residue, leading to activation and altered gene expression or protein function. Quorum sensing involves sensing and responding to population density (Box 2). Global regulators, such as CRP [44, 45] and sigma factors RpoS and RpoN [32], play a role in biofilm formation in the vibrios, but they will not be discussed here.
Figure 3. Regulation of biofilm formation in Vibrio spp.
Biofilm formation in V. cholerae is positively regulated by the regulators VpsR and VpsT. The magnitude of transcriptional control of vps genes by VpsR is greater than that of VpsT. Expression of vps genes and the regulators VpsR and VpsT are negatively controlled by the HapR regulator. In V. parahaemolyticus expression of cps genes is negatively regulated by a homolog of VpsT, CpsS. In the absence of CpsS, OpaR and CpsR (HapR and VpsR homologs, respectively) positively control cps gene expression and biofilm formation. In V. fischeri transcription of syp genes and biofilm formation are positively controlled by the histidine kinases RscS and/or SypF which act through response regulator SypG. SypF also positively regulates production of cellulose (cel), acting though the VpsR homolog of V. fischeri. The roles of VpsR and SypE as inhibitors are poorly understood and thus these are omitted from this figure.
Box 2. Quorum sensing
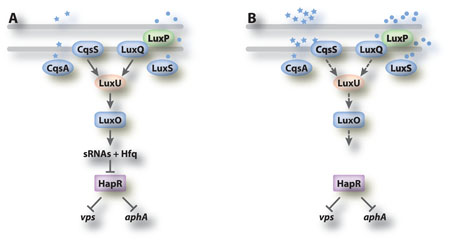
Quorum sensing is a mechanism that cells use to determine their population density. Signal synthases produce a diffusible molecule (autoinducer (AI)) that can accumulate in the extracellular environment. When the cell population is sufficiently high, and enough signal accumulated, this molecule binds to and activates cellular receptors. In Vibrio species, one consequence of this signaling is a phosphorelay that ultimately controls the synthesis of a global regulator and phenotypes that are more useful to a group of cells, such as luminescence, rather than to individuals [50]. Since biofilm formation is a behavior that depends upon a group of cells, it makes sense for bacteria to rely on quorum sensing regulators to control biofilm formation, and indeed they do. In V. cholerae, however, the sense of this control is not as might be expected: V. cholerae mutants, which are ‘locked’ into a regulatory state mimicking high cell density, are impaired in biofilm formation [54].
Legend, Box 2 figure:
V. cholerae quorum sensing. V. cholerae synthesizes two autoinducers, CAI-1 (stars) made by CqsA and AI-2 (circles) made by LuxS, that are detected by sensor kinases CqsS and LuxQ (working with periplasmic protein LuxP), respectively, which control a phosphorelay. Low (panel A) and high (panel B) levels of autoinducers alter the signal transduction cascade as described in the text of box 2, ultimately impacting biofillm formation and virulence gene expression.
V. cholerae produces two AIs [50]. CAI-1 ((S)-3-hydroxytridecan-4-one) [73] is synthesized by CqsA and detected by CqsS. AI-2 (the furanosyl borate diester (2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran borate) is synthesized by LuxS and detected by the LuxQP receptor [50]. Information from the sensors is transduced through a phosphorelay, first to the LuxU phosphotransfer protein and then to LuxO, which is an RpoN dependent response regulator. At low cell density, when the concentrations of AIs are low, LuxO is phosphorylated and activates expression of a set of small regulatory RNAs (figure, panel A) [74]. The result is inhibition of expression of the major quorum sensing regulator, HapR. At high cell density, LuxO is dephosphorylated and expression of HapR is increased. Under these conditions, the vps genes and virulence-associated genes (aphA) are repressed (figure, panel B).
This regulation appears to be both direct and indirect as the empirically-defined HapR binding motif was found in the promoter regions of both the vps locus and its regulator vpsT [32]. Mutants in cqsA that cannot produce the CAI-1 signal, a condition that should mimic low cell density, form thicker biofilms [55]. This suggests that CAI-1 signals are critical for repression of biofilm formation via the quorum sensing regulatory circuitry. It is thought that quorum sensing ensures development of ‘normal’ biofilm structures that permit rapid dispersion of bacteria from the biofilm, thereby facilitating transmission.
Two-component regulators
VpsR and VpsR-like regulators
In V. cholerae, the RR VpsR is a key regulator of biofilm formation. VpsR promotes transcription of the vps genes and formation of typical 3-D biofilm structures [32, 46]. Mutations that alter the residue predicted to be phosphorylated (D59) either inactivate this protein (D59A) or increase its activity (D59E) [14], thus supporting that phosphorylation on this residue is required for activation. However, the HK responsible for phosphorylation of VpsR has not been identified, as it is not physically linked to vpsR. Identifying the cognate HK will facilitate understanding the regulatory network controlling vps gene expression and how environmental signals regulate VPS production.
Under specific genetic conditions, V. parahaemolyticus also produces high levels of CPS and thus rugose colonies. Disruption of the vpsR homolog, cpsR, yields smooth colonies and decreases transcriptional activity of cpsA fused to a lacZ reporter in the rugose background [34]. However, CpsR is not required for basal levels of cps expression in TR or OP strains, as disruption of cpsR in these strains had no effect on cps transcription [34]. This contrasts with the role of VpsR, which is essential for transcription of vps genes in all forms of V. cholerae. It remains to be determined whether CpsR directly regulates transcription of cps genes.
V. fischeri also encodes a VpsR homolog. Disruption of vpsR leads to formation of mucoid colonies, which are indicative of enhanced polysaccharide production, suggesting that VpsR might be a repressor [35]. When overexpressed, however, VpsR induces biofilm formation, indicating that it could also be an activator. Surprisingly, VpsR-induced biofilms depended not on a vps-like locus but on a putative cellulose biosynthesis cluster not found in other characterized vibrios (Figure 2C) [35]. Thus VpsR appears to have a novel role in V. fischeri.
VpsT and VpsT-like
In V. cholerae, a second positive regulator is VpsT, a member of the UhpA (FixJ) family of transcriptional regulators [47]. VpsT shows homology to CsgD, which positively controls curli and cellulose production in Salmonella enterica. Disruption of vpsT yields smooth colonies and reduces vps expression, and biofilm formation [47]. Although the putative phosphorylation site in the receiver domain of VpsT is conserved, it is not necessary for the in vivo function of VpsT (J. Meir and F. Yildiz, unpublished). Thus, whether VpsT functions as a canonical RR remains unclear.
Characterization of vpsT and vpsR mutants in V. cholerae revealed different roles for these regulators in determining biofilm architecture. The vpsT mutant is still able to form biofilms (albeit distinct from its rugose parent), whereas vpsR and vpsT vpsR mutants produce only single cells or small microcolonies attached to the substratum [48]. The VpsR and VpsT regulons are largely identical, although VpsR exerts a larger impact on expression [48]. Thus, VpsR is essential for VPS production and biofilm formation, while VpsT appears to play an accessory role, possibly by increasing the level or activity of VpsR. Whether VpsT and VpsR serve as direct regulators of vps remains unknown.
A VpsT homolog, CpsS, also controls biofilm formation in V. parahaemolyticus, but as a negative regulator [34]. Introduction of a cpsS mutation into TR or OP strains derepresses cpsA transcription, resulting in rugose colonies. In TR strains, derepression is mediated through CpsR, while in OP strains, CpsR plays an accessory role [34]. Thus, V. cholerae and V. parahaemolyticus use similar proteins, but they function in the opposite sense and to different degrees: CpsS is the dominant negative regulator in V. parahaemolyticus whereas VpsT is a positive co-regulator in V. cholerae.
Other two-component regulators
In V. vulnificus, the RR NtrC contributes to biofilm formation through its transcriptional control of gmhD [49]. The GmhD protein, an ADP-|-glycero-D-manno-heptose-6-epimerase, is required for LPS and EPS production and biofilm formation [49]. The relative roles of LPS and EPS in biofilm formation, as well as the identity of the EPS involved in this process, have yet to be determined.
In V. fischeri, two-component regulators play critical roles in the control of biofilm formation, primarily through their activation of the syp polysaccharide locus. For example, the orphan HK RscS, when overexpressed, induces syp transcription, wrinkled colony and pellicle formation, and symbiotic biofilm formation [11]. RscS acts upstream of SypG, a syp-encoded RR that is proposed to be the direct activator of syp transcription [18].
Surprisingly, overexpression of SypG induced syp transcription but not the formation of wrinkled colonies or strong pellicles. However, overexpression of SypG in a sypE mutant permitted wrinkled colony and pellicle formation [18]. Thus, SypE, a putative RR that is not predicted to bind DNA, appears to play an inhibitory role. RscS might therefore both activate SypG and inactivate SypE. Homologs of SypG are present in V. vulnificus and V. parahaemolyticus, but RscS and SypE are unique to V. fischeri.
Overexpression of a third syp-encoded regulator, SypF, also induces biofilm formation in V. fischeri [35]. This regulator appears to function upstream of both SypG and VpsR: mutation of both regulators was necessary to eliminate all the biofilm phenotypes induced by SypF overexpression. The SypF-VpsR path appears to promote cell-surface attachment, while the SypF-SypG path appears responsible for cell-cell attachment [35].
Quorum sensing regulators
Vibrio species use quorum sensing (Box 2) to control numerous traits, including luminescence, virulence, and biofilm formation. Best studied in Vibrio harveyi, where the endpoint regulator is termed LuxR (reviewed in Ref. [50]), similar pathway components are found in all other vibrios studied to date.
The V. parahaemolyticus LuxR homolog, OpaR, positively regulates opacity, cps gene expression and biofilm formation [51]. Disruption of opaR in an OP strain yielded TR colonies, and overexpression of opaR in a TR strain increased cps expression and colony opacity [37]. A similar phenomenon is seen in both V. vulnificus and V. fischeri: mutants defective for the LuxR homologs, SmcR [52] and LitR [53], respectively, form TR colonies instead of the parental OP colonies; in the case of V. vulnificus, this disruption is associated with decreased biofilm formation. However, the molecular mechanisms causing decreases in colony opacity and biofilm formation and their connections to CPS or EPS production by V. vulnificus and V. fischeri are unknown.
In V. cholerae, the story is different (Figure 3). Mutants of the LuxR homolog, HapR, exhibit increased rugosity and increased vps expression [32, 54, 55]. These data indicate that HapR is a negative regulator of biofilm formation. HapR can directly bind DNA and repress expression of vpsT [56]. It can also control vpsR expression in some strains [32]. Thus, quorum sensing control of cell surface properties and biofilm formation is opposite in V. cholerae relative to the other vibrios. The ecological importance of this regulation is yet to be determined.
C-di-GMP signaling and biofilm formation
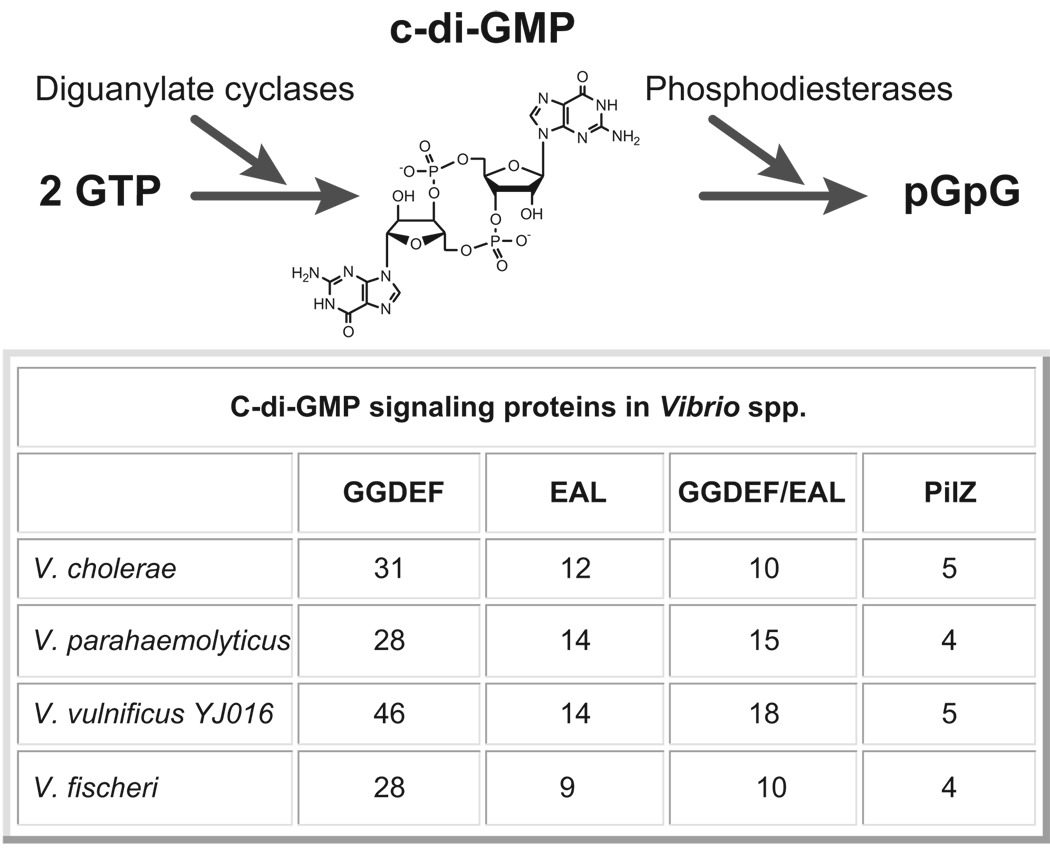
C-di-GMP is a ubiquitous second messenger that controls the transition from a free-living, motile lifestyle to a biofilm lifestyle in many bacteria (reviewed in Ref. [57]), including vibrios [58–62]. Increased c-di-GMP levels tend to promote biofilm formation and/or inhibit flagellar motility. C-di-GMP production and degradation is controlled by diguanylate cyclases (DGCs) and phosphodiesterases (PDEs), respectively [57]. Overexpression of these regulators tends to cause global effects. Intriguingly, the vibrios contain much larger numbers of DGCs and PDEs than other bacteria [63]. The abundance of enzymes controlling synthesis and degradation of c-di-GMP in vibrios indicates the importance of c-di-GMP signaling to the biology of vibrios. Since different types of sensory domains are found in proteins predicted to function as DGCs or PDEs [63], one possibility is that cells adjust their c-di-GMP levels in response to environmental and intracellular signals and that c-di-GMP signaling plays an important role in adaptation of vibrios to different environments.
In V. cholerae, c-di-GMP increases biofilm formation by stimulating transcription of vps genes and the positive transcriptional regulators vpsR and vpsT [60, 64]. Mutants of the known or putative PDE genes, mbaA, rocS, cdgC, cdpA, and vieA, exhibit enhanced biofilm formation, presumably due to increased c-di-GMP levels [65, 66].
Recently, c-di-GMP also has been linked to the natural capacity of V. cholerae to generate rugose variants. For example, the prototype rugose strain A1552 expresses elevated c-di-GMP levels caused by a single amino acid change in a DGC protein, VpvC, relative to the smooth variant [67]. Disruption of vpvC in this rugose variant reduces overall c-di-GMP levels and causes cells to become similar to the smooth variant with respect to biofilm formation and vps transcription. Rugosity can also be generated by deletion of the master quorum sensing regulator hapR. This effect occurs through CdgA, a DGC whose mRNA abundance is increased in the hapR mutant; this increased CdgA presumably increases cellular c-di-GMP. Deletion of cdgA decreased vps transcription and restored smooth colony formation to the rugose hapR mutant [48]. Subsequent studies revealed that HapR serves as a direct regulator of cdgA [56].
Increased c-di-GMP level leads to a decrease in motility. In V. cholerae, as expected mutations in DGC genes cdgD [60], cdgH [68] and vpvC [67] lead to an increase, while mutations in PDE genes vieA, rocS, cdgC and mbaA lead to a decrease in motility relative to wild-type, when tested on LB soft agar motility plates [66].
In V. parahaemolyticus, increases in cellular c-di-GMP levels prevent swarming motility and promote biofilm formation. Two genes involved in c-di-GMP control, scrG and scrC, have been extensively characterized [58, 59, 69]. ScrG functions as a PDE: null mutants increase c-di-GMP and exhibit increased cps and decreased lateral flagellar gene (laf) expression and thus, enhanced biofilm formation and reduced swarming motility [59]. Null mutants defective for the scrABC operon behave similarly, while overexpression of scrABC yields the opposite results [58]. Interestingly, however, overexpression of scrC in the absence of scrAB (encoding putative pyridoxal-phosphate-dependent and extracellular solute-binding proteins, respectively) induces cps, not laf, expression [58]. Subsequent work revealed that ScrC is a bifunctional protein that functions as a DGC to synthesize c-di-GMP, but when co-produced with ScrAB, functions as a PDE and degrades c-di-GMP. Epistasis analysis indicates that ScrG and ScrABC act in the same regulatory circuitry and that scrG and scrABC double mutants show a cumulative effect at the level of laf and cps gene expression [58].
Relatively little is known about c-di-GMP and biofilm formation in V. vulnificus and V. fischeri. In V. vulnificus, expression of the DGC DcpA converted translucent colonies of an acapsular mutant into OP colonies, but did not impact motility [61]. Overexpression of dcpA induced production of an EPS that was structurally distinct from the CPS, rugose colony formation and biofilm formation [61]. In V. fischeri, overexpression of the putative DGC MifA promotes cellulose biosynthesis and biofilm formation, suggesting that c-di-GMP is a player in biofilm formation in this microbe as well [70].
How are c-di-GMP levels sensed by the cell? One protein domain that binds c-di-GMP is the PilZ domain. Of the five PilZ domain proteins in V. cholerae, two of these, PlzC and PlzD, have been recently shown to bind c-di-GMP and are known to regulate biofilm formation and/or motility [71]. Thus, PilZ domain proteins can function as c-diGMP receptors and regulate c-di-GMP-dependent processes in V. cholerae and likely in other vibrios.
It is becoming clear that although Vibrios share common regulatory proteins and signaling systems, the biofilm regulatory circuitry is unique to each vibrio spp. Differences in regulation might reflect the importance of the biofilm life style to each vibrio spp during their in vivo and ex vivo life cycles, differences in niche occupation, differences in environmental parameters they respond to, and/or parameters driving evolution of the pathogens and symbionts.
Concluding remarks
Biofilm formation, particularly on a biotic, possibly nutritional surface, seems likely to provide a substantial survival advantage to aquatic organisms such as Vibrio species. That these organisms use similar traits and regulators to solve the problem of biofilm formation is not unexpected. That they use such diversity in approaches—the relative importance of the traits and regulators, and even the sense (positive or negative) of control—is surprising and thus has the potential to provide great insights into the peculiar lifestyles of these microbes. Some outstanding questions are listed in Box 3. Because biofilm formation is also part of the pathogenic lifestyles of Vibrio spp, elucidation of the molecular mechanisms and regulation of biofilm formation will provide the foundation for developing novel treatments and prevention strategies against Vibrio-associated illnesses.
Box 3. Questions for future research
What combination(s) of polysaccharides are being produced under various laboratory and environmental conditions?
What are the constituents (protein or DNA) of biofilm matrices under laboratory, environmental, and/or disease conditions?
What other structural and regulatory factors are involved in biofilm formation?
What are the environmental conditions that promote formation and dissolution of biofilms?
What are the stimuli sensed by two-component systems regulating biofilms formation?
What is the mechanism of c-di-GMP signaling and how is it connected to the regulatory network controlling biofilm formation?
Do differences in regulation of biofilm formation reflect the importance of the biofilm lifestyle to each Vibrio spp. during their in vivo and ex vivo life cycles?
Figure 4. C-di-GMP signaling proteins in Vibrio spp.
Cyclic di-guanosine-monophosphate (c-di-GMP) controls cell surface structures and biofilm formation in a diverse group of microorganisms. C-di-GMP is created from GTP (guanosine-5’-triphosphate) by diguanylate cyclase proteins that bear a GGDEF amino acid motif and degraded to the dinucleotide pGpG by phosphodiesterase proteins with EAL domains. C-di-GMP can be sensed by proteins with a PilZ domain. Numbers of genes encoding GGDEF, EAL, dual GGDEF/EAL or PilZ domain proteins in different Vibrio species are shown.
Acknowledgements
We thank Emily Yip and Cindy DeLoney-Marino for photos of V. fischeri aggregates, Linda McCarter for Vibrio parahaemolyticus pictures, and Kivanc Bilecen, Sinem Beyhan, and Ates Gurcan for figure preparation. We also thank members of our labs, Karen Ottemann, and Alan Wolfe for critical reading of the manuscript. Work in our laboratories investigating biofilm formation in Vibrio species was supported by NIH grants AI055987 to FHY and GM59690 to KLV.
References
- 1.Faruque SM, et al. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nair GB, et al. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin Microbiol Rev. 2007;20:39–48. doi: 10.1128/CMR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gulig PA, et al. Molecular Pathogenesis of Vibrio vulnificus. J Microbiol. 2005;43(Spec No):118–131. [PubMed] [Google Scholar]
- 4.Visick KL, Ruby EG. Vibrio fischeri and its host: it takes two to tango. Curr Opin Microbiol. 2006;9:632–638. doi: 10.1016/j.mib.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watnick PI, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci U S A. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque SM, et al. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc Natl Acad Sci U S A. 2006;103:6350–6355. doi: 10.1073/pnas.0601277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colwell RR, et al. Reduction of cholera in Bangladeshi villages by simple filtration. Proc Natl Acad Sci U S A. 2003;100:1051–1055. doi: 10.1073/pnas.0237386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nyholm SV, et al. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yip ES, et al. The symbiosis regulator rscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol. 2006;62:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Toole G, et al. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 13.Watnick PI, et al. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol Microbiol. 2001;39:223–235. doi: 10.1046/j.1365-2958.2001.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauriano CM, et al. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J Bacteriol. 2004;186:4864–4874. doi: 10.1128/JB.186.15.4864-4874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawagishi I, et al. The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol Microbiol. 1996;20:693–699. doi: 10.1111/j.1365-2958.1996.tb02509.x. [DOI] [PubMed] [Google Scholar]
- 16.Enos-Berlage JL, et al. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol Microbiol. 2005;55:1160–1182. doi: 10.1111/j.1365-2958.2004.04453.x. [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, et al. Role of flagellum and motility in pathogenesis of Vibrio vulnificus. Infect Immun. 2004;72:4905–4910. doi: 10.1128/IAI.72.8.4905-4910.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussa EA, et al. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J Bacteriol. 2008;190:4576–4583. doi: 10.1128/JB.00130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiavelli DA, et al. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl Environ Microbiol. 2001;67:3220–3225. doi: 10.1128/AEM.67.7.3220-3225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meibom KL, et al. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci U S A. 2004;101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reguera G, Kolter R. Virulence and the environment: a novel role for Vibrio cholerae toxin-coregulated pili in biofilm formation on chitin. J Bacteriol. 2005;187:3551–3555. doi: 10.1128/JB.187.10.3551-3555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watnick PI, et al. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moorthy S, Watnick PI. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol Microbiol. 2004;52:573–587. doi: 10.1111/j.1365-2958.2004.04000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsiao A, et al. Post-transcriptional cross-talk between pro-and anti-colonization pili biosynthesis systems in Vibrio cholerae. Mol Microbiol. 2008;67:849–860. doi: 10.1111/j.1365-2958.2007.06091.x. [DOI] [PubMed] [Google Scholar]
- 25.Shime-Hattori A, et al. Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiol Lett. 2006;264:89–97. doi: 10.1111/j.1574-6968.2006.00438.x. [DOI] [PubMed] [Google Scholar]
- 26.Paranjpye RN, et al. The type IV leader peptidase/N-methyltransferase of Vibrio vulnificus controls factors required for adherence to HEp-2 cells and virulence in iron-overloaded mice. Infect Immun. 1998;66:5659–5668. doi: 10.1128/iai.66.12.5659-5668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paranjpye RN, Strom MS. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect Immun. 2005;73:1411–1422. doi: 10.1128/IAI.73.3.1411-1422.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paranjpye RN, et al. Role of type IV pilins in persistence of Vibrio vulnificus in Crassostrea virginica oysters. Appl Environ Microbiol. 2007;73:5041–5044. doi: 10.1128/AEM.00641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruby EG, et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci U S A. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stabb EV, Ruby EG. Contribution of pilA to competitive colonization of the squid Euprymna scolopes by Vibrio fischeri. Appl Environ Microbiol. 2003;69:820–826. doi: 10.1128/AEM.69.2.820-826.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fong JC, Yildiz FH. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol. 2007;189:2319–2330. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yildiz FH, et al. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol. 2004;53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 33.Grau BL, et al. Further characterization of Vibrio vulnificus rugose variants and identification of a capsular and rugose exopolysaccharide gene cluster. Infect Immun. 2008;76:1485–1497. doi: 10.1128/IAI.01289-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guvener ZT, McCarter LL. Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus. J Bacteriol. 2003;185:5431–5441. doi: 10.1128/JB.185.18.5431-5441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darnell CL, et al. The putative hybrid sensor kinase SypF coordinates biofilm formation in Vibrio fischeri by acting upstream of two response regulators, SypG and VpsR. J Bacteriol. 2008;190:4941–4950. doi: 10.1128/JB.00197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith AB, Siebeling RJ. Identification of genetic loci required for capsular expression in Vibrio vulnificus. Infect Immun. 2003;71:1091–1097. doi: 10.1128/IAI.71.3.1091-1097.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enos-Berlage JL, McCarter LL. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J Bacteriol. 2000;182:5513–5520. doi: 10.1128/jb.182.19.5513-5520.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yip ES, et al. A novel, conserved cluster of genes promotes symbiotic colonization and sigma-dependent biofilm formation by Vibrio fischeri. Mol Microbiol. 2005;57:1485–1498. doi: 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- 39.Bush CA, et al. Classification of Vibrio vulnificus strains by the carbohydrate composition of their capsular polysaccharides. Anal Biochem. 1997;250:186–195. doi: 10.1006/abio.1997.2219. [DOI] [PubMed] [Google Scholar]
- 40.Nakhamchik A, et al. Identification of a Wzy polymerase required for group IV capsular polysaccharide and lipopolysaccharide biosynthesis in Vibrio vulnificus. Infect Immun. 2007;75:5550–5558. doi: 10.1128/IAI.00932-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chatzidaki-Livanis M, et al. Genetic variation in the Vibrio vulnificus group 1 capsular polysaccharide operon. J Bacteriol. 2006;188:1987–1998. doi: 10.1128/JB.188.5.1987-1998.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joseph LA, Wright AC. Expression of Vibrio vulnificus capsular polysaccharide inhibits biofilm formation. J Bacteriol. 2004;186:889–893. doi: 10.1128/JB.186.3.889-893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kierek K, Watnick PI. The Vibrio cholerae O139 O-antigen polysaccharide is essential for Ca2+dependent biofilm development in sea water. Proc Natl Acad Sci U S A. 2003;100:14357–14362. doi: 10.1073/pnas.2334614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fong JC, Yildiz FH. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J Bacteriol. 2008;190:6646–6659. doi: 10.1128/JB.00466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang W, et al. The cyclic AMP receptor protein modulates colonial morphology in Vibrio cholerae. Appl Environ Microbiol. 2007;73:7482–7487. doi: 10.1128/AEM.01564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yildiz FH, et al. VpsR, a Member of the Response Regulators of the Two-Component Regulatory Systems, Is Required for Expression of vps Biosynthesis Genes and EPS(ETr)-Associated Phenotypes in Vibrio cholerae O1 El Tor. J Bacteriol. 2001;183:1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casper-Lindley C, Yildiz FH. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J Bacteriol. 2004;186:1574–1578. doi: 10.1128/JB.186.5.1574-1578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beyhan S, et al. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J Bacteriol. 2007;189:388–402. doi: 10.1128/JB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim HS, et al. Role of NtrC in biofilm formation via controlling expression of the gene encoding an ADP-glycero-manno-heptose-6-epimerase in the pathogenic bacterium, Vibrio vulnificus. Mol Microbiol. 2007;63:559–574. doi: 10.1111/j.1365-2958.2006.05527.x. [DOI] [PubMed] [Google Scholar]
- 50.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 51.McCarter LL. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J Bacteriol. 1998;180:3166–3173. doi: 10.1128/jb.180.12.3166-3173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JH, et al. Identification and functional analysis of Vibrio vulnificus SmcR, a novel global regulator. J Microbiol Biotechnol. 2007;17:325–334. [PubMed] [Google Scholar]
- 53.Fidopiastis PM, et al. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol Microbiol. 2002;45:131–143. doi: 10.1046/j.1365-2958.2002.02996.x. [DOI] [PubMed] [Google Scholar]
- 54.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 55.Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell. 2003;5:647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- 56.Waters CM, et al. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol. 2008;190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Romling U, Amikam D. Cyclic di-GMP as a second messenger. Curr Opin Microbiol. 2006;9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 58.Ferreira RB, et al. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J Bacteriol. 2008;190:851–860. doi: 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim YK, McCarter LL. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J Bacteriol. 2007;189:4094–4107. doi: 10.1128/JB.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lim B, et al. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol Microbiol. 2006;60:331–348. doi: 10.1111/j.1365-2958.2006.05106.x. [DOI] [PubMed] [Google Scholar]
- 61.Nakhamchik A, et al. Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus. Appl Environ Microbiol. 2008;74:4199–4209. doi: 10.1128/AEM.00176-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galperin MY, et al. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 64.Beyhan S, et al. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J Bacteriol. 2006;188:3600–3613. doi: 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamayo R, et al. Role of cyclic Di-GMP during El tor biotype Vibrio cholerae infection: characterization of the in vivo-induced cyclic Di-GMP phosphodiesterase CdpA. Infect Immun. 2008;76:1617–1627. doi: 10.1128/IAI.01337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yildiz FH, Kolter R. Genetics and Microbiology of Biofilm Formation by Vibrio cholerae. Caister Academic Press; 2008. pp. 123–139. [Google Scholar]
- 67.Beyhan S, Yildiz FH. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol Microbiol. 2007;63:995–1007. doi: 10.1111/j.1365-2958.2006.05568.x. [DOI] [PubMed] [Google Scholar]
- 68.Beyhan S, et al. Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J Bacteriol. 2008;190:7392–7405. doi: 10.1128/JB.00564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boles BR, McCarter LL. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J Bacteriol. 2002;184:5946–5954. doi: 10.1128/JB.184.21.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O’Shea TM, et al. Diguanylate cyclases control magnesium-dependent motility of Vibrio fischeri. J Bacteriol. 2006;188:8196–8205. doi: 10.1128/JB.00728-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pratt JT, et al. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem. 2007;282:12860–12870. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grau BL, et al. High-frequency phase variation of Vibrio vulnificus 1003: isolation and characterization of a rugose phenotypic variant. J Bacteriol. 2005;187:2519–2525. doi: 10.1128/JB.187.7.2519-2525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Higgins DA, et al. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 74.Lenz DH, et al. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol. 2005;58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]