Abstract
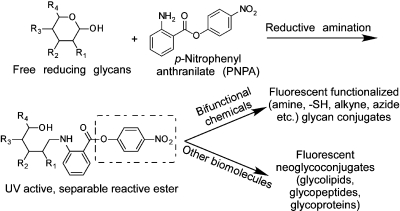
A facile preparation of neoglycoconjugates has been developed with a commercially available chemical, p-nitrophenyl anthranilate (PNPA), as a heterobifunctional linker. The two functional groups of PNPA, the aromatic amine and the p-nitrophenyl ester, are fully differentiated to selectively conjugate with glycans and other biomolecules containing nucleophiles. PNPA is efficiently conjugated with free reducing glycans via reductive amination. The glycan−PNPA conjugates (GPNPAs) can be easily purified and quantified by UV absorption. The active p-nitrophenyl ester in the GPNPA conjugates readily reacts with amines under mild conditions, and the resulting conjugates acquire strong fluorescence. This approach was used to prepare several fluorescent neoglycoproteins. The neoglycoproteins were covalently printed on activated glass slides and were bound by appropriate lectins recognizing the glycans.
Introduction
Glycoconjugates including glycoproteins, glycolipids, and proteoglycans play important roles in many biological systems (1). Neoglycoconjugates, including neoglycolipids and neoglycoproteins as artificial mimics of natural glycoconjugates (2−6), have found many applications in various areas. One of the major applications is the development of carbohydrate-based vaccines and drugs (7−11).
To prepare neoglycoconjugates, and specifically neoglycoproteins, usually one or both of the molecules (glycan and protein) are derivatized with an appropriate functional group and then covalently linked. Theoretically, the linkages could be built through many different chemical reactions. Practically, however, derivatization is limited by several critical requirements. First, since glycans from natural sources or expensive synthetic approaches are often available only in small quantities, the glycan incorporation reaction should be highly efficient. Second, the reaction conditions should ideally be in aqueous buffer under physiological pH to avoid altering the glycan or denaturation of proteins, which might affect the solubility and immunogenicity of the conjugate. Third, the linkage itself should not decrease the immunogenicity of the conjugate and should be of minimal toxicity and antigenicity. There have been numerous methods developed for protein−carbohydrate conjugation, or neoglycoprotein synthesis, based on heterobifunctional or homobifunctional cross-linkers (12,13). Stowell et al. used 2-imino-2-methoxyethyl 1-thioglycosides to link glycans to proteins (14,15). Diethyl squarate has been used for efficient conjugation of proteins and carbohydrates; however, the potential immune response of the linker itself has impeded its application in vaccine development (16). Click chemistry (17) as well as a number of other protein− or peptide−carbohydrate conjugation methods have been employed for carbohydrate bioconjugation (18−21). Activated dicarboxylic acids can be used as homobifunctional linkers, but usually, no intermediate glycan derivatives can be separated. Wu et al. utilized bis-p-nitrophenylesters (22) to conjugate glycans and proteins. The intermediates can be separated and purified; however, the glycans need to be premodified to possess an active amine. Reductive amination has also been used in the conjugation of free reducing glycans (23) or ninhydrin-treated glycopeptides (24) to proteins; however, the reaction conditions are relatively harsh: they require large quantities of glycan and often the yields are very low.
Here, we describe a simple and efficient method to conjugate free reducing glycans with proteins using a commercially available chemical linker (Figure 1). The linker, p-nitrophenyl anthranilate (PNPA)1 , has two different functional groups: an aryl amine that can react with free reducing glycans by reductive amination and an active p-nitrophenyl ester that can react with nucleophiles. We show that this bifunctional linker is a useful reagent for preparing fluorescent neoglycoproteins from naturally occurring complex glycans. Moreover, the neoglycoconjugates acquire fluorescence upon linking, thus providing a ready means of following the reaction and quantifying the products.
Figure 1.
General strategy for using p-nitrophenyl anthranilate (PNPA) as a heterobifunctional linker to make neoglycoconjugates and other potential applications.
Experimental Procedures
Free reducing glycans were purchased from V-LABS and stored at −20 °C until use. All chemicals were purchased from Sigma-Aldrich and used without further purification. HPLC solvents were purchased from Fisher Scientific. An Ultraflex-II TOF/TOF system from Bruker Daltonics was used for MALDI-TOF mass spectrometry analysis of glycan conjugates.
Glycan−PNPA Conjugation
The conjugation of glycan with PNPA was carried out using the common reductive amination procedure for free glycan labeling with modifications on sample purification. Briefly, to a free reducing glycan (0.1 to 1 mg), freshly prepared PNPA solution (0.35 M in DMSO/AcOH = 7:3 (v/v), 25−50 μL) and an equal volume NaCNBH3 solution (1 M in DMSO/AcOH = 7:3 (v/v)) were added. The mixture was heated at 65 °C for 2 h. The reaction mixture was quenched by addition of acetonitrile (0.5 to 1 mL). The mixture was cooled at −20 °C for 2 h and centrifuged at 10 000 g for 5 min. The supernatant was discarded and the pellet subjected to either C18 Sep-pak for desalting or C18 HPLC for direct purification. For reaction with ethylenediamine, LNnT-PNPA (0.1 to 1 mg) was dissolved in 20−200 μL 10% ethylenediamine, and the solution was kept at room temperature for 30 min and subjected to HPLC analysis.
HPAEC-PAD and High-Performance Liquid Chromatography (HPLC)
High-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) analysis was carried out with a Dionex ICS-3000 system with a Carbpac PA-100 column. The eluent gradient was set to 0−125 mM sodium acetate over 50 min in sodium hydroxide (100 mM).
A Shimadzu HPLC CBM-20A system was used for HPLC analysis and separation of glycan−PNPA conjugates. For reverse-phase HPLC, it was coupled with a Vydec C18 HPLC column and a UV detector SPD-20A. UV absorptions at 330 and 280 nm were used to detect and quantify GPNPAs and BSA−GPNPA conjugates. The mobile phase was acetonitrile and water with 0.1% trifluoroacetic acid (TFA). The concentration of acetonitrile increased from 1% to 90% in 30 min with a linear gradient. For SEC-HPLC, a Biobasic-60 SEC column was used. The eluent was ammonium acetate (10 mM) at pH 4.5. The flow rate was set at 1 mL/min for all HPLC runs.
Conjugation of Proteins with GPNPAs
Protein conjugation with GPNPAs was carried out in various buffers as described above. Protein concentration was kept at 5 mg/mL. Aqueous buffer with 1% DMSO was used. The protein conjugates were purified by SEC by collecting the eluent with strong absorption at UV 280 nm. Lyophilized fractions were dissolved in water or suitable buffer for MALDI-TOF characterization and printing.
Printing, Binding Assay, and Scanning
The printing of protein−glycan conjugates on NHS-activated slides and epoxy slides was carried out according to previous procedure (25). Biotinylated lectins were used in the binding assay, and the bound lectins were detected by a secondary incubation with cyanine 5-streptavidin. The slides were scanned with a Perkin-Elmer ProScanArray microarray scanner equipped with 4 lasers covering an excitation range from 488 to 633 nm. The scanned images were analyzed with the ScanArray Express software. For cyanine 5 fluorescence, 649 nm (Ex) and 670 nm (Em) were used. All the images obtained from the scanner were in grayscale and colored for easy discrimination.
Results
Derivatization of Free Reducing Sugars with PNPA by Reductive Amination
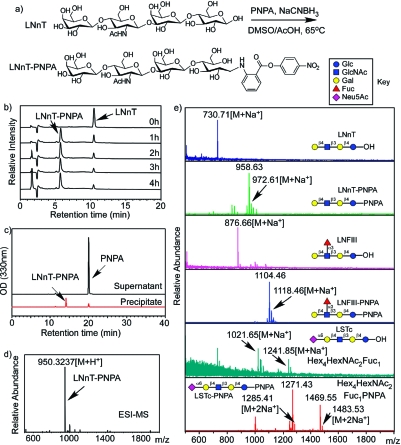
The preliminary study of PNPA conjugation with free reducing glycans was carried out using lactoneotetraose (LNnT) (Galβ1−4GlcNAcβ1−3Galβ1−4Glc) as the model compound. The conjugation reaction is shown in Figure 2a. The reaction time was optimized by analyzing free LNnT and the product mixture after various reaction times using HPAEC-PAD (Figure 2b). The conversion of LNnT did not significantly increase after 2 h of heating at 65 °C, when >80% yield was obtained. The reaction mixture was precipitated in 10 volumes of acetonitrile, and the supernatant and precipitate were analyzed using RP-HPLC (Figure 2c). Most of the excess PNPA remained in the supernatant, and the LNnT−PNPA conjugate was precipitated in high yield (>90%). The major product peak was collected from RP-HPLC and analyzed by ESI-MS (Figure 2d), which showed an expected mass at 950.3237 [M+H+] (calc. 950.3248) and confirmed the successful derivatization of the reducing glycan. Interestingly, the behavior of the LNnT-PNPA conjugate on MALDI-TOF-MS was quite different from ESI-MS. Although the expected molecular ion peak at 972.61 [M+Na+] (calc. 972.3068) is evident in the spectrum, the most intense peak occurs at 958.63, along with other fragmentation peaks at 956.62 and 942.63 (Figure 2e). The fragmentation ions of −14, −16, and −30 Da caused by radical reactions on the nitro group (26) further confirmed the installation of PNPA. The pattern is highly reproducible and serves as an indicator for all glycan−PNPA conjugates in mass spectrometry analysis (Figure 2e).
Figure 2.
The PNPA derivatization of LNnT: (a) the chemical reaction equation; (b) Dionex profiles of LNnT−PNPA conjugation over a time course; (c) RP-HPLC profiles of supernatant and precipitate after precipitation of the LNnT−PNPA conjugation with acetonitrile; (d) ESI-MS of the HPLC purified LNnT−PNPA showing the expected mass; (e) MALDI TOF of the starting material and the product of several compounds, showing the expected masses and fragmented major peaks.
Reaction of Glycan−PNPA (GPNPA) Conjugates with Nucleophiles
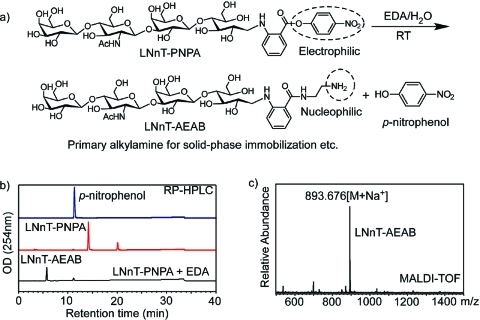
The reactivity of the GPNPAs toward nucleophiles was tested with ethylenediamine. This mild reaction at room temperature transforms the electrophilic p-nitrophenyl ester into a nucleophilic amino group (Figure 3a). Addition of 10% ethylenediamine at room temperature for 30 min quantitatively converted LNnT−PNPA to the expected LNnT−AEAB as shown by RP-HPLC (Figure 3b). The major product was purified by HPLC and characterized by MALDI-TOF (Figure 3c). This confirmed the formation of LNnT−AEAB conjugate, and therefore demonstrated the general reactivity of p-nitrophenyl ester toward alkylamines. The minor peak that eluted at 11.2 min with the reaction products matched the p-nitrophenol standard, confirming the PNPA conjugation with glycans and its reactivity toward amines (nucleophiles) as an active ester.
Figure 3.
(a) The reaction of LNnT−PNPA with ethylenediamine; (b) HPLC profile of p-nitrophenol, LNnT−PNPA, and its product after reaction with ethylenediamine; (c) MALDI-TOF of HPLC purified product of LNnT−PNPA and ethylenediamine.
Conjugation of Proteins with GPNPAs
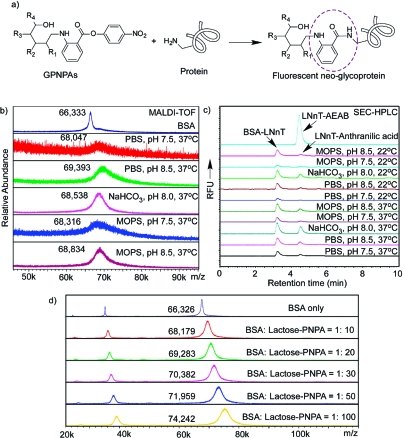
The conjugation of GPNPA with proteins was evaluated with BSA and LNnT−PNPA. To optimize the conjugation, various buffers and temperatures were tested (Figure 4 and Table 1). Figure 4a is a schematic representation of the reaction between GPNPA and proteins. Lysine residues in proteins can attack the active p-nitrophenol ester to form a stable amide bond. In this reaction, glycan and protein are thus linked through a small fluorescent anthranilamide moiety. Figure 4b shows the MALDI-TOF analyses that compare the conjugation of LNnT−PNPA with BSA in various buffers at 37 °C. The starting molar ratio of LNnT−PNPA and BSA was 10:1. While significant conjugation occurred in all buffers, reactions were faster in more basic conditions (pH 8.5 over pH 7.5). There was no significant conjugation difference seen in phosphate, MOPS, and carbonate buffers based on analysis by mass spectrometry. The conjugation efficiencies were estimated to be 2−3.5 glycans/protein molecule based on the MALDI-TOF peak values, indicating a yield of 20−35%. Figure 4c shows the fluorescent SEC-HPLC profiles of the conjugations in different buffers. The protein−glycan conjugates are fluorescent as expected (Ex 330 nm, Em 420 nm), and the intensity of the fluorescence can be directly correlated to the conjugation efficiency. Apparently, a relatively higher temperature (37 °C over 22 °C) and pH (8.5 over 7.5) significantly increased the conjugation efficiency. More interestingly, the fluorescence of the LNnT−BSA conjugate generated in carbonate buffer is stronger than those in PBS or MOPS buffer, suggesting a faster reaction. It is worthwhile to note that another fluorescent peak eluting at the same time as LNnT−AEAB is observed. This is presumably the hydrolysis product of LNnT−PNPA (LNnT-anthranilic acid, MW 828.77), which has a similar size to LNnT−AEAB (870.85). This peak is of significantly lower intensity than the protein peak, indicating that hydrolysis of GPNPA under these conditions is not seriously affecting the protein−glycan conjugation. To further evaluate the conjugation of GPNPAs with proteins, we tested the conjugation of lactose−PNPA (Galβ1−4Glc-PNPA) with BSA at different molar ratios (Figure 4d and Table 1). The increased amount of lactose−PNPA, as expected, increased the average number of lactose−PNPA conjugated per BSA molecule. At 100:1 ratio, ∼17−18 lactose/BSA were conjugated.
Figure 4.
BSA conjugation with LNnT−PNPA: (a) the general equation of protein conjugation with GPNPAs; (b) overlay of MALDI-TOF profiles of BSA conjugation with LNnT−PNPA at 37 °C in various buffers; (c) fluorescent SEC-HPLC profiles of BSA conjugation with LNnT−PNPA at different temperatures and buffers; (d) overlay of MALDI-TOF profiles of BSA conjugation with lactose−PNPA at different ratios.
Table 1. Conjugation of BSA with Glycan−PNPA Conjugates in Different Buffers, pH, and Molar Ratios.
| glycan PNPA | temp (°C) | buffer | pH | starting molar ratio (glycan/BSA) | product molar ratio (glycan/BSA) |
|---|---|---|---|---|---|
| LNnT | 37 | PBS | 7.5 | 10 | 2.1 |
| LNnT | 37 | PBS | 8.5 | 10 | 3.8 |
| LNnT | 37 | NaHCO3 | 8.0 | 10 | 2.7 |
| LNnT | 37 | MOPS | 7.5 | 10 | 2.4 |
| LNnT | 37 | MOPS | 8.5 | 10 | 3.1 |
| LNnT | 22 | PBS | 7.5 | 10 | 0.5 |
| LNnT | 22 | PBS | 8.5 | 10 | 2.2 |
| LNnT | 22 | NaHCO3 | 8.0 | 10 | 2.0 |
| LNnT | 22 | MOPS | 7.5 | 10 | 0.0 |
| LNnT | 22 | MOPS | 8.5 | 10 | 1.5 |
| lactose | 37 | NaHCO3 | 8.0 | 10 | 4.1 |
| lactose | 37 | NaHCO3 | 8.0 | 20 | 6.6 |
| lactose | 37 | NaHCO3 | 8.0 | 30 | 9.1 |
| lactose | 37 | NaHCO3 | 8.0 | 50 | 12.6 |
| lactose | 37 | NaHCO3 | 8.0 | 100 | 17.7 |
Printing and Recognition of Glycan−BSA Conjugates on Microarray Slides
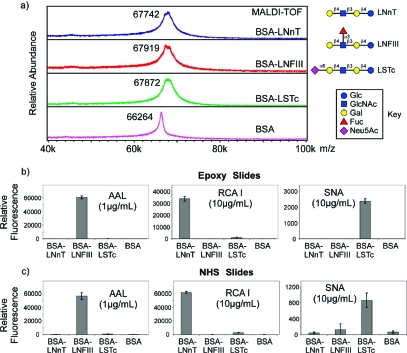
To validate the structural integrity of glycans after conjugation with proteins, we conjugated several GPNPAs (LNnT, LNFIII, and LSTc) with BSA using a 5:1 molar ratio of glycan/protein. The conjugates were purified by SEC-HPLC. The MALDI-TOF profiles in Figure 5a show the mass shifts compared to BSA alone, indicating that 1.5−2 glycans/protein molecule were conjugated. The purified BSA−glycan conjugates were printed on epoxy and NHS-activated glass slides and interrogated with several plant lectins. All three lectins showed expected bindings on both epoxy and NHS-activated slides (Figure 5b). AAL, a fucose binding lectin (27), bound to BSA−LNFIII conjugates. RCA I, a plant lectin that recognizes β1,4-linked galactose residues (28) bound to BSA−LNnT conjugates and also to BSA−LSTc conjugates, weakly, since α2,6-sialylation does not fully abolish its binding to Galβ1,4-R moieties. SNA, a plant lectin that binds α2,6-linked sialic acid residues (29), bound to BSA-LSTc conjugates, as expected.
Figure 5.
(a) MALDI-TOF of conjugates of BSA with LNnT−PNPA, LNFIII−PNPA, and LSTc−PNPA. (b,c) The conjugates of BSA−LNnT, −LNFIII, and −LSTc, and BSA alone printed on epoxy slides (b) and NHS slides (c) and interrogated with the lectins AAL, RCA I, and SNA. The glycan structures are indicated along with a key to the symbols.
Discussion
p-Nitrophenyl ester is known to be an active ester due to the strong electronegativity of the nitro group. This active ester, however, is well-tolerated by the aromatic amine group in PNPA, which is a nucleophile. The existence of two opposing but tolerant functional groups makes PNPA a promising heterobifunctional linker under finely tuned conjugation conditions. Reductive amination of PNPA with free reducing glycans does not interfere with the p-nitrophenyl ester, as shown by HPAEC, HPLC, and MALDI-TOF (Figure 2b−d). Although this reaction does not go to completion under the commonly used conditions, it is still highly efficient (>80% yield) and nonselective toward many glycans, as shown by LNnT, LNFIII, and LSTc conjugation profiles (Figure 2e). Furthermore, GPNPAs can be easily separated from unconjugated free glycans by C18 Sep-pak due to the strong hydrophobicity of the linker. The high conjugation efficiency, nonselectivity, and easy separation make this approach appropriate for extracting natural glycans as active esters for neoglycoconjugate preparation. We have noticed that, by direct reductive amination with glycans, the reducing end ring structure is opened. This is of concern in situations where the whole glycan structures are considered epitopes, such as conjugation of mono- and disaccharides or structures with epitopes close to the reducing end. In most cases, we are more interested in the terminal moieties as epitopes, which should not be affected by this strategy. Under these circumstances, the ability to utilize complex natural glycans directly without laborious synthesis is of more importance than the reducing end structural variation. On the other hand, this strategy could also be applied to any biomolecules with an aldehyde group, which could be installed at the glycan reducing end without breaking the ring structures (30).
It is worthwhile to note the interesting behavior of GPNPAs on MALDI-TOF. While ESI-MS clearly showed the molecular ion, MALDI-TOF of GPNPAs showed a reproducible fragmentation pattern besides the molecular ion. This radical reaction related pattern is specific to the nitro group. While it might complicate mass spectra to a certain extent, it could also serve as a fingerprint for confirming glycan−PNPA conjugates in mass spectrometry analysis. Nevertheless, this does not interfere with the reactivity of GPNPAs for conjugation with other molecules, as shown in Figure 3. LNnT−PNPA quickly reacts with ethylenediamine to form LNnT−AEAB. The electrophile (ester) of LNnT−PNPA is easily transformed to a nucleophile (amine), enabling its direct conjugation with electrophilic molecules such as epoxy and NHS esters. We have demonstrated the use of AEAB conjugation with glycans for generation of fluorescent natural glycan arrays (31), providing an alternative for this application. Although AEAB and PNPA are all heterobifunctional linkers that can be used to conjugate free reducing glycans with other biomolecules or solid surfaces, they are complementary in terms of the reactivity. GAEABs have a nucleophilic alkylamine, while GPNPAs have a good leaving group that is reactive toward nucleophiles. Application of both conjugations offers more flexibility in the design and preparation of bioconjugates.
Protein−carbohydrate conjugation has become an important route for generating glycan-related antibodies. Many methods have been developed, most of which focus on conjugating synthetic glycans through bifunctional linkers. In our effort to develop carbohydrate-based vaccines against schistosomiasis and other pathogens (32), we sought to conjugate complex glycans extracted from natural sources, which are usually very complicated to synthesize with appropriate linkers attached. Therefore, we needed an effective method to efficiently conjugate free reducing glycans with protein carriers. Although there are a number of methods developed for protein−carbohydrate conjugations, most of them are not targeting naturally occurring complex glycans, which are difficult to obtain through synthesis. Although direct coupling of glycans with proteins by reductive amination has been used, the yields are often disappointing. To obtain homogeneous natural glycans is also a major challenge. PNPA, as a heterobifunctional linker, can react efficiently and selectively with glycans and proteins. GPNPAs can be easily prepared and purified from natural sources based on their UV absorbance. The GPNPAs can be efficiently conjugated with BSA, as shown by MALDI-TOF and HPLC (Figure 4). MALDI-TOF is the most common method used to determine the protein−carbohydrate conjugation efficiency based on the mass shift before and after conjugation. However, with more glycans and linkers added, the MALDI-TOF peak broadens quickly so that the peak value does not accurately represent the average molecular weight of protein conjugates. The protein−glycan conjugates prepared with the GPNPA strategy are fluorescent, which can greatly facilitate the quantification of conjugated glycans even at a minimal level (Figure 4c). Furthermore, with appropriate UV range fluorescence detection methods, this fluorescence could be used to detect small amounts of protein in purification steps and bioassays. On the basis of the mass and fluorescence of the conjugates, we confirmed that various glycans such as lactose, LNnT, and LSTc can be reproducibly conjugated to proteins. The molar ratio of glycans versus glycan−protein conjugates can be driven by increasing the molar ratio of glycans to proteins, reaching 17−18 glycans/conjugate at 100:1 glycan/protein molar ratio.
Upon conjugation of glycans to proteins, it is difficult to validate that the structures of glycans are not affected during the glycan−protein conjugation process. Therefore, we conjugated and printed several glycan−protein conjugates on activated glass slides and interrogated them with various lectins. The conjugation of LNnT, LNFIII, and LSTc GPNPAs to BSA showed similar mass shifts (Figure 5a), indicating the general applicability of this approach to different glycans. This is especially useful for sialylated structures, as the sialic acid is not compatible with many conjugation approaches relying on carboxylic acid activation. With p-nitrophenyl ester incorporated onto the glycan reducing end by reductive amination, sialylated glycans can be conjugated to proteins as easily as other glycans. When the BSA conjugates of LNnT, LNFIII, and LSTc were printed on activated glass slides (epoxy and NHS) and assayed with three lectins, AAL, RCA I, and SNA, expected specific binding was observed for each lectin. These results show that the glycan structures are presented after the conjugation process in a manner that is consistent with their predicted recognition, which implies that such conjugates can be used to explore biological functions of glycans. It is also worth noting that this approach provides an alternative platform for natural glycan microarray printing. While suitably derivatized glycans (25,31,33,34) can be directly printed on NHS or epoxy slides for successful carbohydrate binding protein screening, printing of protein−glycan conjugates presents the glycan structures in the context of protein. This somewhat addresses concerns about nonspecific surface interactions related to microarray slide presentation and may provide a desirable alternative presentation strategy under certain circumstances. The fact that the protein−glycan conjugates are fluorescent is also a major advantage when quantifiable microscale material is used in microarray printing.
One general issue in using protein−carbohydrate conjugates as potential vaccines is that the linker itself is sometimes immunogenic or alters immunogenic potential of the glycan. While no testing is yet available on the immunogenicity of anthranilamide derivatives, the fluorescent anthranilamide is generally considered safe as a chemical, and the closely related anthranilic acid is the precursor of the amino acid tryptophan. Thus, while not yet tested in vivo, we expect this linker to have minimal immunogenicity and toxicity.
In conclusion, we have developed a novel approach utilizing a commercially available chemical as a convenient fluorescent linker for protein−carbohydrate conjugation. The ease and efficiency, the ability to utilize natural glycans, and the acquired fluorescence of the final conjugates make this approach very promising in the development of carbohydrate antibodies and carbohydrate-based vaccines.
Acknowledgments
Supported in part by a Collaboration Planning Grant from the Georgia Research Alliance and NIH Grant GM085448 (RDC, DFS). We thank Dr. Jamie Heimburg-Molinaro for manuscript editing and review.
Funding Statement
National Institutes of Health, United States
Footnotes
Abbreviations: AAL, Aleuria aurantia lectin; AEAB, 2-amino-N-(2-aminoethyl)-benzamide; GBP, glycan binding protein; GPNPA, Glycan-PNPA conjugate; HPLC, high performance liquid chromatography; LNFIII, lacto-N-fucopentaose III; LNnT, lacto-N-neotetraose; LSTc, lacto-N-sialyltetraose c; NHS, N-hydroxysuccinimide; PNPA, p-nitrophenyl anthranilate; RCA I, Ricinus communis Agglutinin I; RFU, relative fluorescence unit; SNA, Sambucus nigra lectin.
References
- Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. (2009) Essentials of Glycobiology, 2nd ed., Cold Spring Harbor Press, New York. [PubMed] [Google Scholar]
- Wong S. Y. C. (1995) Neoglycoconjugates and their applications in glycobiology. Curr. Opin. Struct. Biol. 5, 599–604. [DOI] [PubMed] [Google Scholar]
- Lee R. T.; Lee Y. C. (1997) Neoglycoconjugates. Glycosciences 55–77. [Google Scholar]
- Bovin N. V. (2003) Neoglycoconjugates as probes in glycobiology. NATO Sci. Seri., II 129, 207–225. [Google Scholar]
- Davis B. G. (2002) Synthesis of glycoproteins. Chem. Rev. 102, 579–602. [DOI] [PubMed] [Google Scholar]
- Pratt M. R.; Bertozzi C. R. (2005) Synthetic glycopeptides and glycoproteins as tools for biology. Chem. Soc. Rev. 34, 58–68. [DOI] [PubMed] [Google Scholar]
- Slovin S. F.; Keding S. J.; Ragupathi G. (2005) Carbohydrate vaccines as immunotherapy for cancer. Immunol. Cell. Biol. 83, 418–28. [DOI] [PubMed] [Google Scholar]
- Ouerfelli O.; Warren J. D.; Wilson R. M.; Danishefsky S. J. (2005) Synthetic carbohydrate-based antitumor vaccines: challenges and opportunities. Expert Rev. Vaccines 4, 677–85. [DOI] [PubMed] [Google Scholar]
- Lloyd K. O. (2000) Carbohydrate vaccines for the immunotherapy of cancer. Drug News Perspect. 13, 463–70. [PubMed] [Google Scholar]
- Ada G.; Isaacs D. (2003) Carbohydrate-protein conjugate vaccines. Clin. Microbiol. Infect. 9, 79–85. [DOI] [PubMed] [Google Scholar]
- Roy R. (2004) New trends in carbohydrate-based vaccines. Drug Discovery Today: Technologies 1, 327–336. [DOI] [PubMed] [Google Scholar]
- Stowell C. P.; Lee V. C. (1980) Neoglycoproteins: the preparation and application of synthetic glycoproteins. Adv. Carbohydr. Chem. Biochem. 37, 225–81. [DOI] [PubMed] [Google Scholar]
- Macmillan D.; Bertozzi C. R. (2000) New directions in glycoprotein engineering. Tetrahedron 56, 9515–9525. [Google Scholar]
- Stowell C. P.; Lee Y. C. (1982) Preparation of neoglycoproteins using 2-imino-2-methoxyethyl 1-thioglycosides. Methods Enzymol. 83, 278–88. [DOI] [PubMed] [Google Scholar]
- Stowell C. P.; Lee Y. C. (1980) Preparation of some new neoglycoproteins by amidination of bovine serum albumin using 2-imino-2-methoxyethyl 1-thioglycosides. Biochemistry 19, 4899–904. [DOI] [PubMed] [Google Scholar]
- Mawas F.; Niggemann J.; Jones C.; Corbel M. J.; Kamerling J. P.; Vliegenthart J. F. (2002) Immunogenicity in a mouse model of a conjugate vaccine made with a synthetic single repeating unit of type 14 pneumococcal polysaccharide coupled to CRM197. Infect. Immun. 70, 5107–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Chan T. R.; Hilgraf R.; Fokin V. V.; Sharpless K. B.; Finn M. G. (2003) Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 125, 3192–3. [DOI] [PubMed] [Google Scholar]
- Crich D.; Yang F. (2008) Synthesis of neoglycoconjugates by the desulfurative rearrangement of allylic disulfides. J. Org. Chem. 73, 7017–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubler-Kielb J.; Pozsgay V. (2005) A new method for conjugation of carbohydrates to proteins using an aminooxy-thiol heterobifunctional linker. J. Org. Chem. 70, 6987–90. [DOI] [PubMed] [Google Scholar]
- Wong S. Y.; Guile G. R.; Dwek R. A.; Arsequell G. (1994) Synthetic glycosylation of proteins using N-(beta-saccharide) iodoacetamides: applications in site-specific glycosylation and solid-phase enzymic oligosaccharide synthesis. Biochem. J. 300, Pt 3843–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M. C.; Chen L. M.; Wold F. (1990) Complex neoglycoproteins. Methods Enzymol. 184, 653–9. [DOI] [PubMed] [Google Scholar]
- Wu X.; Ling C. C.; Bundle D. R. (2004) A new homobifunctional p-nitro phenyl ester coupling reagent for the preparation of neoglycoproteins. Org. Lett. 6, 4407–10. [DOI] [PubMed] [Google Scholar]
- Gildersleeve J. C.; Oyelaran O.; Simpson J. T.; Allred B. (2008) Improved procedure for direct coupling of carbohydrates to proteins via reductive amination. Bioconjugate Chem. 19, 1485–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencke A. J.; Cheung D. T.; Wold F. (1987) Attachment of oligosaccharide-asparagine derivatives to proteins: activation of asparagine with ninhydrin and coupling to protein by reductive amination. Methods Enzymol. 138, 409–13. [DOI] [PubMed] [Google Scholar]
- Song X.; Xia B.; Lasanajak Y.; Smith D. F.; Cummings R. D. (2008) Quantifiable fluorescent glycan microarrays. Glycoconjugate J. 25, 15–25. [DOI] [PubMed] [Google Scholar]
- Ueda K.; Katagiri T.; Shimada T.; Irie S.; Sato T. A.; Nakamura Y.; Daigo Y. (2007) Comparative profiling of serum glycoproteome by sequential purification of glycoproteins and 2-nitrobenzensulfenyl (NBS) stable isotope labeling: a new approach for the novel biomarker discovery for cancer. J. Proteome Res. 6, 3475–83. [DOI] [PubMed] [Google Scholar]
- Yamashita K.; Kochibe N.; Ohkura T.; Ueda I.; Kobata A. (1985) Fractionation of L-fucose-containing oligosaccharides on immobilized Aleuria aurantia lectin. J. Biol. Chem. 260, 4688–93. [PubMed] [Google Scholar]
- Green E. D.; Brodbeck R. M.; Baenziger J. U. (1987) Lectin affinity high-performance liquid chromatography. Interactions of N-glycanase-released oligosaccharides with Ricinus communis agglutinin I and Ricinus communis agglutinin II. J. Biol. Chem. 262, 12030–9. [PubMed] [Google Scholar]
- Shibuya N.; Goldstein I. J.; Broekaert W. F.; Nsimba-Lubaki M.; Peeters B.; Peumans W. J. (1987) The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(alpha 2−6)Gal/GalNAc sequence. J. Biol. Chem. 262, 1596–601. [PubMed] [Google Scholar]
- Munoz F. J., Perez J., Rumbero A., Santos J. I., Canada F. J., Andre S., Gabius H. J., Jimenez-Barbero J., Sinisterra J. V., Hernaiz M. J. (2009) Glycan tagging to produce bioactive ligands for a surface plasmon resonance (SPR) study via immobilization on different surfaces, Bioconjugate Chem. 20, 673−682. [DOI] [PubMed] [Google Scholar]
- Song X., Xia B., Lasanajak Y., Stowell S. R., Smith D. F., Cummings R. D. (2009) Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem. Biol. 16, 36−47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyame A. K.; Kawar Z. S.; Cummings R. D. (2004) Antigenic glycans in parasitic infections: implications for vaccines and diagnostics. Arch. Biochem. Biophys. 426, 182–200. [DOI] [PubMed] [Google Scholar]
- Blixt O.; Head S.; Mondala T.; Scanlan C.; Huflejt M. E.; Alvarez R.; Bryan M. C.; Fazio F.; Calarese D.; Stevens J.; Razi N.; Stevens D. J.; Skehel J. J.; van Die I.; Burton D. R.; Wilson I. A.; Cummings R.; Bovin N.; Wong C. H.; Paulson J. C. (2004) Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 17033–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer A. R.; Hokke C. H.; Deelder A. M.; Wuhrer M. (2007) General microarray technique for immobilization and screening of natural glycans. Anal. Chem. 79, 8107–13. [DOI] [PubMed] [Google Scholar]