Abstract
RNA editing changes the coding/decoding information relayed by transcripts via nucleotide insertion, deletion, or conversion. Editing of tRNA anticodons by deamination of adenine to inosine is used both by eukaryotes and prokaryotes to expand the decoding capacity of individual tRNAs. This limits the number of tRNA species required for codon-anticodon recognition. We have identified the Arabidopsis thaliana gene that codes for tRNA adenosine deaminase arginine (TADA), a chloroplast tRNA editing protein specifically required for deamination of chloroplast (cp)-tRNAArg(ACG) to cp-tRNAArg(ICG). Land plant TADAs have a C-terminal domain similar in sequence and predicted structure to prokaryotic tRNA deaminases and also have very long N-terminal extensions of unknown origin and function. Biochemical and mutant complementation studies showed that the C-terminal domain is sufficient for cognate tRNA deamination both in vitro and in planta. Disruption of TADA has profound effects on chloroplast translation efficiency, leading to reduced yields of chloroplast-encoded proteins and impaired photosynthetic function. By contrast, chloroplast transcripts accumulate to levels significantly above those of wild-type plants. Nevertheless, absence of cp-tRNAArg(ICG) is compatible with plant survival, implying that two out of three CGN codon recognition occurs in chloroplasts, though this mechanism is less efficient than wobble pairing.
INTRODUCTION
tRNAs are key components of gene expression in every living organism. They serve as adaptor molecules helping to convert nucleic acid–based information into chains of amino acids: each RNA codon is recognized by the anticodon harbored by the corresponding tRNA, according to the wobble rules proposed by Francis Crick (Crick, 1966). However, several genetic systems, such as organelles and some bacterial parasites, encode fewer tRNAs than theoretically required for the translation of all codons (Shinozaki et al., 1986; Osawa et al., 1992).
This lack can be compensated for by import of cytosolic tRNAs as demonstrated for mitochondria of plants, fungi, and protozoa (Salinas et al., 2008), but import is unlikely to explain all cases of incomplete tRNA sets. For example, no import of cytosolic tRNA has been demonstrated in chloroplasts or in human mitochondria (Lung et al., 2006), which according to wobble rules lack complete tRNA sets. This suggests that exceptions to the wobble rules must exist in some genetic systems.
Two mechanisms have been postulated to explain how translation occurs with a reduced tRNA set: two out of three and superwobble decoding. Two out of three decoding postulates that a tRNA pairing with only the first two codon bases (usually a G and a C) can be sufficient for translation and that any base can occur at the third (wobble) codon position (opposite position 34 of the tRNA; Figure 1). This mechanism is supported by several examples (Lagerkvist, 1986). It has also been suggested to occur in chloroplasts (Shinozaki et al., 1986) and supported by in vitro experiments using wheat (Triticum aestivum) germ extracts and bean (Phaseolus vulgaris) chloroplast tRNAs (Pfitzinger et al., 1990). Further analysis showed that two out of three critically depends on the stability of the second base pair formed between the codon and anticodon and that this stability is influenced by the structure of the tRNA anticodon loop (Lehmann and Libchaber, 2008).
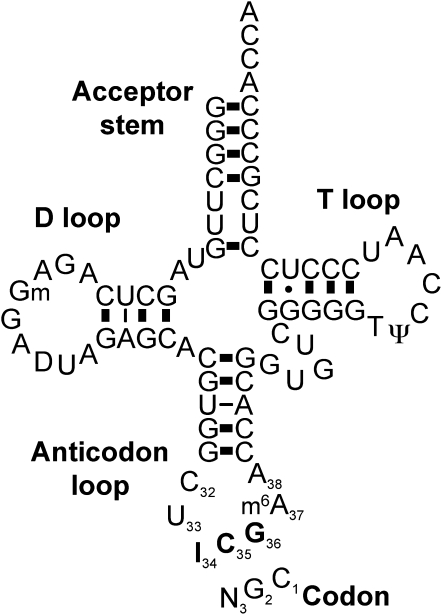
Figure 1.
Representation of cp-tRNAArg(ICG) Binding to Its Corresponding Codon.
cp-tRNAArg(ICG) depicted as a typical cloverleaf structure. The numbering of the anticodon loop residues is according to the convention recommended by Sprinzl et al. (1996). The modifications shown are those identified in other cp-tRNAArg(ICG) that are expected to be conserved in Arabidopsis (Francis et al., 1989; Pfitzinger et al. 1990). I, inosine; N, A, C, U, or G; m6A, N6-methyl A; D, dehydro U; Gm, 2-O-methyl G; T, 5-methyl U; Ψ, pseudo U.
The so-called superwobble, or extended wobble, is a mechanism in which an unmodified uridine at the first anticodon position (position 34 of the tRNA; Figure 1) can read all four nucleotides at the wobble codon position. This mechanism has also been suggested to occur in chloroplasts (Shinozaki et al., 1986; Pfitzinger et al., 1990) and was demonstrated in tobacco (Nicotiana tabacum) when one of the two chloroplast (cp)-tRNAGly genes was deleted (Rogalski et al., 2008). In this study, two out of three was ruled out for decoding of Gly codons, but superwobble was also shown to be inefficient. In every case, exceptions to wobble rules only concern the third codon position (Figure 1), whereas proper Watson-Crick pairing at codon positions 1 and 2 is always required.
tRNAs are the most heavily modified RNA molecules, and numerous examples of posttranscriptional modifications, notably at uridine 34 or purine 37, have been shown to expand the ability of tRNAs to read additional codons (Agris et al., 2007). Modifications at purine 37, outside the anticodon, are very important to prevent hydrogen bonding within the anticodon loop (Dao et al., 1994) and to increase base stacking (Grosjean et al., 1998), strengthening the codon–anticodon interaction. Deamination of adenosine 34 to inosine is a widespread modification expanding the set of codons read by a particular tRNA (Figure 1). This deamination is performed by specific tRNA adenosine deaminases (TADs or ADATs), which are essential in prokaryotes and eukaryotes (Schaub and Keller, 2002). The enzymes mediating tRNA adenosine deamination in bacteria and yeast contain conserved motifs present in cytidine deaminases, suggesting an evolutionary link between the two reactions. The genomes of higher-plant chloroplasts encode two Arg tRNAs: cp-tRNAArg(ACG), thought to be required for decoding the codons CGA/U/G/C (CGN), and cp-tRNAArg(UCU), thought to be required to read the codons AGA/G. It has been shown that cp-tRNAArg(ACG) is deaminated to cp-tRNAArg(ICG) in bean (Pfitzinger et al., 1990), which would be expected to allow efficient translation of CGC and CGA codons. This tRNA modification was inherited from the cyanobacterial ancestor of chloroplasts and is well conserved in prokaryotes where it is essential for cell survival (Wolf et al., 2002). However, as opposed to prokaryotes, which translate the Arg CGG codon using a specific tRNAArg(CCG), neither of the two cp-tRNAArg can form a wobble pair with the CGG codon. The simplest explanation is that chloroplast CGG codons are translated by two out of three recognition by cp-tRNAArg(ICG) (Pfitzinger et al., 1990). However, this mechanism was recently ruled out in tobacco chloroplasts for the decoding of Gly codons (Rogalski et al., 2008), casting doubts on the occurrence of this mechanism in vivo in chloroplasts.
In this work, we identified the Arabidopsis thaliana tRNA adenosine deaminase arginine (TADA) gene that codes for a deaminase responsible for the editing of the adenosine at the wobble position of cp-tRNAArg(ACG). A mutation in TADA leads to slower chloroplast translation, causing profound effects on chloroplast function and plant development. However, as opposed to the case in prokaryotes, it is not lethal, adding weight to the hypothesis that a two out of three mechanism can occur in chloroplasts.
RESULTS
TADA Is a Nuclear Gene Encoding a Protein Containing a Conserved Deaminase Domain
Adenosine-to inosine (A-to-I) editing of the wobble anticodon position of several eukaryote and prokaryote tRNAs is catalyzed by enzymes of the TAD/ADAT protein family (Schaub and Keller, 2002), which contain the conserved cytidine/deoxycytidylate deaminase motif (InterPro: IPR002125). This motif comprises three His and Cys residues involved in the coordination of a zinc ion and an essential Glu residue that is required for the hydrolytic reaction. The single chloroplast tRNA known to be edited by A-to-I deamination is cp-tRNAArg(ACG), which has been directly sequenced in the alga Codium fragile and in bean (Francis et al., 1989; Pfitzinger et al., 1990). While searching for candidate genes that might be implicated in chloroplast RNA editing, we searched the Arabidopsis predicted proteome for sequences containing the characteristic deaminase motif. Fifteen genes were selected (see Supplemental Figure 1 online). Among them eight correspond to the CDA family of cytidine deaminases and were discounted because they lack the requirements for an organellar RNA editing enzyme, namely, organellar targeting sequences, affinity for RNA, deaminase activity on RNA substrates, and conservation in other plant species (Faivre-Nitschke et al., 1999; S.E. Faivre-Nitschke and J.M. Gualberto, unpublished data). Of the remaining candidate genes, only two code for proteins with predicted organellar targeting sequences: At4g20960, which probably codes for the chloroplast riboflavin deaminase; and At1g68720, which codes for a large protein predicted by PREDOTAR (Small et al., 2004) and TargetP (Emanuelsson et al., 2000) to be possibly plastid or mitochondrial respectively. By sequence homology, we identified At1g68720 as likely to code for TADA.
The Arabidopsis TADA gene contains only three introns and codes for a large protein of 1307 amino acids with a C-terminal deaminase domain (Figure 2A). The recent identification of a full-size cDNA (accession number AK117889) confirmed the gene model. The presence of an in-frame stop codon in the 5′-untranslated region unambiguously identified the initiation codon. Most of the large 5′-terminal exon codes a sequence (1080 amino acids) that has no identifiable motif or similarities to sequences outside the plant kingdom. A probable ortholog of TADA is found in rice (Oryza sativa; Os06g0489500), which also codes for a large protein (1590 amino acids). Surprisingly, the predicted N-terminal regions of the Arabidopsis and rice proteins are poorly conserved (15% identity), while the region similar to other deaminases is 65% identical (see Supplemental Figure 2 online). Additional plant homologs can be identified in the databases of genomic and EST sequences (see Supplemental Figure 3 online).
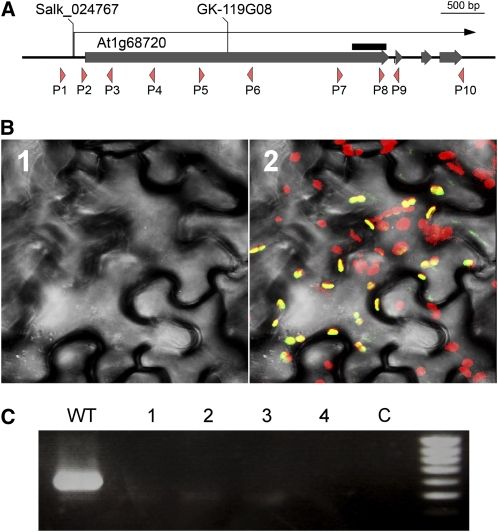
Figure 2.
The Arabidopsis TADA Gene Codes for a Large Protein Targeted to Chloroplasts.
(A) Structure of the Arabidopsis TADA gene. The coding sequences of the exons are indicated by thick gray bars, and the transcribed region is represented by the arrow. The T-DNA insertion sites in the mutants described in this study are shown. The gene sequence selected for expression of a hairpin RNA in the RNAi line Agri-140-69-2-CATMA1a58100 is represented by the black bar. Primers described in the text are indicated by orange arrowheads.
(B) Localization of a protein-GFP fusion in epidermal cells of N. benthamiana. 1, Visible image; 2, merged fluorescence and visible images; green, GFP fluorescence; red, natural fluorescence of chloroplasts; yellow, colocalization of green and yellow fluorescence.
(C) RT-PCR analysis of TADA transcripts using primer P5 and P6 in four independent tada-1 homozygous plants. C, negative control without reverse transcriptase.
TADA Is Targeted to the Chloroplast
To determine the subcellular localization of the TADA protein, several green fluorescent protein (GFP) fusions were prepared. The corresponding plasmids were biolistically transformed into Nicotiana benthamiana leaves, and the localization of the fluorescent fusion proteins observed by confocal microscopy. A full-length TADA:GFP fusion showed no GFP fluorescence. However, the first 259 amino acids of the TADA protein efficiently targeted GFP into chloroplasts (Figure 2B). We observed no fluorescence in mitochondria, cytoplasm, or in the nucleus. Thus, the TADA protein contains an N-terminal targeting sequence that can unambiguously target the protein into chloroplasts.
TADA Is Required for the Specific Editing of Chloroplast tRNAArg(ACG)
We obtained several T-DNA insertions lines for TADA. We could only confirm T-DNA insertions for two lines. Line SALK_024767 has a T-DNA insertion whose left border is located just eight nucleotides upstream of the predicted transcription initiation site (Figure 2A). However, plants homozygous for the insertion showed no decrease in TADA transcript abundance compared with wild-type Columbia-0 (Col-0) plants and showed no visible phenotype differences either. It is likely that promoters internal to the T-DNA efficiently drive expression of the downstream gene (Ulker et al., 2008). Line GK-119G08 contains a T-DNA insertion that was mapped to the first exon of the gene, 1597 nucleotides downstream of the initiation codon. The T-DNA insertion was accompanied by the deletion of 25 nucleotides at the insertion site. In homozygous plants, no transcript could be detected spanning the region containing the T-DNA insertion (Figure 2C). Thus, if a transcript is generated and translated from that locus, the resulting truncated protein would lack most of the N-terminal sequence, including the chloroplast targeting sequence. Line GK-119G08 was therefore identified as mutant tada-1.
We have also characterized several lines transformed with an RNA interference (RNAi) construct that we obtained from the AGRIKOLA collection (Hilson et al., 2004). Plants from line Agri-140-69-2-CATMA1a58100 accumulate no or very little amounts of TADA mRNA (see Supplemental Figure 4B online).
We tested if TADA is involved in cp-tRNAArg(ACG) editing. The tRNA was amplified by RT-PCR, and the cDNA was sequenced. Inosine base pairs with cytosine during reverse transcription, so A-to-I deamination leads to an apparent change in sequence from A to G. In wild-type plants, the sequence profiles suggest that there are almost equimolar amounts of edited and unedited tRNA, as evaluated by the height of the A and G peaks at the position corresponding to the anticodon wobble nucleotide (Figure 3). However, no trace of cp-tRNAArg(ICG) could be found in tada-1 plants. The same result was obtained with RNA extracted from five independent homozygous plants. In RNAi plants, amplification and sequencing of cp-tRNAArg(ACG) confirmed that silencing of TADA correlates with loss of tRNA editing: in a plant where no TADA mRNA could be detected, there was no evidence of tRNA editing, while in two plants showing partial TADA suppression, a residual G is visible at the position corresponding to the tRNA wobble nucleotide. Unaffected plants had equivalent amounts of edited and unedited tRNA, as in wild-type plants (see Supplemental Figure 4C online).
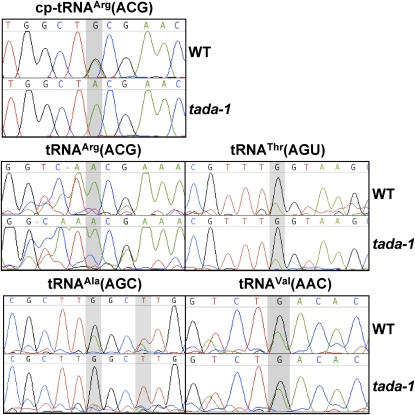
Figure 3.
The TADA Enzyme Is Specifically Required for Editing of cp-tRNAArg(ACG).
The sequences of the cp-tRNAArg(ACG) and of the indicated cytosolic tRNAs were amplified by RT-PCR, ligated to plasmid vector pGEM-T, and reamplified using plasmid and tRNA-specific primers. Direct sequence of the uncloned PCR fragments show that cp-tRNAArg(ACG) is no longer edited in the tada-1 mutant (presence of a G in the sequence at the position 34 [shaded] of the tRNA), while editing of cytosolic tRNAs is not affected. The presence of T instead of A at position 37 (shaded) in the sequences of tRNAAla(ACG) shows that editing of this position into I that is further modified into m1I is also not compromised in tada-1.
Apart from chloroplast tRNAArg(ACG), several cytosolic Arabidopsis tRNAs are also expected to be edited by A-to-I deamination (http://www.inra.fr/internet/Produits/TAARSAT/table.html). We therefore tested the hypothesis that TADA is also responsible for editing of cytosolic tRNAs. Primers were designed to amplify cytosolic tRNAAla(AGC), tRNAVal(AAC), tRNAThr(AGU), and tRNAArg(ACG). Our primers were designed to amplify only a subpopulation of tRNAArg(ACG), for which there are several variants encoded by eight different genes in Arabidopsis. Both in the wild type and in tada-1 we found evidence for efficient editing of cytosolic tRNAAla(AGC), tRNAVal(AAC), and tRNAThr(AGU) (Figure 3). Surprisingly, we found no evidence that cytosolic tRNAArg(ACG) is targeted for editing, either in wild-type Col-0 or in tada-1.
TADA is also not involved in editing of nucleotide A37 of tRNAAla(AGC) which, in all eukaryotic systems studied, is edited to I and further modified to m1I. In yeast and humans, TAD1 is the deaminase involved (Gerber et al., 1998). In the sequence of the RT-PCR products of tRNAAla(AGC), we find a T at position 37, both in the wild type and tada-1, consistent with the presence of m1I.
Thus, TADA seems to be exclusively involved in editing of the prokaryote-type cp-tRNAArg(ACG), and other deaminases must be responsible for editing of cytosolic tRNAs. No other chloroplast tRNA can be a substrate for TADA because no other chloroplast tRNA has an A at the wobble position. Regarding mitochondria, no tRNA adenosine deaminase is theoretically required by the mitochondrial translation system, as there are no mitochondrially encoded tRNAs with an A at the wobble position, and all tRNAs that should require editing for function are imported (presumably preedited) from the cytosol (Duchene et al., 2008).
We also considered the hypothesis that TADA is responsible for C-to-U editing of chloroplast and/or mitochondrial mRNAs that, by analogy to animal APOBEC and AID proteins involved in C-to-U editing, might be catalyzed by enzymes containing the same characteristic cytidine/deoxycytidylate deaminase domain (Conticello, 2008). However, the plant TADA is apparently not involved in organellar C-to-U editing; regions of the ndhB, ndhD, rps12, and rps14 chloroplast transcripts containing 11 editing sites were analyzed by RT-PCR and sequence, but no differences were found between mutant and wild-type plants (see Supplemental Figure 5 online). We conclude that TADA is not required for C-to-U chloroplast mRNA editing. We have also tested if editing of mitochondrial transcripts is affected, but none of 82 editing sites in the transcripts of ccmC, ccmFc, cox3, rps12, nad3, and nad5 is impaired in editing in the tada-1 mutant (see Supplemental Figure 6 online).
The Deaminase Domain of TADA Is Structurally Similar to Other Prokaryotic TADA Enzymes
We constructed a homology model of the TADA deaminase domain based on the crystal structure of Staphylococcus aureus TadA (2B3J) in complex with RNA (Losey et al., 2006). This model shows that most of the residues implicated in interactions with the zinc ion and the ligand are conserved (Figure 4). In particular, the interactions with base 34 of the tRNA are preserved (Figure 4B); Ile-26 is replaced by Val-1131 (as in Escherichia coli and Aquifex aeolicus), while the three other amino acids involved are absolutely conserved (Asn-1148, His-1159, and Ala-1160).
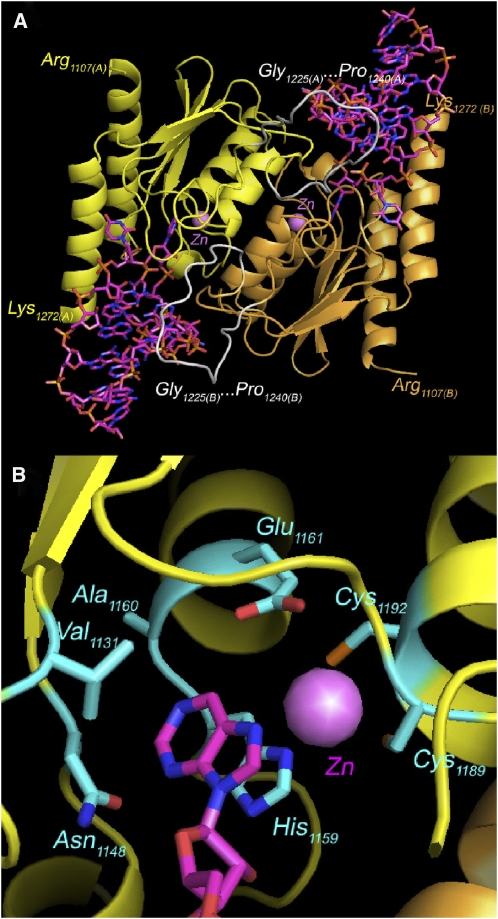
Figure 4.
Model of the Arabidopsis TADA Protein Bound to tRNA Substrate.
(A) Possible structure of an Arabidopsis TADA homodimer bound to two cp-tRNAArg(ACG) substrates. Residues at the C terminus (Arg-1107) and N terminus (Lys-1272) of the represented sequences are indicated. The 16–amino acid insertion (Gly-1225…Pro-1240) is shown in white.
(B) Detail of the active site showing the interactions with A34 of the tRNA anticodon. The coordinated zinc ion is shown.
This allowed us to predict that the N-terminal domain, although not built in our model, is not a hindrance to tRNA recognition, being rather distant from the binding site (Figure 4A). A major difference between bacterial TadAs and plant chloroplast TADAs is an insertion of 16 residues that is present in all plant sequences found in the databases (see Supplemental Figure 3 online), except for the atypical Chlamydomonas reinhardtii protein that also has no predicted N-terminal extension and targeting sequence. This insertion is also poorly conserved in sequence among the different plant TADAs. Finally, our model also proposes a dimer of the same type as in the three bacterial TadA structures known. The portions of the chain involved in contact between the monomers are well conserved, with no sequence insertions and little sequence variation, comparable to that observed between the three known structures.
The Deaminase Domain of TADA Is Sufficient for the Deamination of cp-tRNAArg(ACG) to cp-tRNAArg(ICG) Both in Vitro and in Planta
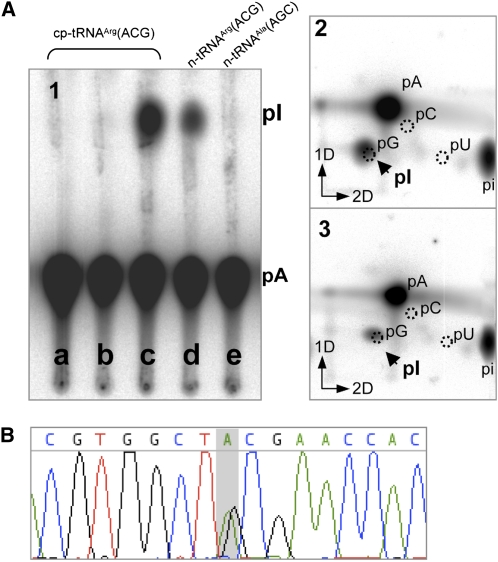
We tested in vitro the activity of the C-terminal part of TADA on the deamination of cp-tRNAArg(ACG). The sequence coding the last 194 amino acids of TADA that comprises the deaminase domain (ΔN-TADA) was expressed in E. coli fused to an N-terminal His-tag, and the activity of the purified soluble protein fraction was tested on in vitro–synthesized cp-tRNAArg(ACG) labeled with [32P]ATP. The presence of inosine was analyzed by thin layer chromatography (TLC) as described (Grosjean et al., 2007). One-dimensional TLC showed a radioactive spot comigrating with inosine monophosphate (IMP), as a consequence of adenosine deaminase activity by ΔN-TADA (Figure 5A1, lane c). No activity was detected in extracts prepared in the same conditions using an unrelated construct cloned in the same expression vector, used as control for contamination by bacterial TadA (Figure 5A1, lane b). The identity of the IMP spot was further confirmed by two-dimensional TLC analysis using different solvent systems (Figure 5A2). To definitely confirm that the activity does not result from bacterial TadA contamination, we also tested the activity of ΔN-TADA purified to homogeneity in denaturing conditions and renatured in vitro. This protein fraction was less active than the protein purified in native conditions but confirmed the deaminase activity of ΔN-TADA on cp-tRNAArg(ACG) (Figure 5A3). We also tested the activity of ΔN-TADA on other tRNA substrates, namely, nuclear encoded cytosolic tRNAArg(ACG) and cytosolic tRNAAla(AGC). Only cytosolic tRNAArg(ACG) was also recognized as substrate by ΔN-TADA (Figure 5A1, lanes d and e, respectively).
Figure 5.
The C-Terminal Domain of TADA Is Sufficient for cp-tRNAArg(ACG) Deamination in Vitro and in Planta.
(A) The C-terminal domain of Arabidopsis TADA (ΔN-TADA) was expressed in E. coli fused to an N-terminal His tag and tested for deamination activity on [32P]ATP-labeled tRNAs synthesized in vitro. The reaction product was digested with nuclease P1 and the resulting 5′-phosphate nucleotides analyzed by one-dimensional (1) or two-dimensional TLC (2 and 3). (1) Deamination activity of ΔN-TADA on cp-tRNAArg(ACG) (lanes a to c), on cytosolic tRNAArg(ACG) and on cytosolic tRNAAla(AGC). a, Control without protein; b, control for bacterial contamination; c to e, plus recombinant ΔN-TADA purified from the E. coli soluble fraction. The position of migration of IMP is indicated (pI). (2) Two-dimensional TLC analysis of ΔN-TADA activity on cp-tRNAArg(ACG). The positions of the four mononucleotides visualized by UV shadowing are indicated. (3) As in 2, with ΔN-TADA purified to homogeneity from the insoluble fraction in denaturing conditions and refolded in vitro.
(B) Deamination of cp-tRNAArg(ACG) is restored to wild-type levels in tada-1 complemented with a ΔN-TADA sequence fused to the chloroplast targeting sequence of RECA1.
We also tested the activity of ΔN-TADA in planta by fusing it to the signal peptide of the chloroplast protein RECA1 (At1g79050) and expressing it in tada-1 under the control of the 35S promoter. ΔN-TADA restored the deamination of cp-tRNAArg(ACG) to cp-tRNAArg(ICG) (Figure 5B), confirming that TADA is responsible for this activity in Arabidopsis and that the large N-terminal extension is dispensable for function both in vitro and in planta.
A Lack of cp-tRNAArg(ICG) Impairs Photosynthesis
Homozygous tada-1 plants show severe phenotypes, including delayed growth, pale-green leaves, and very poor fertility (Figure 6). Leaf cross sections do not show any histological differences compared with young wild-type plants of the same size, and there is no apparent difference in the number of chloroplasts per cell (see Supplemental Figure 7 online). The pale-green color of the mutant is therefore a consequence of reduced chlorophyll content as confirmed by spectrophotometric quantification (see Supplemental Table 1 online). Complementation of the tada-1 mutation by ΔN-TADA targeted to chloroplasts restored a wild-type phenotype (Figure 6), showing that it is the absence of cp-tRNAArg(ICG) that impairs plant growth. RNAi plants showed similar phenotypes of slow growth rate and delay in flowering, which correlated with the accumulation of TADA mRNA (see Supplemental Figure 4A online).
Figure 6.
The tada-1 Mutant Has a Phenotype of Slow Growth and Pale-Green, Round Leaves, Which Is Complemented by ΔN-TADA
(A) Difference in growth between 11-d-old tada-1 plants and wild-type plants grown in vitro.
(B) Difference in size, color, and leaf shape between tada-1 and wild-type plants. The mutant growth phenotype is complemented in tada-1 plants expressing the ΔN-TADA protein.
Different photosynthetic parameters were derived from saturation pulse-induced fluorescence measurements to determine the effects of the tada-1 mutation on photosynthesis and photoprotection (Kramer et al., 2004). The maximum quantum efficiency of photosystem II (PSII) photochemistry (Fv/Fm) of the tada-1 mutant was 60% of that of Col-0 (0.45 ± 0.039 versus 0.77 ± 0.013, respectively). The tada-1 mutation also affected other photosynthetic parameters, such as photochemical and nonphotochemical quenching (NPQ) (Table 1). Measurement of electron transport rates and chlorophyll fluorescence quenching indicated that tada-1 plants dissipated more excitation energy by NPQ than Col-0, as shown by the significantly 2.5-fold higher light-dependent thermal dissipation component of NPQ(ΦNPQ) and lower photochemical efficiency(ΦPSII) (Oxborough, 2004). The coefficient of photochemical quenching, qL, or the fraction of oxidized QA (degree of openness of PSII) was typical for Col-0 (0.59 ± 0.035), while, by contrast, the values for tada-1 plants (0.0) suggest that PSII is closed in the mutant (i.e., reduced QA pool).
Table 1.
Photosynthetic Efficiency
| ΦPSII | ΦNPQ | Φf,D | Σ | |
|---|---|---|---|---|
| Col-0 | 0.51 ± 0.025 | 0.23 ± 0.019 | 0.26 ± 0.013 | 1.0 |
| tada-1 | 0.07 ± 0.077 | 0.58 ± 0.055 | 0.35 ± 0.24 | 1.0 |
The photochemical efficiency, ΦPSII, light-dependent thermal dissipation component of nonphotochemical quenching, ΦNPQ, the sum of fluorescence quenching and light-independent thermal dissipation, Φf,D, and their respective sums, Σ, were determined at normal light. Four 3-week-old plants per line were measured.
A Lack of cp-tRNAArg(ICG) Has Profound Effects on the Chloroplast Transcriptome and Translation
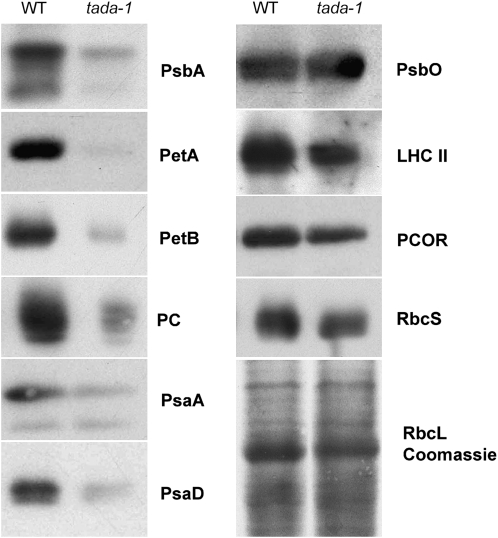
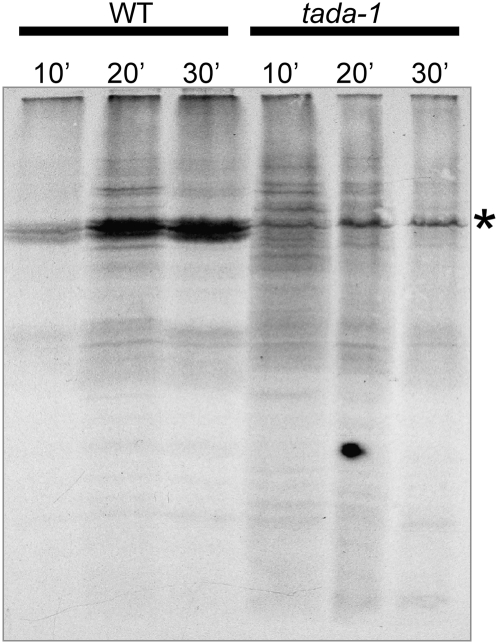
To understand the reasons for the sharp drop in photosynthesis and chlorophyll content, we analyzed the accumulation of several chloroplast proteins by protein gel blots. In tada-1 total protein extracts, we saw a marked decrease of all chloroplast-encoded proteins tested (cytochrome f, cytochrome b6, and D1), whereas most nucleus-encoded proteins were much less affected (Figure 7). Coomassie blue staining of protein gels also showed a decrease in RbcL accumulation. Interestingly, nucleus-encoded PsaD and plastocyanin were also strongly decreased in tada-1. These proteins are associated with the electron transport chain, and their decrease could be a consequence of a deficiency in the assembly of functional photosystems, as shown by the fluorescence measurements. The decrease in plastid-encoded proteins could be related to a decrease in protein synthesis or stability. A pulse-labeling experiment showed that Met incorporation in RbcL is strongly reduced in tada-1, more than fivefold as determined by phosphor-imager quantification (Figure 8). This result does not exclude that the stability of plastid-encoded proteins is also affected but clearly shows that protein synthesis is decreased in tada-1 chloroplasts.
Figure 7.
Reduced Accumulation of Plastid-Encoded Proteins in tada-1.
Protein gel blots of total leaf proteins of wild-type Col-0 and tada-1 plants were analyzed by immunodetection with antibodies recognizing plastid-encoded and nucleus-encoded chloroplast proteins. Plants were grown in vitro in agar plates without sucrose. PsbA, D1 protein; PetA, cytochrome f; PetB, cytochrome b6; PC, plastocyanine; PsaA, photosystem I reaction center subunit A; PsaD, idem subunit D; PsbO, oxygen-evolving enhancer protein 1; LHCII, 26-kD protein from light-harvesting complex II; PCOR, protochlorophyllide oxidoreductase; RbcS, small subunit of Rubisco; RbcL, large subunit of Rubisco.
Figure 8.
Reduced Synthesis Rate of RbcL in tada-1.
Leaf discs of tada-1 and Col-0 were labeled with [35S]Met and incubated for 10, 20, or 30 min in the presence of cycloheximide to inhibit cytosolic translation. Total proteins were then fractionated by SDS-PAGE and autoradiographed. The asterisk indicates the RbcL band.
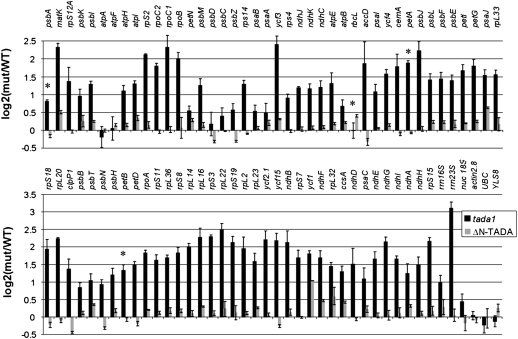
The decrease in the amounts of all chloroplast-encoded proteins tested as well as the decrease in RbcL synthesis suggests a global slowdown of chloroplast protein synthesis. This could be due to a decrease in chloroplast mRNA accumulation or translation. To discriminate between the two possibilities, we analyzed the accumulation of every chloroplast mRNA by quantitative RT-PCR (qRT-PCR) (Figure 9). Surprisingly, the tada-1 mutant overaccumulates most chloroplast transcripts compared with the wild type. This is a consequence of the lack of cp-tRNAArg(ICG), as expression of ΔN-TADA restores a transcription profile that does not significantly deviate from the wild type. In tada-1, transcripts of petA, petB, and psbA (respectively encoding cytochrome f, cytochrome b6, and protein D1) are 3.7, 2.5, and 1.7 times more abundant in tada-1 compared with the wild type respectively, while the rbcL transcript is unchanged. In all cases, there is no correlation between the decrease in protein products and the accumulation of the corresponding transcripts that remain at least at the same levels as in wild-type plants. This further supports the hypothesis that tada-1 plants are impaired in chloroplast translation.
Figure 9.
Accumulation of Chloroplast Transcripts in tada-1 and ΔN-TADA.
Transcript abundances of all protein-encoding and rRNA genes of the chloroplast genome from tada-1 (black bars) and tada-1 complemented with ΔN-TADA (gray bars) were compared with the wild type by qRT-PCR and normalized against a set of nuclear housekeeping genes. The genes are sorted according to their physical location on the chloroplast chromosome. Error bars are standard deviations based on three biological replicates for tada-1 and technical triplicates for tada-1 complemented with ΔN-TADA. Asterisks mark transcripts for which the corresponding protein has been analyzed by protein gel blots (Figure 7). Values are given in Supplemental Table 3 online.
Furthermore, RNAs transcribed by the nucleus-encoded RNA polymerase, such as rpo, accD, and rpl transcripts, accumulate more than those transcribed by the plastid-encoded RNA polymerase (PEP), such as rbcL, psa, and most psb transcripts (Figure 9). Although the quantitative variations are different, this pattern is similar to the chloroplast transcript profiles of Arabidopsis PEP-deficient mutants, such as clb19 and ptac2 (Chateigner-Boutin et al., 2008), suggesting a partial inhibition of PEP activity in the tada-1 mutant. This inhibition is consistent with a decrease in chloroplast translation, as all four of the PEP subunits are encoded in the plastid genome.
tada-1 Can Still Properly Synthesize Chloroplast Proteins
In the absence of cp-tRNAArg(ICG), if CGC and CGA codons could not be recognized by unmodified cp-tRNAArg(ACG), plant survival should depend on misreading of these codons by a noncognate tRNA. This would result in incorporation of amino acids other than Arg at positions coded by CGC or CGA. We checked this possibility by studying the tryptic digest profile of RbcL. Trypsin is a protease cleaving specifically after Arg or Lys residues. If CGC and CGA codons are translated incorrectly, peptides corresponding to cleavage at these particular sites should no longer be detected. After fractionation of total tada-1 proteins by SDS-PAGE and trypsin digestion of the RbcL band, we could unambiguously identify 31 peptides derived from plastid-encoded proteins, namely, RbcL, AtpA, and AtpB (Table 2). Among these peptides, two correspond to cleavage at two out of the eight potentially mistranslated Arg residues in RbcL, three correspond to cleavage at three out of the eight potentially mistranslated Arg residues in AtpB, and two correspond to cleavage at two out of the eight potentially mistranslated Arg residues in AtpA. Therefore, although we cannot exclude that mistranslation occurs in tada-1 chloroplasts, our results show that Arg residues are accurately incorporated at positions corresponding to CGC and CGA codons.
Table 2.
Tryptic Digest of RbcL from tada-1
| m/z Observed | z | Mr Calc | Sequence | Ion Score | |
|---|---|---|---|---|---|
| RbcL | 511.2719 | +2 | 1020.524 | DTDILAAFR41 | 59 |
| 625.3409 | +3 | 1248.6714 | ESTLGFVDLLR350 | 75 | |
| AtpA | 626.8563 | +2 | 1251.6935 | VINALANPIDGR119 | 49 |
| 737.8978 | +2 | 1473.7787 | ASSVAQVVTSLQER216 | 52 | |
| AtpB | 978.0159 | +2 | 1954.016 | (R109)IFNVLGEPVDNLGPVDTR127 | 94 |
| 736.3841 | +2 | 1470.7541 | VGLTALTMAEYFR261 | 89 | |
| 1031.0219 | +2 | 2060.0248 | GIYPAVDPLDSTSTMLQPR378 | 69 |
After mass spectrometry analysis of the RbcL band of tada-1, 31 peptides derived from RbcL, AtpB, and AtpA were unambiguously identified. Only peptides corresponding to cleavage at potentially mistranslated Arg residues (in bold) are listed in this table. Ions score is −10*Log(P), where P is the probability that the observed match is a random event. Ions scores >29 indicate identity or extensive homology (P < 0.05). Mr Calc, relative mass calculated from the mass-to-charge (m/z) ratio. The full list of ions observed during this experiment is available in the Supplemental Data Set 1.
DISCUSSION
TADA Is a tRNAArg Adenosine Deaminase
The chloroplast translation system is derived from its cyanobacterial ancestor and retains predominant prokaryotic characteristics. The recruitment of proteins from the symbiont host has had limited influence on the evolution of the plastid translation system because a significant proportion of its components are still encoded by the plastid genome. These include all rRNAs and tRNAs. Chloroplasts have not acquired exogenous gene sequences during evolution and do not import external tRNAs like plant mitochondria, which have evolved a translation system able to cope with tRNAs that originate from three different genomic compartments (Maréchal-Drouard et al., 1993). cp-tRNAs retain all the typical characteristics of prokaryotic tRNAs. Among these is the editing of cp-tRNAArg(ACG) into cp-tRNAArg(ICG), a deamination reaction that we show is catalyzed by the TADA protein.
The Arabidopsis TADA gene encodes a large protein of which only the C-terminal part resembles bacterial tRNA adenosine deaminases. We modeled this C-terminal domain because the high sequence similarity with bacterial TadA made three-dimensional structure modeling relatively straightforward. In the three bacterial structures that have been determined (2B3J, 1Z3A, and 1WWR), the active site pocket is formed by residues from two different chains. Therefore homodimerization is an absolute requirement for the assembly of the biologically active unit. We identified the positions putatively involved in dimer formation in plant TADAs by comparing the accessible surface of the 2B3J dimer and an isolated monomer (chain 2B3J:A). A third of the residues in those positions is conserved compared with the three known structures. Residues of chain B involved in tRNA binding in pocket A are also well conserved (see Supplemental Figure 3 online): Arg-1176 and Arg-1200 are kept, and at position 1247 an Arg is replaced by a Lys, preserving its basic nature. However, positions 1178 and 1245 are less conserved: Glu is replaced by Ala-1178 (although the E. coli protein has an Ile at this position), and Asn is replaced by His-1245 in all plant sequences. Taken together, despite these small differences, our model suggests that the active form of plant TADA is likely to be a dimer, as in 2B3J, 1Z3A, or 1WWR.
We have shown that the TADA deaminase domain (ΔN-TADA) is sufficient for the deamination of cp-tRNAArg(ACG) to cp-tRNAArg(ICG) both in vitro and in planta (Figure 5). Interestingly, ΔN-TADA was also able to deaminate the cytosolic tRNAArg(ACG) but not tRNAAla(AGC). Because chloroplast and cytosolic tRNAArg(ACG) share little sequence identity, apart from the anticodon loop bases, this suggests that the anticodon loop nucleotides are necessary and sufficient for tRNA recognition by TADA. Mutagenesis studies and the determination of TadA structures showed that each one of the anticodon nucleotides is required for the recognition of the tRNA by bacterial TadA (Wolf et al., 2002; Kuratani et al., 2005; Losey et al., 2006; Luo and Schramm, 2008). The bacterial TadA is probably the ancestor of the eukaryotic ADAT2/TAD2 that, in yeast, forms a heterodimer with ADAT3/TAD3 and catalyzes deamination of seven tRNA species (Gerber and Keller, 1999). However, the bacterial TadA is unable to deaminate substrates of ADAT2/TAD2 in vitro, with the exception of yeast tRNAArg. It is therefore not surprising that the plant TADA, which is phylogenetically related to bacterial TadA, is not involved in the deamination of other tRNAs. In vivo, chloroplast targeting of TADA probably prevents it from deaminating the cytosolic tRNAArg(ACG). The functional homolog of ADAT2/TAD2 in plants remains to be identified.
All higher-plant TADA sequences display large N-terminal domains, whose possible function(s) remain mysterious. They comprise the chloroplast targeting peptide, which in Arabidopsis is contained in the first 259 amino acids (Figure 2B), but the remaining N-terminal sequences contain no identifiable motifs, have no similarities to non-plant proteins and are poorly conserved within the plant kingdom (see Supplemental Table 2 online). The Arabidopsis N-terminal domain is dispensable for the activity and specificity of TADA in vitro (Figure 5A). In vivo, the C-terminal domain alone is sufficient to restore the deamination of cp-tRNAArg(ACG) to levels comparable to the wild type (Figure 5B), reestablishing normal plant growth (Figure 6).
The Absence of cp-tRNAArg(ICG) Slows Down Chloroplast Translation, Which Affects Photosynthesis
Contrary to the situation in bactweria, where TadA is essential for growth, knockout of the Arabidopsis TADA gene is compatible with plant survival. Nevertheless, the mutation has profound effects on the efficiency of chloroplast translation (Figures 6 to 9). In particular, the levels of key components of both photosystems (protein D1 and PsaA) and of the thylakoidal electron transport chain (cytochrome b6f complex subunit VI and cytochrome f) are substantially lower. Although levels of LHCII, a PSII light-harvesting chlorophyll a/b binding protein of the antenna system (Jansson, 1999), are not as affected as chloroplast-encoded proteins, total chlorophyll content is only 40% of wild-type levels in tada-1 plants (see Supplemental Table 1 online). The combination of inefficient energy transfer from the antenna complexes, reduced QA pool, and impaired electron transport chain performance could result in a dramatically lower ratio of photons absorbed by PSII being channeled through photochemical processes (Fv/Fm; Kramer et al., 2004) and the more than doubled amount of nonphotochemical quenching, ΦNPQ, in the tada-1 plants (Table 1).
A priori, the reduced protein synthesis in tada-1 chloroplasts could be due to any of the following defects: (1) reduced levels of translatable mRNA, (2) reduced rates of translation initiation, (3) reduced rates of protein synthesis (chain elongation), and (4) increased rates of protein turnover. We excluded the first of these possible explanations as we found that tada-1 mRNA levels are at least equivalent to those found in wild-type plants. Given that the primary defect in tada-1 plants is the loss of cp-tRNAArg(ICG), the logical assumption is that chloroplast translation is impaired at the elongation step.
When stalled at particular sites, ribosomes can dissociate and/or induce cleavage of the mRNA (reviewed in Dreyfus, 2009). However, slower elongation rates have been associated with the accumulation of heavier polysomes both in yeast and mammals (Hovland et al., 1999; Shenton et al., 2006; Sivan et al., 2007). As a result, mRNAs densely covered with ribosomes might be protected from degradation by nucleases, becoming more stable (Deana and Belasco, 2005; Dreyfus, 2009). This phenomenon, also proposed by Pfalz et al. (2009), could explain the accumulation of plastid transcripts in tada-1, despite the expected reduction in PEP synthesis. A positive effect of reduced chloroplast translation on chloroplast mRNA accumulation was also found for the svr1 Arabidopsis mutant (Yu et al., 2008) but, surprisingly, not in transplastomic tobacco plants impaired in translation by deletion of one of the two cp-tRNAGly genes (Rogalski et al., 2008). This could be because different compensatory mechanisms may be in place. However, in the latter study, only three plastid mRNAs were monitored, including the rbcL transcript whose steady state level is also not affected in tada-1.
Arg Codon Usage Is Biased in Arabidopsis Chloroplasts
The absence of cp-tRNAArg(ICG) in tada-1 plants theoretically prevents translation of two out of the six Arg codons (CGA and CGC) in Arabidopsis chloroplasts. However, the mutation is not lethal, and Arg is still properly inserted at protein positions coded by CGA and CGC codons (Table 2), implying that normal wobble pairing rules can be infringed in chloroplasts. Even in wild-type plants, neither cp-tRNAArg(ACG) nor cp-tRNAArg(ICG) can form a wobble pair with CGG codons, which must be read by a nonstandard decoding mechanism. This might give a clue as to why a lack of cp-tRNAArg(ICG) does not completely block plastid translation.
A detailed analysis of the codon usage of Arabidopsis chloroplast genes shows that the group formed by rbcL, psb, and psa transcripts contains a significantly smaller proportion of CGG codons than other chloroplast transcripts (see Supplemental Table 4 online; Z-test, P value < 0.01). rbcL and psbA transcripts do not contain any CGG codons at all, suggesting that this particular codon is not compatible with high translation rates.
cp-tRNAArg(ACG) and cp-tRNAArg(ICG) have overlapping repertoires according to the wobble rules (both should read CGU codons efficiently), which raises the question of why only ∼50% of cp-tRNAArg(ACG) is apparently deaminated. The Arg codon usage shows a significant bias toward CGU codons compared with CGA and CGC codons in highly translated mRNAs, such as those encoding subunits of ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco), PSI, and PSII (Z-test, P value < 0.001). This might explain the retention of cp-tRNAArg(ACG), as it is expected to be more efficient than cp-tRNAArg(ICG) at translating CGU codons.
These data therefore suggest that CGN codon usage of chloroplast genes is under selection for optimal translation efficiency, given the presence of both cp-tRNAArg(ACG) and cp-tRNAArg(ICG), and that the preferred codon of the CGN series for highly translated RNAs is CGU. This codon bias, which minimizes the necessity for cp-tRNAArg(ICG), is undoubtedly a major reason for why the tada-1 mutation is not lethal.
Two Out of Three Decoding Probably Occurs in Chloroplasts
A complete impairment of chloroplast translation is embryo-lethal in Arabidopsis and tobacco, as demonstrated by knockout mutants either of chloroplast ribosomal proteins (Ahlert et al., 2003; Rogalski et al., 2006; http://www.seedgenes.org), of aminoacyl-tRNA synthetases (Berg et al., 2005), or of some tRNAs (Rogalski et al., 2008). Therefore, the lack of cp-tRNAArg(ICG) is partially compensated for by some other mechanism in tada-1 chloroplasts. According to the superwobble rules, an unmodified uridine at the first anticodon position (position 34 of the tRNA; Figure 1) can read all four nucleotides at the wobble codon position. The possibility for a uridine to accommodate any nucleotide in the codon is restricted to position 34. For this reason, the translation of any of the CGN Arg codons by cp-tRNAArg(UCU) is most unlikely because this would involve superwobbling to occur at both positions 34 and 36 of the tRNA.
Most probably, the translation of CGN codons in tada-1 chloroplasts occurs via two out of three decoding using cp-tRNAArg(ACG). This tRNA lacks a uridine at position 34 required for superwobble but can form the strong Watson-Crick pairing at positions 35 and 36 (Figure 1) required to stabilize codon-anticodon pairing when there is a mismatch at the third position (Lehmann and Libchaber, 2008). However, tobacco cp-tRNAGly(GCC) is apparently unable to decode by two out of three, despite that positions 2 and 3 of the anticodon are cytidines (Rogalski et al., 2008). This could be because, in cp-tRNAGly(GCC), U32 and A38 can form a strong Watson-Crick pairing, whereas C32 and A38 in cp-tRNAArg(ACG) can only form a single weak hydrogen bond (Auffinger and Westhof, 1999). To allow two out of three, the N32-N38 pair has to be non-Watson-Crick to allow the flexibility of the anticodon loop required for proper stacking of the bases (Lehmann and Libchaber, 2008).
These arguments are in favor of two out of three decoding occurring in plastids, even in wild-type Arabidopsis plants, as the Arg CGG codon cannot form a canonical wobble paring with either cp-tRNAArg(ACG) or cp-tRNAArg(ICG). However, the decrease of translation activity in tada-1 clearly shows that cp-tRNAArg(ACG) cannot read all CGN Arg codons efficiently, demonstrating that in this particular case, two out of three pairing is much less effective than wobble pairing. From this perspective, the deamination of cp-tRNAArg(ACG) to cp-tRNAArg(ICG) enhances plastidial translation by replacing inefficient two out of three by canonical wobble codon-anticodon pairing.
METHODS
Plant Materials and Growth Conditions
All experiments used the Col-0 strain of Arabidopsis thaliana as genetic background. Plants were grown on soil under long days (16 h light/8 h dark) at 22°C. For in vitro growth experiments, seeds were surface sterilized and placed on Murashige and Skoog agar plates with or without 1% sucrose. Plates were incubated in the dark at 4°C for 4 d to synchronize germination and subsequently were in a controlled environment chamber (22°C; 8 klux continuous light). T-DNA insertion lines were obtained from the SALK (line SALK_024767) and Gabi-Kat collections (line GK-119G08; Rosso et al., 2003). The RNAi line targeting TADA was obtained from the AGRIKOLA project (line Agri-140-69-2-CATMA1a58100; Hilson et al., 2004).
Genotyping of mutant lines was performed by PCR using gene- and T-DNA–specific primers. For SALK_024767, primers P1 and P3 were used to amplify the wild-type fragment. The mutant allele was detected with primer P3 and the T-DNA left border primer. For GK-119G08, primers P5 and P6 were used to amplify wild-type sequence. The mutant allele was amplified with primer P6 and the T-DNA left border primer. Expression of the mRNAs was tested by amplification of a cDNA fragment spanning the insertion site of the T-DNA, using primers P5 and P6. Total plant RNA and genomic DNA were extracted with TRIzol and DNAzol (Invitrogen), respectively, as described (Zaegel et al., 2006).
tRNA Deaminase Activity Test in Vitro
The C-terminal part of TADA (ΔN-TADA) containing the last 194 amino acids, including the deaminase domain, was amplified with primers P8+P10 and cloned in expression vector pRSET (Invitrogen) in frame with an N-terminal His tag and expressed in Escherichia coli strain ROSETTA(DE3) (Novagen). The soluble protein fraction was purified by zinc-chelating chromatography on Ni-NTA resin (Qiagen). The protein was dialyzed against purification buffer (40 mM Tris-HCl and 300 mM NaCl) containing 50% glycerol and stored at −20°C. Insoluble protein was purified in denaturing conditions in the presence of 6 M urea and renatured in vitro by stepwise dialysis during 3 d against the same buffer containing decreasing concentrations of urea and increasing concentrations of glycerol plus 0.1% Triton and 0.1 mM ZnCl, until a final concentration of 50% glycerol and no urea. The renatured soluble protein was virtually pure, as estimated by Coomassie Brilliant Blue staining.
The activity of the protein was tested on in vitro–synthesized tRNAs labeled with [32P]ATP. The cp-tRNAArg(ACG), cytosolic tRNAArg(ACG), and cytosolic tRNAAla(AGC) sequences were amplified with primers P53+P54, P55+P56, and P57+P58 directly fused to a T7 RNA polymerase promoter including a BstNI site at the 3′ terminus to correctly generate CCA 3′-ends. After BstNI digestion, PCR fragments were used as templates for transcription using the RiboMAX RNA production system (Promega) in the presence of [α-32P]ATP. Incubation conditions were as described (Wolf et al., 2002). Afterwards, the tRNA substrates were digested with nuclease P1 to generate 5′-monophosphate nucleosides that were analyzed by one- or two-dimensional TLC as described (Grosjean et al., 2007).
Complementation of the tada-1 Mutant
The genomic fragment coding for the last 280 amino acids of TADA (ΔN-TADA) was amplified with Gateway primers P47+P48 and recombined into pDONR221 by BP cloning (Invitrogen). Similarly the signal peptide of chloroplast protein RECA1 (At1g79050) was amplified with primers P49 and P50 and recombined by BP cloning into pDONRP4P1R. They were fused into the binary vector pB7m24GW35S under the control of the 35S promoter by LR cloning (Invitrogen). The segregation of the complemented tada-1 mutant was checked as mentioned above, and the presence of the ΔN-TADA construct was confirmed by PCR with primers pairs P51+P52 and P53+P54 amplifying the 5′ and 3′ borders of the construct, respectively.
Protein Gel Blots
Samples of 15 μg of proteins extracted from leaves of 3-week-old plants were fractionated by SDS-PAGE and blotted onto Immobilon-P membranes (Millipore). Antibodies against protein D1, PCOR, PsaD, plastocyanin, PsaA, PsbO, and cytochrome b6 were purchased from Agrisera and used following the manufacturer's recommendations. Antibodies against LHCII, Rubisco small subunit, and cytochrome f were a kind gift of Geraldine Bonnard (Institut de Biologie Moléculaire des Plantes). Proteins recognized by the antibodies were revealed by ECL (GE Healthcare) and autoradiography.
Pulse Labeling of Chloroplast-Encoded Proteins
Pulse labeling experiments were conducted essentially as described (Takahashi et al., 2007). Leaf discs of 19.6 mm2 from young leaves of 3 week-old plants were vacuum infiltrated for 30 s in 0.5 mL of 1 mM KH2PO4 pH6.3, 0.1% Tween 20, 100 μCi of [35S]methionine (specific activity >1000 Ci/mmol). Cycloheximide (100 μg/mL) was added to the infiltration buffer to inhibit cytoplasmic translation. After infiltration, leaf discs were washed in 10 mL water and floated in water plus 100 μg/mL cycloheximide. Discs were exposed to light at 25°C and immediately frozen in nitrogen at the indicated times (10, 20 and 30 min, four leaf discs for each time point). Total proteins were extracted, fractionated by SDS-PAGE and blotted onto Immobilon-P membranes (Millipore) before autoradiography. Quantifications were performed with a Fuji BAS 1000 Imager and MacBAS software version 2.1.
Analysis of RbcL by Mass Spectrometry
Total proteins from leaves of tada-1 were fractionated on a 12% SDS-PAGE gel. After Coomassie Brilliant Blue staining, the band corresponding to RbcL was excised, destained in acetonitrile 50% 10 mM NH4CO3, and trypsin digested overnight. Peptides were extracted from the gel with 50% acetonitrile and 0.5% TFA, dried by vacuum, and analyzed with an 1100 Series HPLC coupled with a 6510 Q-TOF mass spectrometer (Agilent).
Observation of GFP Fluorescence
For in vivo intracellular localization, the sequences encoding the full-length and the first 259 amino acids of TADA were amplified with primers P2+P10 and P2+P4 and cloned in plasmid pCK-GFP3A to express protein:GFP fusions under the control of a 35S promoter as described (Vermel et al., 2002). The resulting plasmids were Nicotiana benthamiana leaves by bombardment, and images were obtained 24 h after transfection with a Zeiss LSM510 confocal microscope with ×60 objectives. The excitation wavelength for GFP detection was 488 nm. Chloroplasts were visualized by the natural fluorescence of chlorophyll. Organelle targeting predictions were determined with programs Predotar and TargetP (http://urgi.versailles.inra.fr/predotar/predotar.html and www.cbs.dtu.dk/services/TargetP/).
Analysis of Chloroplast Transcripts
tada-1 mutants were grown under continuous light in soil, and the analysis of the steady state levels of chloroplast transcripts by qRT-PCR was conducted on three independent biological replicates as described (Chateigner-Boutin et al., 2008). Total Arabidopsis RNA was extracted with the RNeasy plant mini kit (Qiagen), and genomic DNA was removed using RNase-free DNase (DNA-free; Ambion). The absence of DNA was confirmed by PCR prior to reverse transcription with random hexanucleotide primers and Superscript III reverse transcriptase (Invitrogen). Real-time PCR analysis was conducted in 384-well plates with a LightCycler 480 (Roche) using primer pairs described by Chateigner-Boutin et al. (2008) and the LightCycler 480 SYBR I Master mix (Roche). Primer pairs actin2.8F + actin2.8R, Q18SF + Q18SR, UBC-1 + UBC-2, and YLS8-1 + YLS8-2 were used to measure the accumulation of nucleus-encoded transcripts. The specificity of each primer pair was checked by sequencing the PCR product and subsequently by melting curve analysis. For each primer pair and each comparison between mutant and wild-type Col-0, a standard curve based on serial dilutions of the Col-0 cDNA was included. The accumulation of each transcript was analyzed in triplicate and, as the total set of primers was analyzed over more than one PCR plate, primer pairs QC16S, QC23S, and QC18S were included in each plate to correct for variations between plates. The raw data were analyzed using the LightCycler 480 software release 1.5 (Roche) and crossing points determined by second derivative maximum analysis. The accumulation of chloroplast transcripts was normalized by setting the average ratio of nuclear transcripts to 1.
Analysis of tRNA and mRNA Editing
For analysis of tRNA editing by deamination, chloroplast cp-tRNAArg(ACG) and cytosolic tRNAAla(AGC), tRNAVal(AAC), tRNAThr(AGT), tRNALeu(AAG), tRNAIle(AAT), and tRNAArg(ACG) were amplified by RT-PCR with primers P11+P12, P13+P14, P15+P16, P17+P18, P19+P20, P21+22, and P23+P24, respectively. Because the amplified cDNA fragments are too small for direct sequence analysis, they were ligated to plasmid vector pGEM-T (Promega) to obtain DNA fragments long enough for sequencing. Ligated DNA products were reamplified using universal sequence primer (vector-specific) and the reverse tRNA primer. The uncloned cDNA sequences were purified and directly sequenced using universal primer. A-to-I deamination was revealed by the presence of G at the place of A because reverse transcriptase will incorporate a C in place of an I.
Amplification and direct sequence of chloroplast and mitochondrial gene transcripts were done using primers P25 to P46 (see Supplemental Table 5 online).
Fluorescence Analysis and Chlorophyll Content
Fluorescence induction kinetics at room temperature (22°C) were measured using a pulse amplitude modulation fluorometer (PAM101; H. Walz) as described (Giraud et al., 2008) on 3-week-old seedlings grown on soil at 22°C under continuous light at 100 μmol m−2 s−1. Fluorescence measurements were made on four plants for each of the Col-0 and tada-1 lines.
The chlorophyll content of 4-week-old tada-1 and wild-type plants was determined according to Porra (2002) in 80% acetone and 2.5 mM NaH2PO4, pH 7.8.
Modeling of TADA Structure
A search in the Protein Data Bank with the TADA predicted sequence was performed with National Center for Biotechnology Information-BLAST. Homologous proteins belonging to various families were identified: cytosine, guanine, deoxycytidylate and cytidine deaminases, riboflavin biosynthesis proteins, and tRNA adenosine deaminases. Three structures of this last family were found in the library: 1Z3A, 2B3J, and 1WWR, corresponding to sequences from E. coli, Staphylococcus aureus, and Aquifex aeolicus. Their respective scores and E-values are 127 and 2e-29; 124 and 2e-28; and 100 and 2e-21. The 2B3J structure was chosen as scaffold for modeling because although the 1Z3A and 2B3J structures are almost identical (root mean square deviation of 1.2A; Losey et al., 2006), the 1Z3A structure lacks the RNA molecule.
Sequences were aligned using ClustalX (Thompson et al., 1997), and the model was built using Modeler (Sali and Blundell, 1993; http://salilab.org/modeller/about_modeller.html). A large insertion of 17 residues (GGEGNGSEASEKPPPPV) in the Arabidopsis TADA sequence has no equivalent in 2B3J, 1Z3A, or 1WWR and therefore cannot be modeled accurately. No constraint was imposed onto it, and it was modeled as a loop.
The tRNA fragments were modeled as rigid bodies after hand-editing to adjust the sequence to the anticodon stem loop from cp-tRNAArg.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SALK_024767; GK-119G08 (AL757232); Agrik line-140-69-2-CATMA1a58100 (N205329 from the Nottingham Arabidopsis Stock Centre); and TADA, At1g68720 (NM_105544). Accession numbers for other genes used in this study are listed in Supplemental Table 3 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Cytidine/Deoxycytidylate Deaminase Motif Found in Several Arabidopsis Predicted Proteins.
Supplemental Figure 2.Alignment of the Arabidopsis, Rice, and E. coli TADA Proteins Highlights the Poor Conservation of the Large N-Terminal Domains of the Plant TADA.
Supplemental Figure 3. Alignment of the C-Terminal Active Domains of Plant TADA Sequences Found in Genomic and EST Databases Compared with Bacterial TADAs.
Supplemental Figure 4. RNAi Plants Deficient in TADA Expression Are Also Affected in cp-tRNAArg Editing.
Supplemental Figure 5. TADA Is Not Involved in Chloroplast C-to-U Editing.
Supplemental Figure 6. TADA Is Not Involved in Mitochondrial C-to-U Editing.
Supplemental Figure 7. tada-1 Plants Have Normal Leaf Structure and Equivalent Number of Chloroplasts per Cell as Wild-Type Plants.
Supplemental Table 1. Chlorophyll Content of tada-1 Plants.
Supplemental Table 2. Similarities between Plant TADA N-Terminal and C-Terminal Domains.
Supplemental Table 3. Accumulation of Chloroplast Transcripts in tada-1 and tada-1 Complemented with ΔN-TADA Compared with Col-0.
Supplemental Table 4. Frequencies of the CGN Codons in Arabidopsis Chloroplast-Encoded Proteins.
Supplemental Table 5. Primers Used in This Study.
Supplemental Data Set 1. Mass Spectrometry Analysis of the RbcL Band in tada-1.
Supplementary Material
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique, the French Ministry of Research, the Australian Research Council Centre of Excellence in Plant Energy Biology (Grant CEO561495), the State Government of Western Australia, and by an Australian Research Council Linkage International grant (LX0776042). We thank Geraldine Bonnard for antibodies and to Laurence Maréchal-Drouard for helpful discussions.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Etienne Delannoy (edelanno@gmail.com) and José M. Gualberto (jose.gualberto@ibmp-ulp.u-strasbg.fr).
Online version contains Web-only data.
References
- Agris, P.F., Vendeix, F.A., and Graham, W.D. (2007). tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 366 1–13. [DOI] [PubMed] [Google Scholar]
- Ahlert, D., Ruf, S., and Bock, R. (2003). Plastid protein synthesis is required for plant development in tobacco. Proc. Natl. Acad. Sci. USA 100 15730–15735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger, P., and Westhof, E. (1999). Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J. Mol. Biol. 292 467–483. [DOI] [PubMed] [Google Scholar]
- Berg, M., Rogers, R., Muralla, R., and Meinke, D. (2005). Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J. 44 866–878. [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin, A.L., Ramos-Vega, M., Guevara-Garcia, A., Andres, C., de la Luz Gutierrez-Nava, M., Cantero, A., Delannoy, E., Jimenez, L.F., Lurin, C., Small, I., and Leon, P. (2008). CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56 590–602. [DOI] [PubMed] [Google Scholar]
- Conticello, S.G. (2008). The AID/APOBEC family of nucleic acid mutators. Genome Biol. 9 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick, F.H. (1966). Codon-anticodon pairing: the wobble hypothesis. J. Mol. Biol. 19 548–555. [DOI] [PubMed] [Google Scholar]
- Dao, V., Guenther, R., Malkiewicz, A., Nawrot, B., Sochacka, E., Kraszewski, A., Jankowska, J., Everett, K., and Agris, P.F. (1994). Ribosome binding of DNA analogs of tRNA requires base modifications and supports the “extended anticodon”. Proc. Natl. Acad. Sci. USA 91 2125–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana, A., and Belasco, J.G. (2005). Lost in translation: The influence of ribosomes on bacterial mRNA decay. Genes Dev. 19 2526–2533. [DOI] [PubMed] [Google Scholar]
- Dreyfus, M. (2009). Killer and protective ribosomes. Prog. Nucleic Acid Res. Mol. Biol. 85 423–466. [DOI] [PubMed] [Google Scholar]
- Duchene, A.M., Pujol, C., and Marechal-Drouard, L. (2008). Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr. Genet. 55 1–18. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Faivre-Nitschke, E., Grienenberger, J.M., and Gualberto, J.M. (1999). A prokaryotic-type of cytidine deaminase in Arabidopsis thaliana: Cloning and functional characterization. Eur. J. Biochem. 263 1–10. [DOI] [PubMed] [Google Scholar]
- Francis, M.A., Suh, E.R., and Dudock, B.S. (1989). The nucleotide sequence and characterization of four chloroplast tRNAs from the alga Codium fragile. J. Biol. Chem. 264 17243–17249. [PubMed] [Google Scholar]
- Gerber, A., Grosjean, H., Melcher, T., and Keller, W. (1998). Tad1p, a yeast tRNA-specific adenosine deaminase, is related to the mammalian pre-mRNA editing enzymes ADAR1 and ADAR2. EMBO J. 17 4780–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber, A.P., and Keller, W. (1999). An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286 1146–1149. [DOI] [PubMed] [Google Scholar]
- Giraud, E., Ho, L.H., Clifton, R., Carroll, A., Estavillo, G., Tan, Y.F., Howell, K.A., Ivanova, A., Pogson, B.J., Millar, A.H., and Whelan, J. (2008). The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 147 595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean, H., Droogmans, L., Roovers, M., and Keith, G. (2007). Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 425 55–101. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., Houssier, C., Romby, P., and Marquet, R. (1998). Modulatory role of modified nucleotides in RNA loop-loop interaction. In Modification and Editing of RNA, H. Grosjean and R. Benne, eds (Washington, DC: ASM Press), pp. 113–133.
- Hilson, P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res. 14 2176–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovland, R., Eikhom, T.S., Proud, C.G., Cressey, L.I., Lanotte, M., Doskeland, S.O., and Houge, G. (1999). cAMP inhibits translation by inducing Ca2+/calmodulin-independent elongation factor 2 kinase activity in IPC-81 cells. FEBS Lett. 444 97–101. [DOI] [PubMed] [Google Scholar]
- Jansson, S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4 236–240. [DOI] [PubMed] [Google Scholar]
- Kramer, D.M., Johnson, G., Kiirats, O., and Edwards, G.E. (2004). New fluorescence parameters for the determination of q(a) redox state and excitation energy fluxes. Photosynth. Res. 79 209–218. [DOI] [PubMed] [Google Scholar]
- Kuratani, M., Ishii, R., Bessho, Y., Fukunaga, R., Sengoku, T., Shirouzu, M., Sekine, S., and Yokoyama, S. (2005). Crystal structure of tRNA adenosine deaminase (TadA) from Aquifex aeolicus. J. Biol. Chem. 280 16002–16008. [DOI] [PubMed] [Google Scholar]
- Lagerkvist, U. (1986). Unconventional methods in codon reading. Bioessays 4 223–226. [DOI] [PubMed] [Google Scholar]
- Lehmann, J., and Libchaber, A. (2008). Degeneracy of the genetic code and stability of the base pair at the second position of the anticodon. RNA 14 1264–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losey, H.C., Ruthenburg, A.J., and Verdine, G.L. (2006). Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 13 153–159. [DOI] [PubMed] [Google Scholar]
- Lung, B., Zemann, A., Madej, M.J., Schuelke, M., Techritz, S., Ruf, S., Bock, R., and Huttenhofer, A. (2006). Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 34 3842–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., and Schramm, V.L. (2008). Transition state structure of E. coli tRNA-specific adenosine deaminase. J. Am. Chem. Soc. 130 2649–2655. [DOI] [PubMed] [Google Scholar]
- Maréchal-Drouard, L., Weil, J.H., and Dietrich, A. (1993). Transfer RNAs and transfer RNA genes in plants. Annu. Rev. Plant Physiol. 44 13–32. [Google Scholar]
- Osawa, S., Jukes, T.H., Watanabe, K., and Muto, A. (1992). Recent evidence for evolution of the genetic code. Microbiol. Rev. 56 229–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxborough, K. (2004). Imaging of chlorophyll a fluorescence: theoretical and practical aspects of an emerging technique for the monitoring of photosynthetic performance. J. Exp. Bot. 55 1195–1205. [DOI] [PubMed] [Google Scholar]
- Pfalz, J., Bayraktar, O.A, Prikryl, J., and Barkan, A. (May 7, 2009). Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. http://dx.doi.org/10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed]
- Pfitzinger, H., Weil, J.H., Pillay, D.T., and Guillemaut, P. (1990). Codon recognition mechanisms in plant chloroplasts. Plant Mol. Biol. 14 805–814. [DOI] [PubMed] [Google Scholar]
- Porra, R.J. (2002). The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 73 149–156. [DOI] [PubMed] [Google Scholar]
- Rogalski, M., Karcher, D., and Bock, R. (2008). Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 15 192–198. [DOI] [PubMed] [Google Scholar]
- Rogalski, M., Ruf, S., and Bock, R. (2006). Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res. 34 4537–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Sali, A., and Blundell, T.L. (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234 779–815. [DOI] [PubMed] [Google Scholar]
- Salinas, T., Duchene, A.M., and Marechal-Drouard, L. (2008). Recent advances in tRNA mitochondrial import. Trends Biochem. Sci. 33 320–329. [DOI] [PubMed] [Google Scholar]
- Schaub, M., and Keller, W. (2002). RNA editing by adenosine deaminases generates RNA and protein diversity. Biochimie 84 791–803. [DOI] [PubMed] [Google Scholar]
- Shenton, D., Smirnova, J.B., Selley, J.N., Carroll, K., Hubbard, S.J., Pavitt, G.D., Ashe, M.P., and Grant, C.M. (2006). Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 281 29011–29021. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 5 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan, G., Kedersha, N., and Elroy-Stein, O. (2007). Ribosomal slowdown mediates translational arrest during cellular division. Mol. Cell. Biol. 27 6639–6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, I., Peeters, N., Legeai, F., and Lurin, C. (2004). Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4 1581–1590. [DOI] [PubMed] [Google Scholar]
- Sprinzl, M., Steegborn, C., Hubel, F., and Steinberg, S. (1996). Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 24 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, S., Bauwe, H., and Badger, M. (2007). Impairment of the photorespiratory pathway accelerates photoinhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiol. 144 487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker, B., Peiter, E., Dixon, D.P., Moffat, C., Capper, R., Bouche, N., Edwards, R., Sanders, D., Knight, H., and Knight, M.R. (2008). Getting the most out of publicly available T-DNA insertion lines. Plant J. 56 665–677. [DOI] [PubMed] [Google Scholar]
- Vermel, M., Guermann, B., Delage, L., Grienenberger, J.M., Marechal-Drouard, L., and Gualberto, J.M. (2002). A family of RRM-type RNA-binding proteins specific to plant mitochondria. Proc. Natl. Acad. Sci. USA 99 5866–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, J., Gerber, A.P., and Keller, W. (2002). tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J. 21 3841–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., Liu, X., Alsheikh, M., Park, S., and Rodermel, S. (2008). Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell 20 1786–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaegel, V., Guermann, B., Le Ret, M., Andres, C., Meyer, D., Erhardt, M., Canaday, J., Gualberto, J.M., and Imbault, P. (2006). The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell 18 3548–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.