Abstract
Soil salinity affects large areas of cultivated land, causing significant reductions in crop yield globally. The Na+ toxicity of many crop plants is correlated with overaccumulation of Na+ in the shoot. We have previously suggested that the engineering of Na+ exclusion from the shoot could be achieved through an alteration of plasma membrane Na+ transport processes in the root, if these alterations were cell type specific. Here, it is shown that expression of the Na+ transporter HKT1;1 in the mature root stele of Arabidopsis thaliana decreases Na+ accumulation in the shoot by 37 to 64%. The expression of HKT1;1 specifically in the mature root stele is achieved using an enhancer trap expression system for specific and strong overexpression. The effect in the shoot is caused by the increased influx, mediated by HKT1;1, of Na+ into stelar root cells, which is demonstrated in planta and leads to a reduction of root-to-shoot transfer of Na+. Plants with reduced shoot Na+ also have increased salinity tolerance. By contrast, plants constitutively expressing HKT1;1 driven by the cauliflower mosaic virus 35S promoter accumulated high shoot Na+ and grew poorly. Our results demonstrate that the modification of a specific Na+ transport process in specific cell types can reduce shoot Na+ accumulation, an important component of salinity tolerance of many higher plants.
INTRODUCTION
Soil salinity affects large areas of cultivated land in more than 100 countries (Rengasamy, 2006). Increased soil salinity negatively affects the growth of many crop plants, and the continued salinization of arable land provides an increasing threat to global crop production, especially in irrigated systems (Munns and Tester, 2008). Increasing the salinity tolerance of crop plants will provide an important contribution to the maintenance of crop yields.
The Na+ toxicity of many crop plants is correlated with overaccumulation of Na+ in the shoot (Munns, 1993, 2002; Tester and Davenport, 2003; Møller and Tester, 2007). Na+ is taken up from the soil by the plant root system and transported to the shoot in the transpiration stream (Tester and Davenport, 2003). Shoot Na+ accumulation is the net result of distinct Na+ transport processes occurring in different organs and cell types (Tester and Davenport, 2003), and each of these processes contributes to the salinity tolerance of a plant.
Such Na+ transport processes include passive influx of Na+ into the root system, which is likely to be mediated by non-selective cation channels (Davenport and Tester, 2000; Demidchik et al., 2002; Demidchik and Tester, 2002), with cyclic nucleotide-gated channels and Glu receptors being likely candidate gene families encoding these proteins (Demidchik and Tester, 2002; Roy et al., 2008). The HKT family of ion transporters originally was named for the high-affinity potassium transporter properties of the first member of this family that was isolated, but it is more complex than originally realized, as the affinity and selectivity of many proteins encoded by members of this gene family are different to that indicated by the name. As has been shown for HKT2;1 and HKT2;2 in rice (Oryza sativa; Horie et al., 2001; Garciadeblas et al., 2003), the HKT family of ion transporters might also be involved in Na+ influx in some species, although these do not appear to be major contributors to Na+ transport in saline conditions.
Influx of Na+ into the root system is counteracted by efflux of Na+ to the soil solution, an active process likely to occur in antiport with H+. In Arabidopsis thaliana, this process might involve the Salt Overly Sensitive pathway in which SOS3, a calcium binding protein, recruits SOS2, a Ser/Thr protein kinase, to the plasma membrane where it activates the Na+/H+ antiporter, SOS1, by phosphorylation (Qiu et al., 2002). Uncertainty remains about the role of this specific process in Na+ efflux from roots, as this system appears to be preferentially expressed in the stele of mature roots. Other as yet uncharacterized Na+/H+ antiporters might also be involved in the efflux of Na+ from the root system and might be encoded by the SOS1-like gene at locus At1g14660 in Arabidopsis as well as in the cation/H+ exchanger gene family (Pardo et al., 2006).
Radial movement of Na+ across the root occurs via the apoplast and/or the symplast. Flow of Na+ through the apoplast past the endodermal Casparian strip has been found to be substantial in rice (Yeo et al., 1987), but does not contribute significantly to shoot Na+ accumulation in Arabidopsis (Essah et al., 2003).
Na+ is transported from the root to the shoot via the transpiration stream, and Arabidopsis SOS1 is thought to be involved in the loading of Na+ from the xylem parenchyma cells into the xylem tracheids (Shi et al., 2002). The HKT transporters have been shown to be involved in the retrieval of Na+ from the xylem in several species (Ren et al., 2005; Sunarpi et al., 2005; Byrt et al., 2007; Davenport et al., 2007).
Apart from the level of Na+ accumulation in the shoot, another component of plant salinity tolerance is the ability of the tissue to tolerate Na+ (Munns and Tester, 2008). Tissue tolerance to Na+ involves the storage of Na+ in vacuoles, involving Na+/H+ antiporters from the Na+/H+ exchanger gene family and the vacuolar pyrophosphatase (Apse et al., 1999; Gaxiola et al., 1999, 2001). Further aspects of tissue tolerance have recently been reviewed by Munns and Tester (2008).
For glycophytes, growth in saline soils results in an overaccumulation of Na+ in the leaf tissue causing premature leaf senescence. For several crop plants, it has been empirically observed that a significant component of salinity tolerance is the ability to exclude Na+ from the shoot (Munns, 1993, 2002; Tester and Davenport, 2003; Møller and Tester, 2007). Notably, this correlation between salinity tolerance and exclusion of Na+ from the shoot has been found in several wheat (Triticum spp) species, barley (Hordeum vulgare), rice, rye (Secale cereale), and triticale (× Triticosecale) (Tester and Davenport, 2003; Møller and Tester, 2007 and references therein), although exceptions to this have been documented in bread wheat (Triticum aestivum) (Genc et al., 2007) and rice (Yeo et al., 1990). For Arabidopsis, the relationship between Na+ exclusion and salinity tolerance appears complex, and often there is no correlation observed (Møller and Tester, 2007). Thus, Arabidopsis is a good model for studying and manipulating Na+ transport processes to gain an understanding of how shoot Na+ exclusion might be achieved, but how whole plant salinity tolerance compares in Arabidopsis and cereals is less certain (Møller and Tester, 2007).
Drawing upon the current understanding of plant salinity tolerance, it appears likely that modification of specific root Na+ transport processes might improve Na+ exclusion from the shoot and result, at least for some plants, in an increase in salinity tolerance. For example, initial influx of Na+ from the soil could be decreased in the outer cell layers of the root, or efflux of Na+ from these cells to the apoplast or soil solution could be increased. In the stelar cells surrounding the vasculature, the loading of Na+ into the xylem vessels could be decreased or retrieval of Na+ from the transpiration stream increased. Accordingly, at the cellular level, Na+ transport processes need to be modified in opposite directions in the inner and outer parts of the root to minimize Na+ accumulation in the shoot. Consequently, plasma membrane Na+ transport processes in the root need to be altered in a cell type–specific manner.
Several findings have indicated that cell type–specific processes contribute to plant salinity tolerance. These include cell type–specific expression patterns of genes involved in Na+ transport, such as SOS1 (Shi et al., 2002), HKT1;1 (Mäser et al., 2002a), sodium/myoinositol symporters and K+ transporters from Mesembryanthemum crystallinum (Chauhan et al., 2000; Su et al., 2002; Jou et al., 2004), and particular H+-ATPases in Atriplex nummularia (Niu et al., 1996) and Arabidopsis (Vitart et al., 2001). Accordingly, electrophysiological studies have established that cells from different cell types in roots possess different properties (e.g., Roberts and Tester, 1995; Kiegle et al., 2000a; Gilliham and Tester, 2005). Furthermore, the way in which ions might be partitioned between organs, cell types, and organelles within a cell has been described (Tester and Leigh, 2001), and, in a recent study, such partitioning was observed in roots of durum wheat (Triticum turgidum ssp durum) lines. These lines transported different amounts of Na+ to the shoot (Davenport et al., 2005; James et al., 2006) and were also found to differ in the pattern of localization of Na+ and K+ in specific root cells (Läuchli et al., 2008). However, to our knowledge, cell type–specific changes in Na+ transport have not previously been used to generate transgenic plants with reduced shoot Na+ accumulation (and, hence, potentially increased salinity tolerance).
Members of the HKT family of transport proteins have been implicated in Na+ transport in Arabidopsis (Mäser et al., 2002a; Berthomieu et al., 2003; Rus et al., 2004; Sunarpi et al., 2005; Davenport et al., 2007), rice (Ren et al., 2005; Horie et al., 2007), and wheat (Davenport et al., 2005; James et al., 2006; Byrt et al., 2007). The Arabidopsis genome contains a single member of this gene family, HKT1;1 (Platten et al., 2006), which encodes a plasma membrane protein (Sunarpi et al., 2005) that mediates Na+ influx into cells when heterologously expressed in Xenopus laevis oocytes and Saccharomyces cerevisiae (Uozumi et al., 2000). Using HKT1;1 promoter fusions with the β-glucuronidase (GUS) reporter gene as well as immunoelectron microscopy, it was found that HKT1;1 is expressed in stelar cells of the root and within the vascular tissues in leaves (Mäser et al., 2002a; Berthomieu et al., 2003; Sunarpi et al., 2005). The function of HKT1;1 in planta has been elucidated by studies of hkt1;1 mutant plants, which have higher shoot Na+ concentrations and lower root Na+ compared with wild-type plants (Mäser et al., 2002a; Berthomieu et al., 2003; Gong et al., 2004; Rus et al., 2004; Davenport et al., 2007). HKT1;1 is important for plant salinity tolerance through its involvement in regulating the distribution of Na+ between root and shoot by mediating retrieval of Na+ from the transpiration stream (Sunarpi et al., 2005; Davenport et al., 2007); consequently, the hkt1;1 mutants analyzed have been found to be salt sensitive (Mäser et al., 2002a; Berthomieu et al., 2003; Rus et al., 2004; Sunarpi et al., 2005). Using a series of grafting experiments, it was shown that HKT1;1 expressed in the root, not in the shoot, regulates Na+ accumulation in the shoot of Arabidopsis and that HKT1;1 is not expressed in the root in all Arabidopsis accessions (e.g., Ts-1 and Tsu-1; Rus et al., 2006).
Although some members of the HKT gene family, primarily in subfamily 2, encode proteins that can transport K+ in some conditions (Platten et al., 2006), there is no evidence in eukaryotic systems for significant K+ transport capacity by the Arabidopsis gene product. There is also some evidence in planta for the high selectivity of Arabidopsis HKT1;1 for Na+ over other Group I alkali cations: Davenport et al. (2007) found effects of an hkt1;1 mutation on 22Na+ transport but not on that of 86Rb+.
In this study, we have employed a targeted genetic modification approach to express the Arabidopsis HKT1;1 gene, which encodes a Na+ transporter, in specific cell types in the mature root of Arabidopsis to increase the retrieval of Na+ from the transpiration stream. Plants in which HKT1;1 was constitutively overexpressed using the cauliflower mosaic virus 35S promoter (Pro35S) were also produced to allow a comparison of the effect of cell type–specific and constitutive overexpression of HKT1;1 on Na+ accumulation and salinity tolerance. We show that, in contrast with plants constitutively overexpressing the gene, overexpression of HKT1;1, specifically in the root stele of Arabidopsis, significantly decreases shoot Na+ and consequently increases the salinity tolerance of these plants.
RESULTS
Constitutive Overexpression of HKT1;1 Results in Overaccumulation of Na+ in the Shoot and Na+ Toxicity
Plants were produced with constitutive overexpression of HKT1;1 using Pro35S. Primary transformants were grown in a low-nutrient soil-like mix for 5 weeks and watered with a nutrient solution containing 2 mM NaCl. The effects of constitutive overexpression of HKT1;1 were deleterious, as Pro35S:HKT1;1 plants displayed stunted growth and chlorosis (see Supplemental Figure 1 online). Fully expanded leaves were examined using inductively coupled plasma–atomic emission spectroscopy (ICP-AES). The accumulation of Na+ was 3.6-fold higher on average in the leaves of primary transformants compared with controls (438 ± 43 [n = 25 primary transformants] compared with 122 ± 6 [n = 53 Columbia-0 (Col-0) control plants] mg Na+·kg−1 FM (fresh mass); P < 0.001).
As the hkt1;1 mutant phenotype is also characterized by overaccumulation of Na+ in the shoot, the overexpression of HKT1;1 in each of the high Na+ Pro35S:HKT1;1 plants was tested by quantitative RT-PCR (12,400 ± 4600 for Pro35S:HKT1;1 [n = 15] compared with 47 for Col-0, when normalized to Arabidopsis cyclophilin). This was to ensure that the high shoot Na+ phenotype was indeed due to HKT1;1 overexpression rather than to gene silencing. No silencing was observed in any of the plants tested.
Enhancer Trap Lines Driving Cell Type–Specific Gene Expression in the Stelar Cells of the Mature Root
In contrast with the constitutive expression of HKT1;1 reported above, plants were also produced that had high levels of expression of HKT1;1 in tightly defined cell types in the root. The aim was to specifically increase Na+ transport into the stelar root cells to increase Na+ retrieval from the transpiration stream. Accordingly, an enhancer trap system was exploited to drive cell type–specific expression of HKT1;1. This enhancer trap system was developed in Arabidopsis by Jim Haseloff and is based on the use of the yeast transcription activator GAL4 and the GAL4 upstream activation sequence (UASGAL4) as previously described (Haseloff, 1999) (Figure 1A). The system can be used to drive the specific and high level of expression of a gene of choice upon transformation with another DNA construct including the UASGAL4 sequence and the coding sequence of the gene of choice (Figure 1B). The expression of the gene of choice is thereby driven by the trapped enhancer or promoter that also drives the expression of a gene encoding a modified green fluorescent protein targeted to the endoplasmic reticulum (mGFP5-ER).
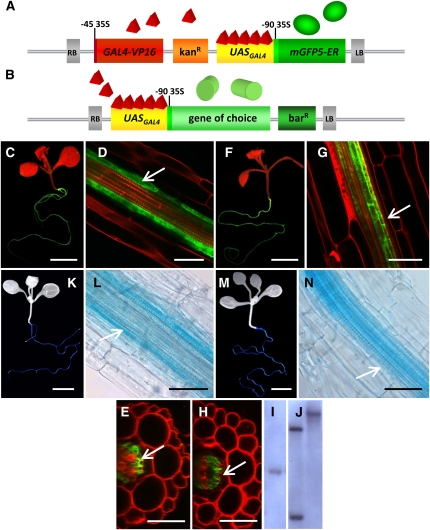
Figure 1.
Characterization of Enhancer Trap Lines with Cell Type–Specific Expression of mGFP5-ER in the Root Stele.
(A) Outline of the enhancer trap T-DNA.
(B) Outline of the transactivation T-DNA in which the coding sequence of a gene of choice is placed downstream of UASGAL4.
(C) to (E) mGFP5-ER expression in whole J2731* plants and longitudinal and transverse section view of mGFP5-ER expression in J2731* roots. Arrows point to pericycle cells.
(F) to (H) Same as (C) to (E), respectively, but with E2586.
(I) DNA gel blot of J2731* genomic DNA restricted with HindIII determined the presence of a single enhancer trap T-DNA insert.
(J) DNA gel blot of E2586 genomic DNA restricted with HindIII (left panel) or HpaI (right panel). Two bands were observed using HindIII, with the lower band of identical size to the T-DNA insert of 6.4 kb. Using HpaI, a single band was detected, indicating that a single concatameric T-DNA insert is present in E2586.
(K) and (L) GUS activity in whole J2731*UASGAL4:uidA plants and longitudinal view of root.
(M) and (N) Same as (K) and (L), respectively, but with E2586 UASGAL4:uidA. RB, right T-DNA border; LB, left T-DNA border; kanR, neomycin phosphotransferase II gene; barR, phosphinotricin acetyl transferase gene; UASGAL4, upstream activation sequence of GAL4-VP16; red triangles, GAL4-VP16 protein; green sphere, mGFP5-ER protein; green cylinder, protein encoded by gene of choice.
Bars = 0.5 cm for (C), (F), (K), and (M) and 40 μm in (D), (E), (G), (H), (L), and (N).
Two Arabidopsis enhancer trap lines were identified that had strong and stable mGFP5-ER expression patterns specific, in roots, to the stele. In line J2731* (in a C24 background), mGFP5-ER expression was localized within the pericycle of main and lateral roots (Figures 1C to 1E). In E2586 (in a Col background), expression of mGFP5-ER was detected in the main and lateral roots specifically in cells within the vascular bundle, excluding the pericycle cells (Figures 1F to 1H). Importantly, in neither line was mGFP5-ER expression observed in the developing root or root tip. No developmental or salinity stress-induced change was observed in the expression pattern of mGFP5-ER, which was stable in the five generations analyzed. The original line J2731 generated by Jim Haseloff had several inserts and was backcrossed until a single insert line was obtained, termed J2731*. Thus, both lines used in this work contained a single site of T-DNA insertion, this being a concatamer in E2586 (Figures 1I and 1J). The two sites were in intergenic regions on the short (J2731*) and long (E2586) arms of chromosome 1 (see Supplemental Figure 2 online).
The GAL4 UASGAL4 enhancer trap system was tested for its ability to drive cell type–specific gene expression using the GUS reporter gene (uidA). Following transformation of the two selected enhancer trap lines J2731* and E2586 with the DNA construct UASGAL4:uidA, the cell type specificity of expression of uidA was tested by GUS staining of 2-week-old seedlings. GUS activity was detected in the same cell types as mGFP5-ER (Figures 1K to 1N). Similar results for the cell type–specific expression of a yellow fluorescent protein-aequorin fusion using the UASGAL4 system have been published previously (Kiegle et al., 2000a; Dodd et al., 2006).
Construction of Plants Overexpressing HKT1;1 Specifically in the Root Stele
In an attempt to increase retrieval of Na+ from the xylem in the root and thus reduce Na+ transfer to the shoot, the Na+ transporter HKT1;1 was overexpressed in specific cell types in the stele using the characterized enhancer trap lines. Lines J2731* and E2586 were transformed with a T-DNA in which the HKT1;1 coding sequence was placed downstream of the UASGAL4 sequence.
To avoid the risk of altering the function of the protein, as has been reported previously (Kato et al., 2003), the encoded HKT1;1 protein was not modified to include an immuno-tag or fluorescent marker. To test the cell type specificity of the overexpression of HKT1;1, functional antibodies were raised to a synthetic peptide corresponding to the region of HKT1;1 targeted by Sunarpi et al. (2005). However, it was found that reliable immunolocalization of the full-length protein in Arabidopsis overexpressing the HKT1;1 gene was not possible using this antibody.
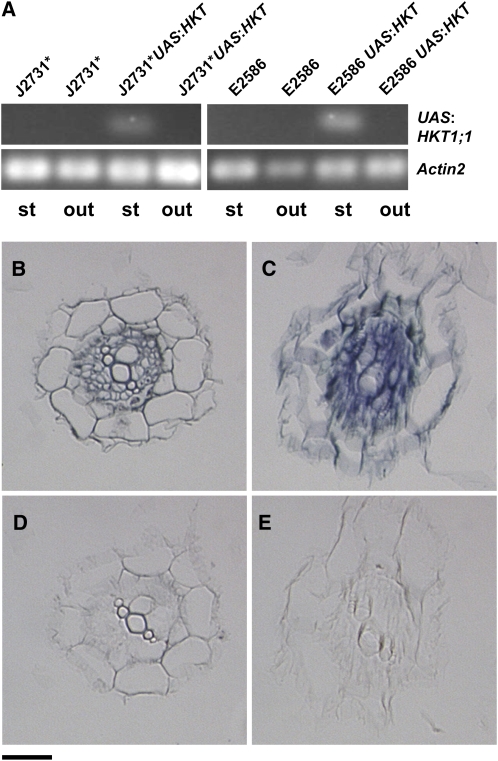
Instead, the cell type specificity of the expression pattern of the HKT1;1 transgene was tested using two independent approaches. Laser dissection allowed the separation of stelar cells from the cortical and epidermal cell layers of transverse sections of roots from J2731* UASGAL4:HKT1;1, E2586 UASGAL4:HKT1;1, and parental J2731* and E2586 plants (see Supplemental Figure 3 online). These laser dissected samples were used for RT-PCR–based detection of the transgene mRNA, which was found only in stelar cells of the UASGAL4:HKT1;1 lines (Figure 2A). Cell autonomy of overexpression of HKT1;1 was further confirmed using mRNA in situ hybridization (Figures 2B to 2E).
Figure 2.
Cell Type–Specific Expression of HKT1;1 Driven by the Enhancer Trap System.
(A) Transgene HKT1;1 message detected specifically in stelar cells using RT-PCR on laser-dissected samples of outer (out) and stelar (st) parts of roots. Three technical replicates were performed on sections taken from one plant.
(B) to (E) HKT1;1 message detected by mRNA in situ localization with an antisense ([B] and [C]) and sense ([D] and [E]) HKT1;1 riboprobe on cross sections of roots from J2731* ([B] and [D]) and J2731*UASGAL4:HKT1;1 ([C] and [E]). Bar = 20 μm.
Na+ Influx Capacity Enhanced in Stelar Root Cells Overexpressing HKT1;1
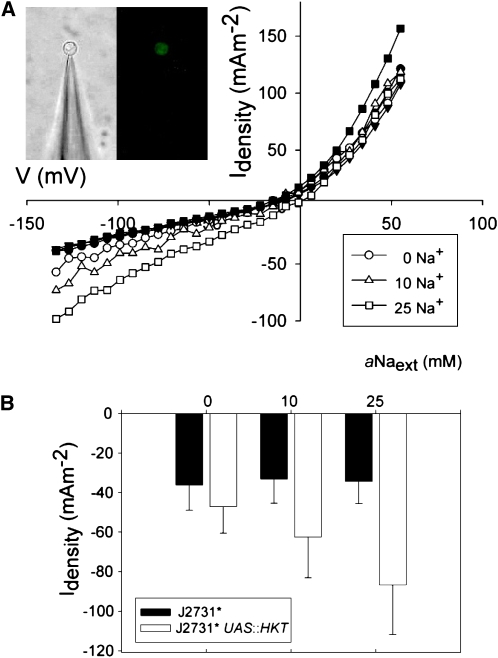
A direct assay of the increased capacity of pericycle cells to take up Na+ from the apoplast upon overexpression of HKT1;1 was conducted using patch clamp electrophysiology of mGFP5-ER–expressing protoplasts isolated from root tissue of J2731*UASGAL4:HKT1;1 and control J2731* plants (n = 5 each) (Figure 3, inset). Inward currents increased in magnitude with an increase in [Na]ext only across the plasma membrane of fluorescent protoplasts from J2731*UASGAL4:HKT1;1 plants (Figure 3A). At −125 mV, a commonly used hyperpolarized potential indicative of an average in planta membrane potential, the mean inward current density of fluorescent protoplasts from J2731* did not significantly vary when [Na+]ext was varied and was similar to that of fluorescent protoplasts from J2731*UASGAL4:HKT1;1 in 0 mM [Na+]ext. By contrast, in fluorescent protoplasts isolated from J2731*UASGAL4:HKT1;1, an increase in [Na+]ext to 10 and 25 mM resulted in a positive shift of the reversal potential of measured currents consistent with the positive shift in ENa and an increase in inward current (Figures 3A and 3B). If currents were due to the movement of Cl−, then on addition of NaCl, the observed reversal potential would move in the opposite direction to the equilibrium (or Nernst) potential for Na+ and toward that of Cl−. However, the observed reversal potential for these currents moves in the same direction as the equilibrium potential for Na+, providing good evidence that the current associated with the overexpression of HKT1;1 was due primarily to the movement of Na+ and not of Cl−. The difference between the actual reversal potential and the theoretical equilibrium potential for Na+ is most likely due to native anion currents contaminating the overall whole-cell currents. These are clearly significant, as the change in reversal potential with increasing salt in control protoplasts follows the change in reversal potential for Cl−, opposite to that observed in protoplasts expressing HKT1;1. This observation of HKT1;1-mediated currents that are consistent with Na+ influx into plant cells is in accordance with the findings of previous heterologous expression studies (Uozumi et al., 2000; Mäser et al., 2002a). From these results, we conclude that overexpression of HKT1;1 leads to the overexpression of a functional protein, which increases Na+ transport into cells.
Figure 3.
HKT1;1-Catalyzed Na+ Inward Transport into Stelar Root Cells.
(A) Enzymatically isolated mGFP5-ER–expressing protoplasts originating from the pericycle of J2731* UASGAL4:HKT1;1 (filled) and J2731* (hollow) roots were examined. Each protoplast was irrigated with solutions containing 0, 10, and 25 mM Na+. Inward current density was plotted against voltage. Photographs showing an mGFP5-ER–expressing protoplast sealed to the glass micropipette viewed in bright field (left) and excited with a mercury arc lamp using a standard GFP filter set (right) are shown in inset.
(B) Mean inward current density at −120 mV across the plasma membrane of J2731* UASGAL4:HKT1;1 (hollow) and J2731* (filled) protoplasts in the same solutions as outlined in (A). Values presented are means ± se. n = 5 for each genotype.
Overexpression of HKT1;1 in Root Stele Decreases Root-to-Shoot Transfer of 22Na+, while Unidirectional 22Na+ Influx Is Unaffected
Using the radioisotope 22Na+, we were able to obtain an insight into how the overexpression of HKT1;1 specifically in the root stele or constitutively affects Na+ influx into the root system and transfer of Na+ from the root to the shoot. Unidirectional influx of 22Na+ into roots was not significantly affected by stelar-specific expression of HKT1;1 (P = 0.6 in J2731* and P = 0.8 in E2586 background), whereas the fraction of 22Na+ transferred to the shoot in intact plants was greatly reduced (Table 1) (P < 0.01 for J2731* and P < 0.001 for E2586). By contrast, constitutive overexpression of HKT1;1 in Col-0 increased unidirectional influx into the root (2.53 ± 0.19 [n = 19] compared with 1.92 ± 0.14 [n = 23] μmol·g−1 FM·min−1; P < 0.05). The transfer of Na+ from root to shoot was also increased following constitutive overexpression of HKT1;1 (33 ± 2 [n = 21] compared with 16 ± 2 [n = 24] percent of 22Na+ translocated to the shoot relative to total plant 22Na+; P < 0.001). These results are consistent with the expected expression patterns of HKT1;1 in these lines and the function of HKT1;1 in Na+ influx into plant cells, in that root stele–specific expression of HKT1;1 should not change Na+ influx in epidermal and cortical cells but should increase Na+ retrieval from the transpiration stream in the root (thereby reducing the transfer of Na+ to the shoot). Constitutive overexpression of HKT1;1 might result in expression in the epidermal and cortical cells, thereby increasing uptake of Na+ from the environment. Although constitutive overexpression might also increase Na+ retrieval from the transpiration stream, these results suggest that such an effect was overcome by the higher Na+ influx into the roots of these lines.
Table 1.
Root Stele–Specific Overexpression of HKT1;1 Reduces Transfer of 22Na+ from Root to Shoot and Changes the Distribution of Na+ and between Shoot and Root
| Genotype | 22Na+ Root Influx (μmol·g−1 FM·min−1) | 22Na+ Translocated to Shoot (%) | Shoot [Na+] (mg·kg−1 FM) | Root [Na+] (mg·kg−1 FM) | Shoot [K+] (mg·kg−1 FM) | Root [K+] (mg·kg−1 FM) |
|---|---|---|---|---|---|---|
| J2731* | 1.62 ± 0.15 (n = 10) | 29 ± 2 (n = 47) | 3039 ± 450 (n = 12) | 751 ± 35 (n = 12) | 2377 ± 177 (n = 12) | 4935 ± 85 (n = 12) |
| J2731* UAS:HKT | 1.74 ± 0.13 (n = 14) | 21 ± 2 (n = 11) | 1278 ± 73 (n = 12) | 1548 ± 61 (n = 12) | 3043 ± 75 (n = 12) | 4060 ± 59 (n = 12) |
| E2586 | 1.61 ± 0.18 (n = 16) | 29 ± 3 (n = 40) | 1009 ± 153 (n = 12) | 1536 ± 60 (n = 12) | 2786 ± 60 (n = 12) | 3463 ± 79 (n = 12) |
| E2586 UAS:HKT | 1.55 ± 0.16 (n = 20) | 12 ± 2 (n = 16) | 632 ± 111 (n = 12) | 1815 ± 79 (n = 12) | 3104 ± 108 (n = 12) | 3176 ± 66 (n = 12) |
Measurements of unidirectional flux of 22Na+ into excised roots and percentage of 22Na+ that was translocated to the shoot in intact plants of J2731* UASGAL4:HKT1;1, E2586 UASGAL4:HKT1;1, and parental controls are shown. Plants used were from the T3 generation. Average shoot and root Na+ and K+ concentrations measured by ICP-AES of 5-week-old T4 plants grown hydroponically and subjected to salt stress for 1 week prior to harvest. The salt stress consisted of 10 mM increases in NaCl every 12 h until 50 mM NaCl was reached. CaCl2 was supplemented at every addition of NaCl to maintain constant Ca2+ activity. Values presented are means ± se.
Overexpression of HKT1;1 Specifically in Root Stele Decreases Shoot Na+ Accumulation
The effect of overexpression of HKT1;1 and thereby increased Na+ uptake capacity of stelar root cells on the elemental composition of the shoot was examined with ICP-AES. Nineteen independent J2731* UASGAL4:HKT1;1 lines and 13 independent E2586 UASGAL4:HKT1;1 lines were studied. From each of these 32 independent lines, 19 segregating T2 plants, together with five parental control plants, were grown in a low-nutrient soil-like mix for 5 weeks and watered with a nutrient solution containing 2 mM NaCl. Each individual T2 plant was genotyped and, accordingly, the T2 plants were divided into two groups: those containing the UASGAL4:HKT1;1 transgene insertion and those that did not (null segregants). This enabled us to investigate the effect of the presence of the UASGAL4:HKT1;1 transgene on the elemental composition of the shoot. The average and standard error of the mean of the shoot Na+ concentration of J2731* UASGAL4:HKT1;1 plants were 217 ± 6 mg Na+·kg−1 FM (n = 285), null segregants were 605 ± 22 mg Na+·kg−1 FM (n = 71), and parental control plants were 605 ± 17 mg Na+·kg−1 FM (n = 93). Thus, the pericycle-specific expression of HKT1;1 in J2731* significantly decreased shoot Na+ accumulation by 64% of the corresponding null segregants (P < 0.001).
Similarly, stelar-specific expression of HKT1;1 in E2586 significantly decreased shoot Na+ accumulation by 47% of the corresponding null segregants (P < 0.001) (see Supplemental Table 1 online). The shoot Na+ concentrations of J2731* and E2586 plants transformed with a control plasmid without the HKT1;1 sequence were not significantly different from the respective null and parent plants (P > 0.4 for J2731* and P > 0.3 for E2586).
The shoot Na+ accumulation in J2731* was found to be three to four times higher than in E2586, which reflects the different ecotype backgrounds involved, as a similar difference is routinely measured in our laboratory for C24 and Col-0 (Møller and Tester, 2007).
Overexpression of HKT1;1 specifically in the root stele resulted not only in a reduction in shoot Na+ but also in an elevated K+ concentration in the shoots compared with controls (P < 0.01) (see Supplemental Table 1 online). The proportional change in K+ was much smaller than the decrease in Na+. It is likely that the increase in shoot K+ is a pleiotropic consequence of the reduced shoot Na+ arising from the stelar-specific expression of HKT1;1 and not a result of HKT1;1-mediated K+ transport. That K+ changes are an indirect effect of the cell type–specific overexpression of HKT1;1 is supported by the observation that the knockout of HKT1;1 affected neither influx nor root-to-shoot transfer of the K+ analog 86Rb+ (Davenport et al., 2007).
Cell type–specific overexpression of HKT1;1 had no other consistent effects on shoot elemental composition (see Supplemental Table 1 online). Constitutive expression of HKT1;1 did not lead to a significant change in the concentration of K+ (4480 ± 72 [n = 25 primary transformants] compared with 4337 ± 106 [n = 53 Col-0 control plants] mg K+·kg−1 FM; P = 0.27).
Na+ Accumulates in Roots upon Overexpression of HKT1;1 Specifically in the Root Stele
To test the heritability of the low Na+ shoot phenotype and allow the measurement of Na+ accumulation in the roots, homozygous UASGAL4:HKT1;1 T4 plants were grown hydroponically for 5 weeks with a gradual increase in salt stress during the last week. Reductions in shoot Na+ concentrations in plants overexpressing HKT1;1 in the root stele (Table 1) were similar to those measured in soil-grown plants (see Supplemental Table 1 online). Concomitant increases in root Na+ concentrations were observed, in accordance with a function for HKT1;1 in the root stele of Na+ retrieval from the xylem. In shoots and roots of both lines, K+ concentrations changed reciprocally with Na+.
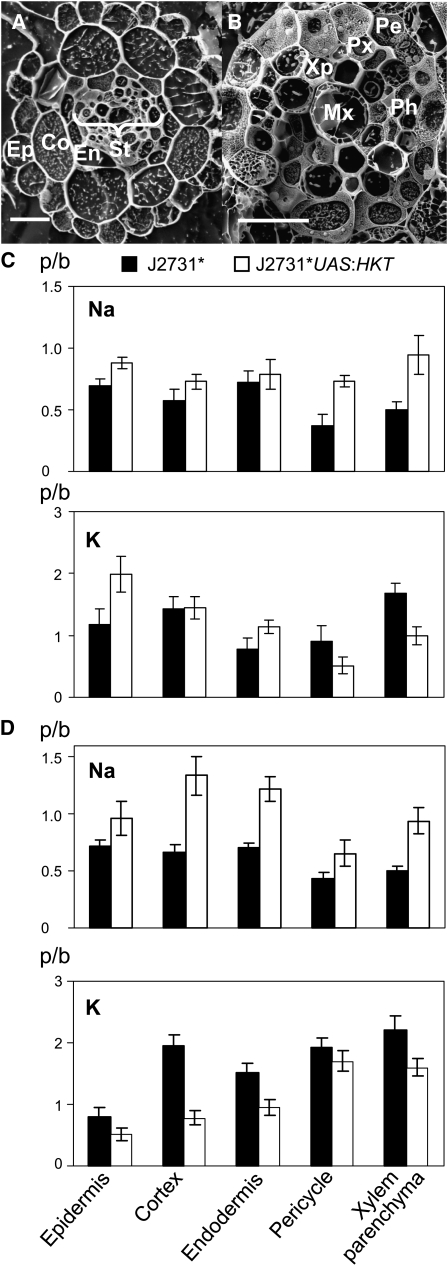
To investigate root cell–specific Na accumulation, x-ray microanalysis of transverse sections of roots was performed (Figures 4A and 4B). As the difference in total root Na+ was larger between control and UASGAL4:HKT1;1 plants in the J2731* background compared with those in the E2586 background, only J2731* plants were used in this more detailed analysis. Plants with a simple root system of a main root with few short lateral roots were used to enable consistent sampling of root tissue from the same position in the root system (20 mm from the main root tip).
Figure 4.
Overexpression of HKT1;1 in the Root Stele Results in Increased Na+ Accumulation in Specific Cell Types in the Root.
(A) and (B) Scanning electron micrographs of a transverse plane of J2731* roots with annotation of cell types. Ep, epidermis; Co, cortex; En, endodermis; St, stele; Pe, pericycle; Xp, xylem parenchyma; Px, protoxylem; Mx, metaxylem; Ph, phloem. Bars = 20 μm.
(C) and (D) X-ray microanalysis was used to measure Na+ and K+ accumulation in different root cell types in J2731* and J2731* UASGAL4:HKT1;1 plants grown in vertical Petri dishes (C) or hydroponics (D).
(C) An average of nine cells per cell type were analyzed in two plants of each genotype.
(D) An average of 21 cells per cell type were analyzed in a total of eight J2731* control plants and six J2731* UASGAL4:HKT1;1 plants. The data are combined from four separate experiments. The results are presented as signal peak:background (p/b) ratios for each element. Values presented are means ± se.
In all plants analyzed, whether grown in Petri dishes or hydroponically, it was found that the concentration of Na+ in J2731*UASGAL4:HKT1;1 roots was higher or similar to the J2731* control in all the cell types analyzed (Figures 4C and 4D). However, the pattern of partitioning of Na+ between the different cell types was dependent on the growth conditions of the plants. In plants grown in Petri dishes, Na+ accumulated particularly in the pericycle and xylem parenchyma cells (Figure 4C), directly reflecting the increased uptake of Na+ into these cells as a consequence of overexpression of HKT1;1 in the root stele. In plants grown hydroponically, the greatest difference in Na+ concentrations was in the cortical cells (Figure 4D). The cortical cells are notably much more vacuolated than stelar cells (see Supplemental Figure 4 online) and thus are better able to store the additional Na+ retrieved from the xylem stream.
The elemental composition of the whole shoot and root system of plants grown in hydroponics under the conditions used for x-ray microanalysis was examined with ICP-AES, and similar differences were found between J2731*UASGAL4:HKT1;1 and J2731* plants for Na+ and K+ (see Supplemental Table 2 online), as was seen previously (Table 1). Both within a specific cell type and within a whole tissue, it was found that K+ concentrations were altered in the opposite direction to Na+ (Figures 4C and 4D; see Supplemental Table 2 online).
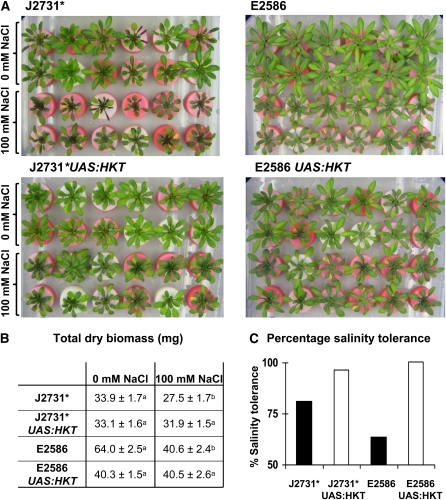
Overexpression of HKT1;1 in Root Stele Improves Whole-Plant Salinity Tolerance
To test the effects on salinity tolerance of stelar-specific expression of HKT1;1, plants were grown hydroponically for 5 weeks, at which point half of the plants were stressed with 100 mM NaCl for 5 d. At the end of the salt stress treatment, the fresh and dry biomass of shoots and whole root systems was recorded. The large decrease in shoot Na+ concentration by stelar-specific expression of HKT1;1 (Table 1) was reflected in an increase in salinity tolerance that was visibly apparent (Figure 5A) and measured as the total dry mass of salt-treated plants relative to control plants (Figure 5C). In both J2731*UASGAL4:HKT1;1 and E2586UASGAL4:HKT1;1 plants, the total dry biomass was not significantly affected by the imposed salt stress (P > 0.5) in contrast with the 19 to 37% decrease in total dry biomass of the two parent lines, which was statistically significant (P < 0.05) (Figure 5B). The smaller stature of the UASGAL4:HKT1;1 plants in the E2586 background compared with the respective parental plants might result from a growth penalty from overexpressing the HKT1;1 transgene, although the physiological basis for this is not known.
Figure 5.
Salinity-Tolerant Plants Overexpressing HKT1;1 Specifically in the Root Stele.
Homozygous T4 plants from lines J2731* UASGAL4:HKT1;1 and E2586 UASGAL4:HKT1;1, together with parental control plants, were grown in hydroponics for 5 weeks and subjected to 5 d of salt stress consisting of 25 mM increases in NaCl every 12 h until 100 mM NaCl was reached. CaCl2 was supplemented at every addition of NaCl to maintain constant Ca2+ activity.
(A) Photograph of plants at the end of the salt stress treatment. The plants were grown in a random arrangement throughout the experiment and were arranged as seen immediately before harvest for the purpose of this photograph.
(B) The total dry masses of plants subjected to control treatment or salt stress. Values that were found to be significantly different using a two-sample two-tailed Student's t test (after use of an F test to determine equality of variance) with P < 0.05 are indicated with different letters. Values presented are means ± se (n = 12 to 14).
(C) The percentage of salinity tolerance of each genotype was measured as the total dry mass of plants grown in saline conditions relative to total dry mass of plants in control treatment.
DISCUSSION
Cell Type–Specific Overexpression of HKT1;1 in Root Stele Was Necessary to Engineer Shoot Na+ Exclusion and Salinity Tolerance
Several strategies to minimize Na+ accumulation in the shoots of plants through modification of Na+ transport processes in specific cell types of the root were previously envisaged (Tester and Davenport, 2003). Here, the strategy of engineering shoot Na+ exclusion by increasing retrieval of Na+ from the transpiration stream and, hence, improving salinity tolerance was examined.
A targeted genetic modification approach was employed to express the gene HKT1;1 in specific stelar cell types in the root of Arabidopsis to increase the retrieval of Na+ from the transpiration stream. To target HKT1;1 to the root stele, an enhancer trap system was used. Enhancer trap lines J2731* and E2586 were chosen because of their high levels of GFP expression with defined expression patterns specific to the mature (and not developing) stelar cells of the entire root system. Such an expression pattern would conceivably be well suited for the cell type–specific overexpression of HKT1;1 to allow maximal retrieval of Na+ from the transpiration stream throughout the mature root system. Pericycle-specific expression of HKT1;1 using the J2731* enhancer trap line decreased shoot Na+ accumulation by 58 to 64% (see Supplemental Table 1 online). Similarly, stelar-specific expression of HKT1;1 in the E2586 enhancer trap line decreased shoot Na+ accumulation by 37 to 47% (Table 1; see Supplemental Table 1 online). This led to a significant increase in salinity tolerance, as the HKT1;1 expressing plants were unaffected by 100 mM NaCl, in contrast with the reduction in growth of 19 to 37% that was observed for the parental lines (Figure 5).
In contrast with the reduction in shoot Na+ observed following overexpression of HKT1;1 specifically in the root stele, constitutive overexpression of HKT1;1 was found to be deleterious. Constitutive overexpression resulted in a 3.6-fold increase in shoot Na+ compared with the approximate halving of shoot Na+ following stelar-specific expression of HKT1;1 in E2586. The Pro35S:HKT1;1 plants were very salt sensitive compared with control Col-0 plants (see Supplemental Figure 1 online) when grown in hydroponics and subjected to a salinity stress treatment of 50 mM NaCl and supplementary CaCl2 under conditions similar to those used in the experiments presented in Figure 5. The overaccumulation of shoot Na+ and resulting sensitivity of these plants was likely to be due to the increased unidirectional influx of 22Na+ into roots of the Pro35S:HKT1;1 plants. These results indicate that, to improve salinity tolerance, cell type–specific overexpression of HKT1;1 is not just sufficient, it is essential.
Importance of Specific Cell Type and Level of HKT1;1 Overexpression for Engineering Na+ Exclusion
HKT1;1 has previously been overexpressed using 2 kb of the native HKT1;1 promoter sequence (ProHKT1;1). The resulting plants were found to accumulate shoot Na+ concentrations similar to Col-0 when grown in transpiring conditions in Turface (Rus et al., 2004) but were reported to exhibit NaCl stress symptoms (salinity tolerance was only quantified as a decrease in primary root length of plants grown in Petri dishes; Rus et al., 2004). Previous studies of plants transformed with ProHKT1;1:uidA reporter gene DNA constructs, using 837 bp (Mäser et al., 2002a) and 2.3 kbp (Berthomieu et al., 2003) of the promoter sequence, showed that the ProHKT1;1 was active in the stelar root tissue and the shoot vasculature (Mäser et al., 2002a; Berthomieu et al., 2003). Therefore, it would be anticipated that the ProHKT1;1::HKT1;1 from Rus et al. (2004) and the E2586:UASGAL4:HKT1;1 plants of this study would have similar phenotypes as both were generated in Col backgrounds and the E2586 enhancer trap-driven expression pattern of HKT1;1 was similar to the expression pattern driven by the native HKT1;1 promoter in roots. However, this was not so, quite likely as a consequence of the higher levels of overexpression of HKT1;1 when driven by the enhancer trap system in this work compared with the native promoter. This result indicates the need for both specific and high levels of gene expression in order to engineer Na+ exclusion from the shoot.
Slight differences in patterns of root stelar expression in this work were found to lead to similar changes in shoot Na+ accumulation; shoot Na+ exclusion phenotypes were found when overexpressing HKT1;1 in the pericycle or in the stelar cells immediately surrounding the vasculature (Table 1; see Supplemental Table 1 online). This indicates that subtle differences in specificity of gene expression are unlikely to cause the differences observed in Na+ accumulation. Although root stele–specific overexpression of HKT1;1 was found to be crucial for engineering Na+ exclusion, the particular stelar cell type used did not appear to be important. Therefore, it is likely that what is important is the level of HKT1;1 overexpression in the root stele. A 7- to 12-fold increase in HKT1;1 transcript abundance was found with quantitative RT-PCR of whole root tissue in E2586UASGAL4:HKT1;1 compared with E2586 (17,400 ± 6500 for E2586UASGAL4:HKT1;1 [n = 3] compared with 1800 for E2586, when normalized to A. cyclophilin). It is worth noting that the grafting experiments of Rus et al. (2006) indicate that the shoot expression driven by the HKT1;1 promoter is unlikely to be a significant element of the shoot Na+ exclusion function of HKT1;1.
In this context, it is also worth noting that the relative effects of the stelar overexpression were different in the two backgrounds. In line E2586 (in Col background), HKT1;1 overexpression resulted in a smaller reduction in shoot Na+ than in line J2731* (in C24 background). It is proposed that this is due to a higher native level of expression of HKT1;1 in Col than in C24 (10- to 20-fold; S.J. Roy, D. Jha, J. Sundstrom, and M. Tester, unpubl results), the overexpression consequently having a smaller relative effect. Consistent with this is the lower shoot Na+ and higher root Na+ in E2586 compared with J2731* (Table 1; see Supplemental Table 1 online). Similar differences among Arabidopsis ecotypes in shoot Na+ accumulation and expression of HKT1;1 in the root have been observed previously for Col-0, Tsu-1, and Ts-1 (Rus et al., 2006).
It was interesting that under transpiring conditions, in plants where all evidence would point to Na+ transport being altered in the stele, there was an additional increase in Na+ within cortical and epidermal root cells (Figure 4D). This is in contrast with the results obtained for plants grown on plates, in a condition of low transpiration, where Na+ was increased and K+ reduced solely in the same cell type in which HKT was overexpressed (Figure 4C). In transpiring conditions with the plants acquiring greater amounts of Na+, it is likely that the Na+ retrieved in the stelar cells is moved back to the cortical cells; these cells are highly vacuolated compared with other cell types within the stele and therefore have greater ion storage potential. The observed difference between transpiring and nontranspiring (plate-grown) plants might be due to the fact that the total Na+ concentration in the roots of the transpiring plants is higher than in the plate-grown plants (Møller, 2008), creating a greater demand for Na+ storage. This greater need for Na+ storage can best be met by the large vacuoles of the cortical cells.
These findings support the proposition that the level of transpiration markedly changes whole-plant ion transport (Flowers, 2004; Møller and Tester, 2007). Furthermore, these observations highlight the plasticity of the ion transport processes occurring in the plant, especially in response to changes in rates of transpiration, and illustrate the importance of undertaking whole-plant studies of ion transport and related processes in transpiring conditions.
Na+ Transport by HKT1;1
HKT1;1 has been characterized as a Na+ transporter when heterologously expressed in S. cerevisiae and Xenopus oocytes but was also found to complement an Escherichia coli mutant with a deficient K+ uptake system (Uozumi et al., 2000). It has been proposed that HKT1;1 facilitates Na+ homeostasis in Arabidopsis and, through that function, modulates K+ nutrient status (Rus et al., 2004). The mechanism by which the levels of Na+ and K+ are coordinated, however, remains unknown. There does not appear to be any evidence in the existing literature for the direct transport of K+ by HKT1;1 in planta.
In this study, the only elements that were consistently found to have changed in the plant shoot following cell type–specific overexpression of HKT1;1 in the root stele were Na+ and K+. The change in concentration of K+ in the shoot as a consequence of HKT1;1 overexpression in the root is likely to be a secondary effect of the change in shoot Na+ accumulation, rather than a direct effect of HKT1;1 overexpression. If HKT1;1 were a protein with transport affinity for both Na+ and K+ and catalyzed the influx of both ions, as has been found for some HKTs from other plant species (Horie et al., 2001; Mäser et al., 2002b; Platten et al., 2006), then the accumulation of K+ in the shoot would have been expected to decrease as a result of increased retrieval of K+ from the transpiration stream. However, the opposite was observed, there being a strong reciprocal relationship between shoot Na+ and shoot K+ concentrations (Table 1; see Supplemental Table 1 online). That HKT1;1 does not mediate K+ transport is also supported by the observation that the knockout of HKT1;1 affected neither influx nor root-to-shoot transfer of 86Rb+, a K+ analog (Davenport et al., 2007). Thus, although the expression of HKT1;1 has been found to relieve a K+ uptake–deficient mutant of E. coli (Uozumi et al., 2000), it seems unlikely that the change in K+ concentration in the shoot of the UASGAL4:HKT1;1 plants is the result of HKT1;1-mediated K+ transport.
During patch clamp experiments, it was found that inward currents increased in magnitude with an increase in the external concentration of Na+ only across the plasma membrane of fluorescent protoplasts from J2731*UASGAL4:HKT1;1 plants (Figure 3). This report of direct measurements of HKT1;1-mediated currents that are consistent with Na+ influx into plant cells is in accordance with the findings of previous heterologous expression studies (Uozumi et al., 2000; Mäser et al., 2002a). Nevertheless, further electrophysiological experiments are necessary to confirm that HKT1;1 does not mediate K+ transport in planta.
In conclusion, cell type–specific gene expression was employed to engineer Na+ exclusion from the shoot through increased retrieval of Na+ from the transpiration stream and, thus, to increase the salinity tolerance of the plants. These results also provide evidence that is consistent with the hypothesized specific Na+ transport activity of HKT1;1 in planta. We propose that cell type–specific, rather than constitutive, overexpression of plasma membrane Na+ transporters, such as HKT1;1, provides an efficient means to decrease shoot Na+ accumulation and increase salinity tolerance. We propose that cell type–specific manipulation of transport processes can usefully be applied to alter shoot accumulation of many other solutes with implications for human nutrition, plant nutrient use efficiency, and phytoremediation. We are currently applying this technology to commercially relevant plants, such as rice and barley, with the aim of increasing the salinity tolerance of these crops.
METHODS
All chemicals were obtained from Sigma-Aldrich unless otherwise stated. Restriction enzymes were obtained from New England Biolabs.
Plant Material
Arabidopsis thaliana ecotypes C24 and Col-0 seeds were obtained from the Nottingham Arabidopsis Stock Centre. J2731 was isolated from a collection of GAL4-VP16 mGFP5-ER enhancer trap lines generated by root transformation of C24 with the enhancer trap plasmid pET-15 (Haseloff, 1999). Expression of mGFP5-ER appeared to be silenced in homozygous J2731 plants in the T3 generation, which were determined by DNA gel blotting to have multiple enhancer trap T-DNA inserts. Therefore, mGFP5-ER–expressing J2731 T2 plants were backcrossed to C24 wild-type plants in an effort to decrease the number of inserts. DNA gel blotting was performed on the offspring to identify a single-insert plant with mGFP5-ER expression in the root pericycle, which was named J2731*. Homozygous plants were generated by self-fertilization. E2586 was obtained from Scott Poethig (University of Pennsylvania, Philadelphia, PA) from a collection of GAL4-VP16 mGFP5-ER enhancer trap lines generated by transformation of Col wild-type plants by floral dip with pET-15.
Plant Growth
Sterile Culture in Vertical Petri Plates
Seeds were surface sterilized following standard protocols (Weigel and Glazebrook, 2002) and placed on square Petri dishes with 0.5× Murashige and Skoog medium (M5519), pH 5.7, with 1% (w/v) sucrose and 0.3% (w/v) Phytagel. The seeds were vernalized at 4°C for 2 d in the dark before the dishes were positioned vertically in a growth chamber under a 10/14-h light/dark cycle at 21°C with an irradiance of 120 μmol m−2 s−1.
For experiments where plants were subjected to salinity stress in Petri plates, the above protocol was followed with the addition of 50 mM NaCl. CaCl2 was added to maintain the activity of Ca2+ constant in the growth solution, as calculated using Visual Minteq software, version 2.52 (KTH).
Soil-Like Mix
Plants were grown in plastic trays with a soil-like mix (3.6 liters perlite, coarse grade, 2 to 6 mm; 3.6 liters coira; and 0.25 liters sand) with a thin layer of Amgrow Seed Raising Mix (Envirogreen) on top. The soil was drenched with Confidor (Bayer) and VectoBac (Valent Biosciences) overnight in 500 mL nutrient solution [2 mM Ca(NO3)2, 15 mM KNO3, 0.5 mM MgSO4, 0.5 mM NaH2PO4, 15 mM NH4NO3, 25 mM NaFeEDTA, 0.2 mM H3BO3, 1 μM Na2MoO4, 1 μM NiCl2, 2 μM ZnSO4, 4 μM MnCl2, 2 μM CuSO4, 1 μM CoCl2, 2 mM NaCl, 5 μM SrCl2, 0.5 mM Na2SiO2, and 1 μM CdCl2] before Arabidopsis seeds were placed on top. Plants were grown under a 10/14-h light/dark cycle at 21°C with an irradiance of 120 μmol m−2 s−1. Trays were regularly rotated and watered twice a week from below with 300 mL nutrient solution.
Hydroponics
Sterilized seeds were sown on 0.8% (w/v) agar in 1.5-mL centrifuge tubes and vernalized for 48 h at 4°C. The bottom of the tubes was cut off and the tubes were suspended over an aerated growth solution consisting of 1.25 mM KNO3, 0.625 mM KH2PO4, 0.5 mM MgSO4, 0.5 mM Ca(NO3)2 and 0.045 mM FeNaEDTA with the following micronutrients: 0.16 μM CuSO4, 0.38 μM ZnSO4, 1.8 μM MnSO4, 45 μM H3BO3, 0.015 μM (NH4)6Mo7O24, and 0.01 μM CoCl2. The agar contained half-strength growth solution with full-strength micronutrients. The plants were grown in a random arrangement in aerated solution on a 10/14-h light/dark cycle at 21°C with an irradiance of 75 μmol m−2 s−1. For high NaCl treatments, solid NaCl was added to the growth solution to make a final concentration of 100 mM. CaCl2 was added to maintain the activity of Ca2+ constant in the growth solution.
Epifluorescence, Confocal, and Transmission Electron Microscopy
mGFP5-ER expression in 2-week-old J2731* and E2586 plants grown in sterile culture were analyzed with the use of an M7FLIII epifluorescence microscope (Leica) with a GFP2 filter and IM50 software version 1.20 (Leica). mGFP5-ER was seen as green and the autofluoresecence of shoot tissue as red color.
Confocal laser scanning microscopy was performed using a TCS NT/SP microscope (Leica) and Leica Confocal Software version 2.00. For propidium iodide staining of cell walls (red), plants were removed from the growth medium, submerged in 10 μg/mL propidium iodide for 10 min, and whole plants were then mounted in water. To examine mGFP5-ER expression further, a z-series of scans was collected and manipulated using the Leica Confocal Software to obtain a transverse section view. The excitation wavelength for propidium iodide and mGFP5-ER was 488 nm.
Transmission electron microscopy was used to examine transverse sections of roots taken 20 mm from the root tip of J2731* and J2731*UASGAL4:HKT1;1 plants. Plants were grown as detailed for x-ray microanalysis. Root tissue was fixed, dehydrated, and infiltrated with LR White resin (London Resin Company). Sections were cut with an Ultramicrotome (Leica), stained with aqueous uranyl acetate (Sigma-Aldrich), and examined with the use of a Philips CM100 transmission electron microscope (Philips Electron Optics) at a voltage of 80 kV and a beam width of 7 μm.
DNA Gel Blotting
Genomic DNA of J2731* and E2586 was restricted with either HindIII or HpaI. A HindIII restriction site is present in the enhancer trap T-DNA outside the GAL4-VP16 gene, whereas HpaI restriction sites are not present in the enhancer trap T-DNA sequence. DNA gel blotting was performed using the Digoxigenin system (Roche Applied Science) following the manufacturer's recommendations. To produce a probe, a 504-bp PCR product was amplified from the pET-15 plasmid using primers mPPR1-5 (5′-AGGCAAGCTTGGATCCAACAATG-3′) and mPPR1-3 (5′-CCCGGAGCTCGTCCCCCAGGCTG-3′). This PCR was used as the template for a nested PCR using the PCR DIG probe synthesis kit (Roche Applied Science) and the primers GAL4-5 (5′-GACATCTGCCGCCTCAAG-3′) and GAL4-3 (5′-GGTCGAGACGGTCAACTG-3′) to produce a DIG-dUTP–labeled 411-bp probe annealing in the GAL4-VP16 sequence.
Thermal Asymmetric Interlaced PCR
Thermal asymmetric interlaced (TAIL)-PCR was performed to amplify the genomic DNA sequences adjacent to the enhancer trap T-DNA inserts in J2731* and E2586 following the protocols previously published (Liu et al., 1995). The nested T-DNA–specific primers ntTR1 (5′-GGAACAACACTCAACCCTATCTC-3′), ntTR2 (5′-TCGGAACCACCATCAAACAG -3′), and ntTR3 (5′-AGGCGGTGAAGGGCAATCAG-3′) were used together with one of the two short arbitrary degenerate primers AD5 [5′-GT(A/C/G/T)CGA(C/G)(A/T)CA(A/C/G/T)A(A/T)GTT-3′] and AD7 [5′-TG(A/T)G(A/C/G/T)AG(A/T)A(A/C/G/T)CA(C/G)AGA-3′]. The TAIL-PCR programs used were identical to those previously published (Liu et al., 1995). The primary TAIL-PCR reaction mixtures (25 μL) contained 1× PCR buffer (Invitrogen), 1.5 mM MgCl2, 0.4 μM ntTR1, 4 μM AD5 or AD7, 0.4 mM deoxynucleotide triphosphate, and 0.04 units/μL U Platinum Taq DNA polymerase (Invitrogen) as well as 30 ng of genomic DNA as template. One microliter of a 50-fold dilution of the primary TAIL-PCR product was added as template in the secondary TAIL-PCR reaction mixtures (25 μL) of the same composition as above. One microliter of a 20-fold dilution of the secondary TAIL-PCR product was added as template in the tertiary TAIL-PCR reaction mixtures (50 μL) of the same composition as above. For both J2731* and E2586, a single PCR band was obtained using AD5 or AD7. The PCR product decreased in size from the secondary to the tertiary TAIL-PCR as expected. The PCR product was cleaned on a NucleoSpin Extract II column (Mackerey-Nagel) and sequenced.
Cloning of DNA Constructs
Pro35S:HKT1;1
A Gateway destination vector with a dual cauliflower mosaic virus 35S promoter was derived from pJawohl8-RNAi (AF408413) by restriction with HindIII and SpeI to remove the intron and second gateway cassette (2047 bp). The DNA fragment was blunt-ended with Klenow DNA polymerase (New England Biolabs) and religated to produce pTOOL2. The HKT1;1 coding sequence was PCR amplified from pGreenII0229UASGAL4:HKT1;1 using primers AtHKT1F (5′-ATGGACAGAGTGGTGGCA-3′) and AtHKT1R (5′-TTAGGAAGACGAGGGGTA-3′). The resultant 1521-bp PCR product was cloned into the Gateway entry vector pCR8/GW/TOPO (Invitrogen). An LR gateway reaction (Invitrogen) with the vectors pCR8/HKT1;1 and pTOOL2 was conducted to produce the Pro35S:HKT1;1 binary vector.
UASGAL4:HKT1;1
The nopaline synthase terminator was PCR amplified from pBI101 (Clontech) using primers ISM8nos-fwd (5′-GCGGGGCCCGAATTTCCCCGATCGTTCAAAC-3′) and ISM9nos-rev (5′-GCCGGTACCCCGATCTAGTAACATAGATGAC-3′). The resultant 283-bp fragment was cloned into pCR4Blunt-TOPO (Invitrogen) followed by cloning into the binary vector pGreenII0229 (Hellens et al., 2000) using restriction enzymes ApaI and KpnI to generate pGreenII0229nos. The UASGAL4 sequence was PCR amplified from the enhancer trap plasmid pET-15 using primers ISM10UASfwd (5′-GAGCCGCGGGCATGCCTGCAGGTCGGAGTAC-3′) and ISM12UASrev (5′-CTGCCCGGGGTCGACCTGCAGGTCGTCCTC-3′). The resultant 250-bp fragment was subcloned into pCR4Blunt-TOPO (Invitrogen). Using restriction enzymes SacI and XmaI, a 286-bp fragment containing UASGAL4 was cloned into pGreenII0229nos to generate pGreenII0229UASGAL4nos. The open reading frame of HKT1;1 was PCR amplified from cDNA from salt-treated roots of Arabidopsis ecotype Col-0 using primers ISM31AtHKT1fwd (5′-CTCCCCGGGAACAATGGACAGAGTGGTGGCAAAAATAG-3′) and ISM32AtHKT1rev (5′-ATCCTCGAGTTAGGAAGACGAGGGGTAAAGTATC-3′). The resultant 1543-bp fragment was cloned into pCR4Blunt-TOPO (Invitrogen) followed by cloning into pGreenII0229UASGAL4nos using restriction enzymes XmaI and XhoI to generate pGreenII0229UASGAL4:HKT1;1.
UASGAL4:uidA
The uidA coding region was PCR amplified from pBI101 (Clontech) using primers ISM29GUSfwd (5′-TGACCCGGGAACAATGTTACGTCCTGTAGAAACCCCAAC-3′) and ISM30GUSrev (5′-GTTCTCGAGTCATTGTTTGCCTCCCTGCTGCGG-3′). The resultant 1834-bp fragment was cloned into pCR4Blunt-TOPO (Invitrogen) followed by cloning into pGreenII0229UASGAL4nos using XmaI and XhoI to generate pGreenII0229UASGAL4:uidA.
The binary vectors were transformed into Agrobacterium tumefaciens GV3101:pMP90(RK) together with pSOUP in the case of the pGreenII0229-based vectors. Arabidopsis plants were transformed using the floral dip method following standard protocols (Weigel and Glazebrook, 2002). Transformed plants were resistant to phosphinotricin.
Genotyping of Segregating Plants
UASGAL4:HKT1;1 T2 Plants
At 5 weeks, leaf tissue from T2 UASGAL4:HKT1;1 plants analyzed for shoot ion accumulation by ICP-AES was harvested and used for DNA extraction with the ChargeSwitch gDNA plant kit (Invitrogen). To genotype individual segregating T2 plants, extracted DNA was subjected to PCR analysis to amplify a 388-bp amplicon from the HKT1;1 transgene in transgenic plants and a 297-bp amplicon from the Arabidopsis actin2 gene in all plants. To specifically amplify HKT1;1 supplied from the transgene, and not the native HKT1;1 gene, the reverse primer was designed to anneal to the sequence comprising the 3′end of the HKT1;1 coding sequence and the 5′ end of the nopaline synthase terminator sequence. The PCRs were performed using Immolase Taq DNA polymerase (Bioline), which requires heat activation at 95°C for 7 min, and 30 cycles of 95°C for 30 s, 61°C for 30 s, and 72°C for 30 s using primers pISM44 (5′-TGATGATTCCGAAAATGGAAAG-3′) and pISM45 (5′-CCCCTCGAGTTAGGAAGACG-3′) for HKT1;1. Arabidopsis Actin2 was amplified following the same protocol using primers pISM42 (5′-GCCCAGAAGTCTTGTTCCAG-3′) and pISM43 (5′-ACATCTGCTGGAATGTGCTG-3′) and an annealing temperature of 50°C.
Pro35S:HKT1;1 Plants
Primary Pro35S:HKT1;1 transformants were grown on a soil-like mix as described above and selected for phosphinotricin resistance after spraying twice, 7 d apart, with a 200 mg/L phosphinotricin (Duchefa) solution in MilliQ water. At 5 weeks, DNA was extracted from a leaf sample and subjected to PCR using primers 35S FORWARD (5′-GATATCTCCACTGACGTAAGG-3′) and HKT REVERSE (5′- TTAGGAAGACGAGGGGTAAAGTATC-3′) and 35 cycles of 95°C for 30 s, 51°C for 30 s, and 72°C for 2 min with Taq DNA polymerase (Qiagen) to amplify a 1688-bp amplicon.
GUS Staining
Two-week-old J2731*UASGAL4:uidA and E2586UASGAL4:uidA plants grown on vertical Petri plates as described above were assayed for GUS activity according to standard protocols (Weigel and Glazebrook, 2002). Whole plants were incubated in staining buffer with X-Gluc (Progen Biosciences) at 37°C for 1 h and cleared with 70% ethanol overnight before the whole-mount samples in 70% ethanol were examined using a dissecting microscope.
Laser Microdissection and RT-PCR
Root samples from 4-week-old Arabidopsis seedlings grown on Petri plates as described above were fixed in Farmer's fixative at 4°C overnight and dehydrated in a graded series of ethanol:double autoclaved MilliQ water followed by an ethanol:xylene series (Kerk et al., 2003). Approximately 25% (w/v) Paramat Gurr (BDH) was added, and samples were kept at 65°C overnight. While at 65°C, another 25% (w/v) Paramat Gurr was added. After 3 h, the solution was replaced with 100% liquefied Paramat Gurr and replaced at regular interval at least six times over 3 d. The root samples were transferred to room temperature to solidify the paraffin. Using a Leica RM2265 microtome (Leica), 6-μm tissue sections were cut and mounted onto PEN membrane slides (Leica) in a 42°C bath with double-autoclaved MilliQ water and left to air-dry. The samples were deparaffinized in two xylene treatments for 10 min and air-dried.
Using the endodermis as a boundary, the stelar and outer root cells of 25 sections from the root system of one plant were dissected into separate caps of 200-μL PCR tubes (Biozym Scientific) with a Leica ASLMD laser-assisted microdissection microscope (Leica). Three technical replicates were performed on sections cut from the same plant. For two replicates, total RNA was extracted using the RNA Aqueous-Micro Kit (Ambion), following the procedure for RNA extraction from laser capture samples with DNA digested with the DNaseI supplied; for one replicate, RNA was isolated using Trizol (Roy et al., 2008). The isolated RNA was reverse transcribed using the Sensiscript RT kit (Qiagen) with 1 μM 15-mer oligo(dT) primer. PCR amplification was performed using the HotStartTaq PCR kit (Qiagen) in a 25-μL reaction volume with 1 μM forward and reverse primers. All PCRs were run under identical conditions for 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min in the same heat block. Arabidopsis Actin2 was amplified with primers Actin2Fwd (5′-GATCTCCAAGGCCGAGTATG-3′) and Actin2Rev (5′- GGCATCAATTCGATCATC-3′) to give a 306-bp PCR product. The UASGAL4:HKT1;1 transgene was amplified with primers HKT1transFwd (5′-TGAACGGCGTGTGGACATCA-3′) and HKT1transRev (5′-CCCCCCCTCGAGTTAGGAAGA-3′) to give a 187-bp PCR product. PCR products were visualized with the use of ethidium bromide on a 2% (w/v) agarose gel.
mRNA in Situ Localization
A 405-bp HKT1;1 fragment was PCR amplified from Col-0 cDNA using primers XhoIAtHKTprobeF (5′-GACCTCGAGCCGTTGACGGAACAAAAGACGATAGAGA-3′) and ClaIAtHKTprobeR (5′- GACATCGATGCCAGATTTGGCTGTGAACTGCTTAAAC-3′), cloned into pSPT73 (Promega) and sequenced. DIG-labeled antisense and sense HKT1;1 riboprobes were generated with a DIG RNA labeling kit (Roche Diagnostics).
Tissue was fixed in TEM fixative (0.25% [v/v] glutaraldehyde, 4% [v/v] paraformaldehyde, and 4% [w/v] sucrose in 1× PBS), for 2 h at room temperature, washed overnight in 1× PBS at 4°C, dehydrated through an ethanol, and then xylene, series, embedded in paraffin, and sectioned to 7 μm. After dewaxing in Histochoice (Sigma-Aldrich) and sequential rehydration, sections were treated with 20 μg/mL Proteinase K (Sigma-Aldrich), postfixed in 4% (v/v) formaldehyde in 1× PBS, acetylated in 0.5% (v/v) acetic anhydride in 0.1 M triethanolamine-HCl, and dehydrated through an ethanol series. mRNA in situ hybridization was performed overnight at 45°C in hybridization buffer (50% [v/v] formamide, 1× SSC, 10% [v/v] dextran sulfate [Progen Biosciences], 1× Denhardt's solution, and 1 mg/mL tRNA [Roche Diagnostics]). Slides were washed three times in 0.2× SSC for 1 h each at 45°C and then held in 1× PBS at 4°C overnight. Antibody incubation, detection block solution, and buffers were as described in DIG nucleic acid detection kit instructions (Roche Diagnostics), with the exception of 1× PBS wash buffer and color detection with BM Purple (Roche Diagnostics). Slides were made permanent with Crystal Mount (Sigma-Aldrich).
Patch Clamp Electrophysiology
Protoplasts were isolated from roots of 9- to 14-d-old plants, grown on vertical Petri plates as described above, following previously published methods (Demidchik and Tester, 2002) upon incubation in 1.5% (w/v) Onozuka RS cellulase (Yakult Pharmaceutical), 1% (w/v) cellulysin cellulase (Calbiochem, Merck), 0.1% (w/v) pectolyase (P5936; Sigma-Aldrich), 0.1% (w/v) BSA, 10 mM CaCl2, 10 mM KCl, 2 MgCl2, 2 mM MES, pH 5.7 with Tris, 290 to 300 mOsm·kg−1 adjusted with D-mannitol, for 2 h and 45 min. Protoplasts were isolated from the enzyme solution by filtration through 50-μm nylon mesh and two successive pelleting and resuspension steps using 2 mM CaCl2, 5 mM KCl, 1 mM MgCl2, 2 mM MES, 10 mM sucrose, and 10 mM glucose, pH 5.7, with Tris, 290 to 300 mOsm·kg−1 adjusted with D-mannitol, with centrifugation at 4°C (Kiegle et al., 2000b). Protoplasts were kept on ice, in the dark, for up to 12 h before use.
Patch clamp electrophysiology of mGFP5-ER–expressing protoplasts was performed as described previously (Kiegle et al., 2000b; Demidchik and Tester, 2002) with the following modifications. Electrode blanks were filled with a solution containing 5 mM NaCl, 25 mM Na-gluconate, 1.3 mM CaCl2, 3 mM EGTA, and 5 mM HEPES, pH 7.2, with NaOH, 290 to 300 mOsm·kg−1 with D-mannitol. A giga-ohm seal was obtained in an external sealing solution containing 10 mM CaCl2 and 2 mM MES, pH 5.7, with Tris, 290 to 300 mOsm·kg−1 with D-mannitol. Three bath solutions were used with 0, 10, or 25 mM NaCl, 10 mM tetraethylammonium chloride, and 5 mM MES, pH 5.7, adjusted with Tris, 290 to 300 mOsm·kg−1 adjusted with D-mannitol. The activity of solution constituents was calculated with the use of Maxchelator (Stanford University, Palo Alto, CA) and Visual Minteq 2.52 and used to calculate reversal potentials. To reduce activity of non-selective cation channels, the activity of Ca2+ in the external solution was kept at 1.71 mM in all bath solutions by addition of CaCl2 and TEA+ at 8.9 mM. The reversal potentials were as follows (in mV): ECa, +140; ETRIS, infinitely positive and ETEA, infinitely positive, in all solutions; in 0, 10, and 25 mM [NaCl]ext, respectively ECl −20, −23, and −32 and ENa −149, −77, and −36. Data were acquired using a voltage ramp protocol; the membrane was hyperpolarized to +60 mV for 1 s and then ramped to −140 mV over 500 ms, then to −1.5 mV, and VH was −1.5 mV for 4 s between 10 sweeps. Data were acquired and analyzed with Axon Laboratory Clampex 9.12 and Clampfit 10 (Molecular Devices) and Microsoft Excel. Membrane current was converted to current density, and junction potentials were measured and corrected for using standard protocols (Gilliham and Tester, 2005). Data were plotted with the use of Sigmaplot 10 (Systat Software).
Radioactive Tracer Experiments
Unidirectional fluxes of Na+ into excised root systems and transfer of Na+ from roots to shoot in whole plants were measured using the radioisotope 22Na+ (GE Healthcare) following the previously published protocols (Essah et al., 2003; Davenport et al., 2007).
For measurements of Na+ influx into excised roots, 4-week-old seedlings grown in vertical Petri plates as described above were used. Three to four plants were combined, and the entire root systems were excised from the shoots. Excised root systems were pretreated in unlabeled 50 mM NaCl and 0.5 mM CaCl2 for 10 min, blotted gently, and transferred to 15 mL influx solution containing 50 mM NaCl, 0.5 mM CaCl2, and 5.5 μCi of 22Na+ on a gently rotating shaker for 2 min. The roots were rinsed in two ice-cold 50 mM NaCl and 10 mM CaCl2 solutions for 2 and 3 min, respectively. The roots were blotted, weighed, mixed with 4 mL of Ecolume scintillation cocktail (MP Biomedicals), and counted in a LS 6500 scintillation counter (Beckman Coulter).
For measurements of the transfer of Na+ from roots to shoot, plants were grown in hydroponics for 5 weeks, with an additional 50 mM NaCl for 5 d prior to the experiment in which whole plants were used. The entire root system was submerged in influx solution containing 10 mL of 50 mM NaCl, 0.5 mM CaCl2, and 5 μCi of 22Na+ on a gently rotating shaker for 60 min. The roots were rinsed as described above, blotted, and separated from the shoots. The samples were weighed, mixed with 4 mL of Ecolume scintillation cocktail (MP Biomedicals), and counted in a LS 6500 scintillation counter (Beckman Coulter).
ICP-AES Analysis of Ion Accumulation in Shoot and Root Tissue
After 5 weeks, the youngest fully expanded leaves (∼50 mg) (and when using hydroponically grown plants, the entire root system) were harvested using plastic tweezers (Technoplas), weighed, and placed in sterile 50-mL PP Falcon tubes (Greiner Bio-One). Following predigestion overnight in 1.5 mL 69% (v/v) nitric acid (GR grade; Merck) at room temperature, the samples were transferred to a hot block (A.I. Scientific) for digestion at 80°C for 30 min and then 125°C for 2 h followed by addition of 0.3 mL 32% (v/v) hydrochloric acid (GR grade; Merck). The samples were made to 15 mL with MilliQ water at 18.2 mega ohms resistance before analysis using a CIROS radial inductively coupled plasma optical emission spectrometer (Spectro). Acid digestion and ICP-AES analysis were performed by Waite Analytical Services (University of Adelaide, Urrbrae, South Australia).
Cryoscanning Electron Microscopy and X-Ray Microanalysis
Homozygous T3 J2731* UASGAL4:HKT1;1 plants and parental control plants were grown under two different experimental conditions for analysis using cryoscanning electron microscopy and x-ray microanalysis. (1) Seeds were placed on sterile growth medium including 50 mM NaCl, and supplemental CaCl2 and plants were grown in vertical Petri dishes as described above and analyzed 10 d after germination. (2) Seeds were germinated on agar in 10-μL pipette tips, and plants were grown in hydroponics as described above. Seven days after germination, NaCl was added to the growth solution in 10 mM steps every 12 h to a final concentration of 50 mM. Plants were analyzed 14 d after germination.
Plants were lifted out of the Petri dishes or hydroponics tanks, and a 1-cm piece of the main root was cut out 20 mm from the root tip and placed in a drop of growth solution without additional NaCl. Using fine tweezers, in one movement the root pieces were gently and quickly placed in a bundle with the same alignment, and a tiny flat piece of LEIT-C-Plast conductive mounting plastic (SPI Supplies) was carefully placed around the end of the bundle at the end that was closest to the root. The root bundle was carefully lifted by the mounting plastic, which was inserted in a hole in the brass stub supporting the root pieces to stand vertically. Five roots of each genotype were placed in the same stub in each experiment with a drop of growth solution without added NaCl surrounding them and snap frozen in a liquid N2 slush. The handling time for the total preparation was <1 min with two people working simultaneously on the preparation of either the control or J2731* UASGAL4:HKT1;1 sample. The frozen specimen was transferred under vacuum to the preparation chamber of the scanning electron microscope where it was cryoplaned using a cooled microtome blade. The samples were sublimed for 1 min 30 s at −92°C. Samples were recooled to −120°C and sputter coated with platinum for 1 min and 30 s to give a thickness of 5 nm. The coated specimens were then loaded onto the microscope stage (held at a temperature below −150°C) and analyzed in a Philips XL 30 scanning electron microscope (Philips Electron Optics) fitted with a CT1500 HF cryotransfer stage (Oxford Instruments) and an EDAX energy-dispersive x-ray detector (EDAX International). XRMA spectra were recorded using an electron beam of 10 keV, spot size 5, a working distance of 10 mm, 10,000 times magnification, and a raster square of 5 μm on each side in the middle of each cell, where possible, and a data collection time of 100 live seconds, giving counts of 1000 to 1500 counts per second. Spectra were analyzed with eDXi software (EDAX), and results presented were measurements of peak over background. All acquired Na and K p-b ratios were consistently above detection limits. The average net integral values for Na used to calculate p-b ratios were 4.9 and 1.3 (net integral) and 3.4 and 3.6 (background) for high and low salt cells, respectively; average integral errors were 6.8 for high salt cells and 13.7 for low salt cells.
Statistical Analysis
Significance was determined according to the Student's t test using Excel software (Microsoft).
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number At4g10310 (Arabidopsis HKT1;1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Salt-Sensitive Phenotype of Salt-Stressed Pro35S:HKT1;1 Plants.
Supplemental Figure 2. Position of Enhancer Trap T-DNA in the Genome of J2731* and E2586.
Supplemental Figure 3. Laser Dissection of Transverse Root Sections.
Supplemental Figure 4. Electron Micrograph of Transverse Section of J2731*UASGAL4:HKT1;1 Root.
Supplemental Table 1. Shoot Accumulation of 10 Elements in UASGAL4:HKT1;1 Plants.
Supplemental Table 2. Accumulation of Na+ and K+ in Shoot and Roots of 2-Week-Old J2731* UASGAL4:HKT1;1 Plants Grown in Hydroponics.
Supplementary Material
Acknowledgments
We thank M. Henderson for assistance with confocal microscopy, the staff at Adelaide Microscopy for technical assistance, N. Tikhomirov for TAIL-PCR, E. Kalashyan for programming of XRMAplot for semi-automated XRMA data analysis, S. Poethig for plant material, and S. Tyerman for access to equipment. We thank N. Shadiac, D. Plett, A. Jacobs, N. Shirley, U. Baumann, E. Kalashyan, D. Salt, B. Lahner, G. Safwat, N. Hannink, R. Hosking, and R. Davenport for technical support and advice. This work was supported by the Biotechnology and Biological Sciences Research Council (UK) and the Australian Research Council (Australia). I.S.M. was supported by the Danish Research Council.
Author Contributions
I.S.M. performed the experiments, apart from the following: M.G. performed x-ray microanalysis and patch clamp electrophysiology experiments with I.S.M. G.M.M performed the mRNA in situ localizations. D.J. performed experiments with 22Na+ and Pro35S:HKT1;1 plants and the 100 mM salinity tolerance assay. S.J.R. performed laser dissection and related RT-PCR. J.C.C. and J.H. supervised, while I.S.M. was based in J.H.'s laboratory. M.T. conceived and supervised the project. I.S.M., M.G., and M.T. wrote the article. All authors discussed the results and commented on the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Mark Tester (mark.tester@acpfg.com.au).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Apse, M.P., Aharon, G.S., Snedden, W.A., and Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285 1256–1258. [DOI] [PubMed] [Google Scholar]
- Berthomieu, P., et al. (2003). Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 22 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrt, C.S., Platten, J.D., Spielmeyer, W., James, R.A., Lagudah, E.S., Dennis, E.S., Tester, M., and Munns, R. (2007). HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 143 1918–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, S., Forsthoefel, N., Ran, Y.Q., Quigley, F., Nelson, D.E., and Bohnert, H.J. (2000). Na+/myo-inositol symporters and Na+/H+-antiport in Mesembryanthemum crystallinum. Plant J. 24 511–522. [DOI] [PubMed] [Google Scholar]
- Davenport, R.J., James, R.A., Zakrisson-Plogander, A., Tester, M., and Munns, R. (2005). Control of sodium transport in durum wheat. Plant Physiol. 137 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport, R.J., Muñoz-Mayor, A., Jha, D., Essah, P.A., Rus, A., and Tester, M. (2007). The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 30 497–507. [DOI] [PubMed] [Google Scholar]
- Davenport, R.J., and Tester, M. (2000). A weakly voltage-dependent, nonselective cation channel mediates toxic sodium influx in wheat. Plant Physiol. 122 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik, V., Davenport, R.J., and Tester, M. (2002). Nonselective cation channels in plants. Annu. Rev. Plant Biol. 53 67–107. [DOI] [PubMed] [Google Scholar]
- Demidchik, V., and Tester, M. (2002). Sodium fluxes through nonselective cation channels in the plasma membrane of protoplasts from Arabidopsis roots. Plant Physiol. 128 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd, A.N., Jakobsen, M.K., Baker, A.J., Telzerow, A., Hou, S.-W., Laplaze, L., Barrot, L., Poethig, S., Haseloff, J., and Webb, A.A.R. (2006). Time of day modulates low-temperature Ca2+ signals in Arabidopsis. Plant J. 48 962–973. [DOI] [PubMed] [Google Scholar]
- Essah, P.A., Davenport, R.J., and Tester, M. (2003). Sodium influx and accumulation in Arabidopsis. Plant Physiol. 133 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers, T.J. (2004). Improving crop salt tolerance. J. Exp. Bot. 55 307–319. [DOI] [PubMed] [Google Scholar]
- Garciadeblas, B., Senn, M.E., Banuelos, M.A., and Rodriguez-Navarro, A. (2003). Sodium transport and HKT transporters: The rice model. Plant J. 34 788–801. [DOI] [PubMed] [Google Scholar]
- Gaxiola, R.A., Li, J., Undurraga, S., Dang, L.M., Allen, G.J., Alper, S.L., and Fink, G.R. (2001). Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc. Natl. Acad. Sci. USA 98 11444–11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola, R.A., Sherman, A., Grisafi, P., Alper, S.L., and Fink, G.R. (1999). The Arabidopsis thaliana proton transporters, Nhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. USA 96 1480–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genc, Y., McDonald, G., and Tester, M. (2007). Re-assessment of tissue Na+ concentration as a criterion for salinity tolerance in bread wheat. Plant Cell Environ. 30 1486–1498. [DOI] [PubMed] [Google Scholar]
- Gilliham, M., and Tester, M. (2005). The regulation of anion loading to the maize root xylem. Plant Physiol. 137 819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, J.-M., Waner, D.A., Horie, T., Li, S.L., Horie, R., Abid, K.B., and Schroeder, J.I. (2004). Microarray-based rapid cloning of an ion accumulation deletion mutant in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 101 15404–15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff, J. (1999). GFP variants for multispectral imaging of living cells. Methods Cell Biol. 58 139–151. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Horie, T., Costa, A., Kim, T.H., Han, M.J., Horie, R., Leung, H.-Y., Miyao, A., Hirochika, H., An, G., and Schroeder, J.I. (2007). Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 26 3003–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie, T., Yoshida, K., Nakayama, H., Yamada, K., Oiki, S., and Shinmyo, A. (2001). Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 27 129–138. [DOI] [PubMed] [Google Scholar]
- James, R.A., Davenport, R.J., and Munns, R. (2006). Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol. 142 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jou, Y., Chou, P., He, M., Hung, Y., and Yen, H.E. (2004). Tissue-specific expression and functional complementation of a yeast potassium-uptake mutant by a salt-induced ice plant gene mcSKD1. Plant Mol. Biol. 54 881–893. [DOI] [PubMed] [Google Scholar]
- Kato, Y., Hazama, A., Yamagami, M., and Ouzumi, N. (2003). Addition of a peptide tag at the C terminus of AtHKT1 inhibits its Na+ transport. Biosci. Biotechnol. Biochem. 67 2291–2293. [DOI] [PubMed] [Google Scholar]
- Kerk, N.M., Ceserani, T., Tausta, S.L., Sussex, I.M., and Nelson, T.M. (2003). Laser capture microdissection of cells from plant tissues. Plant Physiol. 132 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiegle, E., Gilliham, M., Haseloff, J., and Tester, M. (2000. b). Hyperpolarisation-activated calcium currents found only in cells from the elongation zone of Arabidopsis thaliana roots. Plant J. 21 225–229. [DOI] [PubMed] [Google Scholar]
- Kiegle, E., Moore, C.A., Haseloff, J., Tester, M.A., and Knight, M.R. (2000. a). Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. 23 267–278. [DOI] [PubMed] [Google Scholar]
- Läuchli, A., James, R.A., Huang, C.X., McCully, M., and Munns, R. (2008). Cell-specific localization of Na+ in roots of durum wheat and possible control points for salt exclusion. Plant Cell Environ. 31 1565–1574. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8 457–463. [DOI] [PubMed] [Google Scholar]
- Mäser, P., et al. (2002. a). Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 531 157–161. [DOI] [PubMed] [Google Scholar]
- Mäser, P., Hosoo, Y., Goshima, S., Horie, T., Eckelman, B., Yamada, K., Yoshida, K., Bakker, E.P., Shinmyo, A., Oiki, S., Schroeder, J.I., and Uozumi, N. (2002. b). Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc. Natl. Acad. Sci. USA 99 6428–6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller, I.S. (2008). Na+ Exclusion and Salinity Tolerance in Arabidopsis thaliana. PhD dissertation (Cambridge, UK: University of Cambridge).
- Møller, I.S., and Tester, M. (2007). Salinity tolerance of Arabidopsis: A good model for cereals? Trends Plant Sci. 12 534–540. [DOI] [PubMed] [Google Scholar]
- Munns, R. (1993). Physiological processes limiting plant growth in saline soils: Some dogmas and hypotheses. Plant Cell Environ. 16 15–24. [Google Scholar]
- Munns, R. (2002). Comparative physiology of salt and water stress. Plant Cell Environ. 25 239–250. [DOI] [PubMed] [Google Scholar]
- Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59 651–681. [DOI] [PubMed] [Google Scholar]
- Niu, X., Damsz, B., Kononowicz, A., Bressan, R.A., and Hasegawa, P.M. (1996). NaCl-induced alterations in both cell structure and tissue-specific plasma membrane H+-ATPase gene expression. Plant Physiol. 111 679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo, J.M., Cubero, B., Leidi, E.O., and Quintero, F.J. (2006). Alkali cation exchangers: Roles in cellular homeostasis and stress tolerance. J. Exp. Bot. 57 1181–1199. [DOI] [PubMed] [Google Scholar]
- Platten, J.D., et al. (2006). Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 11 372–374. [DOI] [PubMed] [Google Scholar]
- Qiu, Q.S., Guo, Y., Dietrich, M.A., Schumaker, K.S., and Zhu, J.K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99 8436–8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Z.-H., Gao, J.-P., Li, L.-G., Cai, X.-L., Huang, W., Chao, D.-Y., Zhu, M.-Z., Wang, Z.-Y., Luan, S., and Lin, H.-X. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37 1141–1146. [DOI] [PubMed] [Google Scholar]
- Rengasamy, P. (2006). World salinization with emphasis on Australia. J. Exp. Bot. 57 1017–1023. [DOI] [PubMed] [Google Scholar]
- Roberts, S.K., and Tester, M.A. (1995). Inward and outward K+-selective currents in the plasma membrane of protoplasts from maize root cortex and stele. Plant J. 8 811–825. [Google Scholar]
- Roy, S.J., et al. (2008). Investigating glutamate receptor-like gene co-expression in Arabidopsis thaliana. Plant Cell Environ. 31 861–871. [DOI] [PubMed] [Google Scholar]
- Rus, A., Baxter, I., Muthukumar, B., Gustin, J., Lahner, B., Yakubova, E., and Salt, D.E. (2006). Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet 2 e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus, A., Lee, B.H., Muñoz-Mayor, A., Sharkhuu, A., Miura, K., Zhu, J.-K., Bressan, R.A., and Hasegawa, P.M. (2004). AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 136 2500–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Quintero, F.J., Pardo, J.M., and Zhu, J.K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H., Golldack, D., Zhao, C., and And Bohnert, H.J. (2002). The expression of HAK-Type K+ transporters is regulated in response to salinity stress in common ice plant. Plant Physiol. 129 1482–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarpi, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K, Konomi M, Osumi M, Yamagami M, Schroeder JI, Uozumi N. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 44 928–938. [DOI] [PubMed] [Google Scholar]
- Tester, M., and Davenport, R.J. (2003). Na+ tolerance and Na+ transport in higher plants. Ann. Bot. (Lond.) 91 503–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester, M., and Leigh, R.A. (2001). Partitioning of nutrient transport processes in roots. J. Exp. Bot. 52 445–457. [DOI] [PubMed] [Google Scholar]
- Uozumi, N., Kim, E.J., Rubio, F., Yamaguchi, T., Muto, S., Tsuboi, A., Bakker, E.P., Nakamura, T., and Schroeder, J.I. (2000). The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 122 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitart, V., Baxter, I., Doerner, P., and Harper, J.F. (2001). Evidence for a role in growth and salt resistance of a plasma membrane H+-ATPase in the root endodermis. Plant J. 27 191–201. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Yeo, A., Yeo, M., Flowers, S., and Flowers, T.J. (1990). Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theor. Appl. Genet. 79 377–384. [DOI] [PubMed] [Google Scholar]
- Yeo, A.R., Yeo, M., and Flowers, T. (1987). The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. J. Exp. Bot. 38 1141–1153. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.