Abstract
The eukaryotic nucleolus is multifunctional and involved in the metabolism and assembly of many different RNAs and ribonucleoprotein particles as well as in cellular functions, such as cell division and transcriptional silencing in plants. We previously showed that Arabidopsis thaliana exon junction complex proteins associate with the nucleolus, suggesting a role for the nucleolus in mRNA production. Here, we report that the plant nucleolus contains mRNAs, including fully spliced, aberrantly spliced, and single exon gene transcripts. Aberrant mRNAs are much more abundant in nucleolar fractions, while fully spliced products are more abundant in nucleoplasmic fractions. The majority of the aberrant transcripts contain premature termination codons and have characteristics of nonsense-mediated decay (NMD) substrates. A direct link between NMD and the nucleolus is shown by increased levels of the same aberrant transcripts in both the nucleolus and in Up-frameshift (upf) mutants impaired in NMD. In addition, the NMD factors UPF3 and UPF2 localize to the nucleolus, suggesting that the Arabidopsis nucleolus is therefore involved in identifying aberrant mRNAs and NMD.
INTRODUCTION
The eukaryotic nucleus is highly organized and contains nuclear bodies and domains involved in a variety of aspects of regulation of gene expression (Lamond and Spector, 2003; Misteli, 2005). The most prominent nuclear subcompartment is the nucleolus, which is the site of transcription and processing of precursor rRNAs and the assembly of mature rRNAs and ribosomal proteins into ribosomal subunits (Fatica and Tollervey, 2002; Granneman and Baserga, 2004). Besides the production of ribosomal subunits, the nucleolus is involved in many other RNA metabolism processes, as well as other cellular functions. For example, the nucleolus has roles in the maturation, assembly, and export of ribonucleoprotein particles (RNPs), such as the signal recognition particle and telomerase RNPs, some tRNAs and U6snRNA, and functions in cell cycle control, cell growth, and aging, and cellular stress sensing (Pederson, 1998; Rubbi and Milner, 2003; Olson, 2004; Raška et al., 2006; Boisvert et al., 2007). Recently, some microRNAs and precursor microRNAs have been found to be concentrated in the nucleolus of rat myoblasts (Politz et al., 2006, 2009). The nucleolus is also involved in many animal and plant virus infections, with specific viral proteins interacting with host nucleolar proteins and localizing to the nucleolus (Williams et al., 2005; Boyne and Whitehouse, 2006; Hiscox, 2007). In plants, nucleolar trafficking of the ORF3 protein of Groundnut Rosette Virus involves interactions with Cajal bodies and fibrillarin and is required for viral RNP production and systemic virus spread (Kim et al., 2007a, 2007b). Finally, novel functions in RNA metabolism have been proposed for the plant nucleolus and bodies often associated with the nucleolus (Cajal and D-bodies): production of heterochromatic small interfering RNAs involved in transcriptional gene silencing (Li et al., 2006; Pontes et al., 2006) and maturation of microRNAs (Fang and Spector, 2007; Fujioka et al., 2007; Song et al., 2007). Thus, the nucleolus is multifunctional and is associated with processing of many RNAs and dynamic interactions and trafficking of RNA components between nuclear compartments, the nucleoplasm, and cytoplasm (Lamond and Spector, 2003; Olson, 2004; Gorski et al., 2006; Raška et al., 2006; Boisvert et al., 2007).
Plant nucleoli differ from animal nucleoli in their structural organization. The plant nucleolus contains a much larger proportion of dense fibrillar component (DFC) than animal nucleoli. The DFC is surrounded by the granular component and plant nucleoli often have a nucleolar cavity (Brown and Shaw, 1998; Shaw and Brown, 2004). Nevertheless, the fundamental processes of ribosome biogenesis are very similar in all eukaryotes, as reflected by 70% of the 217 proteins identified in proteomic analyses of Arabidopsis thaliana nucleoli having orthologs in the human nucleolar proteome (Pendle et al., 2005). In addition to nucleolar and ribosome biogenesis proteins, splicing and translation factors as well as six exon-junction complex (EJC) proteins (Y14, Mago, eIF4A-III, RNPS1, UAP56, and REF/Aly) were also identified in the plant nucleolar proteome (Pendle et al., 2005). EJCs are deposited upstream of splice junctions on mRNAs following splicing. They contain proteins involved in splicing, mRNA export, and localization and nonsense-mediated decay (NMD), thereby linking transcription and splicing to export, surveillance, and translation (Maquat, 2004; Tange et al., 2004; Lejeune and Maquat, 2005; Behm-Ansmant et al., 2007b; Chang et al., 2007; Isken and Maquat, 2007). In particular, the plant EJC proteins found in the nucleolus were either orthologs of the core of the EJC complex that binds mRNA or were factors that interact with the TAP export factor (Maquat, 2004; Tange et al., 2004; Lejeune and Maquat, 2005; Andersen et al., 2006; Stroupe et al., 2006; Chang et al., 2007; Isken and Maquat, 2007). We further showed that green fluorescent protein (GFP) fusion proteins of these and other EJC proteins localized to the nucleolus in contrast with their animal orthologs, suggesting that messenger RNPs (mRNPs) may be present in the plant nucleolus (Pendle et al., 2005). In this article, we show that the nucleolus and the nucleoplasm contain different profiles of mRNA transcripts and that aberrantly spliced mRNAs are more abundant in the nucleolus. In addition, at least some of the aberrantly spliced transcripts are substrates for NMD, and the Up-frameshift (UPF) NMD proteins UPF2 and UPF3 are localized in the nucleolus, suggesting that the Arabidopsis nucleolus is involved in recognition of aberrantly spliced mRNAs and the NMD pathway.
RESULTS
The Plant Nucleolus Is Enriched in Aberrantly Spliced mRNAs
To examine whether mRNAs are present in plant nucleoli, we constructed cDNA libraries from poly A+ RNA isolated from whole cells, isolated nuclei, and purified nucleoli from Arabidopsis culture cells. Around 1,000 cDNA clones were completely sequenced (approximately 300 each from the whole cell and nuclear libraries and 400 from the nucleolar library) of which 497 were full-length (Figure 1). The cDNA sequences were grouped into three classes of mRNA: 1) transcripts from single exon genes (which do not contain introns and therefore have not undergone splicing); 2) fully spliced mRNAs where all introns in a transcript have been removed; and 3) “aberrantly” spliced mRNAs (Figure 1). The latter class consists of potentially mis-spliced mRNAs (splicing errors), incompletely spliced mRNAs (where one or more introns have not been removed) and alternatively spliced mRNA transcripts. Given our relatively limited knowledge of alternative splicing events for many plant genes, it is difficult to distinguish among is-splicing, incomplete splicing and alternative splicing.
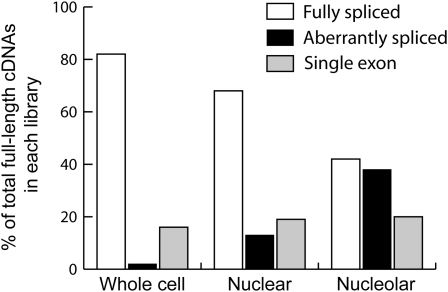
Figure 1.
Distribution and Nature of Transcript Types in cDNA Libraries.
The distribution of fully spliced, aberrantly spliced, and single exon transcripts in whole-cell, nuclear, and nucleolar cDNA libraries expressed as percentage of total full-length cDNAs from the same library (226, 139, and 132 total cDNAs from the nucleolar, nuclear, and whole-cell libraries, respectively). Only full-length, nonredundant cDNAs were scored.
The distribution of mRNA classes in the whole-cell, nuclear, and nucleolar libraries was strikingly different (Figure 1). While the proportion of single exon gene transcripts was similar in the three libraries (ranging from 16 to 20%), the proportion of fully spliced transcripts was highest in the whole-cell library (82%) and less abundant in the nuclear (68%) and nucleolar (42%) libraries (Figure 1). By contrast, aberrantly spliced mRNAs represented only 2% of the transcripts in the whole-cell library, and this proportion rose to 13 and 38% in the nuclear and nucleolar libraries, respectively. The abundance of aberrantly spliced transcripts in the whole-cell library (2%) is similar to that observed generally in plant cDNA libraries and EST collections. The nucleolar preparation used for nucleolar cDNA library construction was the same as that used in the proteomic analysis of Arabidopsis nucleoli (Pendle et al., 2005). The profile of identified proteins and subsequent GFP fusion protein localizations showed this fraction to be highly enriched in nucleolar proteins with ∼10% contamination from nuclear/cytoplasmic proteins (Pendle et al., 2005). Although some contamination is expected in biochemical fractionation, contamination from the nucleoplasm or cytoplasm cannot account for the presence of mRNAs in the nucleolar fraction because of the significantly different composition of mRNA types. If the mRNAs isolated in the nucleolar library were due to nucleoplasmic/cytoplasmic contamination, the proportions of fully spliced, aberrantly spliced, and single exon transcripts in the nucleolus would reflect those found in the whole-cell or nuclear libraries. This is clearly not the case because while the proportion of single exon transcripts remains constant, the level of fully spliced transcripts is halved and that of aberrantly spliced transcripts increases 20- and 3-fold in the nucleolar library over the levels found in whole-cell and nuclear libraries, respectively (Figure 1). Thus, the plant nucleolus contains mRNAs that are enriched in aberrantly spliced transcripts such that almost half of the transcripts from intron-containing genes are aberrantly spliced.
The Majority of Aberrantly Spliced mRNAs Are Potential NMD Substrates
We next examined the nature of the aberrant mRNAs that accumulated in the nucleolus and whether there were implications for gene expression. The most common event was the presence of an unspliced intron, occurring in almost 75% of the aberrantly spliced transcripts (Figures 2A, 2G, and 3A). The remainder exhibited a range of different splicing phenotypes such as the use of cryptic 5′ or 3′ splice sites in intron or exon sequences or exon skipping (Figures 2B to 2F, 2H, 2I, and 3A; see Supplemental Table 1 online). Some transcripts contained more than one aberrant splicing event (e.g., Figure 2D).
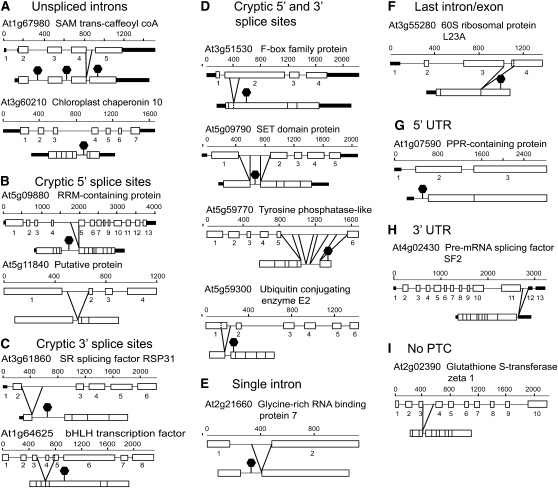
Figure 2.
Schematic Diagrams of Gene Structures of Representative Aberrantly Spliced Transcripts.
The examples demonstrate different splicing phenotypes: unspliced introns (A), use of cryptic 5′ splice sites (B), use of cryptic 3′ splice sites (C), use of both cryptic 5′ and 3′ splice sites (D), aberrant splicing in a single intron gene (no downstream exon-exon junction/EJC) (E), aberrant splicing in the last intron/exon (no downstream EJC) (F), aberrant splicing in a 5′ UTR intron (this example produces a short open reading frame in the UTR) (G), aberrant splicing in a 3′ UTR intron (H), and alternative splicing leaving an intronic fragment in frame (no PTC) (I). The cloned transcript is shown below the exon-intron structure of the gene. The splicing events result in the removal of parts of exon sequences, inclusion of whole or partial intron sequences, or exon skipping. Open boxes, exons; thick black lines, UTRs; solid lines, introns; dashed vertical lines, site of cryptic splice site in exon. Aberrant splicing events are indicated by diagonal lines; black hexagons indicate that PTCs are generated in the transcript.
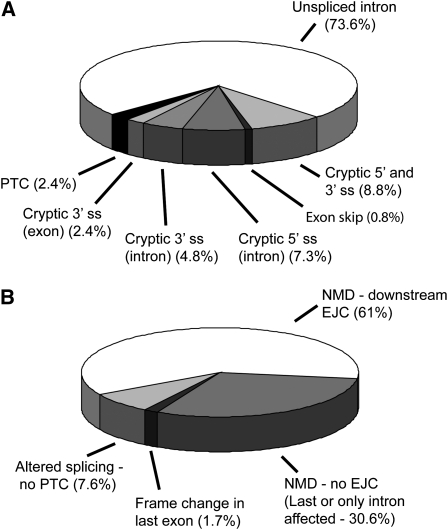
Figure 3.
Distribution of Splicing and NMD Transcript Phenotypes.
(A) Distribution of different splicing phenotypes among 160 aberrantly spliced transcripts from the three cDNA libraries.
(B) Distribution of transcripts by potential consequence of the aberrant splicing event(s). NMD phenotypes were classified as potential NMD substrates with or without a downstream EJC, or non-NMD substrates where no PTC is generated or where the PTC remains close to the 3′ UTR due to a frame change in the last exon. ss, splice site.
By identifying the presence and position of premature termination codons (PTCs), 91% of the aberrantly spliced transcripts were found to be putative NMD substrates on the basis of current models of long 3′ untranslated region (UTR)- and intron-based PTC recognition in plants (Kertész et al., 2006; Schwartz et al., 2006; Hori and Watanabe, 2007; Kerényi et al., 2008) (Figure 3B; see Supplemental Table 1 online). All had an increased distance between the PTC and 3′ UTR, and two-thirds also contained one or more splice junctions (and therefore EJCs) downstream of the PTC (Figures 2 and 3B). The remaining aberrant transcripts (∼9%) were not expected to produce NMD substrates as they caused in-frame changes (Figure 2I) or a change in reading frame in the last exon without significantly affecting the position of the stop codon (Figure 3B). Finally, some aberrant splicing events affected introns in the 5′ or 3′ UTRs (Figures 2G and 2H). In particular, those in the 5′ UTR either had no effect on the reading frame or generated a short open reading frame (ORF) upstream of the normal translation initiation codon that could affect translational efficiency (Meijer and Thomas, 2002) or trigger NMD (He et al., 2003; Mendell et al., 2004) (see Supplemental Table 1 online).
Distribution of mRNAs in Nuclear, Nucleolar, and Nucleoplasmic Fractions
Having shown by sequencing of cDNA libraries that the nucleolus contained mRNAs and was enriched in aberrant mRNAs, we validated these results by RT-PCR of total RNA from isolated nuclei and nucleolar and nucleoplasmic fractions. Initial RT-PCR analyses showed multiple higher molecular weight products for many genes (Figures 4 and 5), making a standard quantitative RT-PCR approach impossible as multiple primer pairs would be required to assess each product separately. Therefore, we used gene-specific primers (designed to the 5′ and 3′ regions of the coding sequence) to amplify all splicing isoforms in the same reaction to compare directly the relative levels of fully spliced and aberrantly spliced transcripts from the same gene. Extensive testing of PCR conditions was performed on a number of genes with known alternatively spliced isoforms and with a range of expression levels. For the majority of genes tested, the ratios of alternatively spliced isoforms remained constant (i.e., in the logarithmic amplification range) over 22 to 24 cycles (see Supplemental Figure 1 online; see Methods).
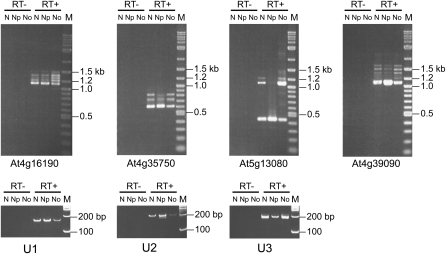
Figure 4.
RT-PCR of mRNA Transcripts in Nuclear, Nucleoplasmic, and Nucleolar RNA.
Products of RT-PCR (RT+) with gene-specific primers from nuclear (N), nucleoplasmic (Np), and nucleolar (No) RNA for different Arabidopsis genes showing enrichment of higher molecular weight products and a reduction in fully spliced product in the nucleolar fraction. M, size markers; RT−, PCR controls without reverse transcriptase. The fractionation was assessed by RT-PCR of U1 snRNA (U1) and U2 snRNA (U2) for the nucleoplasmic and U3 snoRNA (U3) for nucleolar fractions.
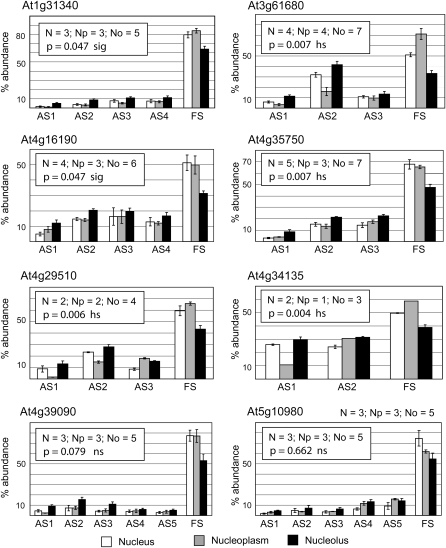
Figure 5.
Relative Abundance of Fully Spliced and Aberrantly Spliced RT-PCR Products for Different Genes in Nuclear, Nucleoplasmic, and Nucleolar RNA Fractions.
Histograms showing the mean and standard errors of the relative abundances (expressed as a percentage) of different higher molecular weight, aberrantly spliced products (AS1-5) and fully spliced (FS) products for the different fractions (white, nuclear; gray, nucleoplasmic; black, nucleolar). The number of different fractions used for RT-PCR is given in the inset for each histogram (N, nuclear; Np, nucleoplasmic; No, nucleolar). The combined means of the aberrantly spliced bands and mean of the fully spliced band were compared between the nuclear and nucleolar preparations by analysis of variance. Probability values for AS>FS in the nucleolus and FS>AS in the nucleoplasm are presented (sig, significant, P < 0.05; hs, highly significant, P < 0.01; ns, not significant).
Gene-specific primers were designed to a number of genes whose transcripts had been cloned in the nucleolar cDNA library either as aberrant transcripts or normal, fully spliced transcripts and to genes that were not represented in any of the libraries (effectively randomly selected). Following an RT reaction with oligo(dT) using total RNA from nuclei, and nucleolar and nucleoplasmic fractions, PCR was performed for 24 cycles. The purity of the nucleoplasmic and nucleolar preparations was assessed using RT-PCR with primers to amplify the spliceosomal small nuclear RNAs, U1 and U2, and the small nucleolar RNA, U3 (Figure 4). U1 and U2 were enriched in the nucleoplasmic fractions, while U3 was enriched in the nucleolar fractions. The presence of some U1 and U2 in plant nucleoli has been shown previously by in situ hybridization (Beven et al., 1995) and localization of U1 and U2 small nuclear RNPs (snRNPs) (Lorković and Barta, 2008). Thus, although some cross-contamination is expected, the fractions are enriched as expected for nucleoplasmic or nucleolar snRNAs.
For virtually all of the genes tested, fully spliced transcripts were more abundant in nucleoplasmic RNA, while the higher molecular weight transcripts (unspliced/aberrantly spliced) were more abundant in nucleolar RNA (Figures 4 and 6, lanes 1 and 2). We showed that the higher molecular weight RT-PCR products represented differently spliced transcripts by cloning and sequencing the products from some genes (see Supplemental Figure 2 online). The enrichment of aberrant, higher molecular weight products and the reduction of fully spliced products in the nucleolus was found consistently in RT-PCR reactions with RNA from numerous different nuclear, nucleoplasmic, and nucleolar preparations from culture cells and was significantly different for most genes tested (Figure 5). Thus, the RT-PCR results corroborate those obtained from cloning and sequencing from cDNA libraries (Figure 1). The relative proportions of higher molecular weight transcripts to fully spliced transcripts in the nucleus were estimated from the relative band intensities in the RT-PCR reactions of nuclear preparations (Figure 4, N lanes) and varied greatly among different genes ranging from between 20 and 50% for the genes studied (Figures 4 and 5). Higher molecular weight aberrant transcripts were detectable for most genes tested in nuclear and, in particular, nucleolar RNA preparations. They were more difficult to detect in RNA from whole cells or seedling material under the PCR conditions used. This reflects the much higher levels of spliced mRNAs derived from the cytoplasm in this material and again is consistent with the 1 to 2% of plant ESTs consisting of transcripts with, for example, unspliced introns (e.g., Figure 1, whole-cell library). Thus, production of aberrant transcripts (whether due to splicing errors or alternative splicing) is a common occurrence likely to require NMD for their turnover, and these mRNA transcripts are enriched in the nucleolus.
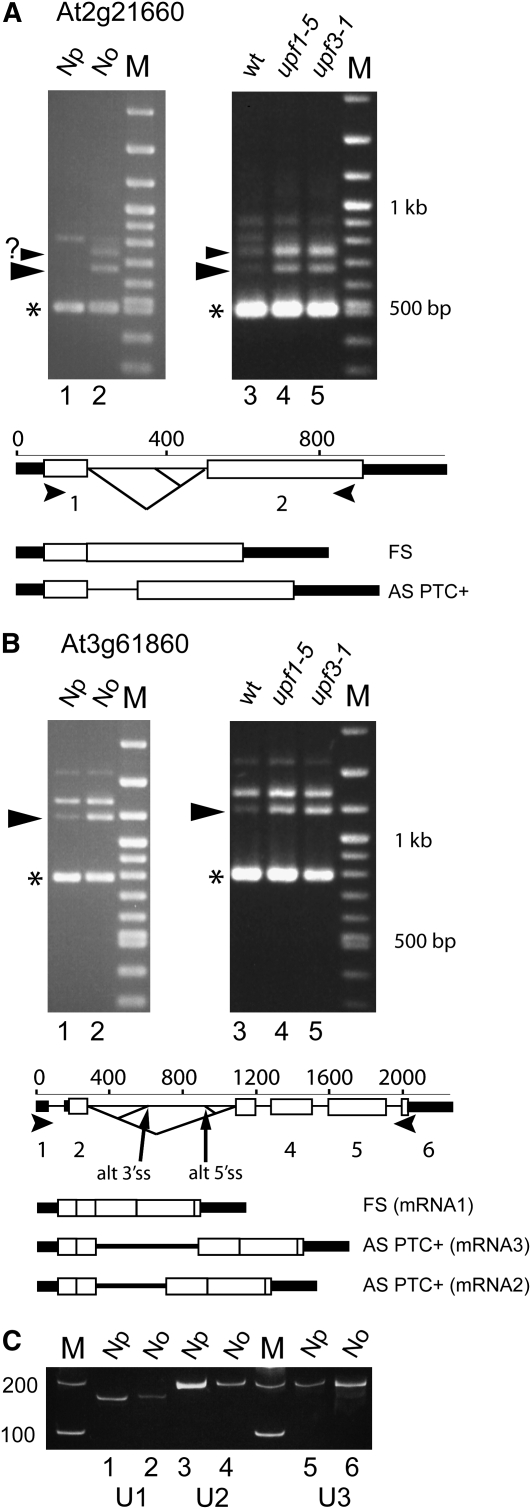
Figure 6.
RT-PCR of mRNA Transcripts in Nuclear, Nucleoplasmic, and Nucleolar RNA and upf Mutants.
(A) and (B) RT-PCR with gene-specific primers for At2g21660 (A) and At3g61860 (B) from RNA from nucleoplasmic (Np) and nucleolar (No) fractions of cell culture cells (lanes 1 and 2) and seedlings of wild-type plants (lane 3), upf1-5 (lane 4), and upf3-1 (lane 5). Transcript structures are shown below the exon/intron structure of the genes based on The Arabidopsis Information Resource (TAIR).
(C) The fractionation was assessed by RT-PCR of U1 snRNA (lanes 1 and 2) and U2 snRNA (lanes 3 and 4) for the nucleoplasmic fraction and U3 snoRNA (lanes 5 and 6) for the nucleolar fraction. M, size markers; large arrowheads, RT-PCR products that are enriched in the nucleolus and in upf mutants; small arrowhead with question mark, unknown product; asterisk, fully spliced product; open boxes, exons; thick black lines, UTRs; solid lines, introns; diagonal lines, splicing events; small arrowheads, primer positions. RT-PCR controls without reverse transcriptase are not shown.
Transcripts Enriched in the Nucleolus Are NMD Sensitive
The majority of aberrant mRNAs from the cDNA libraries contained PTCs and were predicted to be degraded by NMD (Figure 3B; see Supplemental Table 1 online). To demonstrate the link between nucleolar accumulation of particular transcripts and NMD, we performed RT-PCR reactions on total RNA isolated from whole seedlings of wild-type plants and mutant plants of the NMD proteins UPF1 and UPF3. We used two mutants, upf1-5 and upf3-1, which have been shown to be impaired in NMD such that mRNAs that are normally turned over by NMD accumulate in these mutants (Hori and Watanabe, 2005; Arciga-Reyes et al., 2006). RT-PCR was performed using gene-specific primers to At2g21660 and At3g61860 encoding the Gly-rich RNA binding protein, GRP7, and the SR protein, RSp31, respectively. Aberrant transcripts of both these genes were isolated in the nucleolar cDNA library and were detectable in seedling RNA using 24 cycles of PCR. Their alternatively spliced transcripts have also been characterized (Kalyna et al., 2006; Schöning et al., 2007, 2008). In particular, the levels of GRP7 transcripts are regulated by an autoregulatory loop that promotes the production of an alternatively spliced variant that is turned over by NMD (Schöning et al., 2007, 2008). RT-PCR of RNA from the upf mutants showed the accumulation of two higher molecular weight products for At2g21660 in the mutants (Figure 6A, lanes 4 and 5) compared with the wild type (Figure 6A, lane 3). These same products are enriched in the nucleolar fraction compared with the nucleoplasmic fraction (Figure 6A, lanes 1 and 2). The lower product (674 bp) is the alternatively spliced product of GRP7 that is regulated by alternative splicing and NMD; the identity of the larger product is unknown. RT-PCR of At3g61860 also showed two higher molecular weight products, the smaller of which was enriched in both the nucleolar fraction and the upf mutants and is therefore NMD sensitive (Figure 6B). The larger of the two products represents a PTC+ transcript generated by use of alternative 5′ and 3′ splice sites. This transcript is stable and not turned over by NMD. Other naturally occurring NMD transcripts were analyzed in the original descriptions of the upf mutants (Hori and Watanabe, 2005; Arciga-Reyes et al., 2006), and RT-PCR analysis of one such gene, At2g45670, again showed that the PTC+/NMD transcript is enriched in the nucleolus and accumulates in the upf mutants (see Supplemental Figure 3 online). Thus, there is a correlation between the accumulation of aberrant mRNAs in the nucleolus and their turnover by NMD for at least some gene transcripts.
Plant NMD Proteins Are Associated with the Nucleolus
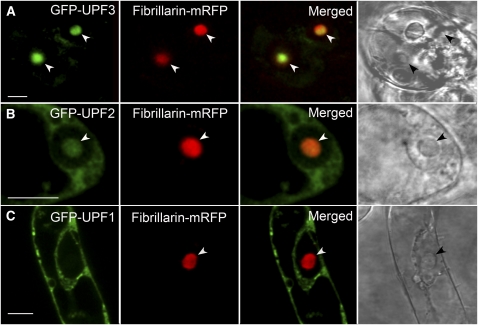
Since most of the aberrant mRNAs in the nucleolus were potential NMD substrates, we asked whether factors required for NMD were also present in the nucleolus. GFP fusion constructs to the NMD proteins UPF1, UPF2, and UPF3 were coexpressed with a fibrillarin-monomeric red fluorescent protein (mRFP) fusion construct (as a marker for the nucleolus) in Arabidopsis suspension culture cells. Although the fusion constructs were expressed from the cauliflower mosaic virus 35S promoter, cells that fluoresced at low levels were imaged to minimize potential problems due to overexpression of GFP fusion proteins (Pendle et al., 2005). GFP-UPF3 was localized predominantly in the nucleolus, GFP-UPF2 was found in both the nucleolus and cytoplasm, while GFP-UPF1 was largely cytoplasmic (Figures 7A to 7C, respectively). GFP-UPF1 exhibited a heterogeneous distribution in the cytoplasm (Figure 7C) with bright fluorescent signal concentrated in many relatively small foci, which might indicate the accumulation of UPF1 in specific compartments, possibly processing bodies (P-bodies) where mRNA decay factors are concentrated and where degradation of aberrant mRNA can occur (Eulalio et al., 2007; Parker and Sheth, 2007). The nucleolar localization of UPF3-GFP was confirmed in stably transformed live seedlings (see Supplemental Figure 4 online).
Figure 7.
Subcellular Localization of NMD Proteins, UPF1, UPF2, and UPF3.
Confocal images of single cells coexpressing GFP-UPF (green signal) and fibrillarin-mRFP (red signal) protein fusions and merged images. GFP-UPF3 (A), GFP-UPF2 (B), and GFP-UPF1 (C). The right-hand panel shows transmission light images. Nucleoli are indicated by arrowheads. Bars = 10 μm.
DISCUSSION
Here, we show the unexpected presence of mRNAs in the plant nucleolus and enhanced levels of aberrantly spliced mRNAs in the nucleolus compared with the nucleoplasm. The majority of the aberrant mRNAs in the nucleolus were potential NMD substrates by virtue of containing PTCs. Our data, using upf mutants impaired in NMD, demonstrate a direct link between aberrantly spliced mRNA transcripts that are enriched in the nucleolus and NMD such that at least some of the nucleolar aberrant mRNAs are targets for NMD. Finally, in addition to the presence of mRNAs in the nucleolus and the nucleolar association of plant EJC proteins (Pendle et al., 2005), we showed that the NMD factors UPF2 and UPF3 also localize to the nucleolus. These observations suggest that the plant nucleolus has a novel function in RNA metabolism by being involved in detection of aberrant mRNAs and the NMD pathway.
The association of mRNAs with the nucleolus has been reported previously. In mammalian cells, evidence for mRNAs in the nucleolus is limited to a few spliced cellular mRNAs (e.g., c-myc), and the reason for their nucleolar localization is unknown (Bond and Wold, 1993; Pederson, 1998; Olson, 2004). The nucleolus has also been implicated in mRNA export in eukaryotes based on the nucleolar accumulation of poly(A)+ RNA in mutants of export components (Pederson, 1998; Ideue et al., 2004; Olson, 2004), although some of these observations may reflect polyadenylation of transcripts undergoing degradation following disruption of export (Carneiro et al., 2007). On the other hand, some viral mRNAs in mammalian cells are exported via the nucleolus. For example, herpes virus saimiri spliced mRNA and human immunodeficiency virus type 1 unspliced and singly spliced mRNAs are bound by the viral proteins ORF57 and Rev, respectively, and transported to the nucleolus. These proteins also cause the redistribution of export factors allowing assembly of export-competent viral RNP complexes in the nucleolus (Williams et al., 2005; Boyne and Whitehouse, 2006; Hiscox, 2007). In the case of unspliced or partially spliced human immunodeficiency virus mRNAs, this export pathway has been suggested to protect the partially spliced mRNAs from the NMD machinery (Hiscox, 2007). More recently, a model has been proposed for nucleolar involvement in the formation of mRNPs that are localized to specific regions of the cytoplasm for translation. Localized mRNAs, such as yeast ASH1 mRNA, enter the nucleolus bound to specific RNA binding proteins. In the nucleolus, translation repressor proteins are loaded onto the mRNA before the mRNP is exported to the cytoplasm and its final destination, where translation is activated (Du et al., 2008; Jellbauer and Jansen, 2008). A similar trafficking pathway may also operate in mammals, for the formation of mRNPs associated with Staufen, an RNA binding protein found in the nucleolus and involved in transport of mRNAs in neurons (Jellbauer and Jansen, 2008). Thus, the eukaryotic nucleolus is involved in the assembly and export of some cellular and viral mRNAs/mRNPs. Here, the levels of aberrant mRNA transcripts that we have observed in the plant nucleolus are unprecedented. They also derive from a wide range of genes, suggesting that their presence in the nucleolus is part of a process to identify aberrant mRNAs, which is not restricted to specific genes or gene sets.
The majority of aberrant mRNA transcripts in the plant nucleolus contain PTCs and are putative or known targets of NMD. These characteristics raise questions of how PTC-containing (PTC+) aberrant mRNAs are identified in plants, how they become enriched in the nucleolus, and what the link to NMD is. NMD identifies PTC+ transcripts that encode truncated proteins that are either nonfunctional or potentially detrimental to cell function and targets them for rapid degradation (Maquat, 2004; Lejeune and Maquat, 2005; Behm-Ansmant et al., 2007b; Chang et al., 2007; Isken and Maquat, 2007; Shyu et al., 2008). NMD is a nucleus-associated and translation-dependent process and, in animal cells, the site of mRNA surveillance is thought to be on the cytoplasmic side of the nuclear pore complex in a pioneer round of translation as mRNPs exit the nucleus, with degradation occurring in the cytoplasm (Maquat, 2004; Lejeune and Maquat, 2005; Behm-Ansmant et al., 2007b; Chang et al., 2007; Isken and Maquat, 2007; Singh et al., 2007; Shyu et al., 2008). The mechanisms of PTC recognition vary among different species but can be summarized as relying on signals downstream of the PTC and, in particular 3′ UTRs and associated proteins, or being splicing and EJC dependent (Bühler et al., 2004, 2006; Amrani et al., 2006; Rehwinkel et al., 2006; Behm-Ansmant et al., 2007a, 2007b; Chang et al., 2007; Isken and Maquat, 2007; Shyu et al., 2008; Stalder and Mühlemann, 2008). In plants, NMD is also triggered by the distance between the PTC and 3′ UTR (long 3′ UTRs) and/or the presence of a downstream exon-exon junction (intron-based NMD) (Kertész et al., 2006; Schwartz et al., 2006; Hori and Watanabe, 2007; Kerényi et al., 2008). These features are evident not only in NMD-sensitive protein-coding gene transcripts, but also in mRNA-like noncoding RNAs, which are also turned over by NMD in Arabidopsis (Kurihara et al., 2009).
Plant orthologs of EJC proteins and the NMD proteins UPF1, UPF2, UPF3, and SMG-7 have been identified (Pendle et al., 2005; van Hoof and Green, 2006; Behm-Ansmant et al., 2007b), and the involvement of plant Mago, Y14, UPF1, UPF2, UPF3, and SMG-7 in the NMD pathway has been shown by the stabilization of PTC+ transcripts either in viable mutants or when the proteins are reduced by silencing (Hori and Watanabe, 2005; Arciga-Reyes et al., 2006; Yoine et al., 2006; Kertész et al., 2006; Wu et al., 2007; Kerényi et al., 2008). In mammals, UPF2 interacts with UPF1 and UPF3, and the order of association of UPF proteins with EJCs is UPF3, UPF2, and finally UPF1, which is important in activating NMD (Kashima et al., 2006). In plants, UPF2 also links UPF3 and UPF1 (Kerényi et al., 2008). Despite these functional similarities, the localization of the Arabidopsis UPF3 and UPF2 to the nucleolus contrasts with that of their mammalian orthologs in that human UPF3 is primarily nuclear, and human UPF2 and UPF1 are primarily cytoplasmic proteins (Lykke-Andersen et al., 2000; Serin et al., 2001). The fact that Arabidopsis UPF1 is cytoplasmic suggests that NMD occurs in the cytoplasm. Although we are, at present, unable to distinguish whether the localization of UPF2 and UPF3 in the nucleolus reflects functional activity in the NMD pathway or some other function, the clear difference in localization of these proteins between plants and animals is unexpected. Thus, while there is some underlying conservation in both mechanisms and components of the NMD pathway between plants and other eukaryotes, our results suggest that there are potentially fundamental differences in the organization of the NMD pathway in plants that involve a function for the nucleolus.
The distinct profiles of fully spliced and aberrantly spliced transcripts in the nucleoplasm and nucleolus suggest that, in plants, different classes of mRNAs are distinguished prior to exit from the cytoplasm and lead to enrichment of aberrant mRNAs in the nucleolus. Aberrant transcripts could be distinguished from normal mRNAs in a number of ways. First, aberrant mRNAs could be discriminated by an NMD-like mRNA surveillance mechanism occurring either in the nucleoplasm to target aberrant mRNAs to the nucleolus or in the nucleolus itself. There is some evidence in favor of nuclear NMD in mammals (Bühler et al., 2002, 2004, 2006). However any mechanism involving the detection of stop codons would presumably have to involve an engaged ribosome, in a nuclear or nucleolar pioneer round of translation, since the ribosome is currently the only known factor that can interpret the reading frame and thus detect stop codons. Evidence has been published for nuclear translation, but this idea is still hotly disputed (Iborra et al., 2001; Dahlberg and Lund, 2004). On the other hand, if mRNA surveillance occurs as mRNPs exit the nuclear pore complex as in animals, the aberrant transcripts would have to be reimported into the nucleus/nucleolus. Although export and reimport of RNAs from the cytoplasm is well established in the biogenesis of small nuclear RNAs and some tRNAs (Hopper and Shaheen, 2008), it is difficult to see the advantage to the cell of exporting aberrant mRNAs to allow cytoplasmic surveillance and reimporting them to the nucleolus.
A second possibility is that aberrant mRNPs are identified by virtue of differences in their mRNP composition. Plant introns are UA rich (up to 80%) with on average 20% lower GC content than exon sequences. U-rich sequences are required for efficient splicing and are thought to bind U-rich binding proteins as an early intron recognition step prior to spliceosome formation (Lorković et al., 2000). Given that the majority of the aberrant nucleolar mRNAs found in our study contained either unspliced introns or intron fragments, U-rich binding proteins could mark the aberrant transcripts and target them to the nucleolus by an unknown mechanism.
Third, normally spliced and aberrantly spliced mRNPs could be distinguished at the level of export, with the export of aberrant mRNPs inhibited through the interaction or lack of interaction between particular proteins on the mRNP and the nuclear pore complex. In yeast, spliced transcripts are discriminated by MLp proteins associated with the nuclear pore complex, which interact with the hnRNP-like protein Nab2p, present on spliced mRNAs, allowing them to be exported (Galy et al., 2004). In animals, splicing commitment factors that recognize splice sites or intronic sequences prevent intron-containing transcripts from being exported (Stutz and Izaurralde, 2003). A similar mechanism of differential export could operate in plants, again dependent on the presence of specific proteins (e.g., U-rich binding proteins) bound to introns or intron fragments in aberrant mRNAs. The latter possibilities would provide a general mechanism for the identification of transcripts containing introns or intron fragments irrespective of which gene they are transcribed from.
How do aberrant transcripts become enriched in the nucleolus and what is the link to NMD? Current ideas of nuclear organization suggest that the dynamic distribution of molecules and complexes relies on overall diffusion with differential accumulation in different compartments depending on mean residence time in the different compartments due to molecular interactions (Gorski et al., 2006; Raška et al., 2006). Thus, while the presence of fully spliced and single exon mRNAs in the plant nucleolus may reflect free diffusion of mRNPs between the nucleoplasm and nucleolus, the enhanced levels of aberrant mRNA transcripts indicate that these mRNAs are retained for longer in the nucleolus. Aberrant mRNPs may reach the nucleolus by targeting due the presence of specific proteins on mRNPs or by diffusion to the nucleolus where they interact with factors that remodel these mRNPs for degradation by the NMD pathway. The localization of UPF3 and UPF2 in the nucleolus suggests a model where the NMD proteins are loaded onto the aberrant mRNPs prior to being exported to the cytoplasm where UPF1 (mainly cytoplasmic) is recruited to activate degradation. The assembly of UPF3/UPF2 onto the aberrant transcripts would parallel the model recently proposed for assembly of translationally repressed, localized mRNPs in yeast and animals, and for export-competent viral mRNPs, in the nucleolus before export to the cytoplasm (Hiscox, 2007; Jellbauer and Jansen, 2008).
Taken together, the presence of EJC, UPF2, and UPF3 proteins and aberrant NMD-substrate mRNAs in the nucleolus suggests that the plant nucleolus is involved in the pathway by which aberrant mRNAs are turned over by NMD. This links the nucleolus to mRNA surveillance and NMD processes and clearly differentiates plants from animals in this respect. That plants have evolved a novel system to deal with aberrant mRNA transcripts may reflect the fundamental differences in plant and animal gene structure and the nature of the transcripts produced by splicing errors. For example, plant introns are UA rich and proteins have been proposed to bind these sequences early in spliceosome assembly (Lorković et al., 2000). In addition, however, plant genes differ significantly from animal genes in intron size. The average size of Arabidopsis introns is ∼170 nucleotides, while that of animals is ∼5.5 kb (see Barbazuk et al., 2008) with some animal introns being tens of thousands of nucleotides long. The effect of this difference is that errors in splicing in animals usually lead to skipping of an exon, requiring translation to detect changes in reading frame and PTCs in the resultant mRNAs. In plants, errors in splicing usually lead to nonremoval of introns (intron retention) and, as we have shown from sequencing, the majority (∼75%) of the aberrant mRNAs contain unspliced introns or intron fragments. Thus, plants may identify intron-containing transcripts and prepare them for degradation by the NMD pathway in the nucleus and nucleolus, while other PTC+ transcripts without intron fragments would be identified on export by the pioneer round of translation as in animals.
METHODS
Plant Materials and Growth Conditions
The upf1-5 and upf3-1 mutants were obtained from Brendan Davies (Hori and Watanabe, 2005; Arciga-Reyes et al., 2006). Wild-type and mutant plants were grown at 25°C in 16-h-light/8-h-dark conditions in a controlled environment cabinet. For nuclei and nucleoli preparation, an Arabidopsis thaliana suspension culture derived from a Landsberg erecta line was used as a source of material. Cultures were grown at 25°C in the light with constant shaking on an orbital incubator at 200 rpm in AT medium (4.4% Murashige and Skoog [MS] salts, 3% sucrose, 0.05 mg/L kinetin, and 0.5 mg/L 1-naphthaleneacetic acid (NAA), pH 5.8).
Purification of Nuclei and Nucleoli from Arabidopsis Culture Cells
Nucleoli were routinely purified from nuclei isolated from an Arabidopsis (Landsberg erecta) suspension culture as described previously (Pendle et al., 2005). The purification was monitored by phase contrast light microscopy, microscopy following staining with pyronin Y, and by enrichment of U3snoRNA in nucleoli by RT-PCR (see below). For preparation of nucleoplasmic and nucleolar RNA fractions for RT-PCR, nuclei were disrupted by sonication, nucleoli harvested by centrifugation, and the supernatant taken as the nucleoplasmic fraction. Fractionation was monitored by RT-PCR of U1 and U2 snRNAs and U3 snoRNAs to demonstrate enrichment in the nucleoplasmic and nucleolar fractions, respectively. Total RNA (250 ng) was primed by 3′-AtU1 (5′-AGGGGCTGCGCGAACGCARRCC-3′), 3′-AtU2 (5′-GYGGGSTGAGCCARGCCCDTGCARTA-3′), or 3′-AtU3 (5′-CCTGTCAGACCCGCCGTGCGACCA-3′) complementary to the 3′ ends of Arabidopsis U1 and U2 snRNAs and U3 snoRNA, respectively. PCR was performed following addition of the 5′-end primers (5′-AtU1, 5′-ACTTACCTGGACGGGGTCWACKCGYGATCA-3′; 5′-AtU2, 5′-ATACCTTTCTCGGCCTTTTGGCTAA-3′; and 5′-AtU3, 5′-TCTGTTCTATAGGCTCGTACCTCTGTTT-3′) using 15 (for U2 and U3) or 20 cycles (for U1). The PCR products were separated an on 8% polyacrylamide gel.
Nucleolar, Nuclear, and Whole-Cell cDNA Libraries
Total RNA was extracted from whole cells, isolated nuclei, and nucleoli using an RNeasy kit (Qiagen), and mRNA was purified from total RNA using Oligotex beads (Qiagen) according to the manufacturer's instructions. cDNA libraries were generated using 100 ng of mRNA prepared from nucleoli, nuclei, and whole cells using the GeneRacer system (Invitrogen) according to the manufacturer's protocols. In brief, total RNA was dephosphorylated prior to decapping of 5′ ends of mRNA by tobacco acid pyrophosphatase and ligation to the RNA oligonucleotide adaptor (5′-CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA-3′) with T4 RNA ligase. This procedure enriched for full-length, capped poly(A)+ RNA. First-strand cDNA was generated using an oligo(dT)-adaptor primer [GCTGTCAACGATACGCTACGTAACGGCATGACAGTG(T)18-24] and Superscript III RT. The second-strand cDNA was generated using primers specific to 3′ and 5′ oligonucleotide adaptor sequences and Platinum Taq polymerase high fidelity (Invitrogen). cDNA fragments >500 bp were enriched by Sepharyl-S500 spin column (Amersham Biosciences) chromatography, inserted into the pCR TA-cloning vector (Invitrogen), and fully sequenced.
Sequence and Gene Structure Analysis of cDNA Clones
cDNA sequences were compared with the current gene models of the Arabidopsis Genome Initiative gene data set containing all Arabidopsis transcription unit sequences with intron and UTR sequences using BLASTN at TAIR (http://www.arabidopsis.org). Resulting alignments were compared with the full-length genomic sequences and gene structure models from The Institute for Genomic Research Arabidopsis thaliana database (http://www.tigr.org/tdb/e2k1/ath1/). Computer-assisted translation of aberrant transcripts with ExPASy (http://www.expasy.ch/tools/dna.html) identified the position of PTCs and ORFs. The analysis accurately identified the exact extent and nature of the aberrant or alternative splicing events.
Expression Analyses of Gene Transcripts
Whole nuclei and nucleolar and nucleoplasmic fractions were extracted with phenol/chloroform and total RNA recovered by ethanol precipitation and treated with RNase-free DNase prior to use. For RT-PCR, 5 μg each of nuclear, nucleoplasmic, or nucleolar total RNA was used for the first-strand cDNA synthesis primed by oligo(dT)25, and one-tenth volume of mixture was amplified by PCR using gene-specific primers. PCR was performed at 24 cycles, which was in the exponential amplification range determined by measuring the levels of RT-PCR products with increasing PCR cycle number (Simpson et al., 2008). The reaction products were fractionated on a 1.2% agarose gel, and band intensities compared using the public domain program ImageJ (Wayne Rasband; http://rsb.info.nih.gov/ij/).
RT-PCR conditions were determined in control experiments using gene-specific primers to genes known to produce alternative splicing isoforms. First-strand synthesis was performed on 5 μg of total RNA of Columbia-0 seedlings using ready-to-go you-prime first-strand beads (Amersham) following incubation at 65°C and chilling on ice for 2 min. Then, 2 μM oligo(dT)18 and beads were added and incubated for 1 min at room temperature followed by 1 h at 37°C. The room temperature reaction was diluted to 100 μL and 1 μL used in gene-specific PCR reactions (25 μL) in 1× PCR buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl, and 3 mM MgCl2), 0.2 mM each of dATP, dGTP, dCTP, and dTTP (Promega), 1.5 μM each of gene-specific primers, and Taq DNA Polymeraase (Roche). The forward primer was labeled with 6-carboxyfluoresceine to visualize RT-PCR products on an ABI3730 capillary sequencing machine. PCR reactions were performed by initial incubation for 30 min at 48°C followed by 20, 22, 24, or 26 cycles of 94°C for 15 s, 50°C for 30 s, 70°C for 1 min, and finally 72°C for 10 min. Primer sequences are given in Supplemental Table 2 online. RT-PCR reactions were purified by miniElute purification (Qiagen), and 1 μL was mixed with 10 μL Hi Di Formamide with 0.05 μL of GeneScan 500 LIZ internal size standards (Applied Biosystems) and separated on an ABI 3730 (Applied Biosystems). Relative fluorescent peak areas for the expected alternatively spliced isoforms were extracted and analyzed using GENEMAPPER software (Applied Biosystems). The relative ratios of the two to three isoforms was calculated for each gene for three technical samples and the means plotted against PCR cycle number.
Construction of GFP and mRFP Fusion Proteins of Nucleolar Components
Cloning was performed using the Gateway system (Invitrogen). The UPF3 and UPF2 full-length ORFs were PCR amplified using Platinum Pfx DNA polymerase (Invitrogen), with sequence-specific primers containing Gateway attB adapters: 5′-AAAAAGCAGGCTCGATGAAGGAACCTTTGCAGAA-3′ and 5′-AGAAAGCTGGGTAACAAGTACCGGATGATGGTT-3′ for UPF3 and 5′-AAAAAGCAGGCTTCATGGATCATCCAGAAGATGAATCCCAC-3′ and 5′-AGAAAGCTGGGTCCTTTCGTCGGGCATGATACGAACCACC-3′ for UPF2. Amplification was performed using cDNA obtained by reverse transcription of RNA from Arabidopsis inflorescence material (isolated using the RNeasy kit [Qiagen]) with Omniscript polymerase (Quiagen) and oligo(dT). The UPF1 ORF region was obtained from the UPF1/pDONR20 clone (kindly provided by Masato Yoine, Nagoya University, Japan). All ORFs were then inserted via BP recombination into a Gateway entry vector pDONR207 (Invitrogen). The inserts were then recombined via LR reaction into the binary plant expression vector GFP-N-bin containing an N-terminal GFP translational fusion. As a marker for the nucleolus, the plasmid, pROK2.mRFPattR, containing a translational fusion with the nucleolar protein, fibrillarin, and mRFP was used (Kim et al., 2007b). The binary expression vectors containing GFP and mRFP fusions were transformed into Agrobacterium tumefaciens and transiently expressed in Arabidopsis cells as described previously (Koroleva et al., 2005).
Arabidopsis plants (Columbia-0 ecotype) were transformed by the floral dip method (Clough and Bent, 1998) and T1 seedlings selected on Petri plates with MS-agar media containing 50 mg/L kanamycin. Kanamycin-resistant seedlings were further selected using fluorescence microscopy. The seedlings were grown vertically on MS-agar plates in a growth room under constant light at 25°C.
Microscopy and Image Analysis
For microscopy, a small volume of cell suspension was placed on a slide and covered with a cover slip. Imaging was performed using a 40× oil lens on Leica SP and SP2 laser scanning confocal microscopes, equipped with Leica software (Leica Microsystems). To overcome potential problems of overexpression of GFP fusion proteins, only cells that fluoresced at low levels were imaged. This approach was used to image >90 nucleolar and EJC proteins showing numerous distinct patterns of subnuclear localization (Pendle et al., 2005). Each image was collected as an average of three sequential scans performed using the excitation wavelengths 488 nm (80%) and 543 nm (20%); for detection of GFP and mRFP signals, channels of 500 to 550 and 630 to 680 nm, respectively, were used. Low levels of UPF3-GFP expression, including in stable transformants, were visualized by wide-field imaging using a Nikon Eclipse 600 epifluorescence microscope equipped with a Hamamatsu Orca-ER cooled CCD camera and a Prior Proscan x–z stage. Image stacks were collected using MetaMorph software (Universal Imaging) and were deconvoluted using AutoDeblur (AutoQuant Imaging). Image processing and preparation of montages were performed using Adobe Photoshop CS2.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: UPF1 (At5g47010), NM_124072; UPF2 (At2g39260), 1009042019; and UPF3 (At1g33980) NM_103120 and in Supplemental Table 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RT-PCR Conditions for Comparison of Relative Abundance of Aberrant and Fully Spliced mRNAs.
Supplemental Figure 2. Aberrantly Spliced Transcripts Sequenced from RT-PCR Reactions.
Supplemental Figure 3. RT-PCR of mRNA Transcripts of At2g45670 in Nucleoplasmic and Nucleolar RNA and RNA from upf Mutants.
Supplemental Figure 4. Root Epidermal Cells from 5-d-Old Arabidopsis Seedling Stably Transformed with UPF-GFP.
Supplemental Table 1. Aberrant cDNA Clones from Nucleolar, Nuclear, and Whole-Cell cDNA Libraries.
Supplemental Table 2. Primers Used in RT-PCR Reactions.
Supplementary Material
Acknowledgments
We thank Brendan Davies for the generous gift of the upf mutants. UPF1 and the GFP-N-bin vector were kind gifts from Masato Yoine (Nagoya University, Japan) and Ben Trevaskis (Max Planck Institute of Molecular Plant Physiology, Golm, Germany), respectively. We are grateful to Gordon Simpson and Steve Hubbard (University of Dundee) for critical reading of the manuscript and statistical analysis, respectively. This work was supported by Scottish Executive Environment and Rural Affairs Department (SCR/909/03), the Biotechnology and Biological Sciences Research Council (BBS/B/13519), and the European Union FP6 Programme Network of Excellence on Alternative Splicing (LSHG-CT-2005-518238).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: John W.S. Brown (j.w.s.brown@dundee.ac.uk).
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Amrani, N., Sachs, M.S., and Jacobsen, A. (2006). Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell Biol. 7 415–425. [DOI] [PubMed] [Google Scholar]
- Andersen, C.B.F., Ballut, L., Johansen, J.S., Chamieh, H., Nielsen, K.H., Oliveira, C.L.P., Pedersen, J.S., Séraphin, B., Le Hir, H., and Andersen, G.R. (2006). Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313 1968–1972. [DOI] [PubMed] [Google Scholar]
- Arciga-Reyes, L., Wootton, L., Kieffer, M., and Davies, B. (2006). UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47 480–489. [DOI] [PubMed] [Google Scholar]
- Barbazuk, W.B., Fu, Y., and McGinnis, K.M. (2008). Genome-wide analysis of alternative splicing in plants: Opportunities and challenges. Genome Res. 18 1381–1392. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant, I., Gatfield, D., Rehwinkel, J., Hilgers, V., and Izzaurralde, E. (2007. a). A conserved role for cytoplasmic poly(A) inding protein (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 26 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant, I., Kashima, I., Rehwinkel, J., Saulière, J., Wittkopp, N., and Izaurralde, E. (2007. b). mRNA quality control: An ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 581 2845–2853. [DOI] [PubMed] [Google Scholar]
- Beven, A.F., Simpson, G.G., Brown, J.W.S., and Shaw, P.J. (1995). The organization of spliceosomal components in the nuclei of higher plants. J. Cell Sci. 108 509–551. [DOI] [PubMed] [Google Scholar]
- Boisvert, F.-M., van Koningsbruggen, S., Navascués, J., and Lamond, A.I. (2007). The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8 574–585. [DOI] [PubMed] [Google Scholar]
- Bond, V.C., and Wold, B. (1993). Nucleolar localisation of myc transcripts. Mol. Cell. Biol. 13 3221–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne, J.R., and Whitehouse, A. (2006). Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. USA 103 15190–15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.W.S., and Shaw, P.J. (1998). Small nucleolar RNAs and pre-rRNA processing in plants. Plant Cell 10 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler, M., Paillusson, A., and Mühlemann, O. (2004). Efficient downregulation of immunoglobulin μ mRNA with premature translation-termination codons requires the 5′-half of the VJD exon. Nucleic Acids Res. 32 3304–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler, M., Steiner, S., Mohn, F., Paillusson, A., and Mühlemann, O. (2006). EJC-independent degradation of nonsense immunoglobulin-u mRNA depends on 3′ UTR length. Nat. Struct. Mol. Biol. 13 462–464. [DOI] [PubMed] [Google Scholar]
- Bühler, M., Wilkinson, M.F., and Mühlemann, O. (2002). Intranuclear degradation of nonsense codon-containing mRNA. EMBO Rep. 3 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro, T., Carvalho, C., Braga, J., Rino, J., Milligan, L., Tollervey, D., and Carmo-Fonseca, M. (2007). Depletion of the yeast nuclear exosome subunit Rrp6 results in accumulation of polyadenylated RNAs in a discrete domain within the nucleolus. Mol. Cell. Biol. 27 4157–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.F., Imam, J.S., and Wilkinson, M.F. (2007). The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76 51–74. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Dahlberg, J.E., and Lund, E. (2004). Does protein synthesis occur in the nucleus? Curr. Opin. Cell Biol. 16 335–338. [DOI] [PubMed] [Google Scholar]
- Du, T.G., Jellbauer, S., Muller, M., Schmid, M., Niessing, D., and Jansen, R.P. (2008). Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep. 9 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio, A., Behm-Ansmant, I., and Izaurralde, E. (2007). P-bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8 9–22. [DOI] [PubMed] [Google Scholar]
- Fang, Y., and Spector, D.L. (2007). Identification of nuclear dicing bodies containing proteins for microRNA biogenesis. Curr. Biol. 17 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica, A., and Tollervey, D. (2002). Making ribosomes. Curr. Opin. Cell Biol. 14 313–318. [DOI] [PubMed] [Google Scholar]
- Fujioka, Y., Utsumi, M., Ohba, Y., and Watanabe, Y. (2007). Location of possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 48 1243–1253. [DOI] [PubMed] [Google Scholar]
- Galy, V., Gadal, O., Fromont-Racine, M., Romano, A., Jacquier, A., and Nehrbass, U. (2004). Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116 63–73. [DOI] [PubMed] [Google Scholar]
- Gorski, S.A., Dundr, M., and Misteli, T. (2006). The road much travelled: Trafficking in the cell nucleus. Curr. Opin. Cell Biol. 18 284–290. [DOI] [PubMed] [Google Scholar]
- Granneman, S., and Baserga, S.J. (2004). Ribosome biogenesis: Of knobs and RNA processing. Exp. Cell Res. 296 43–50. [DOI] [PubMed] [Google Scholar]
- He, F., Li, X., Spatrick, P., Casillo, R., Dong, S., and Jacobson, A. (2003). Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5 ' to 3 ' mRNA decay pathways in yeast. Mol. Cell 12 1439–1452. [DOI] [PubMed] [Google Scholar]
- Hiscox, J.A. (2007). RNA viruses: Hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper, A.K., and Shaheen, H.H. (2008). A decade of surprises for tRNA nuclear-cytoplasmic dynamics. Trends Cell Biol. 18 98–104. [DOI] [PubMed] [Google Scholar]
- Hori, K., and Watanabe, Y. (2005). UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 43 530–540. [DOI] [PubMed] [Google Scholar]
- Hori, K., and Watanabe, Y. (2007). Context analysis of termination codons in mRNA that are recognized by plant NMD. Plant Cell Physiol. 48 1072–1078. [DOI] [PubMed] [Google Scholar]
- Iborra, F.J., Jackson, D.A., and Cook, P.R. (2001). Coupled transcription and translation within nuclei of mammalian cells. Science 293 1139–1142. [DOI] [PubMed] [Google Scholar]
- Ideue, T., Azad, A.K., Yoshida, J., Matsusaka, T., Yanagida, M., Ohshima, Y., and Tani, T. (2004). The nucleolus is involved in mRNA export from the nucleus in fission yeast. J. Cell Sci. 117 2887–2895. [DOI] [PubMed] [Google Scholar]
- Isken, O., and Maquat, L.E. (2007). Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes Dev. 21 1833–1856. [DOI] [PubMed] [Google Scholar]
- Jellbauer, S., and Jansen, R.P. (2008). A putative function of the nucleolus in the assembly or maturation of specialised messenger ribonucleoprotein complexes. RNA Biol. 5 225–229. [DOI] [PubMed] [Google Scholar]
- Kalyna, M., Lopato, S., Voronin, V., and Barta, A. (2006). Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res. 34 4395–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima, I., Yamashita, A., Izumi, N., Kataoka, N., Morishita, R., Hoshino, S., Ohno, M., Dreyfuss, G., and Ohno, S. (2006). Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf phosphorylation and nonsense-mediated decay. Genes Dev. 20 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerényi, Z., Mérai, Z., Hiripi, L., Benkovics, A., Gyula, P., Lacomme, C., Barta, E., Nagy, F., and Silhavy, D. (2008). Inter-kingdom conservation of mechanism of nonsense-mediated decay. EMBO J. 27 1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertész, S., Kerényi, Z., Mérai, Z., Bartos, I., Palfy, T., Barta, E., and Silhavy, D. (2006). Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 34 6147–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H., MacFarlane, S., Kalinina, N.O., Rakitina, D.V., Ryabov, E.V., Gillespie, T., Haupt, S., Brown, J.W.S., and Taliansky, M. (2007. a). Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc. Natl. Acad. Sci. USA 104 11115–11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H., Ryabov, E.V., Kalinina, N.O., Rakitina, D.V., Gillespie, T., Haupt, S., MacFarlane, S., Brown, J.W.S., and Taliansky, M. (2007. b). Cajal bodies, the nucleolus and fibrillarin are required for a plant virus systemic infection. EMBO J. 26 2169–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva, O.A., Tomlinson, M.L., Leader, D.J., Shaw, P.J., and Doonan, J.H. (2005). High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J. 41 162–174. [DOI] [PubMed] [Google Scholar]
- Kurihara, Y., et al. (2009). Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc. Natl. Acad. Sci. USA 106 2453–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond, A.I., and Spector, D.L. (2003). Nuclear speckles: A model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4 605–612. [DOI] [PubMed] [Google Scholar]
- Lejeune, F., and Maquat, L.E. (2005). Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 17 309–315. [DOI] [PubMed] [Google Scholar]
- Li, C.F., Pontes, O., El-Shami, M., Henderson, I.R., Bernatavichute, Y.V., Chan, S.W.-L., Lagrange, T., Pikaard, C.S., and Jacobsen, S.E. (2006). An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126 93–106. [DOI] [PubMed] [Google Scholar]
- Lorković, Z.J., and Barta, A. (2008). Role of Cajal bodies and nucleolus in the maturation of the U1 snRNP in Arabidopsis. PLoS One 3 e3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković, Z.J., Wieczorek-Kirk, D.A., Lambermon, M.H., and Filipowicz, W. (2000). Pre-mRNA splicing in higher plants. Trends Plant Sci. 5 160–167. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Shu, M.D., and Steitz, J.A. (2000). Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103 1121–1131. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. (2004). Nonsense-mediated mRNA decay: Splicing, translation and mRNA dynamics. Nat. Rev. Mol. Cell Biol. 5 89–99. [DOI] [PubMed] [Google Scholar]
- Meijer, H.A., and Thomas, A.M. (2002). Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem. J. 367 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell, J.T., Sharifi, N.A., Meyers, J.L., Martinez-Murillo, F., and Dietz, H.C. (2004). Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36 1073–1078. [DOI] [PubMed] [Google Scholar]
- Misteli, T. (2005). Concepts in nuclear architecture. Bioessays 27 477–487. [DOI] [PubMed] [Google Scholar]
- Olson, M.O.J. (2004). Non-traditional roles of the nucleolus. In The Nucleolus, M.O.J. Olson, ed (Georgetown, USA: Landes Bioscience), pp. 329–342.
- Parker, R., and Sheth, U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25 635–646. [DOI] [PubMed] [Google Scholar]
- Pederson, T. (1998). The plurifunctional nucleolus. Nucleic Acids Res. 26 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendle, A.F., Clark, G.P., Boon, R., Lewandowska, D., Lam, Y.W., Andersen, J., Mann, M., Lamond, A.I., Brown, J.W.S., and Shaw, P.J. (2005). Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol. Biol. Cell 16 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz, J.C.R., Zhang, F., and Pederson, T. (2006). MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl. Acad. Sci. USA 103 18957–18962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz, J.C.R., Hogan, E.M., and Pederson, T. (2009). MicroRNAs with a nucleolar location. RNA (in press). [DOI] [PMC free article] [PubMed]
- Pontes, O., Li, C.F., Nunes, P.C., Haag, J., Ream, T., Vitins, A., Jacobsen, S.E., and Pikaard, C.S. (2006). The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126 79–92. [DOI] [PubMed] [Google Scholar]
- Raška, I., Shaw, P.J., and Cmarko, D. (2006). Structure and function of the nucleolus in the spotlight. Curr. Opin. Cell Biol. 18 325–334. [DOI] [PubMed] [Google Scholar]
- Rehwinkel, J., Raes, J., and Izaurralde, E. (2006). Nonsense-mediated mRNA decay: Target genes and functional diversification of effectors. Trends Biochem. Sci. 31 639–646. [DOI] [PubMed] [Google Scholar]
- Rubbi, C.P., and Milner, J. (2003). Disruption of the nucleolus mediates stabilisation of p53 in response to DNA damage and other stresses. EMBO J. 22 6068–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöning, J.C., Streitner, C., Meyer, I.M., Gao, Y., and Staiger, D. (2008). Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 36 6977–6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöning, J.C., Streitner, C., Page, D.R., Hennig, S., Uchida, K., Wolf, E., Furuya, M., and Staiger, D. (2007). Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 52 1119–1130. [DOI] [PubMed] [Google Scholar]
- Schwartz, A.M., Komarova, T.V., Skulachev, M.V., Zvereva, A.S., Dorokov, Y.L., and Atabekov, J.G. (2006). Stability of plant mRNAs depends on the length of the 3′-untranslated region. Biochemistry (Mosc.) 71 1377–1384. [DOI] [PubMed] [Google Scholar]
- Serin, G., Gersappe, A., Black, J.D., Aronoff, R., and Maquat, L.E. (2001). Identification and characterisation of human orthologues to Saccharomyces cerevisiae Upf2 and Upf3 protein (Caenorhabiditis elegans SMG-4). Mol. Cell. Biol. 21 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, P.J., and Brown, J.W.S. (2004). Plant nuclear bodies. Curr. Opin. Plant Biol. 7 614–620. [DOI] [PubMed] [Google Scholar]
- Shyu, A.B., Wilkinson, M.F., and van Hoof, A. (2008). Messenger RNA regulation: To translate or to degrade. EMBO J. 27 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, C.G., Fuller, J., Maronova, M., Kalyna, M., Davidson, D., McNicol, J., Barta, A., and Brown, J.W.S. (2008). Monitoring changes in alternative pre-mRNA splicing in multiple gene transcripts. Plant J. 53 1035–1048. [DOI] [PubMed] [Google Scholar]
- Singh, G., Jakob, S., Kleedehn, M.G., and Lykke-Andrersen, J. (2007). Communication with the exon junction complex and activation of nonsense-mediated decay by human Upf proteins occur in the cytoplasm. Mol. Cell 27 780–792. [DOI] [PubMed] [Google Scholar]
- Song, L., Han, M.H., Lesicka, J., and Fedoroff, N.V. (2007). Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl. Acad. Sci. USA 104 5437–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder, L., and Mühlemann, O. (2008). The meaning of nonsense. Trends Cell Biol. 18 315–321. [DOI] [PubMed] [Google Scholar]
- Stroupe, M.E., Tange, T.O., Thomas, D.R., Moore, M.J., and Grigorieff, N. (2006). The three dimensional architecture of the EJC core. J. Mol. Biol. 360 743–749. [DOI] [PubMed] [Google Scholar]
- Stutz, F., and Izaurralde, E. (2003). The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 13 319–327. [DOI] [PubMed] [Google Scholar]
- Tange, T.O., Nott, A., and Moore, M.J. (2004). The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 16 279–284. [DOI] [PubMed] [Google Scholar]
- van Hoof, A., and Green, P.J. (2006). NMD in plants. In Nonsense-Mediated mRNA Decay. L. Maquat, ed (New York: Landes Bioscience), pp. 167–172.
- Williams, B.J.L., Boyne, J.R., Goodwin, D.J., Roaden, L., Hautbergue, G.M., Wilson, S.A., and Whitehouse, A. (2005). The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA through the cellular mRNA export pathway. Biochem. J. 387 295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J.Q., Kang, J.H., Hettenhausen, C., and Baldwin, T. (2007). Nonsense-mediated mRNA decay (NMD) silences the accumulation of aberrant trypsin proteinase inhibitor mRNA in Nicotiana attenuata. Plant J. 51 693–706. [DOI] [PubMed] [Google Scholar]
- Yoine, M., Ohto, M., Onai, K., Mita, S., and Nakamura, K. (2006). The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signaling in Arabidopsis. Plant J. 47 49–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.