Abstract
Stable maize (Zea mays) chromosomes were recovered from an unstable dicentric containing large and small versions of the B chromosome centromere. In the stable chromosome, the smaller centromere had become inactivated. This inactive centromere can be inherited from one generation to the next attached to the active version and loses all known cytological and molecular properties of active centromeres. When separated from the active centromere by intrachromosomal recombination, the inactive centromere can be reactivated. The reactivated centromere regains the molecular attributes of activity in anaphase I of meiosis. When two copies of the dicentric chromosome with one active and one inactive centromere are present, homologous chromosome pairing reduces the frequency of intrachromosomal recombination and thus decreases, but does not eliminate, the reactivation of inactive centromeres. These findings indicate an epigenetic component to centromere specification in that centromere inactivation can be directed by joining two centromeres in opposition. These findings also indicate a structural aspect to centromere specification revealed by the gain of activity at the site of the previously inactive sequences.
INTRODUCTION
The centromere is the region of a chromosome that organizes the kinetochore for chromosome movement during mitosis and meiosis. In most multicellular organisms, the centromeric region is composed of species-specific sequence repeats arranged in large tandem blocks (Henikoff et al., 2001; Amor et al., 2004; Ma et al., 2007). However, these arrays can lose their ability to organize a kinetochore and become inactive (Earnshaw and Migeon, 1985). Furthermore, human kinetochores can be formed over entirely unique DNA sequences as neocentromeres (Amor and Choo, 2002; Cleveland et al., 2003) that are perpetuated over cell generations indefinitely. In Drosophila melanogaster, the sites of the centromeres have no sequences in common across chromosomes, and examples of neocentromeres have been described (Williams et al., 1998; Platero et al., 1999; Maggert and Karpen, 2001). These lines of evidence and other observations have led to the idea that centromere specification is epigenetically determined (Karpen and Allshire, 1997; Allshire and Karpen, 2008).
Considerable evidence that an epigenetic component is required for plant centromere specification has been obtained from work with the maize (Zea mays) B chromosome (Han et al., 2006) but also from A chromosomes (Lamb et al., 2007). Furthermore, a neocentromere has been described in barley (Hordeum vulgare) on a chromosome carrying no centromeric repeats (Nasuda et al., 2005). The B or supernumerary chromosome of maize exists only in some lines, and it is neither required nor detrimental unless at a high copy number. It has two major advantages for the study of centromere structure and function. First, the chromosome is basically inert; second, it possesses a specific repeat sequence that is present in and around its centromere with a minor representation at the long arm tip of the chromosome (Alfenito and Birchler, 1993; Jin et al., 2005; Lamb et al., 2005). This second property allows its centromere to be studied independently of all the others in the nucleus (Jin et al., 2005; Lamb et al., 2005).
Because the B chromosome is dispensable, it is maintained in populations by an accumulation mechanism. This mechanism is provided by two properties of the B chromosome (Roman, 1947, 1948). First, the centromere nondisjoins at the second pollen mitosis, which produces the two maize sperm. In microspores that receive one B chromosome, the resulting pollen will usually have one sperm with zero and the other sperm with two B chromosomes. Then the sperm with the B chromosomes will preferentially join with the egg nucleus during the process of double fertilization. The process of nondisjunction requires the tip of the long arm to be present in the same nucleus as the centromere but not necessarily on the same chromosome (Roman, 1947; Lin, 1978). Thus, if the tip of the long arm of the B is missing, the B centromere will disjoin as does every other centromere (Han et al., 2007).
The property of the B chromosome to undergo nondisjunction at the single mitotic division that produces the male gametes has been capitalized upon by maize biologists to produce translocations between the B and the various A chromosomes (Roman, 1947). One such translocation has figured into studies of the B centromere over many years and is referred to as TB-9Sb (Carlson, 1970). It is a reciprocal exchange between the short arm of chromosome 9 (9S) and the B chromosome such that genetic markers on 9S are linked to the B centromere. Carlson (1970) discovered a derivative of TB-9Sb that had suffered a misdivision of the centromere resulting in a chromosome with two copies of 9S. Subsequent studies have continued to recover sequential misdivision derivatives so that now a large collection has been recovered (Kaszás and Birchler, 1996, 1998). Using the centromeric B-specific repeat, it was demonstrated that all of these derivatives had rearranged the B-specific sequences and produced centromeres that were considerably reduced in size relative to the progenitor B centromere in TB-9Sb, which is typical of the normal B centromere (Alfenito and Birchler, 1993; Kaszás and Birchler, 1996, 1998; Jin et al., 2005).
Evidence for inactive plant centromeres came from the study of apparent dicentric minichromosomes that were examined for their centromere behavior (Han et al., 2006). These small chromosomes were originally observed as a byproduct of studies to determine the developmental extent of the chromosomal type of the breakage-fusion-bridge (B-F-B) cycle (Zheng et al., 1999). To examine this issue, TB-9Sb was used by recombining onto it a reverse duplication of the 9S arm originally constructed by McClintock (1939, 1941). The reverse duplication can recombine with itself in prophase of meiosis I and in the process will tie together the B sister centromeres in a dicentric that will separate at meiosis II. When this chromosome is broken at anaphase II, a B-F-B cycle is initiated that continues during the subsequent gametophytic development. At the second pollen mitosis, the B centromere will undergo nondisjunction and can deliver two broken chromosomes to the zygote, which is the condition to establish the chromosome type of B-F-B cycle. Because this chromosome is dispensable, it can continue to break and rejoin over the course of the life cycle. In the process, the size of the B centromere–containing chromosome is gradually diminished until it is stabilized. Stabilization can occur when one centromere fails to rejoin with another centromere or, as recently discovered, by the inactivation of one of the two sets of centromere sequences (Han et al., 2006).
In a collection of minichromosomes recovered in subsequent generations of the B-F-B cycle, five cases were found in which two sets of B centromere sequences were present but only one of the two showed a primary constriction or proceeded to the poles at anaphase (Han et al., 2006). When these chromosomes were examined for molecular correlates of centromere activity, only one of the two sets of centromere sequences was found to be active (Han et al., 2006).
The DNA sequences underlying maize centromeres consist of an array composed of CentC units each ∼156 bp in length (Ananiev et al., 1998; Nagaki et al., 2003). Interspersed among the CentC array are CRM family retrotransposons that are active and transpose almost exclusively to new centromeric sites (Jin et al., 2004; Sharma et al., 2008). The B chromosome centromere is composed of the same elements but also has a B chromosome–specific repeat of canonical length of 1.4 kb that is interspersed with the CentC and CRM elements and continues into pericentromeric regions (Jin et al., 2005).
In addition to specific DNA sequences, three molecular characteristics of centromeres serve as an assay for active versus epigenetically silenced versions (Han et al., 2006). All of the centromeres in maize, including the B centromere, are associated with a specific variant of histone H3 referred to as CENH3 (Zhong et al., 2002). Another basal kinetochore protein is referred to as CENP-C, and the encoding gene has been isolated (Dawe et al., 1999). A third molecular marker of active centromeres is the phosphorylation of Ser-10 of H3 (Houben et al., 2007).
Several observations provided evidence that sequences usually present at the centromeres do not necessarily organize a kinetochore. First, the long arm of the B chromosome was found to contain many sites of CentC hybridizing sequences that in some cases have as great an intensity as present in some primary constrictions of the A chromosomes (Lamb et al., 2005). Nevertheless, these sites do not associate with CENH3 under any known circumstance nor do they foster anaphase movement of the chromosome arm. Secondly, a variant of chromosome 8 was found in which an inversion had occurred that apparently split the centromere sequence array and moved a portion of it (Lamb et al., 2007). The portion left behind is at the canonical position for chromosome 8 as evidenced in heterozygotes of this inversion chromosome with another inbred line of maize. Tests of anaphase movement and the presence of CENH3 indicate that the new site is the active centromere, and the remaining sequences do not organize a kinetochore. These examples indicate that the presence of centromere sequences alone does not necessarily organize an active kinetochore.
While the above evidence points to an epigenetic component to centromere specification, the homogenization of rapidly evolving sequences at centromere sites might argue for a genetic DNA-based property that is also involved. The production of artificial chromosomes in human cells via the introduction of centromere repeats that aggregate to form functional chromosomes might also be taken as evidence of a genetic or structural component (Amor et al., 2004). Here, we report that centromere inactivation can regularly occur when centromeres of different sizes are joined together. An example of the resulting chromosomes shows a high frequency of reactivation of the formerly inactive site, suggesting a structural or topological component is also involved with fostering the specification of centromeric activity.
RESULTS
Inactivation of Centromeres from a Tug of War
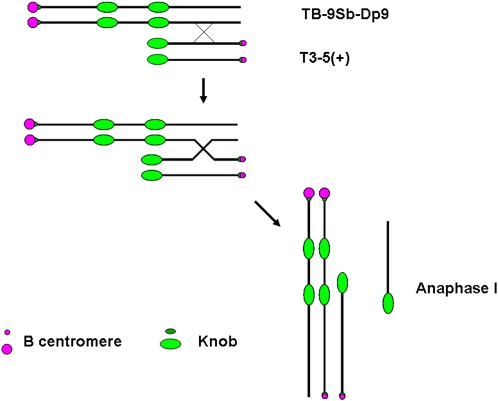
Initially an experiment was set up to test the strength of different sized centromeres. A collection of B chromosome centromeres of reduced size had been recovered in previous work (Kaszás and Birchler, 1996, 1998). Using a translocation of the B chromosome and the short arm of chromosome 9 that has a foldback duplication (TB-9Sb-Dp9), recombination between it and a misdivision derivative also carrying 9S will pit a large and small centromere against each other in anaphase I or allow them to assort to the same pole (Figure 1). The TB-9Sb-Dp9 chromosome is a translocation containing the supernumerary B chromosome centromere with two short arms of maize chromosome 9 but reversed in order (Zheng et al., 1999; Han et al., 2007b). Misdivision derivatives were also derived ultimately from TB-9Sb and have been classified as to their chromosomal constitution (Kaszás and Birchler, 1996, 1998). The derivative Telo 3-5(+) will be the focus of this study. “Telo” refers to its telocentric structure, “3” to the number of misdivisions suffered by the progenitor B centromere sequences to produce the present array, and “(+)” refers to the presence of the small centric heterochromatic knob adjacent to the centromere.
Figure 1.
Centromere Tug of War between a Reduced-Sized (T3-5+) and a Normal B Centromere of TB-9Sb-Dp9.
A reverse duplication of the short arm of chromosome 9 is translocated to the B centromere in TB-9Sb-Dp9. From crosses with misdivision derivative T(3-5)+, which has a greatly reduced sized B centromere, recombination (designated by X) in the indicated orientation will join the two centromeres in a tug of war in anaphase I. The other products of recombination are also shown, including an acentric fragment containing a knob. If the two types of B centromeres do not segregate from each other and assort independently, then the products of recombination can proceed to the same pole during anaphase I. Knobs are blocks of distinct heterochromatin that aid in identifying the chromosome. There is a small knob adjacent to the centromeric B–specific array and a larger knob at the end of chromosome arm 9S, which is internally duplicated in TB-9Sb-Dp9.
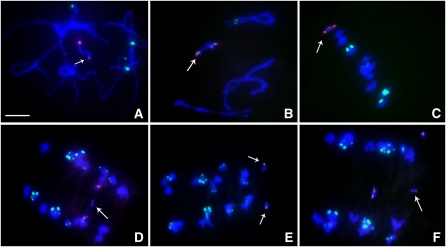
First, we crossed together TB-9Sb-Dp9 and Telo 3-5(+). The plants containing TB-9Sb-Dp9 and Telo 3-5(+) were identified via fluorescence in situ hybridization (FISH) on primary root tip metaphase chromosomes using probes for the B chromosome–specific repeat (ZmBs) and knob heterochromatin (Peacock et al., 1981). The TB-9Sb-Dp9 chromosome contains a large centromere and Telo 3-5(+) has a small one, so the two are easily distinguished from each other (Figure 1). During meiosis I, the TB-9Sb-Dp9 chromosome can pair with Telo 3-5(+) and then recombination can occur between them (Figure 1). If this occurs, the two centromeres can move to different poles and form an anaphase bridge, or they can move to the same pole if they act independently. Five heterozygous plants were analyzed at meiosis. The percentage of pairing between TB-9Sb-Dp9 and Telo 3-5(+) is 60.43%, which can be determined by the association of the large and small B centromere signal. These nuclei formed bridges (54.79%) in anaphase I (see Supplemental Table 1 online). These data suggest that the two centromeres often segregate from each other despite the fact that they are not closely aligned (Figure 2) but also that segregation does not always occur and thus they progress to the same pole.
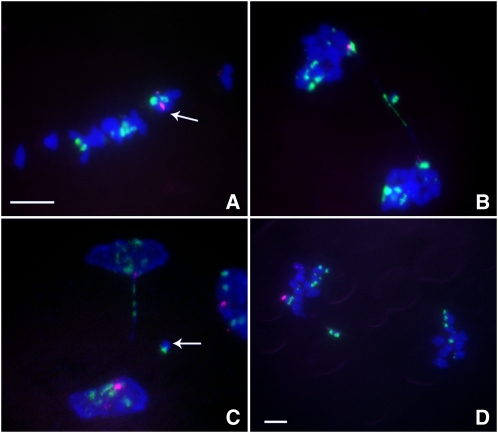
Figure 2.
Cytological Analysis of Plants Containing TB-9Sb-Dp9 and Telo 3-5(+).
ZmBs, which is a B chromosome–specific sequence in and around the centromere, is labeled in magenta. Knob heterochromatin is labeled in green. Chromosomes were counterstained with 4',6-diamidino-2-phenylindole (DAPI) in blue. Bars =10 μm.
(A) Metaphase I. Arrow indicates the pairing between TB-9Sb-Dp9 and Telo 3-5(+).
(B) Anaphase I. When recombination occurs between TB-9Sb-Dp9 and Telo 3-5(+), both centromeres are active and a bridge is formed together with the release of an acentric fragment with knob heterochromatin if the centromeres proceed to opposite poles.
(C) Early telophase I. A bridge was formed and an acentric fragment was released (arrow). The large and small B centromeres (red) are at opposite poles.
(D) Anaphase I. An example of the large and small centromeres proceeding to the same pole is shown. The small centromere appears to be stretched to both poles but with eventual movement to the same pole as the large centromere.
From the progeny of the plants containing one copy each of TB-9Sb-Dp9 and Telo 3-5(+), new dicentric chromosomes were recovered. Different such heterozygotes in various backgrounds produced widely varying numbers of dicentrics in their respective progenies. Nevertheless, all of these dicentric chromosomes had one large B centromere and one small B centromere at opposite ends of the chromosome as predicted from the joining of TB-9Sb-Dp9 and Telo 3-5(+) via recombination. It is not possible to determine whether the resulting chromosomes are the direct product of recombination with progression of the two centromeres to the same pole without further modification or whether an intervening chromosomal breakage occurred before inactivation resulted. All of these dicentrics possess only one primary constriction at the site of the large centromere, suggesting that the small centromere has become inactive. The molecular analysis of the example described below confirmed the inactive state of its small centromere. This example is missing the knob heterochromatin in the middle of the chromosome that would be predicted to be present from recombination with no further modification (Figure 1), suggesting a history of chromosomal breakage and rejoining.
The Inactive Centromere Loses Molecular Features of Normal Activity
From the progeny of plants containing TB-9Sb-Dp9 and Telo 3-5(+), we randomly selected a candidate for centromere activity analysis, which we refer to as Dicentric-15 (Dic-15). From FISH results using probes for CentC, CRM, knob heterochromatin, and B repeat sequences, Dic-15 contains the full complement of DNA sequences at both centromeres. This result confirmed that the chromosome carries a large and small B centromere at opposite ends as predicted from its history (see Supplemental Figure 1 online). Dic-15 appears stable in mitosis as evidenced by no breakage of this chromosome being observed in the many somatic root tip cells examined from individual seedlings in contrast with dicentrics undergoing the B-F-B cycle (Han et al., 2007b).
Antibodies against CENPC, CENH3, and H3 phosphorylated at Ser-10 were applied to Dic-15 and revealed that only the large centromere exhibited signal, whereas none of the three proteins were detected over the smaller centromere (see Supplemental Figure 2 online). The sensitivity of this test was verified by probing a heterozygote of TB-9Sb and Telo 3-5(+) in which the same large and small B chromosome centromeres are unattached and active. Both the large and small centromeres have CENPC, CENH3, and H3 phosphorylated Ser-10 signals that are easily and consistently visualized, illustrating that the failure to detect these proteins on the small centromere in Dic-15 is a reflection of activity loss (see Supplemental Figure 3 online). Indeed, α-tubulin immunostaining of Dic-15 revealed that only the large active centromere was attached to the spindle (see Supplemental Figure 4 online). Thus, the lack of a primary constriction at the small centromere end of the dicentric chromosome is confirmed at the molecular level.
Intrachromosomal Recombination in the Dicentric Can Form New Chromosomes with Only Large or Small Centromeres
When we screened the progeny of plants containing one copy of the Dic-15 chromosome, we observed chromosomes with new structures that contained B centromeres in somatic root tip cells. Because Dic-15 itself is stable from cell to cell as an intact chromosome in root tip somatic spreads, the behavior of Dic-15 in the previous meiosis was suspected of generating these chromosomes. Figure 3 illustrates that the foldback nature of the single chromosome can produce recombinants that would generate new chromosomal structures. Some products of recombination would join the large centromere sisters to themselves as one product and the two small centromere sisters as the reciprocal product. Analysis of the pollen mother cells in tassel samples indicated that Dic-15 does not pair with any of the normal bivalents (Figures 4B and 4C). Dic-15 pairs onto itself, which fosters intrachromosomal recombination (Figures 3 and 4A). In anaphase I, 26% of cells showed new chromosomal structures because the large active centromere and small inactive centromere were separated (Figures 4D and 4E). The large active centromere should exhibit sister chromatid cohesion at this division and proceed to one pole or the other. In some cases, the two newly formed structures moved to the same pole (Figure 4F). The small centromere is a misdivision product of the B centromere, and previous work had suggested that misdivision derivatives are prone to subsequent misdivision, which can result from attachment of a centromere to both poles with further fracture (Kaszás and Birchler, 1998; Kaszás et al., 2002). It is possible that the small centromere attaches to both poles (merotelic orientation) (Cimini, 2008) in these cases, followed by detachment from one pole. This scenario might serve as a potential explanation for the separation of the two centromeres but their eventual progression to the same pole in some meiocytes.
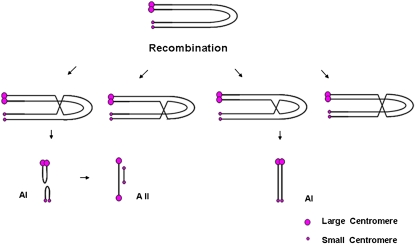
Figure 3.
Intrachromosomal Recombination of Dic-15.
Because of the foldback structure of the chromosome with a large active and a small inactive centromere, recombination can produce new structures depending on the chromatids involved. In the first two recombination events depicted, the large and small sister centromeres become joined together and can be recognized at anaphase I (AI). At anaphase II (AII), active centromeres will form bridges. In the latter two types of recombination events, no change occurs for the structure of the chromosome.
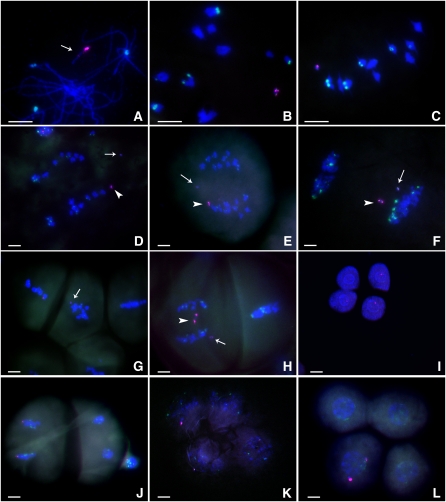
Figure 4.
Cytological Analysis of One Copy of Dic-15.
The ZmBs sequence is labeled in magenta, and the knob is labeled in green. Bars = 10 μm.
(A) Pachynema. The Dic-15 chromosome is undergoing intrachromosomal pairing. The large and small (arrow) centromeres are visible in magenta.
(B) Diakinesis.
(C) Metaphase I. The Dic-15 chromosome has moved to the plate with the other bivalents.
(D) and (E) Anaphase I. The arrowhead indicates the large centromere of Dic-15, and the arrow indicates the small centromere, which has separated from the large one.
(F) Early telophase I. Two chromosomes (one with two large centromeres and another with two small) have moved to the same pole.
(G) Metaphase II. Arrow indicates the smaller centromere. In this case, the two chromosomes separated in anaphase I.
(H) Anaphase II. Arrow indicates the new dicentric chromosome containing two small centromeres, and the arrowhead indicates the chromosome with large centromeres. The large-large centromere chromosome has formed a bridge. The two chromosomes progressed to the same pole in anaphase I.
(I) Tetrad. Three magenta signals in the cells indicate that one chromosome with a small centromere is independent of the large centromeres; this independent segregation requires that the small centromere has been reactivated.
(J) Telophase II. Dic-15 sister chromatids are separated to the two poles (left side), which represents no change in chromosome structure in this case.
(K) Tetrad. Three cells contain magenta signals. In this case, both small centromeres are observed in opposite cells and independent of the large centromere. This observation indicates that at least one small centromere is active. One large centromere is missing, presumably due to loss following anaphase II bridge formation and breakage as shown in (H).
(L) Tetrad. Only one cell contains two magenta signals, representing both the large and small centromeres. This tetrad configuration might result from progression of both new chromosome structures to the same pole or the production of a new dicentric following breakage and fusion.
At meiosis II, we found the new chromosome with only two smaller centromeres can enter metaphase II in some cases (Figure 4G). This finding suggests that the smaller centromere has recovered function. We further found the two new chromosomal structures could be present in the same cell at metaphase II. The large centromeres with its two sister chromatids are attached to the spindle (see Supplemental Figure 5A online) and the chromosome with only smaller centromeres also attracts tubulin (see Supplemental Figure 5B online). A new dicentric chromosome with only smaller centromeres was observed (Figure 4H). Dic-15 also can transmit as an intact chromosome without a change in structure to the next generation (Figure 4J) because not all recombination events change its structure. In tetrads, there is some variation of the B centromere distribution with large and small signals in the same cell. This result would only be found if the smaller inactive centromere recovers its function during meiosis I and survives into the tetrad.
Homologous Pairing of Two Dicentric Chromosomes Reduces Breakage but Also Produces New Dicentric Chromosomes
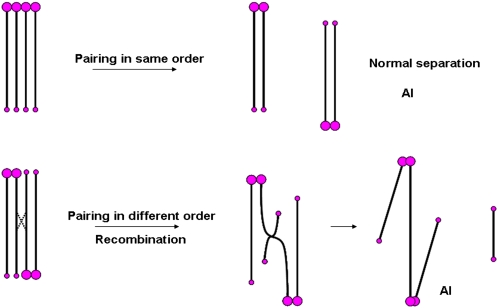
Plants containing two copies of Dic-15 were produced by self-pollination and screening of root tip metaphase spreads for individual seedlings with two intact copies of the chromosome. Because the chromosome is a partial mirror image, there are two ways for homologous pairing to occur (Figure 5). When pairing is in the same order, a large centromere pairs with a large one and the smaller ones are together (Figure 6A). Meiotic examination indicated that the two Dic-15 chromosomes pair with each other in this orientation to form a bivalent in metaphase I (Figures 6A and 6C) and separate normally (Figure 6E). The other pairing scenario occurs in the opposite order: large centromere paired with the smaller centromere at each terminus (Figure 6B), although close examination shows that the centromeres are not perfectly aligned and that a region of asynapsis is present in the middle of the pair. Nevertheless, when recombination occurs between the two Dic-15 chromosomes that are paired in the opposite order, new dicentric chromosomes can be produced at anaphase I. One will have two smaller centromeres together and the other will join the large centromeres that form a bridge at anaphase I (Figures 5, 6D, and 6E; see Supplemental Table 2 online). We also found unpaired Dic-15, which underwent intrachromosomal recombination with the separation of the large and smaller centromeres in anaphase I as described above.
Figure 5.
Chromosome Pairing and Recombination with Two Copies of the Dic-15 Chromosome.
There are two possible orientations for the pairing of two Dic-15 chromosomes: (1) in the same order, there is no bridge formed and the bivalent will be separated in anaphase I; (2) when the pairing occurs in the opposite order, recombination between the two chromosomes forms a bridge, and later in anaphase I, the chromosome containing two small centromeres is released. If the two Dic-15 chromosomes do not pair with each other, there will be some chromosomes separated in anaphase I due to intrachromosomal recombination in Dic-15 as depicted in Figure 3.
Figure 6.
Cytological Analysis of Two Copies of the Dic-15 Chromosome.
ZmBs is labeled in magenta; the knob heterochromatin is labeled in green; DAPI is the counterstain in blue. Bar =10 μm.
(A) Pachynema with the same order pairing. (Both large and both small together at each terminus.) The arrow denotes the small centromere pair in magenta.
(B) Pachynema with the opposite order pairing. (Large with small at each terminus.) Note that in this orientation the large and small centromeres are not perfectly aligned and a region of asynapsis is present. The arrow depicts the Dic-15 bivalent.
(C) Metaphase I. The two Dic-15 chromosomes (arrow) start to separate.
(D) Anaphase I. Arrow indicates the new dicentric chromosome with two small centromeres.
(E) Anaphase I. Normal segregation of the two Dic-15 chromosomes (arrows).
(F) Anaphase I. A bridge is formed between two large centromeres and a chromosome with two small centromeres is released (arrow).
Inheritance of Chromosomes with Newly Reactivated Centromeres
The Dic-15 chromosome as one copy can regularly be transmitted intact to the next generation (see Supplemental Figure 6 online). However, we also found chromosomal fragments with only the large centromere (see Supplemental Figure 6 online) and to a lesser degree with only the smaller centromere. These chromosomes are likely the result of breakage of anaphase II bridges that result from the joining of sister centromeres via intrachromosomal recombination within Dic-15 (Figures 3 and 4). Because the large sister centromeres are active, it is not possible to recover a dicentric with both large centromeres because they form a bridge that is broken in anaphase II. If both small centromeres become reactivated, they too would be fractured at anaphase II, and this possibility could explain the recovery of the fragments with only one small centromere. However, also observed in the progeny were dicentric chromosomes with two small centromeres (Figures 3 and 4). Immunostaining for CENP-C, CENH3, and histone 3 phosphylated at Ser-10 indicated association with only one of the two sets of centromeric sequences (see Supplemental Figure 7 online), which explains their recovery.
The inheritance of chromosomes with only the small centromeres that possess molecular features of functional centromeres provides evidence that the small centromeres have regained activity. Seedlings were selected that possessed a copy of the small-small centromere chromosome and were grown for collection of meiotic tissue. Meiotic analysis of this chromosome with only small centromeres indicated that it was stably inherited throughout the subsequent generation and that cells were observed in which one of the two centromeres showed evidence of activity at anaphase and that the chromosome can progress through meiosis to the tetrad stage (see Supplemental Figure 8 online). These observations indicate that once reactivation occurs, this state can be stably inherited, although the structure of the small-small centromere containing chromosome would predict that it too would be susceptible to intrachromosomal recombination.
Time Course for Centromere Reactivation
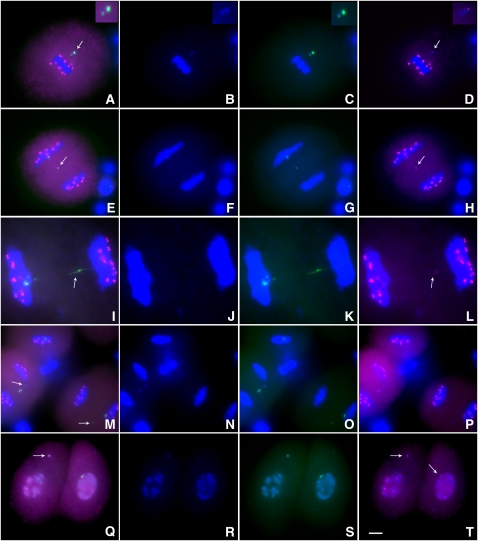
The FISH analysis shows a high frequency of anaphase I separation of the large and small centromeres of Dic-15. In maize, a homolog of mammalian CENP-C has been isolated (Dawe et al., 1999). CENP-C has been confirmed to be present only in the active centromeres (Sullivan and Schwartz, 1995). We investigated the localization of CENP-C in Dic-15 during meiosis using immunocytochemistry and FISH. At metaphase I, CENP-C signals are only detected in the active centromere (Figures 7A to 7D). In anaphase I or early telophase I stages, the smaller centromere can become separated from Dic-15 and CENP-C signals are observed on it (Figures 7E and 7I). However, the frequency of smaller centromeres associated with CENP-C is low (3 to 4%), and most fragments containing the smaller centromere did not exhibit CENP-C signals (Figure 7M). In some telophase cells, the separated centromeres both exhibited CENP-C signals (Figure 7Q). In plants with two copies of Dic-15, we detected very weak CENP-C signals in the newly formed dicentric chromosome at anaphase I (see Supplemental Figure 9 online).
Figure 7.
Immunolocalization Analysis of CENP-C in Meiosis of Plants Containing One Dic-15.
CENP-C signals are magenta, ZmBs is green, and DAPI-stained chromosomes are blue.
(A) to (D) Metaphase I. Arrow indicates that only the large centromere of Dic-15 associates with CENP-C. Insets at the top right show the enlarged Dic-15 chromosome.
(E) and (L) Early telophase I. Arrows indicates that the smaller centromere has gained CENP-C signals. In (I) to (L), the stretched appearance of the small centromeres (arrows) might result from spindle attachment from both poles.
(M) to (P) Early telophase I. Arrows indicate that the smaller centromere does not attract CENP-C proteins in this case.
(Q) to (T) Telophase I. The large and small centromeres moved to different cells and both have CENP-C signals in this case, indicating reactivation of the small centromere. Arrows indicate the two chromosomes. Bar = 10 μm.
DISCUSSION
In the centromere tug of war, when the large and small centromeres were placed in opposition, they usually both functioned to move the ends of the newly formed chromosome to opposite poles in anaphase I. In fewer cases, they moved independently, which can result in their inclusion at the same pole at the end of anaphase I. In still other cases, the smaller misdivision derivative centromere appears to attach to both poles but eventually is included in the same anaphase I pole as the large centromere (Figure 2D). The latter two circumstances can give rise to dicentric chromosomes that may involve breakage and fusion of broken ends and a continuation of the B-F-B cycle. Dicentric chromosomes were recovered in the progeny of the tug of war heterozygotes that contained a large and a small set of B centromeric sequences. In all cases examined in root tip metaphase spreads, a centromeric constriction was only present at the end of the chromosome where the large centromere resides. One example, referred to as Dic-15, was examined in detail. Its structure suggests a history of breakage and fusion because it is missing the two sets of knob heterochromatin in the center of the chromosome that would be predicted from the mere joining of the two chromosomes via recombination (Figure 1). It contains a large active centromere and a small inactive one, which has been inherited in this state through several generations. The analysis of Dic-15 resulted in the discovery of centromere reactivation under conditions that release the inactive centromere from the parent chromosome, indicating that centromere sequences can foster the establishment of centromeric chromatin at their respective sites.
In the plants containing one copy of Dic-15, 26% of the meiocytes showed separation of new structures at anaphase I, but in the plants containing two copies of Dic-15, we only found a very low frequency of the two centromeres separated at this stage (see Supplemental Table 2 online). This result suggests that chromosome pairing reduces intrachromosomal recombination and therefore reduces the frequency of smaller centromeres being separated. When we used CENP-C antibody to examine one copy of Dic-15, it is interesting that only in late anaphase I, a few smaller centromeres associate with CENP-C. Allshire and Karpen (2008) suggested that the timing of CENP-A deposition in new nucleosomes is during telophase through G1. Jansen et al. (2007) suggested that assembly and stabilization of new CENP-A containing nucleosomes is restricted to the subsequent G1 phase in mitosis. In living early Drosophila embryos as revealed by quantitative fluorescence measurements, CENP-A and CENP-C are rapidly incorporated into centromeres during anaphase (Schuh et al., 2007). In Arabidopsis thaliana, the loading of CENH3 in mitosis occurs during G2 (Lermontova et al., 2006). Our results are consistent with CENP-C being incorporated into the formerly inactive centromere during anaphase, although our studies concerned meiosis and unusual circumstances rather than mitosis.
Previous results have shown an epigenetic component to centromere specification (Han et al., 2006). Our results provide further support for this concept. However, the reactivation results provide evidence that an inactivated natural centromere can become reactivated. This result suggests that the underlying DNA repeats can in fact foster centromere specification unless an inherited epigenetic mark of an unknown nature persists in the absence of centromere function. The available data do not discriminate as to whether exactly the same sequences are associated with function in the original active and reactivated centromeres. However, by the criterion of spindle attachment, there is clearly inactivation of the small centromere in Dic-15 and reactivation of a copy of this centromeric region in the newly formed small-small centromere chromosomal derivatives. Taken together, the data indicate that centromere specification can be fostered by the underlying DNA sequence or topology, yet these sequences present on a chromosome will not necessarily condition kinetochore assembly and can be inherited for generations in an epigenetically silent state in the absence of an appropriate trigger for activity.
METHODS
Plant Materials
The construction of TB-9Sb-Dp9 has been previously described (Zheng et al., 1999) as well as the generation of Telo 3-5(+) (Kaszás and Birchler, 1998). TB-9Sb-Dp9 and Telo 3-5(+) seedlings were screened by FISH using probes for the B chromosome–specific repeat and knob heterochromatin sequences. The candidate seedlings were transferred to the greenhouse or field for crosses. Hybrid seeds were screened by FISH, and a number of seedlings containing one copy each of TB-9Sb-Dp9 and Telo 3-5(+) was found. The hybrid seedlings were transferred to the greenhouse for meiotic analysis or crosses. In the progeny, new dicentric chromosomes were scored for copy number by FISH; they were then grown in the greenhouse or the Genetics Farm at the University of Missouri-Columbia. Male inflorescences at the meiotic stage were fixed in ethanol:acetic acid (3:1, v/v) on ice for 2 h and transferred to 70% ethanol and stored at −20°C.
DNA Probe Preparation
For meiotic analysis, the B-specific sequence (Alfenito and Birchler, 1993) was labeled with Texas-red-5-dUTP, and knob-specific sequence (Peacock et al., 1981) was labeled with fluorescein-12-dUTP, both by a modified version of the nick translation method (Kato et al., 2004). CentC (centromeric satellite repeat) was labeled with fluorescein-12-dUTP and CRM (centromeric retrotransposon maize) with Texas-red-5-dUTP as previously described (Han et al., 2006).
Meiotic Analysis
Slides of various stages were collected as described (Gao et al., 1999), UV cross-linked for 2 min, washed in 2× SSC (3 × 5 min), and then rinsed in 70, 95, and 100% ethanol for 5 min each and then air-dried for 30 min. After application of 6 μL probe solution (4 ng/μL of each probe in 2× SSC and 1× TE buffer, previously denatured for 5 min in boiling water and then placed on ice), the slides were heated for 5 min at 100°C and then incubated at 55°C overnight in a humid chamber. After hybridization, the slides were washed in 2× SSC and mounted in Vectashield mounting medium (containing 1.5 μg/mL DAPI; Vector Laboratories). The FISH images were recorded using a Zeiss Universal microscope; images were captured with a Magnafire CCD camera and processed with Photoshop 7.0.
Immunolocalization in Meiotic Cells
Maize (Zea mays) CENH3 and CENP-C antibodies were obtained from Kelly Dawe (University of Georgia); monoclonal rabbit antibody (04-817) raised against histone H3 phosphorylated at Ser-10 and α-tubulin antibodies were obtained from Upstate. Tassels were fixed and stored as described (Han et al., 2007b). Anthers at different stages were collected and then cut open to release the meiocytes into 10 μL of buffer A (80 mM KCl, 20 mM NaCl, 0.5 mM EGTA, 2 mM EDTA, and 15 mM PIPES buffer, pH 7.0) on a glass slide followed by the immediate addition of 5 μL of activated acrylamide stock. The slides were rotated for a few seconds, and a cover glass (18 × 18 mm) was placed on top for 30 min or longer in a moisture box and then removed with a razor blade and transferred to 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4) for 5 min. The slides were treated for 2 to 3 h in 1% Triton X-100 (1× PBS and 1 mM EDTA). The slides were washed twice with 1× PBS, for 5 min each. About 100 μL of diluted antibodies (diluted in 3% BSA, 1× PBS, and 0.1% Tween 20) were added to the pads. The incubation was conducted overnight at room temperature. Samples were then washed in 1× PBS, 0.1% Tween 20, and 1 mM EDTA three times, each for 10 min. The appropriate secondary antibody was added and allowed to bind for 3 to 4 h at 37°C. After washing the slides in 1× PBS three times, each for 5 min, samples were stained with DAPI. Images were obtained using confocal microscopy and image analysis by the Surpass viewer of Imaris version 4.5.2 (Bitplane). Tubulin images were taken as a confocal z-stack, and a flat projection of the three-dimensional image was created with the Surpass viewer of Imaris version 4.5.2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Somatic Chromosome Spread of Dic-15.
Supplemental Figure 2. Immunolocalization Analysis of Dic-15.
Supplemental Figure 3. Immunocolocalization Analysis of a Heterozygous Plant Containing TB-9Sb and Telo 3-5(+).
Supplemental Figure 4. α-Tubulin Immunolocalization on Dic-15 at Meiosis I and for Meiosis II following Separation of Chromosomes with Large or Small Centromeres.
Supplemental Figure 5. α-Tubulin Immunolocalization on Dic-15 at Meiosis II following Separation of Chromosomes with Large or Small Centromeres.
Supplemental Figure 6. Somatic Chromosome Spreads of Progeny Derived from Dic-15.
Supplemental Figure 7. A Chromosome with Two Smaller Centromeres Recovered in the Progeny of a Plant with One Copy of Dic-15.
Supplemental Figure 8. Inheritance and Meiotic Analysis of a Chromosome with a Reactivated Centromere.
Supplemental Figure 9. Immunolocalization Analysis of CENP-C in Meiosis of the Plants Containing Two Dic-15 Chromosomes.
Supplemental Table 1. Meiotic Analysis of Hybrid Plants Containing TB-9Sb-Dp9 and Telo 3-5(+).
Supplemental Table 2. Meiotic Analysis of Newly Formed Dicentric Chromosome #15 (Dic-15).
Supplementary Material
Acknowledgments
We thank Kelly Dawe (University of Georgia) for kindly providing the maize CENH3 and CENP-C antibodies. We also thank the Molecular Cytology Core of University of Missouri-Columbia for imaging assistance. This work was supported by the National Science Foundation (DBI 0421671).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: James A. Birchler (birchlerj@missouri.edu).
Online version contains Web-only data.
References
- Alfenito, M.R., and Birchler, J.A. (1993). Molecular characterization of a maize B chromosome centric sequence. Genetics 135 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire, R.C., and Karpen, G.H. (2008). Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat. Rev. Genet. 9 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor, D.J., and Choo, K.H. (2002). Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71 695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor, D.J., Kalitsis, P., Sumer, H., and Choo, K.H.A. (2004). Building the centromere: From foundation proteins to 3D organization. Trends Cell Biol. 14 359–368. [DOI] [PubMed] [Google Scholar]
- Ananiev, E., Phillips, R.L., and Rines, H. (1998). Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, W.R. (1970). Nondisjunction and isochromosome formation in the B chromosome of maize. Chromosoma 30 356–365. [Google Scholar]
- Cimini, D. (2008). Merotelic kinetochore orientation, aneuploidy and cancer. Biochim. Biophys. Acta 1786 32–40. [DOI] [PubMed] [Google Scholar]
- Cleveland, D.W., Mao, Y., and Sullivan, K.F. (2003). Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112 407–421. [DOI] [PubMed] [Google Scholar]
- Dawe, R.K., Reed, L.M., Yu, H., Muszynski, M.G., and Hiatt, E.N. (1999). A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw, W.C., and Migeon, B.R. (1985). Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma 92 290–296. [DOI] [PubMed] [Google Scholar]
- Gao, Z., Han, F., He, M., Ma, Y., and Xin, Z. (1999). Characterization of genomes and chromosomes in a partial amphiploid of wheat-wheatgrass Zhong 2 using fluorescence in situ hybridization (FISH) and chromosome pairing analysis. Acta Bot. Sin. 41 25–28. [Google Scholar]
- Han, F., Gao, Z., Yu, W., and Birchler, J.A. (2007. b). Minichromosome analysis of chromosome pairing, disjunction, and sister chromatid cohesion in maize. Plant Cell 19 3853–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, F., Lamb, J.C., and Birchler, J.A. (2006). High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. USA 103 3238–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, F., Lamb, J.C., Yu, W., Gao, Z., and Birchler, J.A. (2007). Centromere function and nondisjunction are independent components of the maize B chromosome accumulation mechanism. Plant Cell 19 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H.S. (2001). The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293 1098–1102. [DOI] [PubMed] [Google Scholar]
- Houben, A., Demidov, D., Caperta, A.D., Karimi, R., Agueci, F., and Vlasenko, L. (2007). Phosphorylation of histone H3 in plants—A dynamic affair. Biochim. Biophys. Acta 1769 308–315. [DOI] [PubMed] [Google Scholar]
- Jansen, L.E.T., Black, B.E., Foltz, D.R., and Cleveland, D.W. (2007). Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, W., Lamb, J.C., Vega, J.M., Dawe, R.K., Birchler, J.A., and Jiang, J. (2005). Molecular and functional dissection of the maize B chromosome centromere. Plant Cell 17 1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, W.W., Melo, J.R., Nagaki, K., Talbert, P.B., Henikoff, S., Dawe, R.K., and Jiang, J. (2004). Maize centromeres: Organization and functional adaptation in the genetic background of oat. Plant Cell 16 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen, G.H., and Allshire, R.C. (1997). The case for epigenetic effects on centromere identity and function. Trends Genet. 13 489–496. [DOI] [PubMed] [Google Scholar]
- Kaszás, E., and Birchler, J.A. (1996). Misdivision analysis of centromere structure in maize. EMBO J. 15 5246–5255. [PMC free article] [PubMed] [Google Scholar]
- Kaszás, E., and Birchler, J.A. (1998). Meiotic transmission rates correlate with physical features of rearranged centromeres in maize. Genetics 150 1683–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaszás, E., Kato, A., and Birchler, J.A. (2002). Cytological and molecular analysis of centromere misdivision in maize. Genome 45 759–768. [DOI] [PubMed] [Google Scholar]
- Kato, A., Lamb, J.C., and Birchler, J.A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J.C., Kato, A., and Birchler, J.A. (2005). Sequences associated with A chromosome centromeres are present throughout the maize B chromosome. Chromosoma 113 337–349. [DOI] [PubMed] [Google Scholar]
- Lamb, J.C., Meyer, J.M., and Birchler, J.A. (2007). A hemicentric inversion in the maize line knobless Tama Flint created two sites of centromeric elements and moved the kinetochore-forming region. Chromosoma 116 237–247. [DOI] [PubMed] [Google Scholar]
- Lermontova, I., Schubert, V., Fuchs, J., Klatte, S., Macas, J., and Schubert, I. (2006). Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18 2443–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, B.-Y. (1978). Regional control of nondisjunction of the B chromosome in maize. Genetics 90 613–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., Wing, R.A., Bennetzen, J.L., and Jackson, S.A. (2007). Plant centromere organization: A dynamic structure with conserved functions. Trends Genet. 23 134–139. [DOI] [PubMed] [Google Scholar]
- Maggert, K.A., and Karpen, G.H. (2001). The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics 158 1615–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1939). The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl. Acad. Sci. USA 25 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1941). The stability of broken ends of chromosomes in Zea mays. Genetics 26 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Song, J., Stupar, R.M., Parokonny, A.S., Yuan, Q., Ouyang, S., Liu, J., Hsiao, J., Jones, K.M., Dawe, R.K., Buell, C.R., and Jiang, J. (2003). Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics 163 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasuda, S., Hudakova, S., Schubert, I., Houben, A., and Endo, T.R. (2005). Stable barley chromosomes without centromeric repeats. Proc. Natl. Acad. Sci. USA 102 9842–9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock, W.J., Dennis, E.S., Rhoades, M.M., and Pryor, A.J. (1981). Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc. Natl. Acad. Sci. USA 78 4490–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero, J.S., Ahmad, K., and Henikoff, S. (1999). A distal heterochromatic block displays centromeric activity when detached from a natural centromere. Mol. Cell 4 995–1004. [DOI] [PubMed] [Google Scholar]
- Roman, H. (1947). Mitotic nondisjunction in the case of interchanges involving the B-type chromosome in maize. Genetics 32 391–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman, H. (1948). Directed fertilization in maize. Proc. Natl. Acad. Sci. USA 34 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, A., Schneider, K.L., and Presting, G.G. (2008). Sustained retrotransposition is mediated by nucleotide deletions and interelement recombinations. Proc. Natl. Acad. Sci. USA 105 15470–15474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh, M., Lehner, C.F., and Heidmann, S. (2007). Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 17 237–243. [DOI] [PubMed] [Google Scholar]
- Sullivan, B.A., and Schwartz, S. (1995). Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum. Mol. Genet. 4 2189–2197. [DOI] [PubMed] [Google Scholar]
- Williams, B.C., Murphy, T.D., Goldberg, M.L., and Karpen, G.H. (1998). Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat. Genet. 18 3–4. [DOI] [PubMed] [Google Scholar]
- Zheng, Y.Z., Roseman, R.R., and Carlson, W.R. (1999). Time course study of the chromosome-type breakage-fusion-bridge cycle in maize. Genetics 153 1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C.X., Marshall, J.B., Topp, C., Mroczek, R., Kato, A., Nagaki, K., Birchler, J.A., Jiang, J., and Dawe, R.K. (2002). Centromeric retroelements and satellites interact with maize kinetochore CENH3. Plant Cell 14 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.