Abstract
The apical domain of the embryo is partitioned into distinct regions that will give rise to the cotyledons and the shoot apical meristem. In this article, we describe a novel screen to identify Arabidopsis thaliana embryo arrest mutants that are defective in this partitioning, and we describe the phenotype of one such mutant, bobber1. bobber1 mutants arrest at the globular stage of development, they express the meristem-specific SHOOTMERISTEMLESS gene throughout the top half of the embryo, and they fail to express the AINTEGUMENTA transcript normally found in cotyledons. Thus, BOBBER1 is required to limit the extent of the meristem domain and/or to promote the development of the cotyledon domains. Based on expression of early markers for apical development, bobber1 mutants differentiate protodermis and undergo normal early apical development. Consistent with a role for auxin in cotyledon development, BOBBER1 mutants fail to express localized maxima of the DR5:green fluorescent protein reporter. BOBBER1 encodes a protein with homology to the Aspergillus nidulans protein NUDC that has similarity to protein chaperones, indicating a possible role for BOBBER1 in synthesis or transport of proteins involved in patterning the Arabidopsis embryo.
INTRODUCTION
In angiosperms, the apical half of the globular embryo gives rise to the shoot apical meristem (SAM) and the cotyledon(s). In Arabidopsis thaliana, as in other dicots, the SAM develops between the two cotyledon primordia. This is readily apparent at the heart stage of embryogenesis when the two cotyledons (lobes of the heart) have emerged, flanking the developing SAM. The cotyledon primordia, or seed leaves, are determinate, leaf-like organs with limited growth potential. The SAM, by contrast, has much more extended growth potential. Stem cells within the SAM give rise to the majority of the aboveground portion of the plant.
Genetic and molecular analyses have identified two types of genes involved in development of the apical half of the embryo. The first type includes those genes that are required for the correct development of the cotyledons or the meristem but do not affect the placement or number of either. The second type includes those genes involved in partitioning the SAM into cotyledon and SAM domains.
Examples of the first type are the Arabidopsis WUSCHEL (WUS), CUP-SHAPED COTYLEDON1-3 (CUC1-3), and SHOOTMERISTEMLESS (STM) genes (Barton and Poethig, 1993; Aida et al., 1997, 1999; Mayer et al., 1998; Takada et al., 2001; Vroemen et al., 2003; Hibara et al., 2006). All three are necessary for the formation of the SAM. In their absence, the meristem fails to form normally. WUS is expressed as early as the 16-cell stage of embryo development when it is found in four interior cells of the apical domain. It becomes increasingly limited to a small central domain of the embryo and remains expressed in the L3 layer in a small subset of SAM cells throughout development where it acts to regulate the proliferation of the central zone stem cells (Mayer et al., 1998). CUC and STM are expressed in a band across the top half of the globular embryo that separates the two cotyledons beginning around the late globular to early transition stage (Long and Barton, 1998; Aida et al., 1999). Expression of both of these genes precedes cotyledon outgrowth, with CUC expression preceding STM expression and also being required for STM expression.
The ALTERED MERISTEM PROGRAM (AMP) and PINOID (PID) loci are examples of the second type of gene, one that alters partitioning of the apical domain of the embryo. amp1 and pid mutants typically make three or more cotyledons (Chaudhury et al., 1993; Furutani et al., 2004). The AMP gene encodes an enzyme similar to Glu carboxypeptidases and may act to generate or modulate a small, as yet unknown, signaling molecule (Helliwell et al., 2001). The PID gene encodes a kinase thought to determine the polar localization of the PIN family of auxin transport facilitators (Friml et al., 2004)
Several additional observations have implicated the plant growth hormone auxin in the specification of cotyledon primordia (reviewed in Jenik and Barton, 2005). Based on the pattern of localization of PIN auxin transporters, on the accumulation pattern of auxin inferred from DR5 reporter expression, and on the failure of cotyledons to form in quadruple mutants deficient for four different PIN auxin transporters, a model has been developed in which directional transport of auxin into the incipient cotyledon primordia is required for their formation similar to the mechanism proposed for leaf formation (Benkova et al., 2003; Reinhardt et al., 2003). In support of this, several labs have recently reported that mutants homozygous for both pid and enhancer of pinoid (enp)/macchi bou/npy (an NP3-like protein) fail to form cotyledons (Treml et al., 2005; Cheng et al., 2007; Furutani et al., 2007). These seedlings exhibit spatially limited expression of AINTEGUMENTA (ANT) mRNA, a marker for cotyledon fate, as well as expanded expression of STM mRNA. enp pid double mutant embryos also show disruptions in normal auxin transport as visualized by the accumulation of DR5 reporter expression and by examination of the subcellular location of PIN proteins.
In contrast with amp1 and pid mutants, which make too many cotyledons, topless (tpl) mutant seedlings fail to make both cotyledons and the SAM (Long et al., 2002). The single pin-like structure that forms in tpl mutant embryos fails to express markers for meristem identity, and, instead, the ANT1 transcript, a marker for cotyledon identity, is expressed throughout the apical-most region. Thus, tpl mutant embryos show expanded cotyledon identity along with reduced or absent SAM identity and therefore appear to be defective in apical partitioning. Wild-type TPL function then is required to limit cotyledon fate and promote meristem fate in the apical domain of the embryo. TPL encodes a corepressor that interacts with indoleacetic acid-induced (IAA) proteins and is required for gene repression mediated by auxin response factor/IAA complexes (Szemenyei et al., 2008). Since auxin causes degradation of IAA proteins, auxin may be responsible for lifting the gene repression mediated by TPL in the presumptive cotyledons.
Recently, two receptor-like kinase genes have also been implicated in cotyledon formation and apical partitioning. When both the RECEPTOR-LIKE PROTEIN KINASE1 (RPK1) and TOADSTOOL2 (TOAD2) genes are mutant, embryos lack cotyledons, fail to express the ANT1 cotyledon marker, and show expanded expression of the STM meristem marker (Nodine and Tax, 2008). rpk1 toad2 mutants also fail to establish high local concentrations of auxin in the cotyledon primordia. This may be in part because rpk1 toad2 fails to localize PIN1 transcripts to the cotyledons, instead localizing these transcripts to basal parts of the embryo. Neither the targets for these protein kinases nor the ligands that bind to their extracellular domains are known.
To date, mutants identified in patterning the apical domain of the embryo have been isolated in screens performed at the seedling stage. There are at least two reasons that more mutants have not been found. First, there is likely extensive genetic redundancy as is shown in the case of the rpk1 toad2 mutants (Nodine et al., 2007). Second, some mutants affected in apical partitioning may lead to embryonic arrest. To test the second possibility, we performed a mutant screen in which we first prescreened for embryo arrest mutants. We then scored the mutants for their expression of a marker specific to the SAM. In this article, we describe the isolation, characterization, and molecular cloning of the bobber1 (bob1) mutant allele, a mutant in which the SAM domain is expanded, while the cotyledon domain is missing.
RESULTS
Screen for Embryo Arrest Mutants That Misexpress a Reporter for the STM Gene
With the goal of isolating embryo lethal mutants that are defective in partitioning the apical domain of the embryo, we mutagenized a line carrying a reporter for the STM locus. This reporter expresses the β-glucuronidase (GUS) enzyme marker in the developing SAM (Figure 1; McConnell and Barton, 1998; Jenik et al., 2005). The reporter is not expressed throughout the SAM but is excluded from the central region of the developing meristem (Figure 1D).
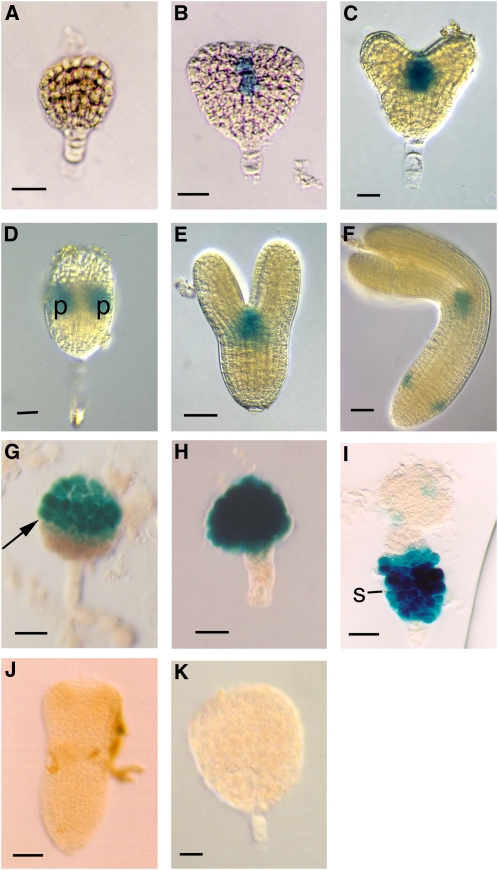
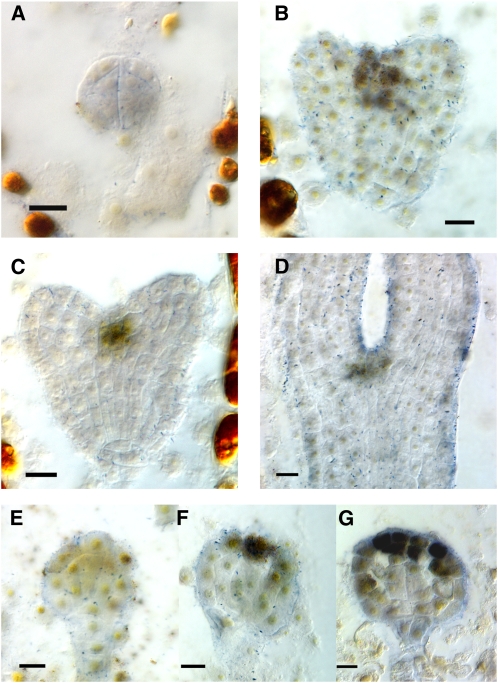
Figure 1.
Expression of the STM:GUS Reporter in Wild-Type and Mutant Embryos.
(A) Late globular stage embryo. Expression is weak to undetectable.
(B) Transition stage embryo showing expression at the site of meristem formation between the developing cotyledons.
(C) Heart stage embryo.
(D) Heart stage embryo with one cotyledon removed and viewed from the side. Note that the reporter expresses most highly in the peripheral regions (p) of the developing meristem. These regions will become the boundaries separating the cotyledon margins.
(E) Torpedo stage embryo.
(F) Walking stick stage embryo. In addition to expression at the apical meristem, expression at the junction of the hypocotyls and root is observed.
(G) Mutant embryo (bob1 mutant) showing expression throughout apical half of embryo. Expression is confined to apical half of embryo. Boundary separating apical and basal halves is indicated with arrow.
(H) Mutant embryo showing expression throughout embryo proper.
(I) Mutant embryo showing ectopic expression in hypertrophied suspensor (s).
(J) Mutant torpedo stage embryo lacking STM expression (sagittal view of embryo with one cotyledon removed).
(K) Globular arrest mutant lacking STM expression.
Bars = 25 μm in (A) to (D), (G) to (I), and (K) and 50 μm in (E), (F), and (J).
Following ethyl methanesulfonate mutagenesis, M2 seeds were harvested from individual M1 plants and planted to soil. A total of 1695 independent embryo arrest mutants, occurring in 819 M2 families, were identified. (Embryo arrest mutants found in the same M2 family were considered independent if individual M2 plants segregated embryos arrested at distinct stages.) Arrested embryos developing in the siliques of M2 plants were dissected out of the ovules when siblings were at heart stage and stained with GUS staining solution. In ∼15% of siliques segregating embryo arrests, neither the wild-type nor the arrested embryos showed GUS expression, presumably reflecting silencing of the transgene.
Among those siliques where the transgene remained active, 69 embryo arrest mutants (4% of total) were found to have ectopic expression of STM-GUS, while 182 embryo arrest mutants (11% of total) failed to express the STM-GUS reporter. In all cases, the wild-type siblings of the arrested embryos expressed STM-GUS in the wild-type fashion. Mutant embryos ectopically expressing STM showed a range of phenotypes with regard both to stage of arrest and to pattern of ectopic expression. Most ectopically expressing mutants formed cotyledons flanking a broader domain of STM expression. A few mutants showed ectopic expression and arrested at the globular stage; examples of such mutant embryos are shown in Figure 1. One type of mutant (represented by one mutation, the bobber1 mutation described below) expressed the STM reporter throughout the top half of the globular embryo (Figure 1G). A second type of mutant (represented by two mutations) expressed the STM reporter throughout the globular embryo (Figure 1H). A third type of mutant (represented by one mutation) expressed the STM reporter in a hypertrophied suspensor (Figure 1I).
Of the mutants that failed to express the STM-GUS reporter, most (154) arrested at the globular stage (Figure 1K). A minority of the nonexpressing class of embryo arrest mutants (26) developed past the globular stage (Figure 1J). Because STM reporter expression does not begin in the wild type until the late globular or transition stage, a globular arrest mutant that fails to express the reporter may do so because it is blocked in its development and has not matured to the stage at which STM is expressed. Alternatively, it may do so due to a patterning defect, such as failure to specify a SAM domain. Because we cannot distinguish between these two possibilities, we chose to focus on an ectopically expressing line for in-depth study rather than a nonexpressing line.
The bob1 Mutant Arrests at the Globular Stage and Shows Ectopic STM Expression
One of the mutants isolated from the screen was bob1, an embryo arrest mutant that fails to develop past the globular stage of embryogenesis (Figures 1G and 2). bob1 mutants arrest consistently at the globular stage of embryogenesis, never morphologically developing to the transition stage (Figure 2). bob1 mutants account for ∼25% of embryos in the self-progeny of heterozygous individuals, consistent with a recessive mutation in a single, nuclear gene (see Supplemental Table 1 online).
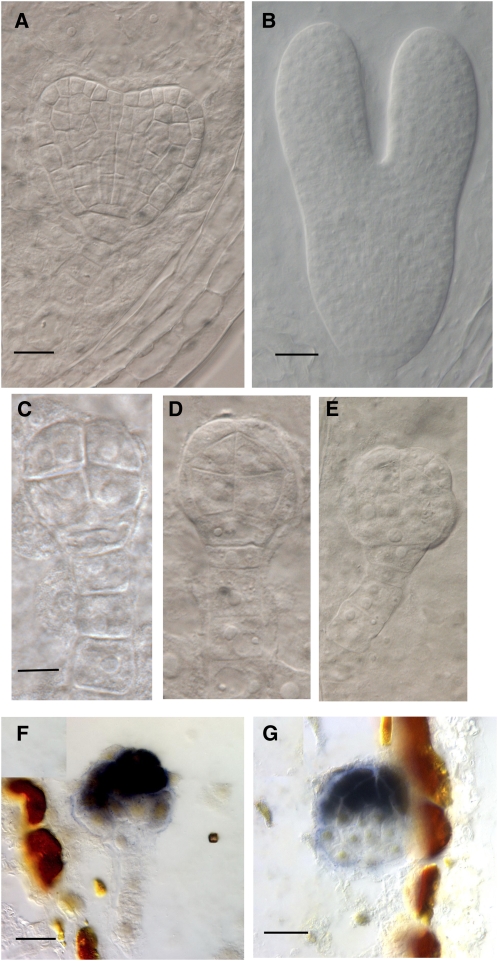
Figure 2.
Development of bob1 Mutant Embryos.
(A) Differential interference contrast (DIC) microscopy image of wild-type heart stage embryo.
(B) DIC microscopic image of wild-type torpedo stage embryo.
(C) DIC microscopic image of eight-cell stage bob1 mutant embryo, sibling to embryo in (A).
(D) DIC microscopic image of 16-cell stage bob1 mutant embryo, sibling to embryo in (A).
(E) DIC microscopic image of ∼32-cell stage bob1 mutant embryo, sibling to embryo in (B).
(F) In situ hybridization to STM mRNA in bob1 mutant embryo, siblings were at heart stage.
(G) In situ hybridization to STM mRNA in bob1 mutant embryo, siblings were at torpedo stage.
Bars = 25 μm in (A), (F), and (G), 50 μm in (B), 5 μm in (C) and (D), and 10 μm in (E).
The initial assay for STM:GUS reporter expression in a bob1 mutant embryo revealed that the expression of STM, normally confined to a few cells in the middle of the apical portion of the embryo, is expressed throughout the apical half of bob1 embryos (Figure 1G). STM expression begins in wild-type embryos at the late globular or transition stage. By the time wild-type siblings have reached early heart stage, bob1 mutant embryos have begun to express the STM:GUS reporter. In situ hybridization experiments on siliques from plants heterozygous for the bob1 mutation using an antisense STM probe confirm that STM is ectopically expressed in bob1 embryos (Figures 2F and 2G). Expression in the entire apical hemisphere of the globular embryo indicates that STM is expressed in cells that would normally be specified as cotyledon tissue.
bob mutant embryos can reliably be distinguished from their wild-type siblings beginning when the wild-type embryos are at heart stage. In siliques of bob1/+ plants, one-quarter of the embryos remain at early globular stages, while none do so in siliques on +/+ sibling plants. (A total of 15/50 embryos in bob1/+ siliques were at the 16-cell stage or earlier; the remainder were beyond the 32-cell stage, with the majority [22] being at the transition stage. For +/+ siliques, 0/43 embryos were at the 16-cell stage or earlier, with the majority [20] being at the transition stage.) The presumed homozygous bob1 embryos have normal division patterns at the 8- and 16-cell stage (Figures 2C and 2D; see also sections of bob embryos in Figures 3 to 5 and 7). In older bob embryos, cells of the epidermal layer are seen to bulge outward (Figure 2E). Additional cell divisions take place in bob embryos (for example, see Figure 3J), but the embryos never make the transition to a heart shape and instead remain globular in shape.
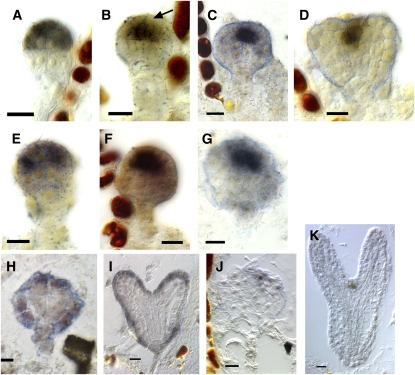
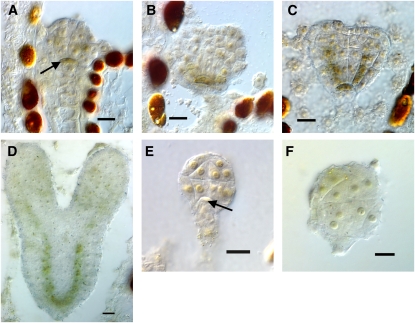
Figure 3.
Expression of CUC2, ATML1, and WUS in Wild-Type and bob Mutant Embryos.
(A) to (D) Wild-type embryos.
(E) to (G) bob1 mutant embryos.
(A) Sixteen-cell stage embryo. CUC2 expression is confined to the apical half of the embryo.
(B) A 32-cell globular stage embryo. CUC2 expression is missing from the protodermis and confined to the subepidermal cells in the apical half of the embryo.
(C) and (D) Late globular embryo (C) and early heart stage embryo (D). CUC2 expression remains in the subepidermal layer(s) as it becomes excluded from the cotyledon primordia.
(E) bob1 16-cell stage mutant embryo showing CUC2 expression principally in the apical half of the embryo similar to the wild type.
(F) and (G) bob1 mutant embryos whose siblings were at heart stage. Expression is confined to subepidermal cells and to the apical half of the embryo. There is little to no evidence of the development of cotyledon primordia.
(H) bob1 mutant embryo expressing ATML1 in the protoderm.
(I) Sibling embryo at heart stage showing ATML1 expression in the protoderm.
(J) bob1 mutant embryo, sibling to wild-type embryo in (K), showing small region of WUS expression slightly offset from center.
(K) Wild-type embryo showing WUS expression in a subset of cells at the presumptive SAM.
Bars = 10 μm.
Figure 4.
Expression of ANT in Wild-Type and bob1 Mutant Embryos.
(A) Wild-type globular embryo. ANT is not expressed at this stage.
(B) Wild-type heart stage embryo showing ANT expression in developing cotyledons.
(C) Wild-type torpedo stage embryo showing persistent expression of ANT in cotyledons.
(D) bob1 mutant embryo, sibling to wild-type embryo in (A).
(E) bob1 mutant embryo, sibling to embryo in (B).
(F) bob1 mutant embryo sibling to wild-type embryo in (C).
Bars = 25 μm.
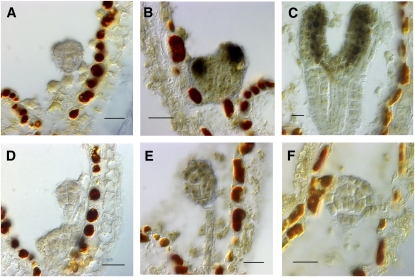
Figure 5.
Expression of UFO in Wild-Type and bob Mutant Embryos.
(A) to (D) Wild-type embryos. Expression begins at the heart stage (B) of development and is limited to the presumptive meristem. By late heart (C), expression is excluded from the protodermal layer (L1 layer) of the apical meristem.
(E) to (G) bob mutant embryos. For (E) and (F), sibling embryos were at the heart stage. For (G), siblings were at the torpedo stage. Expression does not become excluded from the protodermal layer. The number of cells expressing UFO transcript is variable. Note expression in a single cell in (E) and in multiple cells in (G).
Bars = 10 μm.
Figure 7.
SCARECROW Expression in Wild-Type and bob1 Mutant Embryos.
(A) Wild-type globular embryo showing SCARECROW expression in lenticular cell.
(B) Transition stage embryo showing SCARECROW expression in lenticular cell.
(C) and (D) Early heart stage (C) and torpedo stage (D) embryos showing expression in lenticular cell and endodermis.
(E) Early globular stage bob embryo, sibling to transition stage wild-type embryos. Arrow shows properly formed hypophysis cell, not expressing SCARECROW.
(F) bob1 mutant embryo lacking expression of SCARECROW. Siblings were at the torpedo stage.
Bars = 10 μm.
Although bob1 mutant embryos arrest at the globular stage, they do not die at this point. Experiments that rely on the continued ability of bob1 embryos to maintain undegraded RNA (in situ hybridizations) and protein (STM:GUS reporter protein assays) indicate that they remain viable until the time that the wild-type embryos in the silique mature and desiccate. To test if bob1 embryos would recover and restart their development if given enough time, we placed arrested bob1 embryos on three different types of tissue culture media and evaluated their reactions (Murashige and Skoog, 1962; Fischer and Neuhaus, 1996; Li and Thomas, 1998). bob1 embryos were dissected from their seed coats when wild-type siblings were at the bent-cot stage. Although the wild-type siblings placed on Fisher and Li media grew and became wild-type-looking seedlings, none of the bob1 embryos on any of the media ever grew or developed any further.
bob1 Embryos Are Not Rescued by Removing STM Function
To test if ectopic expression of STM is the only defect in bob1 mutant embryos, bob1/+ plants were crossed to stm-11/+ plants. Because stm/stm plants survive through embryogenesis (Barton and Poethig, 1993), if ectopic expression of STM were the only defect in bob1 mutants, bob1/bob1 stm-11/stm-11 double mutants should survive embryogenesis and have the phenotype of stm-11/stm-11 single mutants. In that case, F1 plants heterozygous for both bob1 and stm-11 should segregate 3/16 bob1 embryos (18.75%). If, however, ectopic expression of STM were not the only defect in bob1 mutants, removing STM function should have no impact on the bob1 arrest phenotype; all bob1/+ F1 individuals should segregate 1/4 bob1 embryos, regardless of their STM genotype.
The self-progeny of 16 F1 progeny from four different bob1/+ by stm-11/+ crosses were examined and the number of bob1 and wild-type embryos recorded (see Supplemental Table 1 online). bob1 individuals accounted for 23.5% of the embryos in the siliques of all of the bob1/+ stm-11/+ individuals combined. χ2 testing reveals that the hypothesis of 3/16 segregation of bob1 embryos must be rejected (P < 0.001) but that a 1/4 hypothesis cannot be rejected (P = 0.5). Therefore, removing STM function from bob1 mutant embryos does not rescue the embryo arrest phenotype; the defect in bob1 mutants is not simply the overexpression of STM.
Expression of Markers for Apical Development in bob1 Embryos
CUC2 is required, along with CUC1, for meristem formation and cotyledon separation in the Arabidopsis embryo (Aida et al., 1997). CUC2 mRNA expression precedes STM expression and is required for the accumulation of STM transcript in the embryo (Aida et al., 1999). To determine if the pattern of expression of CUC2 as well as that of STM is affected in bob1 embryos, we examined the expression of CUC2 in bob1 mutant embryos.
Our results indicate that CUC2 is first expressed in the entirety of the apical region of the wild-type 16-cell stage embryo (Figure 3A). This is earlier and more extensive than has been reported previously (Aida et al., 1999). CUC2 expression clears out of the protoderm layer at around the 32-cell stage (Figure 3B), ultimately becoming limited to a stripe across the apical half of the globular embryo. Viewed in frontal section, the expression appears to be confined to the cells of the presumptive meristem (Figure 3C). Viewed in the sagittal plane, however, CUC2 mRNA is present in a band across the apical half of the embryo (Aida et al., 1999). This band of expression represents the portion of the embryo that will become the SAM (central part of the band) and the boundaries separating the cotyledon margins. The areas free of CUC2 expression in the globular embryo represent portions of the apical domain that will grow out as cotyledons (Aida et al., 1999).
In bob1 mutant embryos, CUC2 expression is initially similar to that seen in wild-type embryos; it accumulates throughout the apical domain of the 16-cell stage embryo and then becomes limited to the subepidermal cells of the late globular stage embryo (Figure 3E). However, further limiting of CUC2 to a central stripe separating two cotyledon domains does not appear to occur, and the CUC2 domain of expression may be slightly expanded relative to that of the wild type (cf. Figures 3F and 3G to 3D). In summary, expression of CUC2 is expanded in bob1 mutant embryos but is less affected than the expression of STM.
The exclusion of CUC2 from the epidermis in both wild-type and bob1 mutant embryos indicates that the specification and early development of the protoderm is normal in bob1 mutant embryos. To further test this, we examined the expression of the ARABIDOPSIS THALIANA MERISTEM LAYER L1 (ATML1) mRNA in wild-type and bob1 mutant embryos. The ATML1 gene encodes a transcription factor in the HDZIPIV class (Lu et al., 1996). Some members of this family have been implicated in the establishment and maintenance of the epidermis (Abe et al., 2003). Consistent with this, the ATML1 expression pattern distinguishes the outer cell layer from internal layers beginning at the dermatogen stage (Lu et al., 1996). In bob1 mutant embryos, ATML1 mRNA accumulates normally (Figure 3H). Therefore, formation of the protoderm layer of the embryo does not appear to be affected by the bob1 mutation.
The expanded domain of expression of STM suggests that bob1 embryos are defective in specification of the cotyledon primordia. To test this, we examined expression of the ANT1 mRNA in wild-type and bob1 mutant embryos. The ANT1 gene is expressed in all the lateral organs of the plant, including the cotyledons in the embryo (Elliott et al., 1996). It therefore provides a good marker for cotyledon fate. Wild-type embryos begin expressing the ANT transcript at the late globular stage of embryogenesis (Long and Barton, 1998) in a ring around the periphery of the apical half of the embryo, reflecting the establishment of radial symmetry in the apical region of the embryo. Expression continues throughout embryogenesis, becoming confined to internal tissues of the cotyledons later in embryogenesis (Elliott et al., 1996). The ANT transcript was not detected in bob1 embryos (Figure 4), consistent with a defect in the developmental program that specifies cotyledon fate.
Like STM and CUC2, the WUS gene is required for the normal establishment of the SAM during embryogenesis. However, WUS is expressed in a pattern that is distinct from that of either STM or CUC2. In wild-type embryos, the WUS transcript accumulates from the 16-cell stage throughout embryogenesis, first appearing in all nonprotoderm cells of the apical embryo, then in a progressively smaller region as cell divisions narrow its pattern to a final expression domain that is one to two cells wide and one cell deep in the L3 layer of the SAM in the mature embryo (Mayer et al., 1998).
Accumulation of low levels of the WUS transcript can be seen in bob1 embryos in variable positions. A small region of expression is typically seen somewhere in the apical domain of the embryo (Figure 3J). This region is often off center. It appears that the mRNA accumulates in a slightly different position in each embryo, as if the positional cues WUS needs for correct localization are disrupted or destabilized. However, unlike STM, the number of cells expressing WUS is not increased.
UNUSUAL FLORAL ORGANS (UFO) appears in a subset of meristem cells, beginning around the early heart stage of embryogenesis. Its expression in the embryo is confined to the cells of the presumptive SAM. By the torpedo stage, expression is narrowed to a cup-shaped subdomain within the STM expressing domain (Long and Barton, 1998). Although initially expressed in both protodermal and subepidermal regions, by the late heart stage, expression is restricted to layers below the L1 (Long and Barton, 1998). UFO expression in the embryo requires STM function, and UFO is expressed ectopically in 35S:STM-VP16 transgenic plants (Fernandez, 2002), indicating that UFO is a target, perhaps direct, of STM regulation. While the role of UFO in the embryo, if any, is unknown, it provides a marker for events downstream of STM.
UFO is expressed in bob1 mutants, but in a manner that suggests that the positional cues UFO needs for proper expression are disrupted (Figure 5). The intensity and area of expression are variable from embryo to embryo. Expression is largely, if not entirely, confined to the protoderm in the apical portion of the embryo. Thus, as in the wild type, expression of UFO is limited to a subdomain of STM expressing cells, which is consistent with a requirement for STM plus another factor in activating UFO expression.
Recently, directed transport of auxin into the cotyledon domains has been proposed as a mechanism to explain the early events in specification of cotyledon primordia (for review, see Jenik and Barton, 2005). In support of this, high levels of expression from the auxin-sensitive DR5 reporter are observed in the early cotyledon primordia in wild-type embryos (Figure 6). bob mutant embryos that do not carry the green fluorescent protein (GFP) transgene do not show significant levels of autofluorescence, indicating that the observed fluorescence is not due to alterations in bob mutant physiology. In bob1 mutant embryos, DR5 is expressed throughout the globular stage embryo, suggesting that uniform and high auxin levels are found throughout the embryo (Figure 6). Thus, bob1 mutants are defective in limiting auxin maxima in the apical domain of the embryo to the cotyledon primorida as well as to the root pole of the embryo and show overall higher levels of auxin throughout the globular embryo.
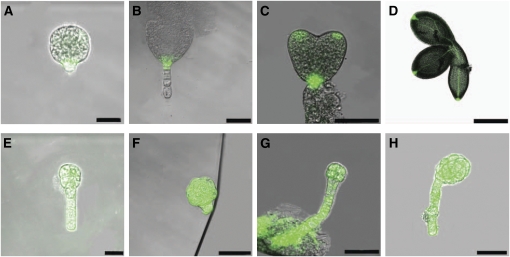
Figure 6.
Expression of DR5:GFP Reporter in Wild-Type and bob1 Mutant Embryos.
(A) to (D) Wild-type embryos. DR5 is expressed in the root pole of wild-type globular (A) and transition (B) embryos. DR5 is also expressed in wild-type cotyledon tips (C) and in the vasculature of mature wild-type embryos (D).
(E) to (H) bob1 mutant embryos. DR5 is expressed throughout both the arrested globular embryo as well as the suspensor in sibling bob1 mutant embryos ([E] to [H]).
Bars = 25 μm in (A) and (E), 50 μm in (B), (C), and (F) to (H), and 250 μm in (D).
Auxin maxima in the embryo have been associated both with the developing cotyledon primordia as well as with the developing root pole of the embryo. To determine if gene expression patterns are abnormal at the root end of the embryo, we analyzed expression of the SCARECROW gene. SCARECROW expression is first seen at the globular stage, where it is found in the lenticular cell (Figure 7). Expression expands to include the endodermis as the embryo progresses through the heart and torpedo stages. By contrast, bob mutant embryos fail to express the SCARECROW transgene. Thus, gene expression associated with the auxin maximum at the root pole of the embryo is also disrupted in bob mutant embryos.
BOB1 Encodes a Gene with Similarity to the Aspergillus nidulans NUDC Gene
Preliminary mapping of the BOB1 locus placed it at the bottom of Arabidopsis chromosome 5, between cleaved-amplified polymorphic sequence (CAPS) markers DFR (87.6 centimorgan [cM] on the recombinant inbred map) and LFY3 (115 cM on the recombinant inbred map) (see Supplemental Figure 1 online). New CAPS markers were developed for fine mapping (see Supplemental Table 2 online), and the BOB1 region was narrowed to a 53-kb region at ∼108 cM on chromosome 5. Two DNA clones overlapped this region: K19E1, a TAC clone; and MYN8, a P1 clone (Sato et al., 2000).
The 53-kb BOB1 region contained 10 predicted genes (see Supplemental Figure 1 online). Predicted genes were amplified from wild-type Landsberg erecta (Ler) genomic DNA and their sequences compared with published sequences from the same open reading frames in wild-type Columbia (Col). Polymorphic loci in the coding sequences of the two ecotypes were identified. Predicted genes were then amplified out of genomic DNA from an F1 plant resulting from a cross between a bob1/+ plant and a wild-type Col plant. Because the bob1 mutation is on a Ler chromosome, the F1 plant contains one Ler chromosome containing the mutation and one wild-type Col chromosome. Amplification reaction products should be 50% from the Ler allele and 50% from the Col allele. Amplification products were cloned into plasmid vectors, and the inserts of several independent clones were sequenced and examined for the presence of a mutation. To ensure that both chromosomes present in the bob1/+ F1 plant had been examined for mutations, the sequences were compared in amplification products containing polymorphisms between Ler and Col sequences.
Sequencing of the MYN8.1 locus (At5g53400) from the Ler chromosome of a bob1-1/+ plant revealed that it contained a C-to-T transition that created a stop codon at amino acid position 196 of the predicted protein (see Supplemental Figure 1 online; Figure 8). To ensure that this was the mutation causing the bob1 phenotype, a 6-kb fragment of MYN8 DNA was amplified and put into binary vector pCAMBIA 1300 for transformation into wild-type Arabidopsis (see Supplemental Figure 2 online). The rescue plasmid was named pREJ14. After transformation, wild-type Ler plants resistant to hygromycin were crossed to bob1/+ plants. The hygromycin-resistant F1 progeny from this cross were allowed to self-fertilize, and the embryos inside self-crossed siliques were examined for segregation of bob1 embryos. bob1 and wild-type embryos in siliques from F1 plants that segregated bob1 embryos were counted (see Supplemental Table 3 online). A total of 5.6% of these embryos displayed the bob1 phenotype, which is in agreement with 1/16 bob1 embryos, the expected ratio for self-progeny segregating from a parent heterozygous for the bob1 mutation and hemizygous for a rescuing plasmid (χ2 testing; P = 0.5).
Figure 8.
Alignment of BOB1 Homologs.
BOB1 homologs contain a highly conserved C-terminal NudC domain (underlined) and a C terminus that is highly conserved within, but not between, phylogenetic groups. The location of the bob1-1 lesion (R197*) is indicated by one asterisk, the location of the bob1-2 T-DNA insertion (I117) is indicated by two asterisks. Species are Arabidopsis (A.t.), Oryza sativa (O.s.), Homo sapiens (H.s.), Danio rerio (D.r.), Schizosaccharomyces pombe (S.p.), and A. nidulans (A.n.).
Finally, a second allele (bob1-2) containing a T-DNA insertion in the N terminus of bob1 at Ile-117 (Salk_001125) was obtained from the Salk T-DNA collection. Self-pollinated bob1-2 siliques contain embryos arrested at the globular stage in a 1:3 ratio. To show that these aborted embryos were due to a lesion in BOB1, crosses between bob1-1 and bob1-2 heterozygous plants were performed. bob1-1 failed to complement the bob1-2 aborted embryo phenotype (see Supplemental Table 4 online). These genetic and molecular complementation experiments demonstrate that the bob1-1 embryo arrest phenotype is due to mutations in the MYN8.1 (At5g53400) gene.
To demonstrate that the STM mislocalization phenotype observed in bob1-1 arrested embryos is due to the bob1-1 lesion as opposed to a separate linked mutation, we further characterized bob1-2. Plants heterozygous for bob1-2 were crossed to the STM:GUS reporter. Arrested embryos in the siliques of the self-pollinated progeny of this cross displayed STM expression, which was similar to the mislocalization observed in bob1-1 (see Supplemental Figure 3 online). Since two independent bob1 alleles both display STM mislocalization, we conclude that this phenotype is specifically due to lesions in BOB1.
BOB1 encodes a homolog of the A. nidulans NUDC (for Nuclear Distribution C) gene, and NudC domain containing proteins are found in fungi, animals, and plants (Figure 8). There is one close BOB1 homolog in the Arabidopsis genome that is contained in a BOB1 syntenous region (Freeling et al., 2007). BOB2 (At4g27890) is very similar to BOB1, sharing 62% identity over the whole protein and 81% identity within the conserved C-terminal domain. The stop codon in bob1-1 is near the start of the conserved C-terminal NudC domain of the BOB1 gene (Figure 8). The placement of the mutation and the severity of the bob1 phenotype indicate that this conserved domain is important to the functioning of BOB1, although it is currently unknown what its biochemical function might be.
BOB1 Is Expressed throughout the Early Arabidopsis Embryo
Sectioned siliques from a plant heterozygous for the bob1 mutation were examined by in situ hybridization using an antisense probe made from BOB1 cDNA. The results indicate that BOB1 is expressed throughout the early embryo (see Supplemental Figure 4 online). BOB1 mRNA first accumulates at a point before the eight-cell stage of embryogenesis, and expression remains strong until the transition stage. In transition and heart stage embryos, variable expression could be detected, and after heart stage, no mRNA could be detected. bob1 mutant embryos did not detectably express BOB1 mRNA. Sectioned siliques from bob1/+ plants examined with a control BOB1 sense probe did not show any hybridization.
A BOB1:GUS reporter transgene was constructed and named pREJ19. BOB1 coding sequences with introns and ∼2.4 kb of 5′ sequences were amplified creating an in-frame fusion to GUS (see Supplemental Figure 5 online). Wild-type Ler plants were transformed with pREJ19, and hygromycin-resistant transformants were allowed to self-fertilize. GUS expression in pREJ19 transformed lines agrees with the in situ hybridization findings for BOB1 mRNA:BOB1-GUS is expressed throughout the early embryo and clears away at a variable time after transition stage. The BOB1-GUS reporter seems to be detectable until later in embryogenesis than BOB1 mRNA, perhaps reflecting the fact that the GUS assay detects protein, while the in situ hybridizations detect mRNA. Alternatively, the GUS reporter may be missing a negative controlling element.
The same promoter fragment was used to drive expression of a BOB-GFP fusion protein. A genomic fragment including the promoter plus BOB exons and introns was fused to GFP. This construct was capable of rescuing bob null mutants. Analysis of embryos expressing this construct shows that the BOB protein accumulates throughout the cytoplasm (Figure 9). In heart stage embryos, the protein may be somewhat enriched in epidermal cells. The location of the BOB protein in the cytoplasm is consistent with a predicted role of NUDC-like proteins as chaperones (Faircloth et al., 2009).
Figure 9.
Expression of BOB1 Protein in Embryogenesis.
Expression of a BOB1-GFP fusion protein expressed under the control of the BOB1 promoter.
(A) Four-cell stage embryo.
(B) Eight-cell stage embryo.
(C) Early heart stage embryo.
Bars = 5 μm in (A) and (B) and 10 μm (C).
DISCUSSION
BOB1 Is Required for Establishment of Normal Patterning in the Apical Domain of Arabidopsis Embryos
Early in dicot embryo development, the top half of the globular embryo becomes partitioned into domains that will form the cotyledons and the SAM. To identify genes required for this partitioning, we designed a mutant screen to isolate mutants with altered expression of a meristem-specific marker, the STM:GUS reporter gene. We limited our screen to mutants that arrest as embryos. This was in part because extensive screens for pattern defective mutants have already been performed at the seedling stage. It was also because it is possible that severe disruption of early patterning events may lead to embryo arrest.
Our screen identified one mutant, the bob1 mutant, in which the STM:GUS reporter is expressed throughout the top half of the globular embryo extending into the regions that would normally form the cotyledons. Moreover, bob1 mutants fail to express the organ-specific marker ANT1. Thus, it appears that in bob1 mutants, the meristem domain is expanded at the expense of the cotyledon domains. The wild-type function of the BOB1 gene then is to promote organ fate and to limit meristem fate. BOB1 is expressed throughout the embryo during the early stages of embryogenesis when these developmental domains are being established. Because BOB1 mRNA expression is not itself restricted to any portion of the embryo, and assuming that BOB1 protein is not posttranslationally modified or transported differentially during embryogenesis, the activity of this gene must enable the correct establishment of patterning information without being part of the pattern itself.
Some pattern elements appear to develop normally in bob1 mutant embryos. The epidermis appears to be normally specified, as evidenced by expression of the ATML1 gene and exclusion of the CUC2 gene. This is in contrast with the rpk1 toad2 mutants, which fail to express ATML1 in the protodermal layer of embryos that lack cotyledons (Nodine and Tax, 2008). In mutants lacking both ATML1 and PDF2 function (both genes encode related HDZIPIV transcription factors), cotyledon initiation is often defective (Abe et al., 2003). It may be that the cotyledon initiation defect in rpk1 toad2 embryos is due to failure to establish a normal protoderm layer. Since bob mutants appear to develop normal protoderm with normal ATML1 expression, it would appear that BOB plays a role in a process separate from the one in which the RPK1 and TOAD2 genes are involved.
A second pattern element that appears to develop normally in bob1 mutant embryos is the separation of the embryo proper into distinct apical and basal halves. This is supported by the finding that the CUC2 and STM genes are strictly limited to the apical domain of the embryo in both wild-type and bob1 mutants.
These two normal pattern elements, dividing the globular embryo into an apical and a basal hemisphere and the establishment of an epidermis, both require early, stereotyped cell divisions in the embryo. These early cell divisions appear normal by inspection with DIC optics as well as in many of the sections that were made of bob mutant embryos. Thus, the defects in pattern that we observe are not due to dramatic differences in early embryonic cell division patterns. In fact, changes in SCARECROW and DR5 expression are observed before changes in cell morphology become apparent.
In our screen for mutants that aberrantly express the STM reporter, we also recovered globular arrest mutants expressing the STM reporter throughout both apical and basal halves of the embryo. The identification of these mutants in our screen illustrates that it is possible to abolish this restriction with what are likely to be single gene mutations. It is therefore significant that bob1 does not abolish this aspect of polarity. This is despite the fact that bob1 mutant embryos fail to properly localize auxin, as determined by the expression of the DR5 promoter throughout the embryo. This is a somewhat surprising result given that auxin has been implicated in establishing apical-basal patterning in the early embryo (Friml et al., 2003). Taken at face value, these data suggest that localized auxin is not absolutely required for the development of at least a subset of cellular differences between the apical and basal halves of the embryo proper.
In addition to its role in development of apical/basal polarity in the embryo, auxin has been proposed to play a role in partitioning the apical domain of the embryo (for review, see Jenik and Barton, 2005; Nawy et al., 2008). In particular, higher local concentrations of auxin, as determined by DR5 reporter gene expression, are found in the presumptive cotyledons. In addition, mutant combinations thought to impair polar auxin transport interfere with cotyledon development. Plants homozygous for these mutant combinations have phenotypes somewhat similar to bob1. For instance, pinformed1 (pin1) pid double mutants and enp pid double mutants both fail to make cotyledons and both exhibit expanded STM and CUC2 expression throughout the apical domain of the embryo (Treml et al., 2005). PIN1 encodes an auxin transporter that directs polar auxin transport through its polarized localization in the plasma membrane, and PID is a kinase that appears to directly regulate the localization of PIN1 (Friml et al., 2004; Michniewicz et al., 2007). ENP (also known as MACCHI BOU and NPY) encodes an NPH3-like protein (Cheng et al., 2007; Furutani et al., 2007). However, there are important differences between these double mutants and bob1 mutants. First, enp pid and pin1 pid double mutants are viable and survive past germination, while bob1 mutants arrest at the globular stage. Second, removal of STM1, CUC1, or CUC2 in pin1 pid mutants can rescue the formation of cotyledons (Furutani et al., 2004), demonstrating that the lack of cotyledons is due to ectopic meristematic gene expression in the apical embryo, while removing STM activity from bob1 embryos does not affect their development. Third, there is some morphological evidence of cotyledon formation in the enp pid double mutants, which form protrusions at the sites of cotyledon formation, whereas there is no morphological indication of cotyledon formation in bob1 mutants. Fourth (and perhaps related to difference number three), the cotyledon marker ANT1 is expressed in pin1 pid and enp pid double mutants (although reduced in extent) but is entirely absent in bob1 mutants. Fifth, in both enp pid and pin1 pid double mutants, reduced auxin maxima are seen in the apical domain of the embryo, and normal auxin maxima are established at the root pole of the embryo, whereas in bob1 mutants, auxin levels appear to be uniform and elevated throughout the embryo. The failure to generate a discrete auxin maximum at the root pole of the embryo may be responsible for the failure of bob mutant embryos to express the SCARECROW transcript.
It is possible that BOB1 acts in the same pathway as the auxin transporters and their regulators and that all five of these differences are differences in severity of the mutant phenotype, with bob1 showing the most extreme mutant phenotype. It should be noted that in all of the mutant combinations affected in auxin transport, there are many gene family members that remain intact and can presumably supply activity, so it is reasonable to assume that a complete deficiency in auxin transport has not yet been observed. However, our data do not rule out that BOB1 may play a role in an as yet described independent pathway that is necessary for apical partitioning and is independent from its effect on auxin localization.
Yet another set of observations involving auxin parallel the observations of the bob1 mutant phenotype. Uniformly high levels of auxin have been observed when Brassica juncea and wheat (Triticum aestivum) embryos were exposed to exogenous auxin. Under these conditions, uniformly high levels of auxin are established throughout the developing embryos, and the embryos fail to develop cotyledons. Like bob1 mutants, the treated embryos never develop past a stage that resembles globular stage (Fischer and Neuhaus, 1996; Hadfi et al., 1998).
As in bob1 mutants, mutants homozygous for the tpl mutation also fail to form cotyledons (Long et al., 2002). However, tpl mutant embryos differ from bob1 mutant embryos and from pin pid and enp pid double mutants in that they fail to express STM and instead express ANT1 across the entire apical dome of the embryo. In this sense, the tpl mutation has the opposite effect from bob1 on the apical development of the embryo. There are other differences as well: tpl mutants survive past germination and are able to localize an auxin maximum at the root pole. TPL encodes a protein that acts as a corepressor (Szemenyei et al., 2008). One of the roles of this corepressor is to repress genes that are regulated by auxin; auxin presumably lifts this repression by degrading the IAA protein. In the absence of TPL function, this repression is defective.
The presence of auxin throughout the bob1 embryo is difficult to reconcile with the inability of bob1 embryos to make cotyledons, as high levels of auxin are associated with cotyledon formation in the apical portion of the embryo in wild-type embryos. It is possible that normal cotyledon development requires the establishment of a differential of auxin concentration rather than depending upon the measurement of a threshold amount of auxin. Alternatively, concentrations of auxin, while generally increased throughout the embryo, may not reach sufficiently high levels in bob1 mutants to trigger cotyledon formation.
BOB1 Encodes a Protein Homologous to A. nidulans NudC
BOB1 encodes a 304–amino acid protein containing an N-terminal domain without homology to any known protein domains and a C-terminal NudC homology domain. The Arabidopsis genome contains a highly conserved BOB1 homolog that is located in a syntenous region, and BOB1 homologs are found throughout the plant kingdom. NudC domain–containing genes are also found in animals and fungi. While the NudC domain is highly conserved across phyla, the N-terminal domain is not conserved between plants and animals and is not present in fungi.
The founding member of this conserved gene family, NudC, was identified in a screen for nuclear distribution mutants in A. nidulans (Osmani et al., 1990). In Aspergillus, spores germinate and undergo several rounds of nuclear division in the absence of cytokinesis. The resulting nuclei are distributed along the fungal hyphae after which cell division occurs. The nudA, nudC, nudF, and nudG mutants fail to move nuclei along the growing hyphae, resulting in short, thick hyphae that have several nuclei at their base (Morris and Enos, 1992; Xiang et al., 1999). NudA and NudG encode the heavy-chain and light-chain subunits of cytoplasmic dynein, respectively (Xiang et al., 1994; Beckwith et al., 1998), and NudF encodes a WD repeat protein homologous to the human Lis1 protein that, when mutated, causes lisencephaly (Xiang et al., 1995). NudF has been shown to interact with dynein complex subunits as well as with tubulin (Hoffmann et al., 2004). NudC and NudF physically interact (Helmstaedt et al., 2008); NudF protein levels are reduced in nudC mutants (Xiang et al., 1995); and NudC and Lis1 interact physically and can be coimmunoprecipitated with dynein (Morris et al., 1998; Aumais et al., 2001). These studies have been interpreted as showing that nuclear migration in Aspergillus is probably mediated by dynein microtubule motors. Similarly, work on human cell lines has implicated human NUDC in dynein assembly and/or function (Dawe et al., 2001; Zhang et al., 2002; Aumais et al., 2003; Zhou et al., 2006). The relevance of these dynein interactions in plants is uncertain since the presence of dynein microtubule motors in higher plants is controversial and the Arabidopsis genome does not contain dynein genes (Lawrence et al., 2001; King, 2002).
The molecular functions of NudC proteins are not well understood; however, NudC genes have been shown to be essential in several organisms. The original Aspergillus nudC mutant is a viable temperature-sensitive allele (Morris, 1976), though a deletion allele is lethal and exhibits morphological and cell wall abnormalities. This suggests that NudC has additional functions within the cell apart from its role in nuclear distribution (Chiu et al., 1997). Disruption of the Caenorhabditis elegans homolog NUD-1 results in embryonic lethality associated with defects in pronuclear migration and spindle rotation in the zygote. Our experiments show that the Arabidopsis NudC homolog BOB1 is also essential and results in embryo arrest when defective.
NudC domains are predicted to have a similar three-dimensional structure as the α-crystallin/HSP20 family of small heat shock proteins (Garcia-Ranea et al., 2002). Based on this observation, Faircloth et al. (2009) recently demonstrated that C. elegans NUD-1 exhibits protein chaperone activity in vitro, preventing the aggregation and reducing the thermal inactivation of model substrates. This chaperone activity is likely to be evolutionarily conserved since the Drosophila melanogaster, C. elegans, and rat NudC genes are all able to rescue the phenotype of Aspergillus nudC mutants (Cunniff et al., 1997; Dawe et al., 2001; Morris et al., 1997). The observations that the original nudC mutation in Aspergillus is temperature sensitive (Osmani et al., 1990), that NudF protein levels depend on NudC activity (Xiang et al., 1995), and that in the absence of NudC activity dynein complexes aggregate (Zhou et al., 2006) all support the idea that NudC proteins may act as chaperones in vivo as well. More recently, Perez et al. (2009) have established that Arabidopsis BOB1 can act as a chaperone in vitro.
If BOB1 functions as a protein chaperone in Arabidopsis, then the phenotypes in bob1 mutants might be explained by a decrease or lack of activity of the substrates of BOB1 chaperone activity. These substrates could include proteins required for polar auxin transport or signaling during embryogenesis but could also include other proteins involved in pattern formation in the early Arabidopsis embryo. In animal systems, chaperones such as HSP90 play an important role in signaling by, among other substrates, steroid hormones (for review, see review Richter and Buchner, 2001). The identification and study of proteins that interact with BOB will be an important area of inquiry in the future and should allow a deeper understanding of the mechanism of BOB action in the embryo and ultimately of embryonic patterning.
METHODS
Plant Growth Conditions
Unless otherwise noted, all plants were grown in soil (Scott's Metro-Mix 200) under constant light at 24°C.
Mutagenesis and Screen
Seeds homozygous for the STM-GUS reporter were mutagenized by soaking in 0.4% ethyl methanesulfonate in deionized water for 8 h at room temperature with agitation and rinsed in deionized water. These M0 seeds were planted in soil and allowed to grow into adult plants. M1 seeds were harvested from individual adults and planted in soil. M2 embryos were dissected from their seed coats using etched tungsten dissecting needles from Fine Science Tools (catalog numbers 10130-05 and 10130-10). GUS expression was assayed by placing the embryos in 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 100 mM NaPO4, pH 7, 5 mM FeCN, 10 mM EDTA, pH 7.6, and 0.1% Triton X-100 solution overnight at 37°C. Staining patterns were visualized with a dissecting microscope and a Nikon Optiphot-2 light microscope.
Clearing of Tissue
Embryos were cleared for visualization using Hoyer's solution (to which 50 mL deionized water, 30 g gum arabic crystals, 200 g chloral hydrate, and 20 g glycerol was added). Embryos, in seed coats, were placed on a glass slide in a drop of Hoyer's solution and covered with a cover slip. Slight pressure was applied to break the seed coat. Embryos were allowed to clear for 30 min to 16 h and visualized with a Nikon Optiphot-2 light microscope.
Mapping and Isolation of the BOB1 Gene
DNA extractions were performed using the method described by Edwards et al. (1991). BOB1 was mapped to a chromosome region using PCR-based CAPS markers (Konieczny and Ausubel, 1993). New CAPS markers were developed for fine mapping (see Supplemental Table 2 online).
Amplification of the genes in the BOB1 region for sequencing was done using ExTaq DNA polymerase from Panvera, and PCR products were cloned using pGEM-T Easy T-vectors from Promega. DNA sequencing was performed by University of Wisconsin Biotechnology Center Sequencing Services. Sequences were assembled and manipulated using DNAStar software. Sequence comparisons were done with ClustalX (Jeanmougin et al., 1998), ReadSeq (copyright 1990 by D.G. Gilbert), and MacBoxshade (developed by Michael D. Baron). Percentage identity values were determined using the BLOSUM62 matrix with BLAST (Altschul et al., 1990) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
Gene Expression
MYN8, a P1 clone containing the BOB1 gene, was obtained from the Kazusa Group, Japan. The wild-type BOB1 coding region (At5g53400) and surrounding sequences were amplified for constructs using ExTaq polymerase. Products were cloned first into pGEM-T Easy vector for manipulation. Binary vectors pCAMBIA 1300 (pREJ14) and pCAMBIA 1291Xb (pREJ19) were obtained from the Center for the Application of Molecular Biology to International Agriculture, Canberra, Australia. Agrobacterium tumefaciens strain GV3101 was used to transform Ler plants.
In situ hybridization was performed as described by Long et al. (1996). The STM probe was transcribed from plasmid MeriHB1, as described by Long et al. (1996). The ANT probe was transcribed from plasmid pANT, as described by Long and Barton (1998). The ATML1 probe was transcribed from plasmid pATML1, made with a 517-bp fragment (base pairs 630 to 1217) amplified from genomic DNA using primers ATML1-1 (5′-GAGAACAATAGGTACAAGG-3′) and ATML1-2 (5′-CTCTCTTGAAGCTTCTGATC-3′), cut with RsaI and HindIII, and ligated into pLitmus 29 (New England Biolabs). The CUC2 probe was transcribed from plasmid pCUC2A, made with a 420-bp (base pairs 1187 to 1563) fragment amplified from genomic DNA using primers CUC2-1 (5′-CACCTCGAGGATCTCTAGGGTTTTCC-3′) and CUC2-2 (5′-CACTCTAGACTAGCGGCGGTGGAAG-3′), cut with XhoI and XbaI (engineered restriction sites), and ligated into pBluescript KS− (Stratagene). The UFO probe was transcribed from plasmid pDW22.1, as described by Lee et al. (1997). The WUS probe was transcribed as described by Mayer et al. (1998). The BOB1 probe was transcribed from plasmid pBOB-AS, made with a 358-bp fragment (base pairs 289 to 647) amplified from the BOB1 cDNA using primers BOBPROBE 5′ (5′-ATATATCTAGACCTGTGGAGAAGAAAGCCGAGAAG-3′) and BOBPROBE 3′ (5′-TATATGAGCTCAGTCCGAAGTGTCTAGCCATCTCG-3′) and ligated into pGEM-T Easy (Promega).
For in situ hybridization experiments on bob1 embryos, siliques were collected from bob1/+ plants, fixed, and sectioned. Because siliques were collected from heterozygous individuals, the bob embryos examined were side-by-side on the slide with their wild-type siblings, providing a positive control for hybridization of probes.
Imaging
Images were captured on a SPOT RT Slider CCD camera (Diagnostic Imaging) and manipulated using Adobe Photoshop software (Adobe Systems).
bob1-1 was genotyped using primers bob1-1_dCAPS_F (5′-TGGGACTAAAGCACGGACTGTTG-3′) and bob1-1_dCAPS_R (5′-AGTAGCAGTCATCAGGCTTCACAGACC-3′) followed by digestion with MspI. The resulting PCR products were 200 bp (wild-type) and 226 bp (bob1-1). bob1-2 was amplified in a reaction containing primers bob1-2_F (5′-GACTTTCTCGGAGAACAGAGC-3′), bob1-2_R (5′-CCTCCTGGAGATTCTGGATCC-3′), and LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′). The resulting PCR products were 630 bp (wild type) and 450 bp (bob1-2).
DR5-GFP Imaging
Plants containing a DR5-GFP transgene (Friml et al., 2003) were crossed to bob1-1/+ plants. The F1 plants were allowed to self-fertilize to generate bob1-1/+ plants homozygous for DR5-GFP. Wild-type and bob1 embryos dissected from the same silique were imaged using a Leica SP5 AOBS confocal microscope (Leica Microsystems).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At BOB1, At5g53400, At BOB2, At4g27890; BOB1 homologs: Os, GI:51535412; Hs, GI:62287138; Dr, GI:46249697; Ce NUD-1 GI:9081901; Sp, GI:3150135; and An NUDC, GI:40743172.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Map-Based Cloning Strategy for the BOBBER1 Locus.
Supplemental Figure 2. Composition of Constructs Used to Rescue the bob1 Mutant Phenotype (A) and to Test Localization of BOBBER1 Promoter Activity (B).
Supplemental Figure 3. Homozygous bobber1-2 Mutant Embryo Showing Ectopic Expression of the STM Reporter.
Supplemental Figure 4. BOBBER mRNA Expression in Wild-Type and bobber Mutant Embryos.
Supplemental Figure 5. Expression of pREJ10, a BOBBER-GUS Fusion Protein, in Wild-Type Embryos.
Supplemental Table 1. Segregation of bobr1 Mutant Embryos in bob1-1/+ and bob1-1/+ stm-11/+ Families.
Supplemental Table 2. CAPS Markers Developed for Mapping the bob1 Mutation.
Supplemental Table 3. Complementation of the bob1 Phenotype by the Wild-Type BOBBER1 Transgene.
Supplemental Table 4. Complementation Data for bob1-1 and bob1-2.
Supplementary Material
Acknowledgments
This work was funded by grants from the National Science Foundation. Matthew Evans and Enrico Magnani provided helpful comments on the manuscript. Nicole Newell provided technical assistance with embryo preparations.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: M. Kathryn Barton (kbarton@stanford.edu).
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Abe, M., Katsumata, H., Komeda, Y., and Takahashi, T. (2003). Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130 635–643. [DOI] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., and Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126 1563–1570. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Aumais, J.P., Tunstead, J.R., McNeil, R.S., Schaar, B.T., McConnell, S.K., Lin, S.H., Clark, G.D., and Yu-Lee, L.Y. (2001). NudC associates with Lis1 and the dynein motor at the leading pole of neurons. J. Neurosci. 21 RC187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumais, J.P., Williams, S.N., Luo, W., Nishino, M., Caldwell, K.A., Caldwell, G.A., Lin, S.H., and Yu-Lee, L.Y. (2003). Role for NudC, a dynein-associated nuclear movement protein, in mitosis and cytokinesis. J. Cell Sci. 116 1991–2003. [DOI] [PubMed] [Google Scholar]
- Barton, M.K., and Poethig, R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 119 823–831. [Google Scholar]
- Beckwith, S.M., Roghi, C.H., Liu, B., and Ronald Morris, N. (1998). The “8-kD” cytoplasmic dynein light chain is required for nuclear migration and for dynein heavy chain localization in Aspergillus nidulans. J. Cell Biol. 143 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M., Letham, S., Craig, S., and Dennis, E.S. (1993). amp1: A mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J. 4 907–916. [Google Scholar]
- Cheng, Y., Qin, G., Dai, X., and Zhao, Y. (2007). NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 104 18825–18829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, Y.H., Xiang, X., Dawe, A.L., and Morris, N.R. (1997). Deletion of nudC, a nuclear migration gene of Aspergillus nidulans, causes morphological and cell wall abnormalities and is lethal. Mol. Biol. Cell 8 1735–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunniff, J., Chiu, Y.H., Morris, N.R., and Warrior, R. (1997). Characterization of DnudC, the Drosophila homolog of an Aspergillus gene that functions in nuclear motility. Mech. Dev. 66 55–68. [DOI] [PubMed] [Google Scholar]
- Dawe, A.L., Caldwell, K.A., Harris, P.M., Morris, N.R., and Caldwell, G.A. (2001). Evolutionarily conserved nuclear migration genes required for early embryonic development in Caenorhabditis elegans. Dev. Genes Evol. 211 434–441. [DOI] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, R.C., Betzer, A.S., Huttner, E., Oakes, M.P., Tucker, W.Q., Gerentes, D., Perez, P., and Smyth, D.R. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faircloth, L., Churchill, P., Caldwell, G., and Caldwell, K. (2009). The microtubule-associated protein, NUD-1, exhibits chaperone activity in vitro. Cell Stress Chaperones 14 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, A.G. (2002). Analyses of SHOOTMERISTEMLESS Function and Development of the Shoot Apical Meristem in Arabidopsis thaliana. PhD dissertation (Madison, WI: University of Wisconsin).
- Fischer, C., and Neuhaus, G. (1996). Influence of auxin on the establishment of bilateral symmetry in monocots. Plant J. 9 659–669. [Google Scholar]
- Freeling, M., Rapaka, L., Lyons, E., Pedersen, B., and Thomas, B.C. (2007). G-boxes, bigfoot genes, and environmental response: characterization of the intragenomic conserved noncoding sequences in Arabidopsis. Plant Cell 19 1441–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jurgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 147–153. [DOI] [PubMed] [Google Scholar]
- Friml, J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865. [DOI] [PubMed] [Google Scholar]
- Furutani, M., Kajiwara, T., Kato, T., Treml, B.S., Stockum, C., Torres-Ruiz, R.A., and Tasaka, M. (2007). The gene MACCHI-BOU 4/ENHANCER OF PINOID encodes a NPH3-like protein and reveals similarities between organogenesis and phototropism at the molecular level. Development 134 3849–3859. [DOI] [PubMed] [Google Scholar]
- Furutani, M., Vernoux, T., Traas, J., Kato, T., Tasaka, M., and Aida, M. (2004). PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131 5021–5030. [DOI] [PubMed] [Google Scholar]
- Garcia-Ranea, J.A., Mirey, G., Camonis, J., and Valencia, A. (2002). p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 529 162–167. [DOI] [PubMed] [Google Scholar]
- Hadfi, K., Speth, V., and Neuhaus, G. (1998). Auxin-induced developmental patterns in Brassica juncea embryos. Development 125 879–887. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A., Chin-Atkins, A.N., Wilson, I.W., Chapple, R., Dennis, E.S., and Chaudhury, A. (2001). The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell 13 2115–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedt, K., Laubinger, K., Vosskuhl, K., Bayram, O., Busch, S., Hoppert, M., Valerius, O., Seiler, S., and Braus, G.H. (2008). The nuclear migration protein NUDF/LIS1 forms a complex with NUDC and BNFA at spindle pole bodies. Eukaryot. Cell 7 1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara, K., Karim, M.R.K., Takada, S., Taoka, K., Furutani, M., Aida, M., and Tasaka, M. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postemebryonic shoot meristem and organ boundary formation. Plant Cell 18 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, B., Zuo, W., Liu, A., and Morris, N.R. (2004). The LIS1-related protein NUDF of Aspergillus nidulans and its interaction partner NUDE bind directly to specific subunits of dynein and dynactin and to alpha- and gamma-tubulin. J. Biol. Chem. 279 820. [PubMed] [Google Scholar]
- Jeanmougin, F., Thompson, J.D., Gouy, M., Higgins, D.G., and Gibson, T.J. (1998). Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23 403–405. [DOI] [PubMed] [Google Scholar]
- Jenik, P.D., and Barton, M.K. (2005). Surge and destroy: The role of auxin in plant embryogenesis. Development 132 3577–3585. [DOI] [PubMed] [Google Scholar]
- Jenik, P.D., Jurkuta, R.E., and Barton, M.K. (2005). Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase epsilon. Plant Cell 17 3362–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.M. (2002). Dyneins motor on in plants. Traffic 3 930–931. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Lawrence, C.J., Morris, N.R., Meagher, R.B., and Dawe, R.K. (2001). Dyneins have run their course in plant lineage. Traffic 2 362–363. [DOI] [PubMed] [Google Scholar]
- Lee, I., Wolfe, D.S., Nilsson, O., and Weigel, D. (1997). A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr. Biol. 7 95–104. [DOI] [PubMed] [Google Scholar]
- Li, Z., and Thomas, T.L. (1998). PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10 383–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., and Barton, M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125 3027–3035. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Woody, S., Poethig, S., Meyerowitz, E.M., and Barton, M.K. (2002). Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development 129 2797–2806. [DOI] [PubMed] [Google Scholar]
- Lu, P., Porat, R., Nadeau, J.A., and O'Neill, S.D. (1996). Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons, E., and Freeling, M. (2008). How to usefully compare plant genes and chromosomes as DNA sequences. Plant J. 53 661–673. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815. [DOI] [PubMed] [Google Scholar]
- McConnell, J., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis thaliana. Development 125 2935–2942. [DOI] [PubMed] [Google Scholar]
- Michniewicz, M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130 1044–1056. [DOI] [PubMed] [Google Scholar]
- Morris, N.R. (1976). A temperature-sensitive mutant of Aspergillus nidulans reversibly blocked in nuclear division. Exp. Cell Res. 98 204–210. [DOI] [PubMed] [Google Scholar]
- Morris, N.R., and Enos, A.P. (1992). Mitotic gold in a mold: Aspergillus genetics and the biology of mitosis. Trends Genet. 8 32–37. [DOI] [PubMed] [Google Scholar]
- Morris, S.M., Albrecht, U., Reiner, O., Eichele, G., and Yu-Lee, L.Y. (1998). The lissencephaly gene product Lis1, a protein involved in neuronal migration, interacts with a nuclear movement protein, NudC. Curr. Biol. 8 603–606. [DOI] [PubMed] [Google Scholar]
- Morris, S.M., Anaya, P., Xiang, X., Morris, N.R., May, G.S., and Yu-Lee, L. (1997). A prolactin-inducible T cell gene product is structurally similar to the Aspergillus nidulans nuclear movement protein NUDC. Mol. Endocrinol. 11 229–236. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Nawy, T., Lukowitz, W., and Bayer, M. (2008). Talk global, act local-patterning the Arabidopsis embryo. Curr. Opin. Plant Biol. 11 28–33. [DOI] [PubMed] [Google Scholar]
- Nodine, M.D., and Tax, F.E. (2008). Two receptor-like kinases required together for the establishment of Arabidopsis cotyledon primordia. Dev. Biol. 314 161–170. [DOI] [PubMed] [Google Scholar]
- Nodine, M.D., Yadegari, R., and Tax, F.E. (2007). RPK1 and TOAD2 are two receptor-like kinases redundantly required for Arabidopsis embryonic pattern formation. Dev. Cell 12 943–956. [DOI] [PubMed] [Google Scholar]
- Osmani, A.H., Osmani, S.A., and Morris, N.R. (1990). The molecular cloning and identification of a gene product specifically required for nuclear movement in Aspergillus nidulans. J. Cell Biol. 111 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, D.E., Hoyer, J.S., Johnson, A.I., Moody, Z.R., Lopez, J., and Kaplinsky, N.J. (July 1, 2009). BOBBER1 is a non-canonical Arabidopsis small heat shock protein required for both development and thermotolerance. Plant Physiol. http://dx.doi.org/. [DOI] [PMC free article] [PubMed]
- Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Richter, K., and Buchner, J. (2001). Hsp90: Chaperoning signal transduction. J. Cell. Physiol. 188 281–290. [DOI] [PubMed] [Google Scholar]
- Sato, S., Nakamura, Y., Kaneko, T., Katoh, T., Asamizu, E., Kotani, H., and Tabata, S. (2000). Structural analysis of Arabidopsis chromosome 5. X. Sequence features of the regions of 3,076,755 bp covered by sixty P1 and TAC clones. DNA Res. 7 31–63. [DOI] [PubMed] [Google Scholar]
- Szemenyei, H., Hannon, M., and Long, J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319 1384–1386. [DOI] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128 1127–1135. [DOI] [PubMed] [Google Scholar]
- Treml, B.S., Winderl, S., Radykewicz, R., Herz, M., Schweizer, G., Hutzler, P., Glawischnig, E., and Ruiz, R.A.T. (2005). The gene ENHANCER OF PINOID controls cotyledon development in the Arabidopsis embryo. Development 132 4063–4074. [DOI] [PubMed] [Google Scholar]
- Vroemen, C.W., Mordhorst, A.P., Albrecht, C., Kwaaitaal, M.A.C.J., and deVries, S.C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, X., Beckwith, S.M., and Morris, N.R. (1994). Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 91 2100–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, X., Osmani, A.H., Osmani, S.A., Xin, M., and Morris, N.R. (1995). NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol. Biol. Cell 6 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, X., Zuo, W., Efimov, V.P., and Morris, N.R. (1999). Isolation of a new set of Aspergillus nidulans mutants defective in nuclear migration. Curr. Genet. 35 626–630. [DOI] [PubMed] [Google Scholar]
- Zhang, M.Y., Huang, N.N., Clawson, G.A., Osmani, S.A., Pan, W., Xin, P., Razzaque, M.S., and Miller, B.A. (2002). Involvement of the fungal nuclear migration gene nudC human homolog in cell proliferation and mitotic spindle formation. Exp. Cell Res. 273 73–84. [DOI] [PubMed] [Google Scholar]
- Zhou, T., Zimmerman, W., Liu, X., and Erikson, R.L. (2006). A mammalian NudC-like protein essential for dynein stability and cell viability. Proc. Natl. Acad. Sci. USA 103 9039–9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.