Abstract
Male germline development in angiosperms produces the pair of sperm cells required for double fertilization. A key regulator of this process in Arabidopsis thaliana is the male germline-specific transcription factor DUO POLLEN1 (DUO1) that coordinates germ cell division and gamete specification. Here, we uncover the role of DUO3, a nuclear protein that has a distinct, but overlapping role with DUO1 in male germline development. DUO3 is a conserved protein in land plants and is related to GON-4, a cell lineage regulator of gonadogenesis in Caenorhabditis elegans. Mutant duo3-1 germ cells either fail to divide or show a delay in division, and we show that, unlike DUO1, DUO3 promotes entry into mitosis independent of the G2/M regulator CYCB1;1. We also show that DUO3 is required for the expression of a subset of germline genes under DUO1 control and that like DUO1, DUO3 is essential for sperm cell specification and fertilization. Furthermore, we demonstrate an essential sporophytic role for DUO3 in cell division and embryo patterning. Our findings demonstrate essential developmental roles for DUO3 in cell cycle progression and cell specification in both gametophytic and sporophytic tissues.
INTRODUCTION
The production of twin functional sperm cells is vital for double fertilization in flowering plant reproduction. Sperm cells are produced by the haploid male gametophytes that develop from unicellular microspores (McCormick, 2004). Development of the microspore involves microtubule-dependent migration of the nucleus to produce a highly polarized microspore. An asymmetric mitotic division then results in two differently sized cells with different fates, and the asymmetry of this division is essential in establishing the germline (Eady et al., 1995). The larger vegetative cell exits the cell cycle and eventually produces the pollen tube, while the smaller germ cell establishes the male germline. During development, the germ cell is engulfed in the vegetative cell cytoplasm and divides once to produce the twin sperm cells that are delivered to the embryo sac by the pollen tube.
Recent microarray analysis of the Arabidopsis thaliana sperm cell transcriptome showed that a large number of genes (∼6000) are expressed in the male germline (Borges et al., 2008). Moreover, the promoters of several germline-specific or enhanced genes have been developed as cell fate markers in Arabidopsis. GENERATIVE-CELL SPECIFIC1 (GCS1) is a germline-specific plasma membrane protein that is essential for fertilization in Arabidopsis (Mori et al., 2006; von Besser et al., 2006). GAMETE EXPRESSED2 (GEX2) is a plasma membrane protein of unknown function that is expressed in the male germline as well as in the female gametophyte (Engel et al., 2005; Alandete-Saez et al., 2008). Another characterized male germline-specific protein in Arabidopsis is the histone H3.3 variant MGH3 (HTR10) (Okada et al., 2005; Ingouff et al., 2007).
Recently, several proteins have been identified that either promote male germline cell cycle progression independent of cell specification (Iwakawa et al., 2006; Nowack et al., 2006; Chen et al., 2008; Kim et al., 2008; Gusti et al., 2009) or have a dual role, promoting both germ cell division and gamete specification (Johnston et al., 2008; Brownfield et al., 2009; Chen et al., 2009). Mutations in either the conserved cyclin-dependent kinase gene CDKA;1 or the gene encoding the F-box protein FBL17 that is responsible for the degradation of CDKA;1 inibitory proteins prevent germ cell division and result in mutant pollen containing a single germ cell rather than twin sperm cells (Iwakawa et al., 2006; Nowack et al., 2006; Kim et al., 2008; Gusti et al., 2009). The lack of CDKA;1 activity results in a delayed S-phase with the germ cell failing to complete S-phase by anther dehiscence. Mutations in the germline-specific R2R3 Myb gene DUO POLLEN1 (DUO1) also result in pollen with a single germ cell (Durbarry et al., 2005; Rotman et al., 2005). DUO1 is required for germ cell cycle progression at G2/M, so that unlike cdka;1 and fbl17 mutant germ cells, duo1 germ cells complete S-phase but fail to enter mitosis. Recently, we have shown that entry of male germ cells into mitosis involves DUO1-dependent expression of the CDKA regulatory subunit CYCB1;1 (Brownfield et al., 2009).
Interestingly, the single germ cell present in mutant cdka;1 and fbl17 pollen is capable of fertilization of the egg cell (Iwakawa et al., 2006; Nowack et al., 2006; Kim et al., 2008; Gusti et al., 2009), and the male germline markers GCS1, MGH3, and GEX2 are expressed in cdka;1 germ cells (Brownfield et al., 2009). Thus, germline cell cycle progression and cell specification can clearly be uncoupled. However, pollen deficient in DUO1 differs from cdka;1 and fbl17 pollen in that the single duo1-1 germ cells fail to express germline markers and do not fertilize (Brownfield et al., 2009). Thus, DUO1 has a dual role in male germline development, promoting germ cell specification and division to form twin functional sperm cells. Current data on these regulatory proteins have enabled the formulation of basic models for the regulation of sperm cell production in flowering plants (Borg et al., 2009; Brownfield et al., 2009). The identification of further male germline regulatory proteins and how these may cooperate with known proteins in the established model for sperm cell production is an important goal in plant reproductive biology.
Here, we identify DUO3 as a key regulatory protein that, like DUO1, links control of male germ cell division and sperm cell specification. We show that DUO3 is conserved throughout the land plants and contains motifs conserved in the GONADLESS-4 (GON-4) protein, a cell lineage regulator of gonadogenesis in Caenorhabditis elegans (Friedman et al., 2000). Similar to duo1 mutants, most male germ cells in duo3-1 pollen complete S-phase but fail to enter mitosis. However, unlike duo1-1 germ cells, duo3-1 germ cells express CYCB1;1. We show that DUO3 is a positive regulator of germ cell fate that, like DUO1, is required for the normal expression of germline markers GCS1 and GEX2. Conversely, DUO3 is not required for MGH3 expression, distinguishing the role of DUO3 in sperm cell specification from that of DUO1. We also generated homozygous duo3-1 embryos that show delayed development and abnormal morphogenesis, indicating a wider role for DUO3 in cell cycle control and patterning. Thus, we show the overlapping, but distinct, roles of DUO3 and DUO1 in male germline development.
RESULTS
DUO3 Is Required for Male Germ Cell Division in Arabidopsis
The duo3-1 mutant was identified in a screen for pollen cell division mutants wherein the duo1-1 and duo2-1 mutants were identified (Durbarry et al., 2005). Approximately 40% of the pollen from heterozygous duo3-1 plants contain a dispersed vegetative nucleus and a single germ cell nucleus (Figures 1A and 1B). Ultrastructural analysis shows that the single germ cell in duo3-1 pollen is surrounded by an intact plasma membrane (Figures 1C and 1D). When heterozygous duo3-1 (+/duo3-1) plants are selfed, the progeny segregate 1:1 for wild-type and +/duo3-1 plants (see Supplemental Table 1 online). Reciprocal crosses with wild-type plants showed that the duo3-1 allele is transmitted normally through the female, but there is no male transmission (see Supplemental Table 1 online). When the duo3-1 allele was introduced into a quartet mutant background, in which the four products of meiosis within a tetrad remain associated (Preuss et al., 1994), there were never more than two mutant members in a tetrad, showing that duo3-1 acts postmeiotically (see Supplemental Table 2 online).
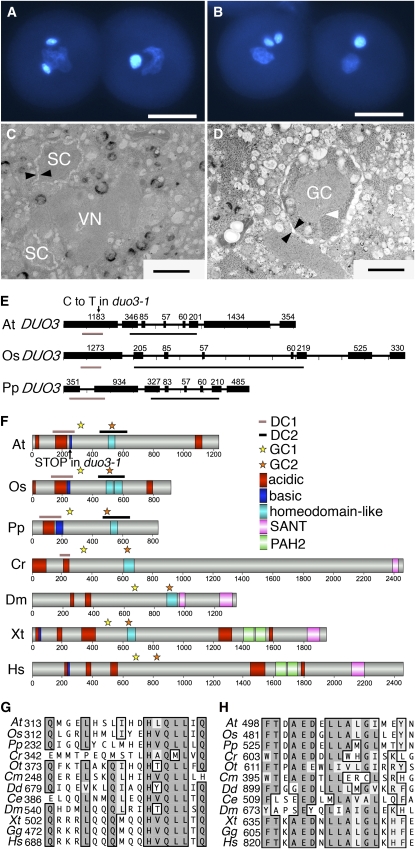
Figure 1.
Phenotype of duo3-1 Pollen and DUO3 Protein Organization.
(A) and (B) Two examples of duo3-1 pollen stained with DAPI. Each shows a wild-type pollen grain on the left, with two brightly stained sperm cells and a diffusely stained vegetative nucleus, and a duo3-1 pollen grain on the right, with a single brightly stained germ cell and a diffuse vegetative nucleus. Bars = 10 μm.
(C) Transmission electron micrograph of a sectioned wild-type pollen grain with two sperm cells (SC) and a vegetative nucleus (VN).
(D) Transmission electron micrograph of a sectioned duo3-1 pollen grain with a single germ cell (GC). Black arrowheads indicate the membranes surrounding the sperm cell (C) or germ cell (D), and the white arrowhead indicates the germ cell nucleus (D). Bars = 1.4 μm.
(E) The genomic structure of the DUO3 gene from Arabidopsis (At DUO3), O. sativa (Os DUO3), and P. patens (Pp DUO3). Exons are represented by black boxes with the size in base pairs of each exon indicated. The site of the duo3-1 mutation is indicated with an arrow, and the regions encoding conserved regions DC1 and DC2 are marked with bars under the gene.
(F) Domains in DUO3 and GON-4L proteins. The conserved regions, DC1 and DC2, are indicated by lines above the protein and the sites of the conserved regions GC1 and GC2 by yellow and orange stars, respectively. Other domains are indicated with colored squares, and the amino acid number is indicated below.
(G) and (H) Alignment of the conserved regions GC1 (G) and GC2 (H) in DUO3 and GON-4L proteins. Conserved residues are boxed, with identical residues colored dark gray and conservative substitutions shaded light gray. Species are as follows: At, Arabidopsis thaliana, Os, Oryza sativa; Pp, Physcomitrella patens; Cr, Chlamydomonus reinhardtii; Ot, Ostreococcus tauri; Cm, Cyanidioschyzon merolae; Dd, Dictyostelium discoideum; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Xt, Xenopus tropicalis; Gg, Gallus gallus; Hs, Homo sapiens.
The DUO3 locus was mapped to a 10-kb region containing two genomic loci, At1g64570 and At1g64580 (see Supplemental Figure 1 online). A genomic fragment containing the promoter and coding region of At1g64570 complemented the bicellular phenotype of duo3-1 pollen (see cDNA complementation in Figure 2C), while a fragment containing At1g64580 did not (data not shown), indicating that At1g64570 is DUO3. The DUO3 gene was sequenced from ecotype No-0 and four independent +/duo3-1 plants. A double chromatogram peak was consistently observed at position 760 nucleotides in the DNA from +/duo3-1 plants, indicating a single base pair substitution (C>T) in the duo3-1 allele. This single base pair substitution results in a codon change from CAG, encoding Gln, to TAG, a stop codon (Figure 1E).
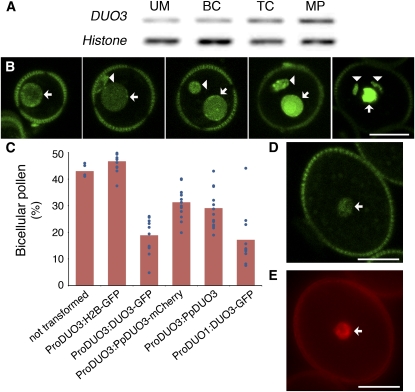
Figure 2.
Analysis of DUO3 Promoter Activity and Functional Complementation of duo3-1.
(A) RT-PCR analysis of DUO3 expression in uninucleate microspores (UM), bicellular (BC), tricellular (TC), and mature (MP) pollen. A Histone H3 gene was used as a control.
(B) Expression of ProDUO3:H2B-GFP during pollen development. Panels (left to right) show five progressive stages: polarized uninucleate microspores and early, mid, and late bicellular and tricellular pollen. GFP is detected in the microspore nucleus (arrow) and in both the vegetative (arrow) and germ cell (arrowhead) nuclei.
(C) Complementation of the germ cell division defect in duo3-1. Pollen was stained with DAPI and the percentage of BC pollen grains determined for untransformed +/duo3-1 plants (n = 5) and for +/duo3-1 plants hemizygous for ProDUO3:H2B-GFP (n = 14 lines), ProDUO3:DUO3-GFP (n = 12 T1 lines), ProDUO3:PpDUO3-mCherry (n = 14 T1 lines), ProDUO3:PpDUO3 (n = 14 T1 lines), or ProDUO1:DUO3-GFP (n = 11 T1 lines). Columns represent the average, with data from individual plants indicated by circles.
(D) and (E) DUO3-GFP (D) and PpDUO3-mCherry (E) fusion proteins are located in the vegetative cell nucleus (arrow).
Bars = 10 μm.
[See online article for color version of this figure.]
The DUO3 cDNA was amplified from floral bud cDNA using primers at the start and stop codons based on The Arabidopsis Information Resource (TAIR) prediction. DUO3 encodes a 1239–amino acid protein with a predicted molecular mass of 137 kD and pI of 4.72. The duo3-1 mutation is predicted to result in a truncated protein of 254 amino acids and is therefore likely to be a null allele (Figure 1F). BLAST searches of the Arabidopsis genome indicate that DUO3 is a unique protein in Arabidopsis. The DUO3 protein has regions rich in acidic or basic amino acids, including an acidic rich region adjacent to a basic region toward the N terminus and an acidic rich region near the C terminus (Figure 1F). DUO3 also contains a homeodomain-like region from amino acids 499 to 550. In other proteins, homeodomain-like regions are involved in binding DNA, and the predicted basic pI (9.13) of this region in DUO3 is consistent with it interacting with DNA.
DUO3 Is Conserved in Land Plants and Shares Features of Gonadless-4-Like Proteins
BLAST searches with DUO3 revealed homologous proteins in a number of flowering plants and in the nonflowering plants Selaginella moellendorffii and Physcomitrella patens (see Supplemental Table 3 online). We obtained the full-length P. patens cDNA and sequenced it. We compared the genomic structure of DUO3 proteins in plants representing dicots (Arabiodpsis, At), monocots (Orzya sativa, Os), and nonflowering plants (P. patens, Pp) (Figure 1E). The coding sequence of both flowering plant sequences begins with a large exon, while this region is interrupted by a single intron in Pp DUO3. The gene structure in the central region of the protein is well conserved but varies at the 3′ end with At DUO3 and Os DUO3 possessing two exons of different lengths and Pp DUO3 a single exon.
In all species, the DUO3 protein is relatively large, although the O. sativa and P. patens proteins (∼100 kD) are smaller than other members (∼130 to 150 kD) (see Supplemental Table 3 online). Interestingly, DUO3 proteins from all the flowering plants and S. moellendorffii are acidic (pI 4.7 to 5.2), while the protein from P. patens has an overall basic pI of 8.76, although it does have local acidic-rich regions. When protein sequences from all the plant homologs were aligned, we identified two regions of high similarity in all plant sequences that we named DUO3 Conserved 1 (DC1; see Supplemental Figure 2 online) and DC2 (see Supplemental Figure 3 online). DC1 is located toward the N terminus of the protein and consists of ∼150 amino acids with ∼30% of amino acids being identical in all proteins. This region contains an acidic-rich region followed by a basic region in all plant proteins (Figure 1F). DC2 is ∼185 amino acids long and is encoded by the central DUO3 gene region that has a conserved genomic structure (Figure 1E). Plant DC2 sequences share ∼50% amino acid identity and at least one predicted homeodomain-like region, although in some species, such as rice, a second homeodomain-like region is also predicted (Figure 1F).
Further BLAST searches using just the DC2 region of DUO3 detected related proteins in the green algae Chlamydomonas reinhardtii and Ostreococcus tauri and the red alga Cyanidioschyzon merolae as well as GON-4L (for Gonadless-4-Like) proteins in animals (see Supplemental Table 4 online). The GON-4 protein in C. elegans is required for normal cell cycle progression in specific lineages during gonadogenesis (Friedman et al., 2000). The GON-4L proteins are relatively large proteins, and like DUO3, they tend to be acidic, especially toward the N terminus (Figure 1F; see Supplemental Table 3 online). There are two short sequences that are highly conserved between DUO3 from plants and GON-4L from animals, which we called GON-4L Conserved 1 (GC1; Figure 1G) and GC2 (Figure 1H). The first of these, GC1, is present shortly after DC1 in the plant proteins, while GC2 occurs within DC2 and includes part of the predicted homeodomain-like region (Figure 1F). DC1 is partially conserved in the C. reinhardtii protein (Figure 1F) but not in O. tauri or any of the animal GON-4L proteins (Figure 1F; see Supplemental Table 4 online). Both the C. reinhardtii protein and the animal GON-4L proteins are distinguished by a predicted SANT or Myb-like domain toward the C-terminal end of the protein that is not present in plant DUO3 proteins (Figure 1F; see Supplemental Table 3 online). These analyses indicate that DUO3 is a conserved regulatory protein with potential for DNA binding that is present throughout the land plants with ancient features conserved in algae and animal proteins.
DUO3 Is Expressed in the Germline and Vegetative Cell in Developing Pollen
RNA was extracted from pollen at various stages of development and DUO3 expression analyzed by RT-PCR (Figure 2A). DUO3 was expressed throughout pollen development from microspores to mature pollen. In microarray studies, DUO3 expression was reliably detected in mature pollen and in isolated sperm cells (Honys and Twell, 2004; Pina et al., 2005; Borges et al., 2008).
We also used the Arabidopsis DUO3 promoter to drive expression of an H2B-green fluorescent protein (GFP) fusion protein, which results in a nuclear GFP signal, enabling germline expression to be distinguished from expression in the surrounding vegetative cell. A relatively weak GFP signal was present in polarized microspores prior to asymmetric division (Figure 2B). In early bicellular pollen, GFP was present in the germ cell nucleus and the vegetative cell nucleus. In late bicellular and tricellular pollen, the GFP fluorescence increased in both the germline and the vegetative cell (indicated by brighter signal and decreased levels of wall autofluorescence; Figure 2B). The vegetative nuclear fluorescence was higher than that in the germline, indicating enhanced expression of DUO3 in the vegetative cell or alternatively increased stability of H2B in the vegetative cell.
We used the cDNA clone of At DUO3 to express a DUO3-GFP fusion protein under the control of the DUO3 promoter. When this construct was introduced into +/duo3-1 plants, the percentage of bicellular pollen decreased compared with nontransformed +/duo3-1 plants or to plants containing the ProDUO3:H2B-GFP construct (Figure 2C). Thus, the DUO3-GFP fusion protein was able to rescue the germ cell division defect in duo3-1. When pollen transformed with ProDUO3:DUO3-GFP was examined by confocal laser scanning microscopy, a relatively weak GFP signal could be detected in the vegetative nucleus of late tricellular and mature pollen (Figure 2D) but not in earlier stages. The detectable signal only in the vegetative cell again indicates higher expression of the DUO3 protein in the vegetative cell than in the sperm cells. The lack of detection of DUO3-GFP in the sperm cells is most likely due to low protein or fluorescence levels, as DUO3 was detected in microarray analysis of isolated Arabidopsis sperm cells (Borges et al., 2008) and the expression of ProDUO3:H2B-GFP was detected in sperm cells. The same DUO3 promoter fragment was used to express the H2B-GFP and DUO3-GFP fusion proteins, yet the intensity of the GFP signal is vastly different (cf. Figure 2B, panel 5, to 2D). This suggests that the coding region of DUO3 may contain posttranscriptional information that regulates translation or protein stability or affects the level of GFP fluorescence.
DUO3 Is Required for Male Germ Cell Cycle Progression
The bicellular phenotype of duo3-1 pollen shows that DUO3 is involved in male germ cell division. Examination of 4',6-diamidino-2-phenylindole (DAPI)-stained mature pollen from +/duo3-1 plants revealed that the proportion of bicellular pollen is ∼40% (Figure 2C, not transformed). As 50% of pollen has the duo3-1 allele, this indicates that ∼20% of duo3-1 pollen complete germ cell division. To further investigate the cell cycle defect in duo3-1 pollen and to determine when these duo3-1 germ cells divide, we examined developing pollen stained with DAPI in wild-type, +/duo3-1, and +/duo1-1 plants. There was no noticeable difference in microspores or pollen from +/duo3-1 plants compared wild-type or +/duo1-1 plants up to mid bicellular pollen stage (data not shown). In late bicellular pollen, the majority of pollen is still bicellular, but a few germ cells had entered mitosis or divided (Figures 3A to 3C). During the next stage, ∼10% of germ cells are in mitosis in pollen from both +/duo1-1 and +/duo3-1 plants, while this number is increased in pollen from wild-type plants.
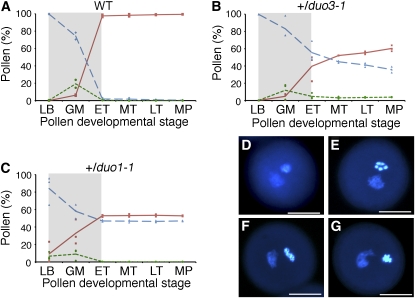
Figure 3.
Abnormal Germ Cell Cycle Progression in duo3-1 Pollen.
(A) to (C) Analysis of pollen development in wild-type (A), +/duo3-1 (B), and +/duo1-1 (C) plants. Pollen from individual flowers was stained with DAPI, and the percentage that was tricellular (continuous line), bicellular (long dashes), or that showed mitotic figures (short dashes) was determined. The stage of development is classified as late bicellular (LB), germ cell mitosis (GM), early (ET), mid (MT), and late (LT) tricellular, and mature pollen (MP). Inflorescences from two wild-type and three +/duo3-1 and +/duo1-1 plants, respectively, were analyzed. Lines and dashes represent the average for each stage, and squares (tricellular), diamonds (bicellular), and circles (mitotic figures) represent individual data points. The approximate developmental window of mitosis is indicated by gray shading.
(D) to (G) DAPI-stained duo3-1 pollen grains showing a single evenly stained mutant germ cell nucleus in BC pollen (D) and three examples of mutant duo3-1 germ cells ([E] to [G]) showing condensed chromosomes. Bars = 10 μm.
[See online article for color version of this figure.]
Corresponding with the mitotic figures, the proportion of tricellular pollen increases, while bicellular pollen decreases as the germ cells divide (Figures 3A to 3C). At early/mid tricellular stage, the proportion of pollen from both +/duo3-1 and +/duo1-1 plants is not significantly different from 50% bicellular and 50% tricellular (χ2 analysis, P < 0.05). As virtually all germ cells in wild-type pollen have divided at the equivalent stage, this indicates that wild-type pollen have divided but the mutant pollen have not. In +/duo3-1 plants, there is an increase in tricellular and a corresponding decrease in bicellular pollen from mid tricellular to mature pollen stages (Figure 3B), such that there is a significant difference in the proportions of tricellular and bicellular pollen at anthesis (χ2 analysis, P < 0.05). This increase in tricellular pollen is not observed in pollen from +/duo1-1 plants (Figure 3C). Thus, it appears that in some duo3-1 pollen, entry into mitosis is delayed rather than completely blocked. This delayed division is observed in ∼20% of the duo3-1 pollen, which are thus tricellular by anthesis with the majority of duo3-1 pollen grains remaining bicellular. A small proportion of pollen from +/duo3-1 plants display germ cell mitotic figures at tricellular and mature pollen stages (Figures 3B and 3D to 3G), but this is not observed in pollen from wild-type or +/duo1-1 plants (Figures 3A and 3C). Since the proportion of tricellular pollen in +/duo3-1 increases through these stages, these mitotic germ cells most likely represent duo3-1 mutant germ cells delayed in entry into mitosis rather than germ cells that have failed to exit mitosis.
In duo1-1 pollen, the G2/M regulator CYCB1;1 is not expressed in mutant germ cells, and germline-specific expression of CYCB1;1 can partially rescue division defects (Brownfield et al., 2009). Therefore, we monitored expression of CYCB1;1 in pollen from +/duo3-1 plants using the pCDGFP vector, containing the CYCB1;1 promoter driving expression of the N terminus of CYCB1;1, which has the mitotic destruction box (CYCB1;1DB) fused to GFP (Figures 4A to 4C). GFP fluorescence was observed in ∼100% of polarized microspores from both wild-type and +/duo3-1 plants and in the vegetative cell soon after asymmetric division (Figure 4A). The fusion protein (CYCB1;1DB-GFP) is rapidly degraded after mitosis as shown by the decrease in the number of pollen with vegetative cell GFP signal by mid bicellular stage (Figure 4A). Prior to germ cell division, GFP is detected specifically in the germ cell in virtually all pollen from both wild-type and +/duo3-1 plants (Figure 4B). Thus, the failure of the majority of duo3-1 germ cells to divide is not due to a failure to express CYCB1;1.
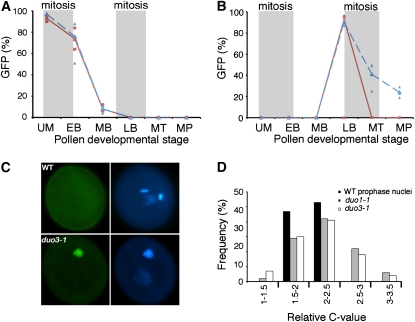
Figure 4.
CYCB1;1 Expression and DNA Content in duo3-1 Germ Cells.
(A) and (B) The frequency of GFP detection in microspores and vegetative cells soon after asymmetric division (A) and in germ cells close to mitosis (B) is similar in wild-type (continuous line) and +/duo3-1 (long dashes) plants transformed with the pCDGFP vector. Stages of pollen development are indicated below each graph, and successive mitoses are indicated by gray shading. Stages are as follows: uninucleate microspores (UM), early (EB), mid (MB), or late (LB) bicellular pollen, mid tricellular pollen (MT), or mature pollen (MP). Analysis was conducted on three wild-type or four +/duo3-1 individuals, with lines and dashes representing the average and data from individual plants plotted as squares (wild type) or triangles (+/duo3-1).
(C) Wild-type (top panels) and duo3-1 (bottom panels) pollen containing the pCDGFP vector at anthesis showing persistent GFP in the undivided duo3-1 germ cell. GFP and DAPI fluorescence is shown in the left and right panels, respectively.
(D) Distribution of the DNA content of wild-type prophase cells (n = 47), undivided duo3-1 germ cells (n = 252), and duo1-1 germ cells (n = 224) measured in a single experiment. The relative C-values were calculated from DAPI fluorescence values normalized to the mean fluorescence of prophase (2C) nuclei.
[See online article for color version of this figure.]
Furthermore, the difference in the population, with ∼20% of duo3-1 germ cells dividing, is not due to differences in CYCB1;1 expression. The parallel accumulation of CYCB1;1DB-GFP in wild-type and duo3-1 pollen indicates that the delay in germ cell division in duo3-1 pollen grains is not related to a delay in S-phase and entry into G2. Rather, it appears that entry into mitosis is delayed and that some duo3-1 germ cells remain in G2 until anthesis. Consistent with this, CYCB1;1DB-GFP persists in ∼70% of the duo3-1 pollen (∼35% of total pollen from heterozygous duo3-1 plants) at tricellular pollen stage, when all CYCB1;1DB-GFP has been degraded in wild-type pollen (Figures 4B and 4C). The proportion of duo3-1 germ cells with persistent CYCB1;1DB-GFP decreases to ∼40% in mature pollen. As ∼20% of duo3-1 germ cells go on to divide and no longer express CYCB1;1DB-GFP, this means that ∼50% of the undivided duo3-1 germ cells contain CYCB1;1DB-GFP at anthesis, while the remaining ∼50% of germ cells degrade CYCB1;1 despite the germ cell failing to divide.
After germ cell mitosis, wild-type sperm cells enter S-phase (Friedman, 1999) and mutant duo1 germ cells also reenter S-phase even though mitosis is blocked (Durbarry et al., 2005; Rotman et al., 2005). To determine if a similar process is occurring in duo3-1 germ cells, we determined the DNA content of single germ cells in mature pollen by measuring DAPI fluorescence. The level of DAPI fluorescence was also measured in germ cell nuclei in prophase to determine the level of 2C fluorescence and in duo1-1 germ cells as a control. The average signal in both duo1-1 and duo3-1 germ cell nuclei was significantly higher than in prophase nuclei (analysis of variance; P = 0.0019). Consistent with previous results (Durbarry et al., 2005), the average DNA content of duo1-1 germ cell nuclei was >2C (2.24C). The average DNA content in single duo3-1 germ cell nuclei was 2.17C, indicating that, like duo1-1 germ cells, duo3-1 germ cells reenter S-phase.
We then investigated the range of DNA content in the mutant germ cells and found that the range of relative DNA contents based on DAPI fluorescence was greater among the undivided duo3-1 germ cell nuclei than among wild-type prophase nuclei (Figure 4D). As all prophase nuclei should have the same DNA content (2C), the range of wild-type values reflects technical variation in the measurement of fluorescence. The greater variation in the duo3-1 germ cells thus likely reflects variation in the amount of DNA in different duo3-1 germ cells. Many duo3-1 germ cells have values close to 2C (the prophase nuclei), while others have values higher than those seen in the prophase nuclei, suggesting that only a subpopulation of duo3-1 germ cells have reentered S-phase. A similar pattern is observed in duo1-1 germ cell nuclei, suggesting that only a portion of mutant duo1-1 germ cells also reenter S-phase (Figure 4D).
As CYCB1;1 inhibits exit from mitosis and entry into S-phase (Weingartner et al., 2004), it is likely that only the population of undivided duo3-1 germ cells that degrade CYCB1;1 reenter S-phase. To confirm this, we measured the DNA content at anthesis in undivided duo3-1 germ cells containing the pCDGFP marker. Those germ cells with GFP, and thus containing CYCB1;1DB-GFP, had an average DNA content close to that of wild-type prophase cells (2.04C, n = 62), indicating that they had not reentered S-phase. Undivided duo3-1 germ cells without GFP, and thus with degraded CYCB1;1DB-GFP, had a higher average DNA content of 3.24C (n = 29), confirming that those duo3-1 germ cells that have degraded CYCB1;1 have reentered S-phase, while those with persistent CYCB1;1 have not.
DUO3 Is Required for Sperm Cell Specification
As ∼20% of duo3-1 pollen grains are tricellular at dehiscence, and thus could be involved in double fertilization, cell cycle defects alone do not account for the complete lack of male transmission of duo3-1 (see Supplemental Table 1 online). This could result from a defect in pollen tube growth or guidance and subsequent failure to deliver the germ cells to the embryo sac. To determine if vegetative cell development is affected in duo3-1 pollen, we used the vegetative cell-specific LAT52 promoter (Twell et al., 1989) to drive expression of an H2B-mRFP1 fusion protein in +/duo3-1 plants. All tricellular pollen and undivided duo3-1 bicellular pollen had a strong red fluorescent protein (RFP) signal, suggesting vegetative cell fate had been correctly specified (Figure 5A). Furthermore, pollen tube germination in vitro occurred at a similar rate in duo3-1 and wild-type pollen (see Supplemental Table 5 online).
Figure 5.
DUO3 Is Required for Male Germ Cell Specification.
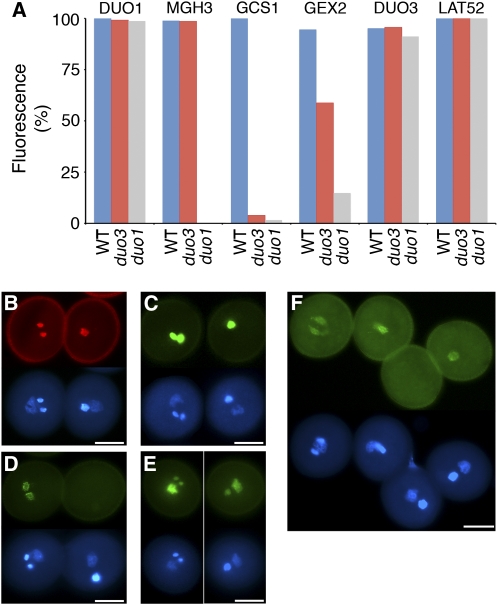
(A) The percentage pollen showing GFP or RFP expression in sperm cells of wild-type pollen or in the single germ cell of duo3-1 and duo1-1 mutant pollen is shown for plants homozygous for various cell fate markers. One to three individual plants were examined, with n denoting the total number of pollen grains examined from +/duo3-1 plants: ProDUO1:H2B-mRFP (DUO1; n = 566), ProMGH3:H2B-GFP (MGH3; n = 1302), ProGCS1:GSC1-GFP (GCS1; n = 589), ProGEX2:GFP (GEX2; n = 764), ProDUO3:H2B-GFP, (DUO3; n = 487) and ProLAT52:H2B-mRFP (LAT52; n = 735). Data for duo1-1 germ cells (Brownfield et al., 2009) are included for comparison.
(B) to (F) Examples of pollen grains from different +/duo3-1 plants homozygous for individual cell fate markers, viewed by fluorescence microscopy. ProDUO1:H2B-mRFP (B), ProMGH3:H2B-GFP (C), ProGCS1:GCS1-GFP (D), ProDUO3:H2B-GFP (E), and ProGEX2:GFP (F). Images in (B) to (E) show wild-type pollen to the left and duo3-1 pollen to the right, while (F) has two wild-type pollen grains to the left and two duo3-1 pollen grains to the right (see bottom DAPI images). Bars = 10 μm.
To determine if duo3 pollen tubes compete with wild-type pollen tubes in reaching the ovules, we examined siliques from selfed +/duo3-1 plants for the presence of undeveloped seeds. As only a single pollen tube enters each ovule, the presence of undeveloped seeds can be due to the arrival of a pollen tube containing germ cells that fail to fertilize or result in embryo abortion. Thus, if duo3 pollen tubes compete with wild-type pollen tubes, failure of fertilization will reduce the number of seeds in siliques fertilized with pollen from +/duo3-1 plants. Siliques from selfed wild-type plants contained an average of 54 ± 6 green seeds (n = 1252), while selfed +/duo3-1 plants had significantly fewer seeds per silique at 40 ± 7 (n = 919) (t test, P < 0.01). There was also a significant increase in the number of undeveloped seeds in siliques from +/duo3-1 plants (t test, P < 0.01) with 22 ± 5 (n = 507) compared with 5 ± 4 (n = 122) in wild-type siliques, indicating that some duo3-1 pollen tubes are successfully guided to ovules. If duo3-1 and wild-type pollen tubes are guided equally to embryo sacs in selfed +/duo3-1 plants, it is expected that there would be close to 50% undeveloped seeds. This however is not observed (χ2 analysis, P < 0.05), with only ∼35% of duo3-1 pollen tubes resulting in undeveloped seeds, indicating a partial role for DUO3 in pollen tube growth and/or guidance.
Microscopy analyses of siliques containing dermatogen stage embryos, which have 16 cells following the first tangential cell divisions, showed that the undeveloped seeds were unfertilized ovules without any embryo development, indicating lack of fertilization, rather than aborted embryos resulting from postfertilization defects. The single germ cells in cdka and fbl17 mutant pollen are capable of fertilization and form early embryos before aborting, showing that single germ cells can fertilize (Iwakawa et al., 2006; Nowack et al., 2006; Kim et al., 2008). Thus, the unfertilized ovules observed in selfed +/duo3 plants indicate that duo3 germ cells are not competent to fertilize.
We then investigated if, like DUO1, DUO3 is also required for sperm cell specification using a range of fluorescent marker constructs. We first investigated if DUO1 is expressed in duo3-1 germ cells using the DUO1 promoter to drive expression of an H2B-RFP fusion protein in +/duo3-1 plants. Fluorescence was detected in nearly all sperm cells in tricellular pollen and in single germ cells in mutant bicellular pollen, indicating that DUO3 is not required for the expression of DUO1 (Figures 5A and 5B). We then introduced three other DUO1-dependent germline markers, ProMGH3:H2B-GFP, ProGCS1:GCS1-GFP, and ProGEX2:GFP, into +/duo3-1 plants. Bright fluorescence from the ProMGH3:H2B-GFP marker was observed in both mutant and wild-type pollen (Figures 5A and 5C), while fluorescence from ProGCS1:GCS1-GFP was absent or very weak in the majority of duo3-1 germ cells (Figures 5A and 5D). The expression of ProGEX2:GFP was reduced in duo3-1 germ cells, with ∼40% of duo3-1 mutant germ cells showing no fluorescence (Figure 5A) and the level of fluorescence in the remaining 60% being highly variable (Figure 5F). Thus, DUO3 is required for the normal germline expression of GCS1 and GEX2, but unlike DUO1, it is not required for the expression of MGH3. We also introduced the ProDUO3:H2B-GFP construct into +/duo3-1 plants to determine if DUO3 is required for its own expression. GFP was detected in both sperm cells and in mutant duo3-1 germ cells, indicating that DUO3 is not essential for its own expression (Figures 5A and 5E). The ProDUO3:H2B-GFP construct was also introduced into +/duo1-1 plants, and GFP was again detected in both wild-type sperm cells and mutant germ cells, showing that DUO1 is not required for the expression of DUO3 (Figure 5A).
Pp DUO3 Can Restore Germ Cell Division and Specification in duo3-1 Pollen
To analyze the functional conservation of DUO3 proteins across land plants, we expressed the moss cDNA, Pp DUO3, in +/duo3-1 plants under the control of the Arabidopsis DUO3 promoter, either alone or as a Pp DUO3-mCherry fusion protein. In pollen from +/duo3-1 plants transformed with either ProDUO3:PpDUO3 or ProDUO3:PpDUO3-mCherry, the percentage of bicellular pollen was reduced compared with untransformed plants or the ProDUO3:H2B-GFP control but was higher than in plants transformed with the DUO3-GFP fusion protein (Figure 2C). This indicates that Pp DUO3 can partially complement the bicellular phenotype in duo3-1 pollen. Similar to the DUO3-GFP fusion protein, Pp DUO3-mCherry could only be detected in the nucleus of the vegetative cell in late tricellular and mature pollen (Figure 2E).
To determine if the presence of Pp DUO3 can restore sperm cell function as well as cell division, we examined if duo3-1 pollen containing Pp DUO3 could transmit the duo3-1 allele. Pollen from three individual T1 plants transformed with ProDUO3:PpDUO3-mCherry and with ∼50% of pollen with fluorescence (indicating a single locus insertion) was crossed to the wild type. Transmission of duo3-1 was observed in each case (Table 1), indicating that Pp DUO3 has restored sperm cell function. The level of transmission was, however, variable, and in two of the three lines, the transmission efficiency was less than the 2:1 ratio expected if Pp DUO3 fully restored transmission (Table 1). This partial restoration of sperm cell function is consistent with the partial rescue of germ cell division by Pp DUO3 (Figure 2C).
Table 1.
Transmission of the duo3-1 Allele through Pollen of Plants in Which the Bicellular Phenotype Has Been Complemented
| Genotypes of Progeny
|
||||
|---|---|---|---|---|
| Male Parent | Wild Type | +/duo3-1 | % (+/duo3-1) | χ2 |
| +/duo3-1; −/− | 447 | 0 | 0 | na |
| +/duo3-1; -/ProDUO3:PpDUO3-mCherry | 33 | 5 | 13 | 4.669* |
| +/duo3-1; -/ProDUO3:PpDUO3-mCherry | 33 | 7 | 18 | 2.984 |
| +/duo3-1; -/ProDUO3:PpDUO3-mCherry | 39 | 1 | 3 | 11.375* |
| +/duo3-1; -/ProDUO1:DUO3-GFP | 25 | 14 | 36 | 0.078 |
| +/duo3-1; -/ProDUO1:DUO3-GFP | 15 | 9 | 38 | 0.125 |
| +/duo3-1; -/ProDUO1:DUO3-GFP | 17 | 11 | 39 | 0.311 |
Pollen from +/duo3-1 plants either not transformed or hemizygous for ProDUO3:PpDUO3-mCherry or ProDUO1:DUO3-GFP was crossed onto a wild-type female. The genotype of progeny was deduced by observing the phenotype of DAPI-stained pollen. A χ2 test was then applied to determine if there was a significant difference from the 2:1 ratio of wild-type to +/duo3-1 plants expected if the complemented pollen competes equally with wild-type pollen. The asterisk indicates a significant difference at P < 0.05. Data are shown for three independent complementing lines for each construct, each with a single locus insertion. na, not applicable.
DUO3 Is Expressed in the Sporophyte and Is Required for Embryo Development
RT-PCR analysis indicated that DUO3 is expressed widely in sporophytic tissues (Figure 6A). To determine the precise locations of DUO3 expression, we used the DUO3 promoter to drive expression of a GFP-β-glucuronidase (GUS) fusion protein and stained various tissues for GUS activity. In seedlings, the DUO3 promoter was active in the vascular tissue of cotyledons, leaves, and roots and in the shoot apical meristem (Figures 6B and 6C). In roots, DUO3 is expressed in a patchy pattern, appearing in short files of cells near the root tip (Figure 6D), in the pericycle, and at sites of lateral root initiation (Figure 6E). Thus, DUO3 expression is enhanced in regions of cell division activity and potential, including meristems and vascular tissues and in nondiving cells, such as pollen vegetative cells.
Figure 6.
DUO3 Is Expressed in Various Sporophytic Cell Types and Is Essential for Embryo Patterning.
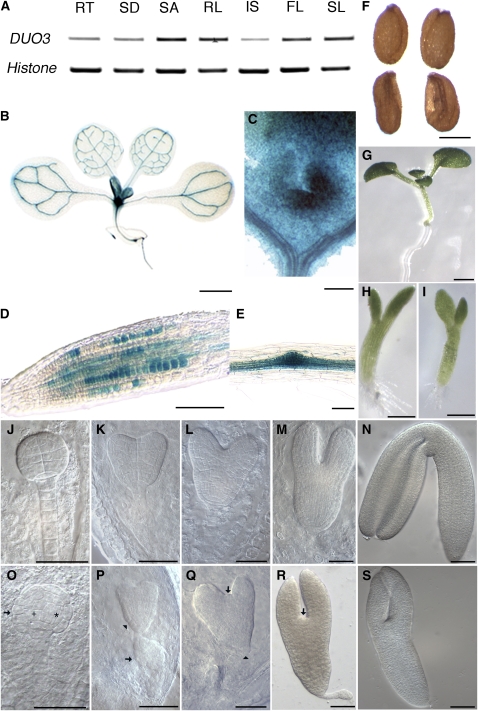
(A) RT-PCR analysis of DUO3 expression in roots (RT), seedlings (SD), shoot apex (SA), rosette leaves (RL), flowers (FL), inflorescence stems (IS), and siliques (SL). Histone H3 was used as a control.
(B) to (E) GUS-stained material from plants transformed with the ProDUO3:GFP-GUS construct.
(B) Whole seedling.
(C) Shoot apical meristem.
(D) Root tip.
(E) Lateral root primordia.
(F) Wild-type seeds (top) and aberrant seeds (bottom) from selfed +/duo3-1 plants hemizygous for ProDUO1:DUO3-GFP.
(G) to (I) Seedlings after 10 d of growth on agar from wild-type (G) or +/duo3-1 ([H] and [I]) plants hemizygous for ProDUO1:DUO3-GFP.
(J) to (S) Embyros from wild-type ([J] to [N]) or +/duo3-1 ([O] to [S]) plants hemizygous for ProDUO1:DUO3-GFP. Embryos are shown at globular ([J] and [O]), early heart ([K] and [P]), late heart ([L] and [Q]), torpedo ([M] and [R]), and bent cotyledon ([N] and [S]) stages of development. Asterisk indicates long cells, plus sign indicates square cells, an arrow indicates radially expanded epidermal cells (O), a swollen suspensor (P), or the prospective site of the shoot meristem ([Q] and [R]), and an arrowhead indicates an uneven epidermis (P) or the prospective site of the root meristem (Q).
Bars = 1 mm in (B) and (G), 50 μm in (C) to (E) and (J) to (S), and 250 μm in (F), (H), and (I).
The study of homozygous mutants can reveal sporophytic functions of gene products. To overcome the lack of male transmission, preventing isolation of duo3-1 homozygotes, we expressed the DUO3∷GFP fusion protein in +/duo3-1 plants under the control of the male germline-specific DUO1 promoter (Rotman et al., 2005). Similar to the ProDUO3:DUO3-GFP construct, no sperm cell GFP was detected in plants transformed with ProDUO1:DUO3-GFP (data not shown), even though fluorescence is detected when the DUO1 promoter is used to express a H2B-RFP fusion protein (Rotman et al., 2005). However, complementation of the bicellular phenotype was observed, with a reduction in the proportion of bicellular pollen similar to that observed when the DUO3 promoter was used (Figure 2C). Thus, germline-specific expression of DUO3 is sufficient to promote germ cell division.
To determine if the complemented pollen was functional, pollen from three lines with a single insertion was used in a cross with wild-type plants. The duo3-1 phenotype was observed in approximately one in three of the offspring, demonstrating full transmission of the duo3-1 allele through the male (Table 1). This indicates that vegetative cell expression of DUO3 is not required for competitive pollen tube growth and guidance.
As the DUO1 promoter is active only in the male germline, the transmission of the duo3-1 allele by mutant pollen containing ProDUO1:DUO3-GFP means that plants homozygous for the duo3-1 allele should arise when complemented +/duo3-1 plants are allowed to self. When such self-seed was examined, we observed an increase in the number of abnormal seeds and seedlings from +/duo3-1 plants compared with wild-type plants (Table 2). There was an average of 12.2% aberrantly shaped seeds from three independent +/duo3-1 plants hemizygous for ProDUO1:DUO3-GFP (Table 2). These seeds were of a similar color and length to wild-type seeds but had shrunken sides (Figure 6F). Seeds were incubated on agar plates and an average of 9.6% of seedlings from +/duo3-1 parents hemizygous for ProDUO1:DUO3-GFP displayed an aberrant phenotype (Table 2). Seedlings had small, thick cotyledons that were commonly unequal in size and failed to expand after germination, the hypocotyls were often fat, and there was limited root growth (Figures 6G to 6I). After 10 d, aberrant seedlings had not initiated leaf development when wild-type plants carrying the construct had two true leaves.
Table 2.
Aberrant Seeds and Seedlings from Selfed +/duo3-1 Plants Hemizygous for ProDUO1:DUO3-GFP or ProDUO3:DUO3-GFP
| Parent Genotype | % Aberrant Seeds (n) | % Aberrant Seedlings (n) |
|---|---|---|
| +/+;-/ProDUO1:DUO3-GFP | 2.0 (152) | 1.6 (191) |
| +/+;-/ProDUO1:DUO3-GFP | 0.7 (140) | 1.1 (185) |
| +/duo3-1;-/ProDUO1:DUO3-GFP | 16.6 (193) | 9.5 (95) |
| +/duo3-1;-/ProDUO1:DUO3-GFP | 8.1 (183) | 11.9 (176) |
| +/duo3-1;-/ProDUO1:DUO3-GFP | 12.0 (167) | 7.5 (160) |
| +/+;-/ProDUO3:DUO3-GFP | 1.4 (346) | 1.7 (181) |
| +/duo3-1;-/ProDUO3:DUO3-GFP | 1.4 (324) | 1.8 (168) |
| duo3-1/duo3-1;+/ProDUO3:DUO3-GFP | 1.7 (477) | 1.1 (176) |
The number of aberrant seeds and seedlings was determined for selfed wild-type plants (two independent lines) or +/duo3-1 plants (three independent lines) hemizygous for ProDUO1:DUO3-GFP and for wild-type, +/duo3-1, and duo3-1/duo3-1 plants hemizygous or homozygous for ProDUO3:DUO3-GFP. Data show the percentage of aberrant seeds or seedlings, with the numbers counted indicated in parentheses.
We also sequenced genomic DNA from aberrant seedlings from three independent lines containing ProDUO1:DUO3-GFP, which confirmed the seedlings were homozygous for the duo3-1 allele. We observed only 1 to 2% of abnormal seeds and seedlings from selfed +/duo3-1 and wild-type plants when hemizygous for ProDUO3:DUO3-GFP. Furthermore, plants homozygous for duo3-1 (confirmed by sequencing genomic DNA) and ProDUO3:DUO3-GFP were generated, and these plants also produced only a low number of aberrant seeds and seedlings (Table 2) and the sporophytic phenotype of these plants were not noticeably different from wild-type plants. Therefore, the presence of DUO3-GFP fully rescues the seed and seedling phenotypes, ruling out the possibility that a mutation linked the duo3-1 allele is responsible for these phenotypes.
The morphology of embryos from two selfed +/duo3-1 and two selfed wild-type plants containing ProDUO1:DUO3-GFP from each of three independent lines was examined. The data for all six +/duo3-1 and wild-type plants are compiled in Table 3. At early stages of development, embryos from +/duo3-1 plants hemizygous for ProDUO1:DUO3-GFP appeared similar to those from wild-type parents. At the heart stage, a few of the embryos from the +/duo3-1 parents were delayed at the preglobular stage. This delay was even more apparent at the torpedo and bent torpedo stages, with some embryos from +/duo3-1 plants still being at globular and heart stages. Interestingly, probable duo3-1/duo3-1 embryos are delayed but not arrested since the majority have progressed to heart stage when the majority of embryos have reached bent torpedo stage. Given full transmission of the duo3-1 allele in pollen containing ProDUO1:DUO3-GFP, it is expected that one embryo in every six will be duo3-1/duo3-1, which is close to the proportion of seeds that show a delay in embryo development.
Table 3.
Embyro Development in Selfed +/duo3-1 Plants Hemizygous for ProDUO1:DUO3-GFP
| Silique Stage
|
||||||
|---|---|---|---|---|---|---|
| Parental Genotpye | Embryo Developmental Stage | Preglobular | Globular | Heart | Torpedo | Bent Torpedo |
| Wild type | Preglobular | 164 | 37 | 1 | 0 | 0 |
| Globular | 30 | 171 | 51 | 0 | 0 | |
| Heart | 0 | 23 | 450 | 37 | 0 | |
| Torpedo | 0 | 0 | 43 | 301 | 28 | |
| Bent torpedo | 0 | 0 | 0 | 23 | 401 | |
| +/duo3-1 | Preglobular | 184 | 36 | 14 | 0 | 1 |
| Globular | 16 | 218 | 92 | 22 | 10 | |
| Heart | 0 | 20 | 343 | 68 | 45 | |
| Torpedo | 0 | 0 | 30 | 213 | 40 | |
| Bent torpedo | 0 | 0 | 0 | 2 | 358 | |
Seeds from siliques of wild-type and +/duo3-1 plants hemizygous for ProDUO1:DUO3-GFP were cleared and the developmental stage of each embryo determined. The stage of the silique was determined based on the morphology of the majority of the embyros. Embryos from two inflorescences from three independent lines were analyzed for both wild-type and +/duo3-1 plants and the data combined. Wild-type embryos progress from preglobular to bent torpedo stage (italics), while some embryos from +/duo3-1 plants are delayed in development (bold).
Not only is there delayed development in embryos from +/duo3-1 plants hemizygous for ProDUO1:DUO3-GFP, there are also abnormal embryos (Figures 6J to 6S). Wild-type embryos follow a defined pattern of cell divisions to produce symmetrical, globular embryos (Figure 6J). Abnormal embryos are not symmetrical with differences in the number and shape of cells in different regions (Figure 6O). Some embryos show delayed longitudinal divisions in central and lower tier cells in one-half of the embryo to produce an embryo with long, thin cells on one side (Figure 6O, asterisk) and square cells on the other (Figure 6O, plus sign). The epidermal layer is also disrupted at globular stage, often with radially expanded cells that give the embryo an uneven surface (Figure 6O, arrow). Wild-type embryos produce symmetrical embryos with a clearly visible epidermis and regular files of cells through heart and torpedo stages (Figures 6K to 6M). In the delayed embryos, the symmetry is often lost, with developing cotyledons being unequal in size from early heart to torpedo stage (Figures 6P to 6R). Also, the epidermal layer is not clearly defined and appears uneven (Figure 6P, arrowhead), and embryos often have disordered and indistinct cell files. In some mutant embryos, the suspensor cells are swollen and sometimes undergo additional cell divisions to form embryo-like outgrowths (Figure 6P, arrow). The region between the cotyledon initials where the shoot apical meristem forms is also disturbed, being wider and flatter than in wild-type embryos (Figures 6Q and 6R, arrows). A defect in the shoot apical meristem is consistent with the lack of true leaves in the germinated seedlings (Figures 6H and 6I). At the bent torpedo stage, the potential duo3-1 mutant embryos are delayed in development, showing reduced elongation along the embryonic axis and irregular cotyledon morphology (Figures 6N and 6S). Thus, a loss of DUO3 leads to a delay in embryo growth and developmental abnormalities, consistent with a delay in cell division, similar to the cell cycle defects observed in the male germline.
DISCUSSION
We identified the Arabidopsis DUO3 gene as a key regulatory factor in plant development. DUO3 is essential for the production of twin functional sperm cells and for normal embryo development. Here, we focused on the well-defined cell lineages in haploid male gametophyte development to explore the dual roles of DUO3 in cell cycle progression and cell specification.
DUO3 Is Required in the Male Germline
Although DUO3 is first expressed in polarized microspores, asymmetric division is not affected in duo3-1 mutant pollen, suggesting that DUO3 is not required for microspore division. After asymmetric division, DUO3 is expressed in both the vegetative cell and the germ cell. Even though DUO3 is expressed in both cell types, it is necessary only in the germ cell, as shown by the complementation of duo3-1 when DUO3 is expressed under control of the male germline-specific DUO1 promoter. These germline-complemented pollen grains compete equally with wild-type pollen grains, even though DUO3 is absent from the vegetative cell. Furthermore, it appears that only a small amount of the DUO3 protein is required for function in the germline due to the low fluorescence levels of both H2B-GFP and DUO3-GFP fusion proteins under the control of the DUO3 promoter.
A DUO3-GFP fusion protein under the control of the DUO3 promoter was detectable only in the nucleus of the vegetative cell. This differs from the DUO3-driven H2B-GFP fusion protein that is detected in both the vegetative and germ cells. A similar situation was observed in sperm cells using the DUO1 promoter, with the ProDUO1:H2B-mRFP fusion being visible in sperm cells (Figure 4B; Rotman et al., 2005) but not the functional ProDUO1:DUO3-GFP fusion. It is possible that DUO3 interferes with the level of GFP fluorescence, although the GFP signal is low with either N-terminal or C-terminal fusions to GFP (data not shown). The difference in the level of GFP fluorescence could also be due to translational repression by the coding region of DUO3 or relative instability of the DUO3-GFP protein. The related YY1AP protein in humans (see below) is also proposed to be unstable (Wang et al., 2004).
DUO3 Regulates the Entry of Germ Cells into Mitosis
The bicellular phenotype of duo3-1 pollen shows that DUO3 has a role in germ cell division. Several other proteins have also been shown to be required for male germ cell division; however, the role of DUO3 is distinct. Mutations in CDKA1 and the F-BOX protein FBL17 both result in delayed S-phase (Nowack et al., 2006; Kim et al., 2008). In duo3-1 mutant germ cells, at least one round of S-phase is completed, as mature germ cells have a DNA content of 2C or more, similar to duo1-1. The timing of S-phase does not appear to be affected in duo3-1 germ cells, as CYCB1;1 expression, which initiates in early G2 and increases strongly at the G2/M transition (Menges and Murray, 2002; Menges et al., 2005), is expressed similarly in duo3-1 and wild-type germ cells. This indicates that DUO3 is unlikely to regulate CDKA or other proteins, such as CYCD family members required for G1/S transition or CYCA family members expressed during S-phase (reviewed in De Veylder et al., 2007).
Initially, duo3-1 and duo1-1 germ cells show similar division defects but subsequently display distinct differences. In contrast with duo1-1 pollen that all remain bicellular at anthesis, ∼20% of duo3-1 germ cells divide to produce tricellular pollen, although the division is delayed in comparison to wild-type germ cells. Mutant duo3-1 and duo1-1 germ cells (Brownfield et al., 2009) also differ in their expression of CYCB1;1, with expression only in duo3-1 germ cells. This is consistent with the ability of some duo3-1 germ cells, but not duo1-1 germ cells, to divide. However, failure of the majority of duo3-1 germ cells to divide indicates a deficiency in other G2/M regulatory factors, which could include other CYCB family members that are also expressed in pollen (Honys and Twell, 2004) or their substrates. The persistence of CYCB1;1DB-GFP in some duo3-1 germ cells at anthesis is consistent with a failure of these germ cells to complete mitosis or progress beyond anaphase when CYCB1;1 is normally degraded.
The duo3-1 germ cells that have not divided by anthesis show two distinct phenotypes, as indicated by the broad range of DNA content in these cells compared with prophase nuclei. Some mutant germ cells have persistent CYCB1;1DB-GFP and a 2C DNA content at anthesis consistent with a failure of these germ cells to enter mitosis or progress beyond anaphase when CYCB1;1 is normally degraded upon activation of the anaphase promoting complex (APC; reviewed in Harper et al., 2002). Persistent CYCB1;1DB-GFP in duo3-1 germ cells could therefore result from failure to enter mitosis and thus to activate APC. Alternatively, DUO3 could have a role in controlling the expression of APC activator proteins (Fülöp et al., 2005). Other undivided duo3-1 germ cells degrade CYCB1;1 and, despite not progressing through mitosis, reenter S-phase as they have a DNA content in excess of 2C. Mutant duo1-1 germ cells also displayed a range of DNA content values, suggesting that there may also be a population of cells that enter S-phase and a population that does not, despite the lack of CYCB1;1 in all duo1-1 germ cells.
Overall, our findings indicate that DUO3 may be required for the coordinated expression of cell cycle regulators or their substrates. Interestingly, the related GON-4 protein in C. elegans is also required for coordinated cell cycle progression in Z1 and Z4 cell lineages during gonadogenesis (Friedman et al., 2000).
DUO3 Has a Role in Sperm Cell Specification
Analysis of germline markers shows that DUO3 is also required for sperm cell specification. Since male germ cell cycle progression and germ cell specification can be uncoupled in Arabidopsis (Iwakawa et al., 2006; Nowack et al., 2006; Kim et al., 2008) and male germline genes are expressed prior to germ cell division (Brownfield et al., 2009), the failure of duo3-1 germ cells to fully express some germline markers is not a consequence of their failure to divide.
Our findings reveal that DUO3 and DUO1 have overlapping but distinct targets (see model in Supplemental Figure 4 online). Both DUO1 and DUO3 are required for the complete expression of GCS1 and GEX2, while DUO1, but not DUO3, is required for the expression of MGH3. The overlap in targets is not due to DUO3 being required for the expression of DUO1. Similarly, DUO1 is not required for the expression of DUO3. How DUO1 and DUO3 cooperate to activate expression of their common targets is unknown. It is possible that DUO3 and DUO1 interact, directly or indirectly, in a complex that activates transcription. Expression of MGH3 in duo3-1 germ cells shows, however, that DUO3 is not always required for DUO1 to activate expression of its targets. Another possibility is that the two proteins act in sequence. As such, DUO3 could have a role in chromatin remodeling, enabling DUO1 access to some of its target gene promoters. Under this model, the variable expression of GEX2 and the low level of expression of GCS1 could be due to DUO1 having some, but not always full, access to these promoter regions in the absence of DUO3.
Many mutant duo3-1 germ cells are delivered to the embryo sac but fail to fertilize. This most likely arises due to incomplete specification and, in particular, a lack of GCS1, which is essential for fertilization in Arabidopsis (Mori et al., 2006; von Besser et al., 2006). Although some duo3-1 pollen tubes are successfully guided to the female gametophyte, they do not compete equally with wild-type pollen tubes. However, mutant duo3-1 pollen tubes are competitive when germline defects have been complemented with DUO1-driven DUO3-GFP. Thus, the germline expression of DUO3 appears to have an impact on pollen tube guidance. The proportion of seed gaps in siliques from +/duo3-1 plants is very similar to that from +/gcs1 plants (von Besser et al., 2006), suggesting that it may be the lack of germline expression of GCS1 that prevents complete guidance of duo3-1 pollen tubes.
DUO3 Has an Essential Role in the Sporophyte during Embryogenesis
As discussed above, DUO3 is required for cell cycle progression and specification in the male germline, and it may have similar roles in the sporophyte. Promoter GUS analysis indicates that DUO3 is expressed in a range of tissues, often at sites of cell division, and mutant duo3-1 embryos show a delay in development that is consistent with a reduced rate and coordination of cell division. Thus, DUO3 may have a general role in regulation of the cell cycle. As the extent of cell division and the level of disturbance differ between embryos, it is possible that like in the male germ cell, DUO3 has a complex role and regulates a suite of cell cycle factors that are necessary to coordinate the rate of cell division in embryos.
DUO3 may also have a wider role in cell specification in the sporophyte since homozygous duo3-1 embryos are viable and germinate but functional shoot and root apical meristems are not established. In some duo3-1 mutant embryos, the suspensors form embryo-like outgrowths, which could arise from a failure to specify suspensor cell fate, a process influenced by correct apical patterning of the embryo (reviewed in Jenik et al., 2007). Other embryo patterning mutants, such as fass, that drastically alter division patterns do not prevent meristem specification and organogenesis (Torres-Ruiz and Jürgens, 1994). This indicates that DUO3 does not only have a role in cell cycle progression but may also have a role in promoting cell specification associated with the formation of functional meristems.
DUO3 Is a Conserved Protein with General Regulatory Functions
Although DUO3 has a specialized role in male germline development, our findings strongly suggest it also has general regulatory functions. Such functions are likely to be conserved throughout land plants as the DUO3 homolog from moss, Pp DUO3, can partially restore male germline division and function to duo3-1 mutant pollen. The sequence similarity shared by At DUO3 and Pp DUO3 mainly lies within the DC1 and DC2 regions. The high degree of conservation in DC1 and DC2, and the ability of Pp DUO3 to partially complement duo3-1, suggests that these regions are important for DUO3 function. The duo3-1 mutation results in a truncated protein missing part of DC1 and all of DC2 and is not likely to be functional. The DC1 region contains a high number of charged residues and may be involved in protein interactions or DNA binding. The DC2 region may bind DNA as it shows some sequence similarity with homeodomain-like domains and has a basic pI.
The plant DUO3 proteins also have sequence similarity to the GON-4L proteins in animals. The GON-4 protein was first identified in C. elegans where it is expressed in somatic gonadal precursor cells (Friedman et al., 2000). Like DUO3, GON-4 is a nuclear protein, and mutations result in a delay in cell cycle progression. GON-4 homologs in other animal species have been called GON-4L, and a function for GON-4L proteins in other animal species is yet to be shown. GON-4L genes are expressed in a wide range of tissues in humans and rats (Kuryshev et al., 2006; Ohtomo et al., 2007, 2008). Interestingly, one of the tissues with the highest expression levels of GON-4L in rats is in spermatogenic cells in the testis (Ohtomo et al., 2008). Thus, members of the DUO3/GON-4L family in Arabidopsis, C. elegans, and rats may all have roles in male gamete development.
The similarity between the plant DUO3 proteins and the animal GON-4L protein is mostly within the two conserved domains GC1 and GC2 and a generally acidic N terminus. GON-4L proteins contain a predicted SANT domain close to the C terminus that may have a role in protein or DNA binding, whereas the C-terminal acidic region in DUO3 proteins has potential for protein or chromatin interactions. Interestingly, the C. reinhardtii DUO3-related protein is intermediate between the plant and animal proteins, having part of the DC1 domain found in the plant proteins and also a C-terminal SANT domain present in the GON-4L proteins. The vertebrate GON-4L proteins also have two PAH2 domains. PAH2 domains in eukaryotic Sin3 proteins bind a number of different proteins and enable the protein to act as a scaffold (Le Guezennec et al., 2006).
Not only do DUO3 and GON4L proteins contain a number of potential protein binding sites, some members have been shown to bind specific proteins. The GON-4L protein from Drosophila melanogaster has been shown to bind D. melanogaster CycD in two-phase yeast two-hybrid analysis (Zhong et al., 2003). While Dm GON-4L binds CycD, it appears that At DUO3 is not involved in S-phase progression, suggesting that DUO3 and GON-4L members may bind different substrates. Also, the YY1AP protein from Homo sapiens binds the transcription factor YY1 through two domains of YY1AP that are homologous to regions of GON-4L (Wang et al., 2004) and include the GC2 region. Although GC2 is conserved in DUO3, protein interactions are likely to differ since YY1 homologs are not present in plants.
There is no predicted biochemical activity for plant DUO3 proteins or the related animal GON-4L proteins, although as they are likely to bind to multiple proteins and DNA, DUO3/GON-4L proteins may act as scaffolding proteins, bringing together proteins to form a complex that subsequently alters transcription. Such a complex may be required directly for transcriptional activation and could include other proteins, such as DUO1 in plant male germ cells. Alternatively, such a complex could be involved in chromatin remodeling with histone-modifying enzymes being among those recruited by DUO3/GON-4L proteins. An alteration in chromatin structure could enable transcription factors, such as DUO1 in the plant male germline, to interact with promoter regions of their target genes. It will be intriguing to determine the role of such a DUO3 complex, whether it acts as a coactivator or is involved in the remodeling of chromatin in preparation for other transcription factors.
METHODS
Plant Material and Transformation
Arabidopsis thaliana plants were grown at 21°C with a 16-h-light and 8-h-dark cycle or with 24 h light (120 to 140 μmol/m2/s), with variable humidity. Experiments were conducted in the +/duo3-1 or the wild-type No-0 backgrounds. The ProAtGCS1:AtGCS1-GFP, ProAtGEX2:GFP, and pCDGFP marker lines are in Columbia-0. Plants were transformed with Agrobacterium tumefaciens (GV3101) using a standard floral dipping method (Clough and Bent, 1998). Transformants were selected either on Murashige and Skoog agar containing 50 μg/mL kanamycin or on soil with 30 μg/mL BASTA (glufosinate ammonium, DHAI PROCIDA) fed by subirrigation.
Vector Construction
Gateway single and multisite construction (Invitrogen) was used to generate most vectors. The DUO3 promoter region (1052 bp upstream from the initiation codon) was amplified from Columbia-0 genomic DNA. Pp DUO3 cDNA was amplified from cDNA clone pph23a09 from the Riken Biological Resource Centre (BRC Resource number pdp06309; Nishiyama et al., 2003) and mCherry from pRSETBmCherry (Shaner et al., 2004). All PCR reactions were performed with high-fidelity Phusion DNA polymerase (Finnzymes) and primers with suitable attachment site (attB) adapters (see Supplemental Table 6 online; attB adapters in italics). Full-length attB sites were added to each fragment in a second high-fidelity PCR. PCR fragments were cloned into pDONR vectors (Invitrogen; pDONR221 for DUO3 and Pp DUO3 cDNAs, pDONRP4P1R and pDONR207 for DUO3 promoter regions, and pDONRP2RP3 for mCherry) via a BP reaction using BP Clonase II (Invitrogen). The product of BP reactions was transformed into α-select chemically competent cells (Bioline) and all clones were verified by sequencing.
Donor clones as described above or by Brownfield et al. (2009) were used in Gateway multipart LR reactions using LR Clonase plus (Invitrogen) and the destination vectors pK7m34GW, pB7m34GW, or pB7m24GW (Karimi et al., 2005) to generate ProDUO3:H2B-GFP, ProDUO3:DUO3-GFP, ProDUO3:PpDUO3-mCherry, ProDUO3:PpDUO3, ProDUO1:DUO3-GFP, and ProLAT52:H2B-mRFP. To generate the ProDUO3:GFP-GUS vector, the DUO3 promoter region in pDONR207 was transformed into pKGWFS7 (Karimi et al., 2002) that contains the GFP-GUS reporter using LR Clonase I.
pCDGFP was constructed by replacing a BamHI-SacI fragment excised from pCDG (Colon-Carmona et al., 1999), which removed the uidA gene and replaced it with a BamHI-SacI fragment corresponding to mGFP5. This fragment was generated by PCR amplification using the primers GFP-1 and GFP-2 (see Supplemental Table 6 online) using a template kindly provided by Jim Haseloff, cloned into pGEM-T easy (Promega), sequence verified, excised with BamHI and SacI, and ligated into BamHI-SacI cut pCDG.
Marker Line Analysis
Established marker lines were used for analysis of DUO1 (Rotman et al., 2005; Brownfield et al., 2009), MGH3 (Brownfield et al., 2009), GCS1 (Brownfield et al., 2009), and GEX2 (Engel et al., 2005) expression in germ cells. As we had not observed any obvious variation in marker expression in wild-type or duo1 mutant pollen, one to three individual +/duo3-1 plants homozygous for each marker were examined to determine if the markers are expressed in duo3-1 germ cells. For newly established markers lines (DUO3 and LAT52), a minimum of 20 primary transformants (mix of wild-type and +/duo3-1 plants) were analyzed, and, as marker expression was similar in a number of lines, representative lines were selected to create homozygous plants for detailed analysis. Marker expression in duo3-1 or duo1-1 germ cells was analyzed in one to three individuals.
RT-PCR Analysis
Pollen from ecotype Landsberg erecta at different stages of development was isolated and RNA extracted as described (Honys and Twell, 2004). For sporophytic tissues, RNA was extracted from frozen samples using the Qiagen RNeasy kit. Samples of 750 ng of total RNA for pollen stages and sporophytic tissues were reverse transcribed in a 20-μL reaction using Superscript II RNase H reverse transcriptase (Invitrogen) and an oligo(dT) primer as per the manufacturer's instructions. For PCR amplification, 1 μL of a 10× diluted cDNA was used in a 25-μL reaction using Biotaq DNA polymerase (Bioline) and 12.5 pmol of each primer (see Supplemental Table 7 online). PCR conditions were as follows: 96°C for 1 min, 30 cycles of 96°C for 30 s, 55°C for 30 s, and 72°C for 40 s followed by 5 min at 72°C.
Microscopy Analyses
Mature pollen was stained with DAPI as described previously (Park et al., 1998). For developmental analysis, pollen from buds at different stages of development was teased out of the anther with a needle and mounted directly in DAPI solution (analysis of cell cycle) or 0.3 M mannitol (analysis of GFP).
Fluorescence and confocal laser scanning microscopy were performed using methods and equipment as described (Brownfield et al., 2009). Transmission electron microscopy was performed as described (Park and Twell, 2001). Histochemical staining for GUS activity was performed as described (Honys et al., 2006) with tissues incubated in a solution containing 1 mM X-gluc (5-bromo-4-chloro-3-indolyl β-d-glucuronide) and 0.5 mM K3Fe[CN]6, at 37°C for 1 d. Material was cleared with 70% ethanol and viewed by bright-field microscopy. For phenotypic characterization of mutant embryos, cleared whole-mount seeds were prepared and viewed with differential interference contrast microscopy as described (Park et al., 2004).
Bioinformatic Analyses
Sequences were analyzed using the MacVector program (MacVector Inc) and alignments created using ClustalW through MacVector with default settings. TBLASTN searches of the Arabidopsis genome were conducted through TAIR (http://www.Arabidopsis.org/Blast/index.jsp) with the TAIR9 Genes (+introns, + UTRs) database using default settings (matrix, blosum 62; genetic code, universal; word size, 3; gap opening penalty, 11; gap extension penalty, 1). Motif predictions used Interproscan (http://www.ebi.ac.uk/interpro/; Zdobnov and Apweiler, 2001). DUO3 homologs in land plants were identified using TBLASTN searches with the entire DUO3 protein sequence and default settings in various databases listed in Supplemental Table 3 online. The animal GON-4L proteins were identified through TBLASTN using just the DC2 region of DUO3 and default setting in the National Center for Biotechnology Information and used the GenBank nonredundant database.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data library under accession numbers FJ461627 (No-0 Arabidopsis DUO3 genomic sequence), FJ461625 (Landsberg erecta Arabidopsis cDNA sequence), NP_191605.1 (Arabidopsis DUO1 protein sequence), and FJ461626 (P. patens DUO3 cDNA sequence) and in the Arabidopsis Genome Initiative database with locus identifiers At1g64570 (Arabidopsis DUO3) and At3g60460 (Arabidopsis DUO1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Representation of Map-Based Cloning of the DUO3 Gene.
Supplemental Figure 2. Alignment of DUO3 Conserved 1 (DC1) Region.
Supplemental Figure 3. Alignment of DUO3 Conserved 2 (DC2) Region.
Supplemental Figure 4. Model of the Roles of DUO3 and DUO1 in Male Germline Development.
Supplemental Table 1. Genetic Transmission of the duo3-1 Allele.
Supplemental Table 2. Tetrad Analysis of the duo3-1 Mutation.
Supplemental Table 3. DUO3 Homologs in Land Plants.
Supplemental Table 4. Characteristics of the DUO3 and GON-4L Proteins from Land Plants, Algae, and Animals.
Supplemental Table 5. The Germination Rate of Wild-Type and duo3-1 Pollen in Vitro.
Supplemental Table 6. Primers Used in Vector Construction.
Supplemental Table 7. Primers Used in RT-PCR Analysis.
Supplementary Material
Acknowledgments
We thank Roger Y. Tsien (Howard Hughes Medical Institute Laboratories, University of California) for providing pRSETBmCherry and Shelia McCormick (Plant Gene Expression Center, Albany, CA) and Toshiyuki Mori (RIKEN) for ProGEX2:GFP and ProGCS1:GCS1-GFP marker lines, respectively. We thank Anthony Wardle (University of Leicester) for construction of plasmid ProDUO3:GUS-GFP and Stefan Hyman and Natalie Allcock (Electron Microscopy Laboratory, University of Leicester) for assistance and advice with electron microscopy. This work was funded by the Biotechnology and Biological Sciences Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: David Twell (twe@le.ac.uk).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Alandete-Saez, M., Ron, M., and McCormick, S. (2008). GEX3, expressed in the male gametophyte and in the egg cell of Arabidopsis thaliana, is essential for micropylar pollen tube guidance and plays a role during early embryogenesis. Mol. Plant 1 586–598. [DOI] [PubMed] [Google Scholar]
- Borg, M., Brownfield, L., and Twell, D. (2009). Male gametophyte development: A molecular perspective. J. Exp. Bot. 60 1465–1478. [DOI] [PubMed] [Google Scholar]
- Borges, F., Gomes, G., Gardner, R., Moreno, N., McCormick, S., Feijo, J.A., and Becker, J.D. (2008). Comparative transcriptomics of Arabidopsis thaliana sperm cells. Plant Physiol. 148 1168–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield, L., Hafidh, S., Borg, M., Sidorova, A., Mori, T., and Twell, D. (2009). A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genet. 5 e10000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Hafidh, S., Shi, H.P., Twell, D., and Berger, F. (2009). Proliferation and cell fate establishment during Arabidopsis male gametogenesis depends on the Retinoblastoma protein. Proc. Natl. Acad. Sci. USA 17 7257–7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Jeanie, L.H., Mathieu, I., Venkatesan, S., and Berger, F. (2008). Chromatin assembly factor 1 regulates the cell cycle but not cell fate during male gametogenesis in Arabidopsis thaliana. Development 135 65–73.18045841 [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Colon-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20 503–508. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., and Inze, D. (2007). The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8 655–665. [DOI] [PubMed] [Google Scholar]
- Durbarry, A., Vizir, I., and Twell, D. (2005). Male germ line development in Arabidopsis: duo pollen mutants reveal gametophytic regulators of generative cell cycle progression. Plant Physiol. 137 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady, C., Lindsey, K., and Twell, D. (1995). The significance of microspore division and division symmetry for vegetative cell-specific transcription and generative cell differentiation. Plant Cell 7 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel, M.L., Holmes-Davis, R., and McCormick, S. (2005). Green sperm. Identification of male gamete promoters in Arabidopsis. Plant Physiol. 138 2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, L., Anna-Arriola, S.S., Hodgkin, J., and Kimble, J. (2000). gon-4, a cell lineage regulator required for gonadogenesis in Caenorhabditis elegans. Dev. Biol. 228 350–362. [DOI] [PubMed] [Google Scholar]
- Friedman, W. (1999). Expression of the cell cycle in sperm of Arabidopsis: Implications for understanding patterns of gametogenesis and fertilization in plants and other eukaryotes. Development 126 1065–1075. [DOI] [PubMed] [Google Scholar]
- Fülöp, K., Tarayre, S., Kelemen, Z., Horváth, G., Kevei, Z., Nikovics, K., Bakó, L., Brown, S., Kondorosi, A., and Kondorosi, E. (2005). Arabidopsis anaphase-promoting complexes: Multiple activators and wide range of substrates might keep APC perpetually busy. Cell Cycle 4 1084–1092. [PubMed] [Google Scholar]
- Gusti, A., Baumberger, N., Nowack, M., Pusch, S., Eisler, H., Potuschak, T., De Veylder, L., Schnittger, A., and Genschik, P. (2009). The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS One 4 e4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.W., Burton, J.L., and Solomon, M.J. (2002). The anaphase-promoting complex: It's not just for mitosis any more. Genes Dev. 16 2179–2206. [DOI] [PubMed] [Google Scholar]
- Honys, D., Oh, S.A., Renak, D., Donders, M., Solcova, B., Johnson, J.A., Boudova, R., and Twell, D. (2006). Identification of microspore-active promoters that allow targeted manipulation of gene expression at early stages of microgametogenesis in Arabidopsis. BMC Plant Biol. 6 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys, D., and Twell, D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5 R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff, M., Hamamura, Y., Gourgues, M., Higashiyama, T., and Berger, F. (2007). Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr. Biol. 17 1032–1037. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H., Shinmyo, A., and Sekine, M. (2006). Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 45 819–831. [DOI] [PubMed] [Google Scholar]
- Jenik, P.D., Gillmor, C.S., and Lukowitz, W. (2007). Embryonic patterning in Arabidopsis thaliana. Annu. Rev. Cell Dev. Biol. 23 207–236. [DOI] [PubMed] [Google Scholar]
- Johnston, A.J., Matveeva, E., Kirioukhova, O., Grossniklaus, U., and Gruissem, W. (2008). A dynamic reciprocal RBR-PRC2 regulatory circuit controls Arabidopsis gametophyte development. Curr. Biol. 18 1680–1686. [DOI] [PubMed] [Google Scholar]
- Karimi, M., De Meyer, B., and Hilson, P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10 103–105. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kim, H.J., Oh, S.A., Brownfield, L., Hong, S.H., Ryu, H., Hwang, I., Twell, D., and Nam, H.G. (2008). Control of plant germline proliferation by SCFFBL17 degradation of cell cycle inhibitors. Nature 455 1134–1137. [DOI] [PubMed] [Google Scholar]
- Kuryshev, V.Y., et al. (2006). An anthropoid-specific segmental duplication on human chromosome 1q22. Genomics 22 143–151. [DOI] [PubMed] [Google Scholar]
- Le Guezennec, X., Vermeulen, M., and Stunnenberg, H.G. (2006). Molecular characterization of Sin3 PAH-domain interactor specificity and identification of PAH partners. Nucleic Acids Res. 34 3929-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, S. (2004). Control of male gametophyte development. Plant Cell 16 (suppl.): S142–S153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges, M., de Jager, S.M., Gruissem, W., and Murray, J.A.H. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41 546–566. [DOI] [PubMed] [Google Scholar]
- Menges, M., and Murray, J.A.H. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30 203–212. [DOI] [PubMed] [Google Scholar]
- Mori, T., Kuroiwa, H., Higashiyama, T., and Kuroiwa, T. (2006). Generative Cell Specific 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8 64–71. [DOI] [PubMed] [Google Scholar]
- Nishiyama, T., Fujita, T., Shin-I, T., Seki, M., Nishide, H., Uchiyama, I., Kamiya, A., Carninci, P., Hayashizaki, Y., and Shinozaki, K. (2003). Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: Implication for land plant evolution. Proc. Natl. Acad. Sci. USA 100 8007–8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack, M.K., Grini, P.E., Jakoby, M.J., Lafos, M., Koncz, C., and Schnittger, A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38 63–67. [DOI] [PubMed] [Google Scholar]
- Ohtomo, T., Horii, T., Nomizu, M., Suga, T., and Yamada, J. (2007). Molecular cloning of a structural homolog of YY1AP, a coactivator of the multifunctional transcription factor YY1. Amino Acids 33 645–652. [DOI] [PubMed] [Google Scholar]
- Ohtomo, T., Horii, T., Nomizu, M., Suga, T., and Yamada, J. (2008). Cloning and expression analysis of YY1AP-related protein in the rat brain. Amino Acids 34 155–161. [DOI] [PubMed] [Google Scholar]
- Okada, T., Endo, M., Singh, M.B., and Bhalla, P.L. (2005). Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 44 557–568. [DOI] [PubMed] [Google Scholar]
- Park, S.K., Howden, R., and Twell, D. (1998). The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125 3789–3799. [DOI] [PubMed] [Google Scholar]
- Park, S.K., Rahman, D., Oh, S.A., and Twell, D. (2004). Gemini pollen 2, a male and female gametophytic cytokinesis defective mutation. Sex. Plant Reprod. 17 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.K., and Twell, D. (2001). Novel patterns of ectopic cell plate growth and lipid body distribution in the Arabidopsis gemini pollen1 mutant. Plant Physiol. 126 899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina, C., Pinto, F., Feijo, J.A., and Becker, J.D. (2005). Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 138 744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, D., Rhee, S., and Davis, R. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264 1458–1460. [DOI] [PubMed] [Google Scholar]
- Rotman, N., Durbarry, A., Wardle, A., Yang, W.C., Chaboud, A., Faure, J.E., Berger, F., and Twell, D. (2005). A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 15 244–248. [DOI] [PubMed] [Google Scholar]
- Shaner, N.C., Campbell, R.E., Steinbach, P.A., Giepmans, B.N.G., Palmer, A.E., and Tsien, R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22 1567–1572. [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz, R.A., and Jürgens, G. (1994). Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development 120 2967–2978. [DOI] [PubMed] [Google Scholar]
- Twell, D., Wing, R., Yamaguchi, J., and McCormick, S. (1989). Isolation and expression of an anther-specific gene from tomato. Mol. Gen. Genet. 217 240–245. [DOI] [PubMed] [Google Scholar]
- von Besser, K., Frank, A.C., Johnson, M.A., and Preuss, D. (2006). Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133 4761–4769. [DOI] [PubMed] [Google Scholar]
- Wang, C.Y., Liang, Y.J., Lin, Y.S., Shih, H.M., Jou, Y.S., and Yu, W.C.Y. (2004). YY1AP, a novel co-activator of YY1. J. Biol. Chem. 279 17750–17755. [DOI] [PubMed] [Google Scholar]
- Weingartner, M., Criqui, M.-C., Mészáros, T., Binarova, P., Schmit, A.-C., Helfer, A., Derevier, A., Erhardt, M., Bögre, L., and Genschik, P. (2004). Expression of a nondegradable cyclin B1 affects plant development and leads to endomitosis by inhibiting the formation of a phragmoplast. Plant Cell 16 643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov, E.M., and Apweiler, R. (2001). InterProScan - An integration platform for the signature-recognition methods in InterPro. Bioinformatics 17 847–848. [DOI] [PubMed] [Google Scholar]
- Zhong, J., Zhang, H., Stanyon, C.A., Tromp, G., and Finley, R.L. (2003). A strategy for constructing large protein interaction maps using the yeast two-hybrid system: Regulated expression arrays and two-phase mating. Genome Res. 13 2691–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.