Abstract
Plants adjust their photosynthetic activity to changing light conditions. A central regulation of photosynthesis depends on the xanthophyll cycle, in which the carotenoid violaxanthin is converted into zeaxanthin in strong light, thus activating the dissipation of the excess absorbed energy as heat and the scavenging of reactive oxygen species. Violaxanthin deepoxidase (VDE), the enzyme responsible for zeaxanthin synthesis, is activated by the acidification of the thylakoid lumen when photosynthetic electron transport exceeds the capacity of assimilatory reactions: at neutral pH, VDE is a soluble and inactive enzyme, whereas at acidic pH, it attaches to the thylakoid membrane where it binds its violaxanthin substrate. VDE also uses ascorbate as a cosubstrate with a pH-dependent Km that may reflect a preference for ascorbic acid. We determined the structures of the central lipocalin domain of VDE (VDEcd) at acidic and neutral pH. At neutral pH, VDEcd is monomeric with its active site occluded within a lipocalin barrel. Upon acidification, the barrel opens up and the enzyme appears as a dimer. A channel linking the two active sites of the dimer can harbor the entire carotenoid substrate and thus may permit the parallel deepoxidation of the two violaxanthin β-ionone rings, making VDE an elegant example of the adaptation of an asymmetric enzyme to its symmetric substrate.
INTRODUCTION
Light is a fundamental source of energy but is also potentially harmful to organisms because chlorophyll has a high yield of triplet states, which can react with molecular oxygen and produce reactive oxygen species (ROS). Plants have evolved a variety of regulatory mechanisms to respond to the naturally varying light conditions (Barber and Andersson, 1992; Niyogi, 2000; Horton et al., 2005) and minimize damage by ROS while maximizing photosynthetic output. Zeaxanthin (Z) is a carotenoid specially synthesized when light is in excess. It plays a fundamental role in photoprotection mechanisms: Z is able to decrease light-harvesting efficiency by enhancing energy dissipation as heat (Demmig-Adams, 1990; Niyogi et al., 1998; Holt et al., 2004, 2005; Ahn et al., 2008; Amarie et al., 2009), and it efficiently scavenges ROS due to its strong antioxidant properties (Havaux and Niyogi, 1999; Demmig-Adams and Adams, 2002; Havaux et al., 2007; Johnson et al., 2007). Z content is regulated by the so-called xanthophyll cycle: in strong light, the energy input exceeds the photosynthetic capacity and leads to overacidification of the thylakoid lumen, when the saturated activity of the ATP synthase, which allows proton efflux, cannot match the rate of inward proton pumping driven by the electron transfer chains. This drop in lumen pH leads to activation of the violaxanthin deepoxidase (VDE), thus shifting the xanthophyll balance from violaxanthin (V; a di-epoxide), which acts as a light-harvesting pigment, toward Z (no epoxide group), which favors dissipation of the excitation energy through heat. This reaction comes with a transient accumulation of antheraxanthin, a mono-epoxide intermediate (Hartel et al., 1996; Frommolt et al., 2001). When the lumen pH increases, Z is converted back to V by a stromal enzyme zeaxanthin epoxidase. This xanthophyll cycle is regulated so as to avoid unnecessary quenching of excitation energy when light is limiting (Havaux and Niyogi, 1999; Demmig-Adams and Adams, 2002; Havaux et al., 2007; Johnson et al., 2007).
Although the physiology of the xanthophyll cycle has been studied in depth, the molecular and structural mechanisms involved are less well known. VDE is a soluble lumenal protein at neutral pH, but it attaches to the thylakoid membrane with a marked cooperativity at acidic pH (Yamamoto and Higashi, 1978; Hager and Holocher, 1994). It uses ascorbate as a cosubstrate with a pH-dependent Km that may reflect a preference for ascorbic acid (for example, see Bratt et al., 1995).
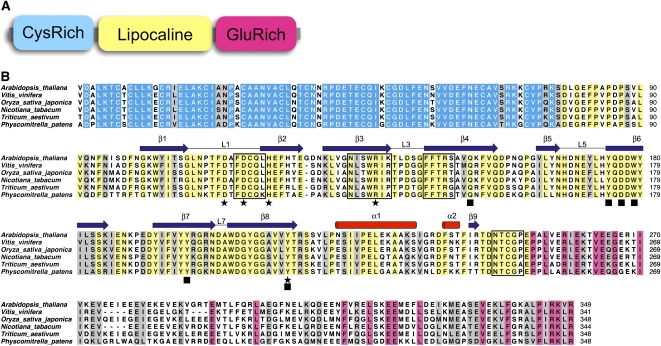
VDE is a single enzyme composed of three domains (Figure 1). The Cys-rich N-terminal domain (∼80 residues) contains 11 of the 13 Cys residues of the mature polypeptide of Arabidopsis thaliana. This domain is not dispensable to the enzymatic activity and is thought to be responsible for the reported inhibition of VDE by reducing agents such as DTT (Hieber et al., 2002). Despite low sequence identity, the central domain was classified as a member of the lipocalin protein family (Bugos and Yamamoto, 1996), although this classification has been challenged (Ganfornina et al., 2000). Lipocalins are a large group of small proteins with a low level of sequence identity, but with remarkably homogeneous three-dimensional structures (Schlehuber and Skerra, 2005; Grzyb et al., 2006). The lipocalin fold consists of an eight-stranded antiparallel β-barrel with a repeated +1 topology enclosing an internal ligand binding site. Initially described as passive transport proteins of hydrophobic ligands (such as retinoic acid or pheromone), some lipocalins were also shown to possess enzymatic activity, expanding their function to the biosynthesis of prostaglandin in animals and jasmonate in plants (Nagata et al., 1991; Hofmann et al., 2006). VDE together with zeaxanthin epoxidase, the other enzyme of the xanthophyll cycle, are two of the first examples of a lipocalin-type enzyme (Bugos et al., 1998; Hieber et al., 2000). The central lipocalin domain was proposed to bind the hydrophobic V substrate and to contain the catalytic residues.
Figure 1.
Sequence Analysis of VDE.
(A) Schematic domain organization of VDE.
(B) Sequence alignment of a range of VDE enzymes. Conserved residues are color coded according to the domain organization. Secondary structure features are shown and labeled above the alignment (cylinders indicate α-helices and arrows β-sheets); residues important for the pH switch are marked with black stars and residues forming the putative active site with black squares. Residues at the dimer interface are boxed.
The C-terminal tail of VDE contains a high number of Glu residues and is predicted to fold as a series of α-helices. Although the C-terminal tail is partly dispensable without affecting enzyme activity, the partial protonation of glutamic residues at pH 5.0 (the optimal pH of VDE) has been proposed to increase the binding of the enzyme to the thylakoid membrane (Hieber et al., 2002).
To elucidate the structure of the enzyme and gain more insight into its catalytic mechanism and regulation, the central lipocalin domain of Arabidopsis VDE (VDEcd), equivalent to about half of the mature protein, was overexpressed in bacteria and purified to homogeneity, and its structure was solved by x-ray crystallography at two different pH values. The structural comparison between the acidic and neutral form of VDEcd shed light on how the enzyme is regulated by pH and how pH may affect its activity.
RESULTS
Structure Determination
The structure of VDEcd was solved by the single-wavelength anomalous diffraction method using a protein crystal grown at pH 5.0 and soaked in a Gadolinium solution (Girard et al., 2003). Subsequently, the structure of VDEcd was solved by molecular replacement using a crystal grown at pH 7. Since, at these two pH values, VDE is either inactive (pH 7) or active (pH 5), our structural study brings novel insights into the pH-dependent activity of VDE.
Structure at Neutral pH
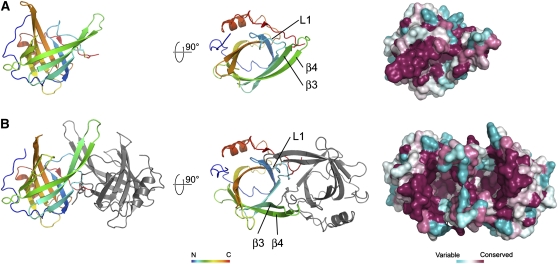
At pH 7, VDEcd adopts a typical lipocalin fold with an eight-stranded antiparallel β-barrel, thus unequivocally confirming its classification as a member of the lipocalin protein family (Figure 2A). The central hydrophobic cavity usually found in lipocalins and necessary for ligand binding is, however, partially occluded by the L1 loop. The bottom of the cavity is hydrophobic, but several charged or polar residues (Gln-153, Tyr-175, Tyr-198, Asp-177, and Tyr-214) colocalize on one side of the barrel, contrasting with most lipocalins, which bind hydrophobic molecules but otherwise have no enzymatic activity. The location of these residues together with their strict conservation in the VDE family strongly suggests that they define the active site of VDE.
Figure 2.
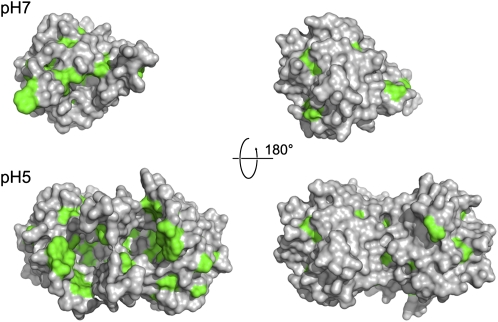
Architecture of the Central Lipocalin Domain of VDE at pH 7 and 5.
Crystal structure of VDEcd at pH 7.0 (A) and pH 5.0 (B) with amino acid sequence variation mapped onto the surface view (right). The opening of the top and side of the barrel at pH 5.0 is visible in this surface view. Membrane anchoring is expected to occur on this side of the molecule due to the abundance of hydrophobic residues (see also Figure 5).
Structure at Acidic pH
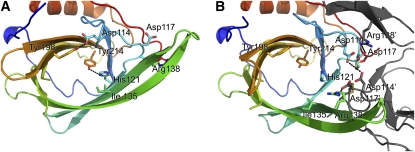
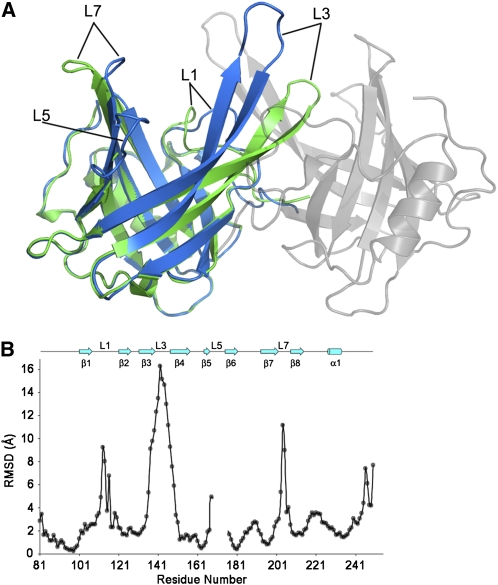
At pH 5, the lipocalin fold of VDEcd is very different from the one observed at neutral pH (Figure 2B). The protein exists as a dimer with a buried surface area on each monomer of ∼1135 Å2. This value is larger than the one measured for two other well-documented dimeric lipocalins, namely, the major horse allergen (1024 Å2; Lascombe et al., 2000) and the bacterial lipocalin blc (750 Å2; Campanacci et al., 2006). In addition, thirteen hydrogen bonds, six salt bridges, and numerous hydrophobic interactions stabilize the dimer in VDEcd, further supporting a physiological dimer at pH 5. Two main structural rearrangements of the lipocalin fold at acidic pH lead to an overall root mean square deviation of 4.4 Å between the monomeric and dimeric conformations of the enzyme (Figures 3 and 4; see Supplemental Movie 1 online for morphing between the two states). Probably of primary importance is that the side of the barrel opens between strands β2 and β3 with a considerable rearrangement of strands β3 and β4 and their connecting loop L3. For example, the distance between the Cα of Leu-129 (in strand β2) and the Cα of Ser-136 (in strand β3) goes from 5.4 Å at pH 7.0 to 13.6 Å at pH 5. The second major rearrangement of the lipocalin fold is the movement of loop L1 from a closed state at pH 7.0 to an open state at pH 5. In the closed state, Asp-114 in loop L1 is hydrogen bonded to Tyr-198 in the barrel cavity, thereby partially blocking accessibility to the internal lipocalin cavity. At pH 5, Asp-114 is involved in a salt bridge with Arg-138 and hydrogen bonded to Asp-114 of the adjoining monomer. Two other structural modifications occur in the pH transition; the polypeptide stretch from residues 168 to 177 is disordered at pH 7.0 but ordered at pH 5, and loop L7 connecting strands β7 and β8 is rearranged (this loop contains the highly conserved Asn-202 to Gly-210 region). Overall, the movements described here involve conserved residues and mostly affect the access to the binding cavity located at the top of the barrel (Figure 4). In this region, a conserved ring of hydrophobic residues, which becomes surface exposed only at pH 5.0 (Figure 5), may interact with the membrane, a requirement for the binding of the liposoluble V substrate.
Figure 3.
pH-Dependent Dimerization and Opening of the VDE Active Sites.
Positions of key conserved residues at pH 7.0 (A) and pH 5.0 (B) are shown. Note the rotation of the side chain of His-121 and the rearrangement of the L1 loop with Asp-114 hydrogen bonded to Tyr-198 at neutral pH, whereas it is clustered with Asp-117, Asp-114, and Asp-117 from the adjoining monomer at acidic pH.
Figure 4.
Structural Comparison of the Open and Closed Forms of VDEcd.
(A) Superposition of the VDEcd structure obtained at pH 7.0 (green) with the VDEcd structure obtained at pH 5.0 (one monomer colored in blue and the other colored in gray and rendered semitransparent).
(B) Root mean square deviation plot between the structures obtained at pH 7 and 5. Secondary structures and their labeling are depicted above the plot.
Figure 5.
Position of Conserved Hydrophobic Residues at Neutral (Top) and Acidic pH (Bottom).
Surface representation of VDEcd with conserved hydrophobic residues (Ala, Val, Ile, Leu, Phe, Tyr, and Trp) colored in green. Being more exposed to the solvent at acidic pH may play a role in the attachment of VDE to the membrane.
Structural Transitions between Inactive and Active Conformations
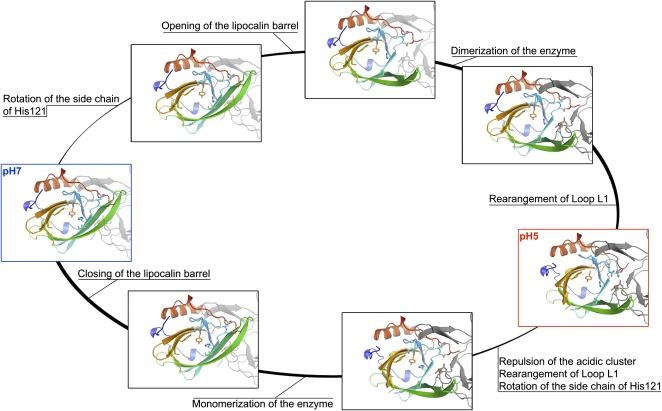
Although we only have snapshots of the closed and open forms, analysis of the VDEcd structures obtained at pH 7.0 and pH 5.0 suggests a plausible sequence of events for the transition between these two states (see Supplemental Movie 1 online; Figure 6). First, the opening of the barrel side would allow the dimer to form, which in turn would promote the rearrangement of loop L1 and the complete opening up of the active sites. Consistent with this sequence, His-121 is ideally positioned to act as a trigger. At pH 7.0, the imidazole ring is uncharged and its Nδ1 position is hydrogen bonded to the Tyr-214 phenolate group. At low pH, the Nδ1 of His-121 is protonated so that it can no longer form this hydrogen bond and is therefore forced to swivel around the Cβ-Cγ bond. This rather small reorientation of the side chain would, however, place the imidazole ring in steric clash with the adjacent Leu-135 on strand β3, hence pushing this strand and ultimately causing the barrel side to open.
Figure 6.
Plausible Transitions between the Closed (pH 7) and Open (pH 5) States of VDEcd.
Residues discussed in the text as being important for the pH transition are represented in stick form. The adjoining monomer is shown in gray and is either transparent or solid depending on the estimated plausibility of the structure. Pauses separate His-121 reorientation, barrel opening, loop rearrangements, and dimer formation. For the neutral to acidic pH transition, the His-121 side chain rotation and barrel opening are probably the first events. Transition in the reverse order (rearrangement of loop L1 followed by dimer formation and strand opening) would be disallowed, as the closed barrel structure would prevent dimer formation. For the acidic-to-neutral pH transition, repulsion within the acidic clusters (Asp-114 and Asp-117 from both monomers) and rearrangement of loop1 are the most likely primary events, as otherwise steric clashes would prevent the barrel from closing before loop1 rearranges.
While His-121 appears important in initiating the transition to the low pH form, Asp-114 is a good candidate for priming the acidic-to-neutral pH transition as it clusters with Asp-117 and Asp-114 Asp-117 from the adjoining monomer. Indeed, this spatial proximity is only possible because at least some of these Asp residues are protonated at low pH. If the pH increases, the deprotonation of these residues will surely destabilize this acidic cluster, therefore promoting the rearrangement of the L1 loop, the closure of the internal cavity, and, finally, the monomerization of the enzyme by the closure of the lipocalin barrel. Interestingly, His residues in a high pH form and acidic residues in a low pH form have also been found to act as pH-sensitive molecular switches in virus fusion (Roche et al., 2007). Thus, very different biological processes appear to rely on similar molecular mechanisms.
Structure-Function Analysis by Site-Directed Mutagenesis
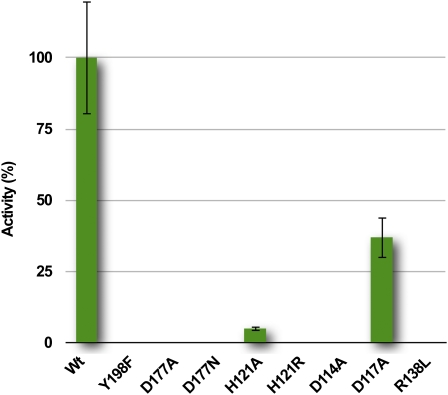
The structural analysis of the pH-dependent conformational transition described above suggests that His-121 should play a key role in initiating the opening of the lipocalin barrel prior to protein dimerization. Since His pKa values are ideal for pH sensor activity in VDE, such a role for His residues has previously been suggested and studied using site-directed mutagenesis and chemical modification (Emanuelsson et al., 2003; Gisselsson et al., 2004). Several pairwise substitutions and only one single mutation (His-124) were performed, revealing a complete loss of VDE activity when all four His residues were substituted with Ala or Arg. We confirmed the key role of His-121 by replacing this residue with Ala or Arg (Figure 7). In both cases, the single mutation drastically affected the activity of the full-length enzyme VDE, consistent with our proposed role of His-121 in acting as a trigger for barrel opening.
Figure 7.
Enzymatic Activity of VDE Mutants.
The enzymatic activity of VDE mutants is indicated as a percentage of that of the wild type and quantified from an increase in absorption at 502 nm (Yamamoto, 1985; Bugos et al., 1999). Data are expressed as the means ± sd of six independent experiments. In the case of inactive mutants, the absence of Z was confirmed by HPLC. Protein concentration was estimated by protein gel blot analysis before testing their activity.
[See online article for color version of this figure.]
Conformational change of the L1 loop is likely critical for VDE activity since it modulates the access to the active site and participates in the stabilization (destabilization) of the dimeric form at acidic (neutral) pH. Indeed, we found that mutating either Asp-114 into Asn or Arg-138 into Leu led to a complete loss of enzyme activity (Figure 7). These two residues are located away from the active site, and the fact that they are critical supports a major role for this loop rearrangement process in enzyme activity.
DISCUSSION
We have determined the crystal structure of VDEcd in its inactive (pH 7) and active (pH 5) conformation and describe the structural transitions that occur between the inactive and active conformations. The structure confirms that VDEcd is a lipocalin protein comprising a single eight-stranded antiparallel β-barrel. The protein exists as a monomer at pH 7, but at acidic pH, it adopts a stable dimeric conformation. In the dimeric state, the barrel adopts an open conformation that facilitates ligand access to the active site.
A Dimeric Model for the Deepoxidation of V
As already mentioned, several charged or polar residues (Gln-153, Tyr-175, Tyr-198, Asp-177, and Tyr-214) are located inside the lipocalin cavity of VDE, whereas this region is highly enriched in hydrophobic residues in the structures of the other members of the family. The location of these residues together with their strict conservation in the VDE family strongly suggests that they define the minimal active site of the protein. These polar residues would be responsible for the binding of the β-ionone ring of V and/or ascorbate. In support of this hypothesis, we found that mutating Asp-177 to Ala and Tyr-198 to Phe completely abolished VDE activity (Figure 7). The identification of a catalytic aspartic residue in VDE is in agreement with the observation that pepstatin A, an inhibitor of aspartic proteases, also inhibits VDE (Kuwabara et al., 1999; Kawano and Kuwabara, 2000).
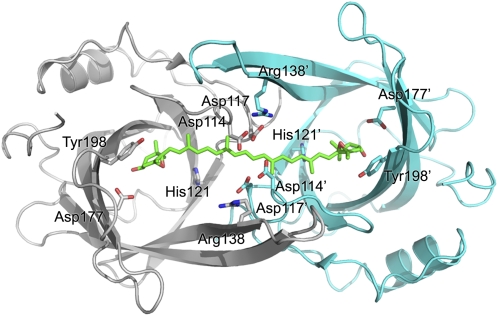
On the basis of the acidic dimeric structure, we propose a model of the VDE enzyme substrate complex. We noticed that the distance between the two putative active sites of the dimer fits the distance between the two heads of V (Figure 8). Since an open channel connects the active sites, there would be no constraint on the polyene chain of the xanthophyll substrate or product. It is therefore possible to relate the dimeric structure of VDEcd to its capability to deepoxidate the two ends of the substrate at once. Previous studies have shown that violeoxanthin (9-cis-violaxanthin) and 9-cis-neoxanthin are not substrates of VDE contrary to their all-trans counterparts (Yamamoto and Higashi, 1978). Authors of this work suggested that VDE is a mono-de-epoxidase with a 30-Å-deep cavity. Our dimeric model of VDE supports an alternative view where VDE is a di-de-epoxidase with two active sites separated by 30 Å. Indeed, the dimer would not be able to deepoxidate cis-substrates such as violeoxanthin because they would not bind the dimeric cavity. In addition, the di-de-epoxidase activity of VDE would explain its ability to produce antheraxanthin: in fact, within the dimer, there are two ascorbate binding sites that provide the reducing power for the deepoxidation of the two epoxy groups. Thus, there is a certain probability that the enzyme binds one V and only one ascorbate molecule, thus producing antheraxanthin instead of Z. This picture is consistent with the observation that ascorbate binding to VDE is a limiting step in the deepoxidation reaction rate (Neubauer and Yamamoto, 1994).
Figure 8.
Model of V Docking on the VDEcd Dimer.
Approximate model of the interaction between VDEcd and V. The V structure was taken from the x-ray structure of the spinach light-harvesting complex LHCII (Liu et al., 2004) without modification and manually docked into the dimeric structure obtained at pH 5. The ascorbate cosubstrate was not modeled, but there is enough space in the central lipocalin cavity for this cosubstrate, even in the presence of V.
Conclusions
The three-dimensional structures of VDEcd in closed and open states, together with site-direct mutagenesis experiments, reveal the molecular basis of the pH transition and how it may affects the xanthophyll cycle. The protonation state of His-121 and Asp-114 will play a central role in modulating the stability of a dimeric enzyme. By allowing the parallel deepoxidation of the two V heads instead of modifying them sequentially, the structure of this dimeric VDEcd at acidic pH is a unique and elegant example of how an asymmetric enzyme can be adapted to its symmetric substrate by dimerization. This pH-dependent conformational transition seen in the crystals substantiates many of the previous studies and gives a molecular and structural basis for the xanthophyll cycle and how it is regulated by the pH of the lumen.
METHODS
Protein Expression and Purification
To overcome the limitations in overexpression of full-length VDE in heterologous systems (Hieber et al., 2000), truncated VDE from Arabidopsis thaliana (residues 78 to 252 of the mature protein) was cloned into the pQE60 vector (from Qiagen) and expressed in Escherichia coli BL21 (CodonPlus-RIL; Stratagene). The protein was purified by nickel affinity chromatography (HisTrap; GE Healthcare) and gel filtration (Superdex 200; GE Healthcare). It was concentrated to 8 mg mL−1 in 50 mM HEPES, pH 7.5, and 50 mM NaCl.
Crystallization
All the crystallization experiments were performed at 293 K using the hanging drop vapor diffusion technique. Crystals were obtained at pH 7.0 by mixing the protein solution (2 μL + 0.2 μL of 20% (w/v) benzamidine hydrochloride hydrate) with 2 μL of a reservoir solution containing 100 mM Hepes (pH 7), 22% PEG3350 and 200 mM MgNO3. The needle-shaped crystals (0.02 × 0.02 × 0.5 mm3), which appeared after two weeks, were soaked in the same solution containing ethylene glycol at concentrations gradually increasing up to 30%, and then frozen in liquid nitrogen. Crystals were obtained at acidic pH by mixing the same amount of protein (2 μL) with a reservoir solution containing 100 mM sodium acetate (pH 5) and 2 M ammonium sulfate. The diamond-shaped crystals (0.3 × 0.3 × 0.5 mm3) that appeared after two months were soaked in paraffin oil and frozen in liquid nitrogen.
Crystal Structure Determination
Diffraction data were collected using beamline ID23-1 at the European Synchrotron Radiation Facility (ESRF) in Grenoble. The structure of the acidic form of the protein was solved by the single-wavelength anomalous diffraction method using crystals soaked for 2 h in the same solution that serves to grow them, but supplemented with 100 mM GdHPDO3, a neutral Gadolinium complex (Girard et al., 2003). Diffraction data up to 2.5 Å (peak) or 2.0 Å (remote) resolution were collected with the FIP beamline (ESRF), reduced using Mosflm (Leslie, 1993) and scaled using Scala (Evans, 1997) from the CCP4 suite (Collaborative Computational Project, Number 4, 1994). The crystal belongs to space group I4122 (a = b = 122.3 Å, c = 158.4 Å). The Solve and Resolve (Terwilliger, 2003) programs were used to determine the position of the three gadolinium atoms in the asymmetric unit and to determine initial phases, using data collected at the peak wavelength (λ = 1.7112 Å). In the resulting electron density map, the two molecules of the asymmetric unit were readily modeled using the Coot (Emsley and Cowtan, 2004) program. The final native model containing two molecules (consisting of amino acids 81 to 251), 192 solvent molecules, and three gadolinium ions was refined using Refmac5 (Murshudov et al., 1997), and data were collected at a remote wavelength (λ = 0.9797 Å; Table 1). Electron density was not detected for the His-tag or the three N-terminal and the C-terminal amino acids. The crystal of neutral pH form belongs to space group P1 with cell dimensions a = 34.6 Å, b = 52.5 Å, c = 54.6 Å, α = 82.5°, β = 76.0°, and γ = 74.5° (see Table 1). To solve the structure, one molecule of the asymmetric unit was used as a search model in molecular replacement using Molrep (Vagin and Teplyakov, 2000). The resulting two solutions were refined using the NCSREF combination of programs to give an interpretable electron density map of the closed conformation. The model was refined using Refmac5 (Murshudov et al., 1997) (Table 1). All images were generated using Pymol, and sequence conservation was mapped onto the surface using the Consurf server (Landau et al., 2005).
Table 1.
Phasing and X-Ray Refinement Statistics
| Closed Form (pH 7) | Open Form (pH 5) | ||
|---|---|---|---|
| Data Collection Statistics | |||
| Wavelength (Å) | 0.9762 | 0.9797 | 1.7112 |
| Resolution range | 30–2.0 (2.11–2.0) | 30–2.0 (2.11–2.0) | 30–2.5 (2.64–2.5) |
| Completness (%) | 96.9 (97.1) | 98.3 (100) | 100 (99.9) |
| I/σI | 6.2 (1.6) | 5.6 (1.3) | 10 (3.2) |
| Rsym | 0.088 (0.340) | 0.070 (0.267) | 0.043 (0.180) |
| Rano | – | – | 0.052 (0.139) |
| Refinement | |||
| Resolution range (Å) | 30–2.0 | 30–2.0 | |
| Number of reflections | 22,270 | 37,869 | |
| Rwork/Rfree (%) | 20.0/27.2 | 20.9/23.4 | |
| Number of atoms | 2,771 | 2,979 | |
| Protein | 2,613 | 2,798 | |
| Ion | – | 3 | |
| Water | 158 | 178 | |
| B-factors | |||
| Protein | 33.4 | 28.7 | |
| Ion | – | 27.1 | |
| Water | 44.0 | 36.9 | |
| Root mean squared deviations | |||
| Bond lengths (Å) | 0.02 | 0.018 | |
| Bond angles (°) | 2.20 | 1.797 |
Mutations and VDE Activity
Wild-type VDE from Arabidopsis, cloned into a modified pQE60 vector as described by Hieber et al. (2002), was expressed using E. coli Origami cells. Site-specific mutants were produced using the QuickChange site-directed mutagenesis kit (Stratagene). VDE activity of protein purified by nickel affinity chromatography was determined by monitoring the change in absorbance at 502 to 540 nm as described (Yamamoto, 1985; Bugos et al., 1999). In the case of inactive mutants, the absence of Z production was confirmed by HPLC (Havaux et al., 2005). The enzyme assay contained 1.5 μM V (purified from spinach by HPLC), 40 μM MGDG (from Lipid Products), 60 mM ascorbate, and 200 mM citrate, pH 5. In each test, around 0.02 enzyme units (equivalent to ∼1 μg of VDE) of wild-type protein were employed. In the case of mutants, the protein amount required to obtain a signal equivalent to the wild type was quantified from protein gel blotting using antibodies recognizing the His-tag.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under accession number At1g08550 (Arabidopsis VDE). Coordinates and structure factors of VDEcd in its closed and open forms have been deposited in the Protein Data Bank (http://www.rcsb.org) with the accession codes 3CQN and 3CQR, respectively.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Movie 1. Morphing and Plausible Transitions between the Closed (pH 7) and open (pH 5) states of VDEcd.
Supplementary Material
Acknowledgments
We thank Prof. H.Y. Yamamoto for the gift of complementary DNA encoding the full-length VDE, and the ESRF staff from beamlines FIP and ID23-1 for their help in data collection. We also thank R. Kahn for providing GdHPDO3 salts and J. Lavergne for comments on the manuscript. T.M. and G.S. thank G.M. Giacometti (Università di Padova) for support.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Tomas Morosinotto (tomas.morosinotto@unipd.it) or David Pignol (david.pignol@cea.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Ahn, T.K., Avenson, T.J., Ballottari, M., Cheng, Y.C., Niyogi, K.K., Bassi, R., and Fleming, G.R. (2008). Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320 794–797. [DOI] [PubMed] [Google Scholar]
- Amarie, S., Laura, W., Tiago, B., Werner, K., Andreas, D., and Josef, W. (2009). Properties of zeaxanthin and its radical cation bound to the minor light-harvesting complexes CP24, CP26 and CP29. Biochimic. Biophys. Acta 1787 747–752. [DOI] [PubMed] [Google Scholar]
- Barber, J., and Andersson, B. (1992). Too much of a good thing: Light can be bad for photosynthesis. Trends Biochem. Sci. 17 61–66. [DOI] [PubMed] [Google Scholar]
- Bratt, C., Arvidsson, P., Carlsson, M., and Akerlund, H. (1995). Regulation of violaxanthin de-epoxidase activity by pH and ascorbate. Photosynth. Res. 45 169–175. [DOI] [PubMed] [Google Scholar]
- Bugos, R.C., Chang, S.H., and Yamamoto, H.Y. (1999). Developmental expression of violaxanthin de-epoxidase in leaves of tobacco growing under high and low light. Plant Physiol. 121 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugos, R.C., Hieber, A.D., and Yamamoto, H.Y. (1998). Xanthophyll cycle enzymes are members of the lipocalin family, the first identified from plants. J. Biol. Chem. 273 15321–15324. [DOI] [PubMed] [Google Scholar]
- Bugos, R.C., and Yamamoto, H.Y. (1996). Molecular cloning of violaxanthin de-epoxidase from romaine lettuce and expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 93 6320–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanacci, V., Bishop, R.E., Blangy, S., Tegoni, M., and Cambillau, C. (2006). The membrane bound bacterial lipocalin Blc is a functional dimer with binding preference for lysophospholipids. FEBS Lett. 580 4877–4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50 760–763. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams, B. (1990). Carotenoids and photoprotection in plants: A role for the xanthophyll zeaxanthin. Biochim. Biophysic. Acta 1020 1–24. [Google Scholar]
- Demmig-Adams, B., and Adams III, W.W. (2002). Antioxidants in photosynthesis and human nutrition. Science 298 2149–2153. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, A., Eskling, M., and Akerlund, H. (2003). Chemical and mutational modification of histidines in violaxanthin de-epoxidase from Spinacia oleacera. Physiol. Plant. 119 97–104. [Google Scholar]
- Emsley, P., and Cowtan, K. (2004). Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60 2126–2132. [DOI] [PubMed] [Google Scholar]
- Evans, P.R. (2006). Scaling and assessment of data quality. Acta Cryst. D. 62 72–82. [DOI] [PubMed] [Google Scholar]
- Frommolt, R., Goss, R., and Wilhelm, C. (2001). The de-epoxidase and epoxidase reactions of Mantoniella squamata (Prasinophyceae) exhibit different substrate-specific reaction kinetics compared to spinach. Planta 213 446–456. [DOI] [PubMed] [Google Scholar]
- Ganfornina, M.D., Gutierrez, G., Bastiani, M., and Sanchez, D. (2000). A phylogenetic analysis of the lipocalin protein family. Mol. Biol. Evol. 17 114–126. [DOI] [PubMed] [Google Scholar]
- Girard, E., Stelter, M., Vicat, J., and Kahn, R. (2003). A new class of lanthanide complexes to obtain high-phasing-power heavy-atom derivatives for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 59 1914–1922. [DOI] [PubMed] [Google Scholar]
- Gisselsson, A., Szilagyi, A., and Akerlund, H. (2004). Role of histidines in the binding of violaxanthin de-epoxidase to the thylakoid membrane as studied by site-directed mutagenesis. Physiol. Plant. 122 337–343. [Google Scholar]
- Grzyb, J., Latowski, D., and Strzalka, K. (2006). Lipocalins - A family portrait. J. Plant Physiol. 163 895–915. [DOI] [PubMed] [Google Scholar]
- Hager, A., and Holocher, K. (1994). Localization of the xanthophyll-cycle enzyme violaxanthin de-epoxidase within the thylakoid lumen and abolition of its mobility by a (light-dependant) pH decrease. Planta 192 581–589. [Google Scholar]
- Hartel, H., Lokstein, H., Grimm, B., and Rank, B. (1996). Kinetic studies on the xanthophyll cycle in barley leaves (influence of antenna size and relations to nonphotochemical chlorophyll fluorescence quenching). Plant Physiol. 110 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux, M., Dall'osto, L., and Bassi, R. (2007). Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol. 145 1506–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux, M., Eymery, F., Porfirova, S., Rey, P., and Dormann, P. (2005). Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 17 3451–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux, M., and Niyogi, K.K. (1999). The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc. Natl. Acad. Sci. USA 96 8762–8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieber, A.D., Bugos, R.C., and Yamamoto, H.Y. (2000). Plant lipocalins: Violaxanthin de-epoxidase and zeaxanthin epoxidase. Biochim. Biophys. Acta 1482 84–91. [DOI] [PubMed] [Google Scholar]
- Hieber, A.D., Bugos, R.C., Verhoeven, A.S., and Yamamoto, H.Y. (2002). Overexpression of violaxanthin de-epoxidase: Properties of C-terminal deletions on activity and pH-dependent lipid binding. Planta 214 476–483. [DOI] [PubMed] [Google Scholar]
- Hofmann, E., Zerbe, P., and Schaller, F. (2006). The crystal structure of Arabidopsis thaliana allene oxide cyclase: Insights into the oxylipin cyclization reaction. Plant Cell 18 3201–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, N.E., Fleming, G.R., and Niyogi, K.K. (2004). Toward an understanding of the mechanism of nonphotochemical quenching in green plants. Biochemistry 43 8281–8289. [DOI] [PubMed] [Google Scholar]
- Holt, N.E., Zigmantas, D., Valkunas, L., Li, X.P., Niyogi, K.K., and Fleming, G.R. (2005). Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307 433–436. [DOI] [PubMed] [Google Scholar]
- Horton, P., Wentworth, M., and Ruban, A. (2005). Control of the light harvesting function of chloroplast membranes: The LHCII-aggregation model for non-photochemical quenching. FEBS Lett. 579 4201–4206. [DOI] [PubMed] [Google Scholar]
- Johnson, M.P., Havaux, M., Triantaphylides, C., Ksas, B., Pascal, A.A., Robert, B., Davison, P.A., Ruban, A.V., and Horton, P. (2007). Elevated zeaxanthin bound to oligomeric LHCII enhances the resistance of Arabidopsis to photooxidative stress by a lipid-protective, antioxidant mechanism. J. Biol. Chem. 282 22605–22618. [DOI] [PubMed] [Google Scholar]
- Kawano, M., and Kuwabara, T. (2000). pH-dependent reversible inhibition of violaxanthin de-epoxidase by pepstatin related to protonation-induced structural change of the enzyme. FEBS Lett. 481 101–104. [DOI] [PubMed] [Google Scholar]
- Kuwabara, T., Hasegawa, M., Kawano, M., and Takaichi, S. (1999). Characterization of violaxanthin de-epoxidase purified in the presence of Tween 20: Effects of dithiothreitol and pepstatin A. Plant Cell Physiol. 40 1119–1126. [DOI] [PubMed] [Google Scholar]
- Landau, M., Mayrose, I., Rosenberg, Y., Glaser, F., Martz, E., Pupko, T., and Ben-Tal, N. (2005). ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33 W299–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascombe, M.B., Gregoire, C., Poncet, P., Tavares, G.A., Rosinski-Chupin, I., Rabillon, J., Goubran-Botros, H., Mazie, J.C., David, B., and Alzari, P.M. (2000). Crystal structure of the allergen Equ c 1. A dimeric lipocalin with restricted IgE-reactive epitopes. J. Biol. Chem. 275 21572–21577. [DOI] [PubMed] [Google Scholar]
- Leslie, A.G.W. (1993). Autoindexing of rotation diffracton images and parameter refinement. In Proceedings of the CCP4 Study Weekend: Data Collection and Processing, L. Sawyer, N. Isaacs, and S. Bailey, eds (Daresbury,UK: SERC Daresbury Laboratory), pp. 44–51.
- Liu, Z., Yan, H., Wang, K., Kuang, T., Zhang, J., Gui, L., An, X., and Chang, W. (2004). Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428 287–292. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53 240–255. [DOI] [PubMed] [Google Scholar]
- Nagata, A., Suzuki, Y., Igarashi, M., Eguchi, N., Toh, H., Urade, Y., and Hayaishi, O. (1991). Human brain prostaglandin D synthase has been evolutionarily differentiated from lipophilic-ligand carrier proteins. Proc. Natl. Acad. Sci. USA 88 4020–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer, C., and Yamamoto, H.Y. (1994). Membrane barriers and Mehler-peroxidase reaction limit the ascorbate available for violaxanthin de-epoxidase activity in intact chloroplasts. Photosynth. Res. 39 137–147. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K. (2000). Safety valves for photosynthesis. Curr. Opin. Plant Biol. 3 455–460. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K., Grossman, A.R., and Bjorkman, O. (1998). Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche, S., Rey, F.A., Gaudin, Y., and Bressanelli, S. (2007). Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315 843–848. [DOI] [PubMed] [Google Scholar]
- Schlehuber, S., and Skerra, A. (2005). Lipocalins in drug discovery: From natural ligand-binding proteins to “anticalins”. Drug Discov. Today 10 23–33. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. (2003). SOLVE and RESOLVE: Automated structure solution and density modification. Methods Enzymol. 374 22–37. [DOI] [PubMed] [Google Scholar]
- Vagin, A., and Teplyakov, A. (2000). An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 56 1622–1624. [DOI] [PubMed] [Google Scholar]
- Yamamoto, H.Y. (1985). Xanthophyll cycles. Methods Enzymol. 110 303–312. [Google Scholar]
- Yamamoto, H.Y., and Higashi, R.M. (1978). Violaxanthin de-epoxidase. Lipid composition and substrate specificity. Arch. Biochem. Biophys. 190 514–522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.