Abstract
Plants have developed numerous mechanisms to store hormones in inactive but readily available states, enabling rapid responses to environmental changes. The phytohormone auxin has a number of storage precursors, including indole-3-butyric acid (IBA), which is apparently shortened to active indole-3-acetic acid (IAA) in peroxisomes by a process similar to fatty acid β-oxidation. Whereas metabolism of auxin precursors is beginning to be understood, the biological significance of the various precursors is virtually unknown. We identified an Arabidopsis thaliana mutant that specifically restores IBA, but not IAA, responsiveness to auxin signaling mutants. This mutant is defective in PLEIOTROPIC DRUG RESISTANCE8 (PDR8)/PENETRATION3/ABCG36, a plasma membrane–localized ATP binding cassette transporter that has established roles in pathogen responses and cadmium transport. We found that pdr8 mutants display defects in efflux of the auxin precursor IBA and developmental defects in root hair and cotyledon expansion that reveal previously unknown roles for IBA-derived IAA in plant growth and development. Our results are consistent with the possibility that limiting accumulation of the IAA precursor IBA via PDR8-promoted efflux contributes to auxin homeostasis.
INTRODUCTION
As sessile organisms, plants rely on a variety of signaling mechanisms to respond to their environment. One key to rapid response may be the availability of hormone storage molecules. The critical phytohormone auxin is stored in a variety of forms that can be metabolized into the active auxin indole-3-acetic acid (IAA; reviewed in Woodward and Bartel, 2005b). Active auxin regulates cell division and elongation to control plant growth and developmental processes, such as embryo and leaf patterning, root and stem elongation, lateral root initiation, and leaf expansion (reviewed in Davies, 2004; Woodward and Bartel, 2005b).
In addition to storage in amide- or ester-linked conjugates with amino acids, peptides, and sugars (reviewed in Woodward and Bartel, 2005b), auxin can be stored in the side chain–lengthened precursor indole-3-butyric acid (IBA). IBA, initially described as a synthetic auxin that induces rooting in woody plant cuttings (Cooper, 1935), is an endogenous plant compound (Blommaert, 1954). Genetic evidence in Arabidopsis thaliana suggests that auxin activity of IBA requires peroxisomal shortening of the four-carbon carboxyl side chain to the two-carbon side chain of IAA (Zolman et al., 2000, 2007). For example, peroxisome biogenesis mutants, such as pex5 and pex7, are resistant to exogenous IBA, but remain sensitive to IAA (Zolman et al., 2000; Woodward and Bartel, 2005a). Additionally, mutants defective in the peroxisomally localized apparent β-oxidation enzymes IBR1, IBR3, or IBR10 are IBA resistant and IAA sensitive, suggesting that the conversion of IBA to IAA takes place in a process similar to peroxisomal fatty acid β-oxidation (Zolman et al., 2007, 2008).
Auxin transport is tightly regulated, controlled by specialized influx and efflux carriers (reviewed in Vieten et al., 2007). Several families of IAA carriers have been identified. IAA influx appears to primarily be controlled by the AUXIN RESISTANT1 (AUX1) family of proteins, which are similar to amino acid permeases (reviewed in Vieten et al., 2007). aux1 mutants are resistant to IAA and 2,4-D (Marchant et al., 1999), which are substrates of the AUX1 transporter (Yang et al., 2006). The IAA efflux carrier PIN-FORMED (PIN) proteins are often localized to a specific face of the cell to direct appropriate polar auxin transport in an organ (reviewed in Vieten et al., 2007). pin mutants have altered local IAA accumulation patterns and can be phenocopied by chemically inhibiting polar auxin transport (Luschnig et al., 1998; Friml et al., 2002b, 2002a, 2003). Additional auxin transporters include members of the MULTIDRUG RESISTANCE/P-GLYCOPROTEIN (PGP) class of ATP binding cassette (ABC) transporters. Members of this ABCB family of auxin effluxers may mediate long-distance IAA transport, providing IAA to the PIN proteins for vectorial transport (Bandyopadhyay et al., 2007; Blakeslee et al., 2007; Bailly et al., 2008; Mravec et al., 2008).
IAA is recognized by TIR1/AFB receptor proteins, components of SCFTIR1/AFB ubiquitin-protein ligases, which when bound to auxin promote degradation of Aux/IAA transcriptional repressors by the 26S proteasome (reviewed in Parry and Estelle, 2006). Degradation of Aux/IAA proteins likely permits auxin-responsive transcription by relieving repression of the activating class of AUXIN RESPONSE FACTOR (ARF) proteins. Mutations in genes encoding or modulating the SCFTIR1/AFB complex, including components of the AXR1/ECR1 pathway, confer resistance to applied and endogenous auxin because Aux/IAA proteins are stabilized and auxin-responsive transcription is reduced (reviewed in Woodward and Bartel, 2005b; Parry and Estelle, 2006). The Arabidopsis iba response5 (ibr5) mutant is defective in a dual-specificity protein phosphatase and is resistant to applied and endogenous auxin (Monroe-Augustus et al., 2003; Lee et al., 2009). Although ibr5, like other auxin response mutants, exhibits decreased basal and auxin-induced levels of the auxin-responsive transcriptional reporter DR5-β-glucuronidase (GUS) (Monroe-Augustus et al., 2003; Strader et al., 2008a), Aux/IAA proteins appear not to be stabilized in ibr5 (Strader et al., 2008a), making ibr5 unique among characterized auxin response mutants.
We have isolated and characterized a set of modifier mutations that restore IBA responsiveness to ibr5 (Strader et al., 2008b). Some of these suppressors restore responsiveness to both IBA and IAA, whereas others restore IBA responsiveness but retain IAA resistance. Here, we report that loss-of-function mutations in PLEIOTROPIC DRUG RESISTANCE8/PENETRATION3/ABCG36 (PDR8/PEN3/ABCG36) specifically restore ibr5 responsiveness to auxins with four-carbon side chains. We demonstrate that pdr8 mutations can suppress the IBA (but not IAA) resistance of additional auxin signaling mutants and that pdr8 in a wild-type IBR5 background is IBA hypersensitive, a phenotype that is suppressed by mutants unable to convert IBA into IAA. Our characterization provides insight into transport of the auxin precursor IBA and is consistent with the possibility that IAA from IBA can promote root hair and cotyledon growth and development.
RESULTS
Mutations in the PDR8/ABCG36 Transporter Gene Suppress a Subset of ibr5 Phenotypes
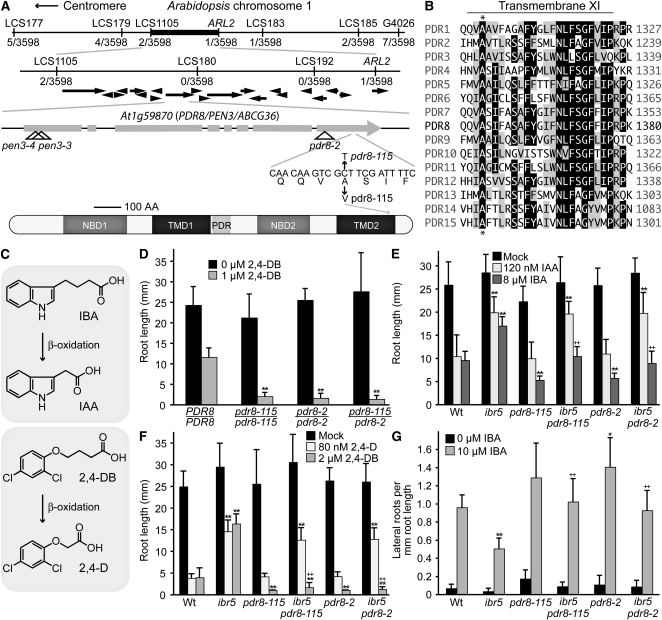
MS115 was isolated as a suppressor that restores ibr5 IBA sensitivity without suppressing ibr5 resistance to IAA (Strader et al., 2008b). The synthetic auxins 2,4-D and 2,4-dichlorophenoxybutyric acid (2,4-DB) are analogous to IAA and IBA; 2,4-D differs from 2,4-DB in that it has a two- rather than four-carbon side chain (Figure 1C). Like IBA (Zolman et al., 2000, 2007), 2,4-DB likely requires peroxisomal carboxyl side-chain shortening to 2,4-D for auxin activity (Hayashi et al., 1998). The MS115 suppressor restores 2,4-DB sensitivity to ibr5 but fails to suppress ibr5 2,4-D resistance (Strader et al., 2008b). We used the phenotype of restored IBA and 2,4-DB responsiveness to map the ibr5-suppressing lesion in MS115 to a 72-kb region on the lower arm of chromosome 1 (Figure 1A). This region contains PDR8/ABCG36, previously identified as the gene defective in pen3, a mutant displaying increased susceptibility to nonhost pathogens (Stein et al., 2006). pdr8 mutants also display altered responses to other pathogens (Kobae et al., 2006; Stein et al., 2006), decreased extracellular accumulation of Flg22-induced callose (Clay et al., 2009), hyperaccumulation of Flg22- or pathogen-elicited 4-methoxyindol-3-ylmethylglucosinolate (Bednarek et al., 2009; Clay et al., 2009), hyperaccumulation of pathogen-elicited camalexin (Bednarek et al., 2009), and increased heavy metal sensitivity (Kim et al., 2007). Although PDR8 had not been implicated in auxin sensitivity, we considered it a candidate gene because mutation of a related ABC transporter, PDR9/ABCG37, restores ibr5 responses to a subset of auxinic compounds (Strader et al., 2008b). We PCR amplified and sequenced PDR8 from MS115 genomic DNA and identified a C-to-T base change at position 4844 (where the A of the ATG is position 1) that causes an Ala-to-Val missense mutation (Figure 1A) in an amino acid conserved in all Arabidopsis ABCG family members (Figure 1B). This mutation falls in the eleventh predicted transmembrane span, which is found in the second transmembrane domain of PDR8 (Figure 1B). We named the ibr5-suppressing mutation in MS115 pdr8-115.
Figure 1.
pdr8 Alleles Suppress a Subset of ibr5 Phenotypes.
(A) Recombination mapping with the indicated PCR-based markers (see Supplemental Table 1 online) localized MS115 to a 72-kb region containing 21 predicted genes (black arrows) between LCS1105 and ARL2 with 2/3598 north and 1/3598 south recombinants. Examination of the PDR8 (At1g59870) gene in this region revealed a C-to-T mutation at position 4844 in MS115 DNA and results in an Ala-1357 to Val substitution. pdr8-2 (SALK_142256) carries a T-DNA insert in the eighth exon of PDR8 (Kobae et al., 2006); pen3-3 (SALK_110926) and pen3-4 (SALK_000578) carry T-DNA inserts in the first exon of PDR8 (Stein et al., 2006). The cartoon shows a PDR8 schematic based on output from the domain-predicting program SMART (Schultz et al., 1998). PDR8 contains two nucleotide binding domains (NBD), two transmembrane domains (TMD) containing six predicted transmembrane spans each, and a PDR signature motif. A thirteenth predicted transmembrane span after TMD2 is not shown.
(B) The pdr8-115 mutation disrupts a conserved Ala in TMD2. The alignment shows the eleventh predicted transmembrane-spanning region of the 15 Arabidopsis PDR/ABCG family members (accession numbers listed in Supplemental Table 4 online). Sequences were aligned using the MegAlign program (DNAStar). Amino acid residues identical in at least eight sequences are boxed in black; chemically similar residues in at least eight sequences are shaded in gray. The position of the pdr8-115 mutation is indicated with an asterisk.
(C) IBA is a naturally occurring IAA precursor; 2,4-DB is a precursor of the synthetic auxin 2,4-D. Both IBA and 2,4-DB are converted to the active auxins IAA and 2,4-D in a process similar to fatty acid β-oxidation.
(D) pdr8-115 is allelic to pdr8-2. Complementation test showing primary root lengths of 8-d-old Col-0 wild-type (PRD8/PDR8), pdr8-115/pdr8-115, pdr8-2/pdr8-2, and pdr8-115/pdr8-2 seedlings grown under yellow-filtered light at 22°C on medium supplemented with ethanol (mock) or 1 μM 2,4-DB (n ≥ 10).
(E) Primary root lengths of 8-d-old Col-0 (Wt), ibr5-1, pdr8-115, ibr5-1 pdr8-115 (MS115), pdr8-2, and ibr5-1 pdr8-2 seedlings grown under yellow-filtered light at 22°C on medium supplemented with 120 nM IAA or 8 μM IBA (n ≥ 12).
(F) Primary root lengths of 8-day-old Col-0 (Wt), ibr5-1, pdr8-115, ibr5-1 pdr8-115 (MS115), pdr8-2, and ibr5 pdr8-2 seedlings grown under yellow-filtered light at 22°C on medium supplemented with 80 nM 2,4-D or 2 μM 2,4-DB (n ≥ 15).
(G) Lateral roots were counted 4 d after transfer of 4-d-old seedlings to medium supplemented with either 0 (ethanol control) or 10 μM IBA (n = 12).
Error bars represent sd of the means. Statistically significant differences from the wild type in two-tailed t tests assuming unequal variance are indicated by single (P ≤ 0.001) and double (P ≤ 0.0001) asterisks. Statistically significant differences from ibr5-1 in two-tailed t tests assuming unequal variance are indicated by single (P ≤ 0.001) and double (P ≤ 0.0001) plus symbols.
To test whether the observed MS115 phenotypes were caused by the pdr8-115 lesion, we isolated the pdr8-115 mutant in a wild-type IBR5 background and then crossed pdr8-115 to the previously described pdr8-2 likely null allele (Kobae et al., 2006) and tested 2,4-DB responsiveness in the pdr8-115/pdr8-2 F1 progeny. We found that pdr8-2, like pdr8-115, was hypersensitive to root growth inhibition by 2,4-DB and that pdr8-2 failed to complement the 2,4-DB hypersensitivity of pdr8-115 (Figure 1D). Because both pdr8-2 (Kobae et al., 2006) and pdr8-115 (data not shown) are recessive, this noncomplementation confirms that the lesion we identified in pdr8-115 confers a loss of PDR8 function.
To determine whether other pdr8 alleles could suppress the same subset of ibr5 phenotypes as pdr8-115, we crossed pdr8-2 to ibr5-1 and compared ibr5 pdr8-2 auxin responses to those of ibr5 pdr8-115. We found that neither pdr8-115 nor pdr8-2 abrogated adult ibr5-1 morphological phenotypes (short stature and small serrated leaves; see Supplemental Figure 1A online). However, pdr8-115 and pdr8-2 similarly restored ibr5 primary root elongation inhibition responses to the auxin precursors IBA and 2,4-DB without affecting the resistance of ibr5 to the active auxins IAA and 2,4-D (Figures 1E and 1F).
In Arabidopsis, IBA promotes lateral root production (Zolman et al., 2000), and ibr5 mutants produce fewer lateral roots than the wild type both with and without auxin treatment (Monroe-Augustus et al., 2003). Like pdr8-115 (Strader et al., 2008b), pdr8-2 restores lateral root production to ibr5 with and without IBA treatment (Figure 1G). We previously reported that MS115 (ibr5-1 pdr8-115) remains resistant (like ibr5) to the auxin transport inhibitor 2,3,5-triiodobenzoic acid (TIBA), the ethylene precursor 1-aminocyclopropane-1-carboxylate (ACC), and the phytohormone abscisic acid (ABA) (Strader et al., 2008b). Similarly, pdr8-2 failed to restore ibr5 sensitivity to TIBA, ACC, or ABA (see Supplemental Figures 1B to 1D online).
Because pdr8-115 responded similarly to pdr8-2 in these assays and because both alleles similarly restored ibr5 responsiveness to IBA and 2,4-DB (Figures 1E to 1G), we concluded that the pdr8-115 lesion reduced PDR8 function and was responsible for suppression of the subset of ibr5 phenotypes that were restored in MS115.
pdr8 Mutants Are Hypersensitive to IBA and 2,4-DB and Exhibit Increased Lateral Root Production
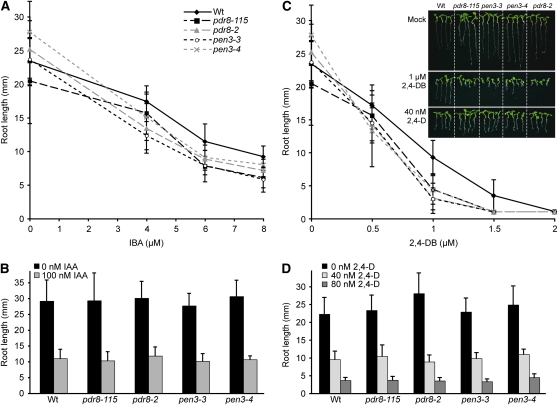
To examine the effects of pdr8 lesions in a wild-type IBR5 background, we compared the phenotypes of three PDR8 T-DNA insertional alleles (Figure 1A), pdr8-2 (Kobae et al., 2006), pen3-3, and pen3-4 (Stein et al., 2006), to those of our pdr8-115 missense allele. We found that all four pdr8 alleles displayed mild hypersensitivity to root elongation inhibition by IBA (Figure 2A) but wild-type IAA sensitivity (Figure 2B). Similarly, each pdr8 allele was hypersensitive to 2,4-DB in root elongation inhibition (Figure 2C) but retained wild-type 2,4-D sensitivity (Figures 2C and 2D).
Figure 2.
pdr8 Alleles Display Hypersensitivity to IBA and 2,4-DB.
(A) Primary root lengths of 8-d-old Col-0 (Wt), pdr8-115, pdr8-2, and pen3-4 seedlings grown under yellow-filtered light at 22°C on medium supplemented with ethanol (0 μM IBA) or various concentrations of IBA (n ≥ 15).
(B) Primary root lengths of 8-d-old Col-0 (Wt), pdr8-115, pdr8-2, and pen3-4 seedlings grown under yellow-filtered light at 22°C on medium supplemented with ethanol (0 nM IAA) or various concentrations of IAA (n ≥ 12).
(C) Primary root lengths of 8-d-old Col-0 (Wt), pdr8-115, pdr8-2, and pen3-4 seedlings grown under yellow-filtered light at 22°C on medium supplemented with ethanol (0 μM 2,4-DB) or various concentrations of 2,4-DB (n ≥ 15). Inset: Photograph of 8-d-old Col-0 (Wt), pdr8-115, pdr8-2, and pen3-4 seedlings grown under yellow light at 22°C on medium supplemented with ethanol (mock), 1 μM 2,4-DB, or 40 nM 2,4-D.
(D) Primary root lengths of 8-d-old Col-0 (Wt), pdr8-115, pdr8-2, and pen3-4 seedlings grown under yellow-filtered light at 22°C on medium supplemented with ethanol (0 μM 2,4-D) or various concentrations of 2,4-D (n ≥ 15).
Error bars represent sd of the means. pdr8-2 and pen3-3 roots were significantly shorter than wild-type roots on 4 μM IBA (P ≤ 0.001), and pdr8-115, pdr8-2, pen3-3, and pen3-4 roots were significantly shorter than wild-type roots on 6 and 8 μM IBA (P ≤ 0.001) in two-tailed t tests assuming unequal variance. pdr8-115, pdr8-2, pen3-3, and pen3-4 roots were significantly shorter than wild-type roots on 1 and 1.5 μM 2,4-DB (P ≤ 0.001) in two-tailed t tests assuming unequal variance.
[See online article for color version of this figure.]
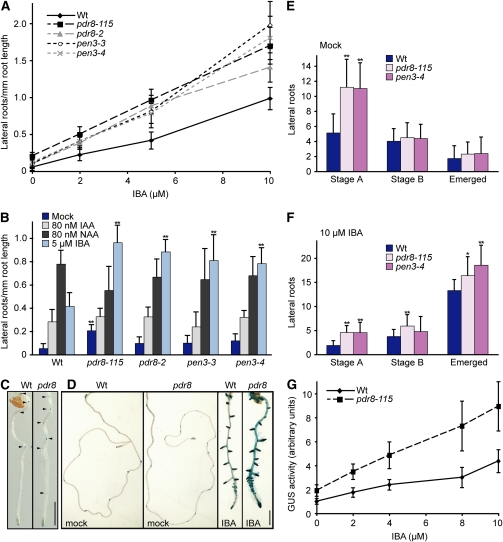
Like pdr8-115, the pdr8 insertional alleles were all hypersensitive to the lateral root–inducing activity of IBA (Figures 3A and 3B) but displayed wild-type sensitivity to IAA and the synthetic auxin 1-naphthylacetic acid (NAA) (Figure 3B). Additionally, the pdr8 alleles all displayed modestly increased lateral root production in the absence of exogenous hormone (Figures 3A and 3B), suggesting that PDR8 normally inhibits lateral root formation. To examine pdr8 lateral root initiation in more detail, we crossed pdr8-115 to the wild type carrying DR5-GUS, a reporter in which GUS expression is driven from a synthetic auxin-responsive promoter (Ulmasov et al., 1997). DR5-GUS is expressed in early lateral root primordia, facilitating the detection of lateral roots prior to emergence. We observed more lateral root primordia (LRP) in 4- and 8-d-old pdr8 than the wild type (Figures 3C and 3D). To quantify differences in LRP, we examined cleared wild-type, pdr8-115, and pen3-4 roots and counted LRP at different developmental stages: LRP of up to three cell layers (stage A), unemerged LRP greater than three cell layers (stage B), and emerged lateral roots. We found that pdr8-115 and pen3-4 exhibited significantly more stage A LRP than the wild type both in the absence and presence of IBA (Figures 3E and 3F) as well as more emerged lateral roots than the wild type following IBA treatment (Figure 3F). In addition to increased LRP, pdr8 DR5-GUS seedlings exhibited more GUS activity in the primary root than the wild type in response to IBA (Figure 3D). Quantitative fluorometric GUS assays showed that GUS activity was higher in pdr8 DR5-GUS than wild-type DR5-GUS with or without IBA treatment (Figure 3G), suggesting that endogenous auxin signaling is increased in pdr8 and demonstrating that the DR5 promoter is more sensitive to IBA in the pdr8 background.
Figure 3.
pdr8 Mutants Exhibit Increased Lateral Root Production.
(A) Lateral roots of Col-0 (Wt), pdr8-115, pdr8-2, and pen3-4 were counted 4 d after transfer of 4-d-old seedlings to medium supplemented with either 0 μM (ethanol control) or various concentrations of IBA (n = 12).
(B) Lateral roots of Col-0 (Wt), pdr8-115, pdr8-2, and pen3-4 were counted 4 d after transfer of 4-d-old seedlings to medium supplemented with either ethanol (Mock) or the indicated concentrations of IBA, IAA, or NAA (n = 12).
(C) Four-day-old Col-0 (Wt) and pdr8-115 seedlings carrying the DR5-GUS construct (Ulmasov et al., 1997) were stained for GUS activity. Arrowheads indicate lateral root primordia.
(D) Four-day-old Col-0 (Wt) and pdr8-115 seedlings carrying the DR5-GUS construct were transferred to medium supplemented with either 0 (ethanol control) or 10 μM IBA, grown under yellow light at 22°C for an additional 4 d, and then stained for GUS activity. Bar = 1 mm.
(E) and (F) Four-day-old Col-0 (Wt) and pdr8-115 seedlings were transferred to medium supplemented with either 0 (Mock) (E) or 10 μM IBA (F), grown under yellow light at 22°C for an additional 4 d, and then cleared and the number and stage of lateral root primordia recorded (n ≥ 30). Stage A spans the first anticlinal division of a pericycle cell to a lateral root primordium with three cell layers. Stage B consists of preemergent lateral roots with more than three cell layers.
(G) Four-day-old Col-0 (Wt) and pdr8-115 seedlings carrying the DR5-GUS construct were transferred to medium supplemented with either 0 (ethanol control) or various concentrations of IBA, grown under yellow light at 22°C for an additional 4 d, and then fluorometrically assayed GUS activity. GUS activity is presented as normalized fluorescence. Data are from 16 replicates of three seedlings each.
Error bars represent sd of the means. Statistically significant differences from the wild type in two-tailed t tests assuming unequal variance are indicated by single (P ≤ 0.001) and double (P ≤ 0.0001) asterisks in (B), (E), and (F). In (A), pdr8-115 exhibited significantly more lateral roots per root length than the wild type in the absence of IBA treatment (P ≤ 0.0001); pdr8-115, pdr8-2, pen3-3, and pen3-4 exhibited significantly more lateral roots per root length than the wild type at all IBA concentrations (P ≤ 0.0001). In (G), GUS activity in pdr8-115 carrying the DR5-GUS construct was significantly higher than in the wild type carrying the DR5-GUS construct in the absence of IBA and at all tested IBA concentrations (P ≤ 0.0001).
pdr8 Affects Subsets of tir1, axr1, and aux1 Auxin Response Defects
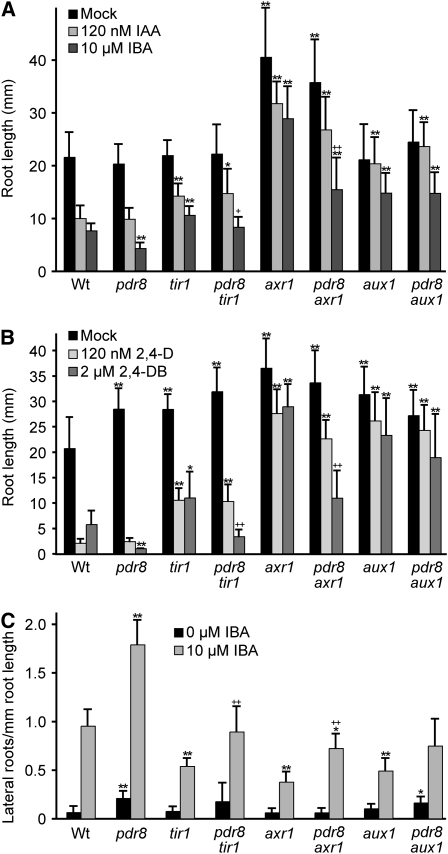
The tir1, axr1, and aux1 mutants, like ibr5 (Monroe-Augustus et al., 2003), are resistant to auxin in primary root elongation inhibition and are deficient in lateral root formation (Maher and Martindale, 1980; Lincoln et al., 1990; Hobbie and Estelle, 1995; Ruegger et al., 1998; Marchant et al., 2002). Because ibr5 differs from tir1 and axr1 in that ibr5 appears to affect auxin responses without stabilizing Aux/IAA proteins (Strader et al., 2008a), and because a previously reported suppressor of axr1 auxin resistance, sar3 (Parry et al., 2006), does not restore auxin responsiveness to ibr5 or tir1 (Strader et al., 2008b), we assessed whether loss of PDR8 could suppress tir1, axr1, or aux1 auxin response phenotypes. We found that pdr8 suppressed the 2,4-DB and IBA-resistant root elongation phenotypes of both tir1 and axr1 (Figures 4A and 4B). Similarly, pdr8 suppressed the IBA-resistant lateral root production phenotype of tir1 and axr1 (Figure 4C). By contrast, pdr8 failed to suppress the IAA or 2,4-D resistance of tir1 and axr1 (Figures 4A and 4B). We concluded that pdr8 generally promotes IBA and 2,4-DB responsiveness and is not specific to the IBR5 pathway.
Figure 4.
pdr8 Suppresses a Subset of Auxin Response Mutant Defects.
(A) Normalized root lengths of 8-d-old Col-0 (Wt), pdr8-115, tir1-1, pdr8-115 tir1-1, axr1-3, pdr8-115 axr1-3, aux1-7, and pdr8-115 aux1-7 seedlings grown under yellow-filtered light at 22°C on medium supplemented with 120 nM IAA or 10 μM IBA (n ≥ 15).
(B) Normalized root lengths of 8-d-old Col-0 (Wt), pdr8-115, tir1-1, pdr8-115 tir1-1, axr1-3, pdr8-115 axr1-3, aux1-7, and pdr8-115 aux1-7 seedlings grown under yellow-filtered light at 22°C on medium supplemented with 120 nM 2,4-D or 2 μM 2,4-DB (n ≥ 13).
(C) Lateral roots were counted 4 d after transfer of 4-d-old seedlings to medium supplemented with either 0 (ethanol control) or 10 μM IBA (n = 15).
Error bars represent sd of the means. Statistically significant differences from the wild type in two-tailed t tests assuming unequal variance are indicated by single (P ≤ 0.001) and double (P ≤ 0.0001) asterisks. Statistically significant differences between the auxin-resistant parent and its corresponding pdr8 double mutant in two-tailed t tests assuming unequal variance are indicated by single (P ≤ 0.001) and double (P ≤ 0.0001) plus symbols.
We also tested whether loss of PDR8 could suppress aux1 auxin resistance phenotypes. The aux1 mutant is resistant to 2,4-D and IAA, which are brought into cells by the AUX1 transporter (Maher and Martindale, 1980; Yamamoto and Yamamoto, 1998; Marchant et al., 1999; Yang et al., 2006) but responds normally to NAA, which is not transported by AUX1 (Yamamoto and Yamamoto, 1998; Marchant et al., 1999; Yang et al., 2006). IBA appears not to be an AUX1 substrate (Rashotte et al., 2003; Yang et al., 2006; Strader et al., 2008b), yet the aux1 mutant is moderately IBA resistant, likely because of resistance to IAA converted from IBA (Zolman et al., 2000). We found that the aux1 mutant and the pdr8 aux1 double mutant displayed similar resistance to root elongation inhibition by IAA, IBA (Figure 4A), 2,4-D, and 2,4-DB (Figure 4B). Thus, unlike its effects on ibr5, tir1, and axr1, pdr8 failed to suppress the IBA or 2,4-DB resistant root elongation phenotypes of aux1. By contrast, pdr8 did appear to suppress aux1 defects in IBA-induced lateral root initiation (Figure 4C).
Mutants Defective in Peroxisome Biogenesis and Import Suppress pdr8
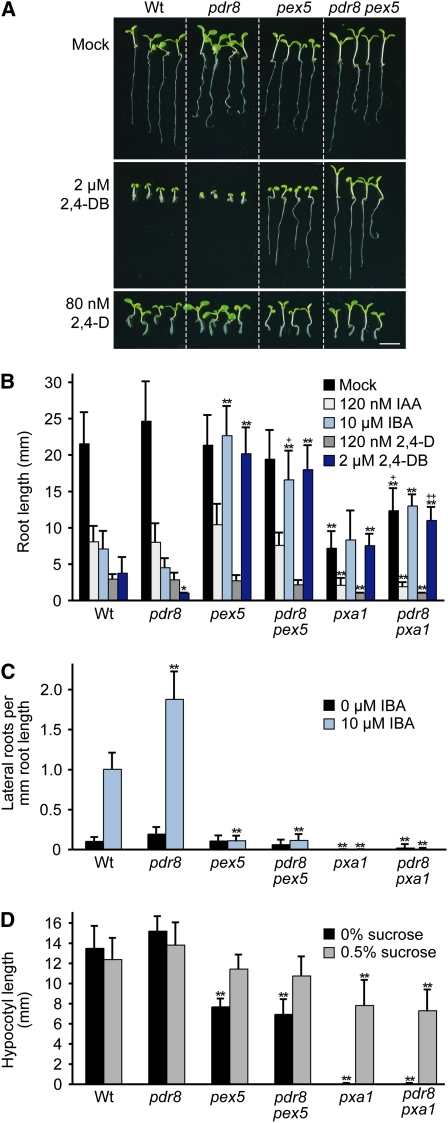
Because pdr8 appears to promote IBA and 2,4-DB responsiveness, but not IAA or 2,4-D responsiveness, we examined the genetic interaction between pdr8 and mutants specifically defective in IBA and 2,4-DB responses. pex5-1 (Zolman et al., 2000) and pxa1-1 (Zolman et al., 2001) are defective in import of peroxisomal enzymes and β-oxidation substrates, respectively. Both pex5 and pxa1 retain wild-type IAA and 2,4-D sensitivity but are resistant to IBA and 2,4-DB, presumably because peroxisomal β-oxidation of IBA to IAA and 2,4-DB to 2,4-D is impaired (Zolman et al., 2000, 2001). The pxa1 mutant forms fewer lateral roots than the wild type, even in the absence of hormone supplementation, suggesting that endogenous IAA from IBA contributes to lateral root development (Zolman et al., 2001). Moreover, pex5 and pxa1 mutants are sucrose dependent during early seedling development, likely because reduced seed storage fatty acid β-oxidation rates result in an energy deficit (Zolman et al., 2000, 2001).
We found that the pex5 and pxa1 mutants fully suppressed the IBA and 2,4-DB root elongation inhibition hypersensitivity of pdr8; the pdr8 pex5 and pdr8 pxa1 double mutant roots were as IBA and 2,4-DB resistant as the pex5 and pxa1 single mutants (Figures 5A and 5B). Interestingly, the pdr8 pex5 double mutant shoots appeared to be even more 2,4-DB resistant than the pex5 single mutant (Figure 5A). In addition, the increased lateral root production of pdr8 both with and without supplemental IBA was fully suppressed by pex5 or pxa1 (Figure 5C). The double mutants retained the wild-type sensitivity to IAA and 2,4-D of the parents (Figures 5A and 5B), and pdr8 had no effect on the sucrose dependence of either pex5-1 or pxa1-1 (Figure 5D). These epistatic relationships suggest that the increased pdr8 response to long side chain auxins requires β-oxidation of these precursors to IAA or 2,4-D in peroxisomes.
Figure 5.
The Heightened Responses of pdr8 to IBA and 2,4-DB Are Suppressed by Peroxisome Biogenesis and Transport Mutants.
(A) Photograph of 8-d-old Col-0 (Wt), pdr8-115, pex5-1, and pdr8-115 pex5-1 seedlings grown under yellow light at 22°C on medium supplemented with ethanol (mock), 2 μM 2,4-DB, or 80 nM 2,4-D. Bar = 1 cm.
(B) Primary root lengths of 8-d-old Col-0 (Wt), pdr8-115, pex5-1, pdr8-115 pex5-1, pxa1-1, and pdr8-115 pxa1-1 seedlings grown under yellow-filtered light at 22°C on medium supplemented with 120 nM IAA, 10 μM IBA, 120 nM 2,4-D, or 2 μM 2,4-DB (n ≥ 11).
(C) Lateral roots were counted 4 d after transfer of 4-d-old seedlings to medium supplemented with either 0 (ethanol control) or 10 μM IBA (n = 12).
(D) Sucrose dependence of Col-0 (Wt), pdr8-115, pex5-1, pdr8-115 pex5-1, pxa1-1, and pdr8-115 pxa1-1. Hypocotyl lengths were measured 4 d after transfer of 1-d-old seedlings to the dark (n ≥ 16).
Error bars represent sd of the means. Statistically significant differences from the wild type in two-tailed t tests assuming unequal variance are indicated by single (P ≤ 0.001) and double (P ≤ 0.0001) asterisks. Statistically significant differences between the resistant parent and its corresponding pdr8 double mutant in two-tailed t tests assuming unequal variance are indicated by single (P ≤ 0.001) and double (P ≤ 0.0001) plus symbols. pdr8 pex5 had significantly fewer lateral roots than pdr8 in both mock- (P ≤ 0.001) and IBA-treated (P ≤ 0.0001) conditions. pdr8 pxa1 had significantly fewer lateral roots than pdr8 in both mock and IBA-treated conditions (P ≤ 0.0001).
Additional pdr8 Developmental Phenotypes
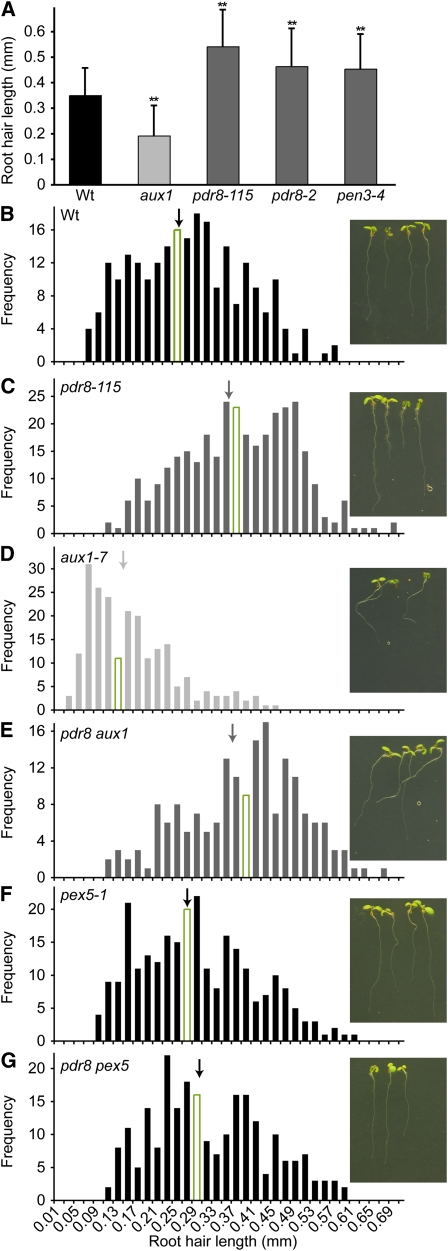
In addition to increased lateral root production (Figure 3), we observed that pdr8 mutants had abnormally long root hairs (Figures 6A and 6C). Root hairs emerge from specialized epidermal cells called trichoblasts. Auxin promotes root hair elongation, and many mutants defective in auxin response or transport display altered root hair growth (reviewed in Grierson and Schiefelbein, 2002). Similar to the pgp4 IAA efflux mutant (Santelia et al., 2005; Cho et al., 2007) and unlike the aux1-7 IAA influx mutant (Figures 6A and 6D; Pitts et al., 1998), pdr8-115, pdr8-2, and pen3-4 displayed longer root hairs than the wild type (Figures 6A and 6C). The long root hairs of pdr8 were not suppressed by impeding IAA influx with the aux1 mutation (Figure 6E). In fact, pdr8 completely suppressed the short root hair phenotype of aux1; pdr8 aux1 root hairs were as long as pdr8 root hairs (Figures 6C and 6E). By contrast, pdr8 did not notably impact the agravitropic root growth of aux1 (Figure 6E, inset). These results suggest that loss of PDR8 can compensate for a lack of AUX1-facilitated IAA movement during root hair elongation but does not compensate for a lack of IAA movement into gravity responsive cells. Furthermore, pdr8 pex5 displayed wild-type root hair length (Figures 6F and 6G), suggesting that conversion of IBA to IAA is required for the lengthened root hairs found in pdr8.
Figure 6.
pdr8 Mutants Have Long Root Hairs.
(A) Lengths of roots hairs of 5-d-old vertically grown Col-0 (Wt), pdr8-115, pdr8-2, and pen3-4 seedling roots were measured using NIH Image software (n ≥ 158 total root hairs from at least 10 seedlings). Error bars represent sd of the means. Statistically significant (P ≤ 0.0001) differences from the wild type in two-tailed t tests assuming unequal variance are indicated by double asterisks.
(B) to (G) Histograms of root hair lengths of 5-d-old vertically grown Col-0 (Wt) (B), pdr8-115 (C), aux1-7 (D), pdr8-115 aux1-7 (E), pex5-1 (F), and pdr8-115 pex5-1 (G) light-grown seedlings. Arrows indicate mean root hair lengths; the bar depicting the median root hair length is outlined. Insets show photographs of seedlings used for root hair measurements, demonstrating the disrupted gravitropism of aux1-7 and pdr8-115 aux1-7 roots.
[See online article for color version of this figure.]
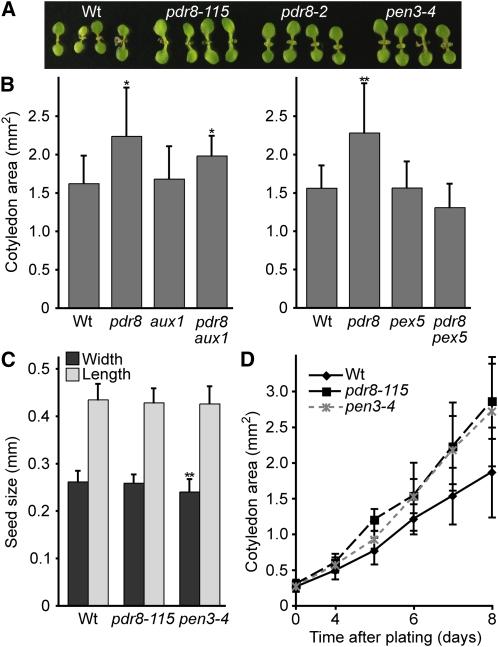
We also found that pdr8 cotyledons were larger than the wild type (Figure 7A). Cotyledons of 7-d-old pdr8 aux1 double mutants were intermediate in size compared with the parents (Figure 7B). pdr8 pex5 displayed wild-type cotyledon size (Figure 7B), suggesting that conversion of IBA to IAA is required for the increased cotyledon expansion found in pdr8. Because cotyledons are formed during embryogenesis, and the bulk of Arabidopsis seeds is composed of these cotyledons, we examined wild-type and pdr8 seeds and found that they were similarly sized (Figure 7C). Indeed, cotyledons of wild-type and pdr8 germinating seedlings were similarly sized upon emergence from the seed coat, but pdr8 mutant cotyledons expanded more rapidly than those of the wild type (Figure 7D).
Figure 7.
pdr8 Mutants Have Enlarged Cotyledons.
(A) Photograph of 7-d-old Col-0 (Wt), pdr8-115, pdr8-2, and pen3-4 light-grown seedlings.
(B) Cotyledon areas of 7-d-old Col-0 (Wt), pdr8-115, aux1-7, pdr8-115 aux1-7, pex5-1, and pdr8-115 pex5-1 light-grown seedlings (n ≥ 18). Separate graphs depict separate experiments.
(C) Seed lengths and widths of Col-0 (Wt), pdr8-115, and pen3-4 (n ≥ 171).
(D) Cotyledon areas of Col-0 (Wt), pdr8-115, and pen3-4 seedlings during early postgerminative growth (n ≥ 23). Measurements were made using NIH Image software.
Error bars represent sd of the means. In (B), statistically significant differences from the wild type in two-tailed t tests assuming unequal variance are indicated by single (P ≤ 0.001) and double (P ≤ 0.0001) asterisks. In (D), pdr8-115 (P ≤ 0.0001) and pen3-4 (P ≤ 0.001) had significantly larger cotyledons than the wild type from 4 to 8 d after plating.
[See online article for color version of this figure.]
pdr8 Mutants Have Reduced IBA Efflux
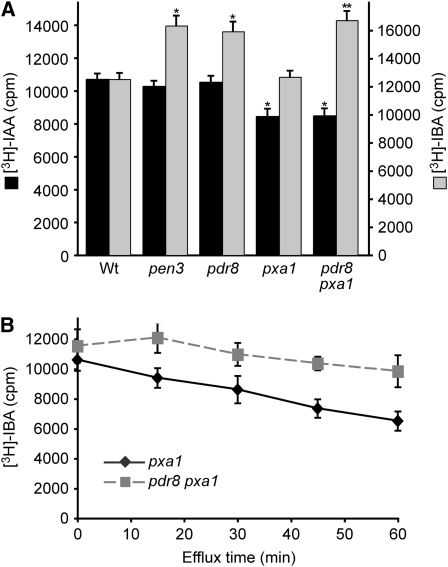
The increased IBA responsiveness of the pdr8 alleles (Figures 2A and 3A), together with the identity of PDR8 as a plasma membrane–localized (Kobae et al., 2006; Stein et al., 2006) ABC transporter (van den Brule and Smart, 2002; Crouzet et al., 2006; Verrier et al., 2008), suggested that PDR8 might function to efflux IBA from cells. To test this possibility, we examined IBA accumulation in pdr8 using an excised root tip auxin transport assay (Ito and Gray, 2006; Strader et al., 2008b). We compared [3H]-IAA and [3H]-IBA accumulation in root tips excised from 8-d-old seedlings. Consistent with wild-type responsiveness to IAA in root elongation inhibition (Figure 2B), pdr8 root tips displayed wild-type [3H]-IAA accumulation in this assay (Figure 8A). The pxa1-1 mutant displayed slightly less [3H]-IAA accumulation (Figure 8A). In contrast with the normal [3H]-IAA accumulation, we found that root tips of both pen3-4 and pdr8-115 hyperaccumulated [3H]-IBA (Figure 8A). pdr8-115 also hyperaccumulated [3H]-IBA in a pxa1-1 background (Figure 8A), which is likely blocked in IBA-to-IAA conversion (Zolman et al., 2001), suggesting that the major radiomolecule assessed in this assay is [3H]-IBA and not [3H]-IAA derived from [3H]-IBA. These results are consistent with the possibility that PDR8 facilitates IBA efflux from root cells.
Figure 8.
pdr8 Mutants Display Increased [3H]-IBA Accumulation and Reduced [3H]-IBA Efflux.
(A) Root tips of 8-d-old Col-0 (Wt), pen3-4, pdr8-115, pxa1-1, and pdr8-115 pxa1-1 seedlings were incubated for 1 h in buffer containing 25 nM [3H]-IAA or 25 nM [3H]-IBA, rinsed three times, and incubated for an additional 20 min in buffer. Root tips were then removed and analyzed by scintillation counting. Data are from eight replicates of assays with five root tips of each genotype.
(B) Root tips of 8-d-old pxa1-1 and pdr8-115 pxa1-1 seedlings were incubated for 1 h in buffer containing 25 nM [3H]-IBA, rinsed three times, and incubated in a large volume of buffer. Root tips were removed at various time points and analyzed by scintillation counting. Data are from six replicates of assays with five root tips of each genotype.
Error bars represent sd of the means. Statistically significant differences from the wild type in two-tailed t tests assuming unequal variance are indicated in (A) by single (P ≤ 0.001) and double (P ≤ 0.0001) asterisks. In (B), pdr8 pxa1 root tips retained significantly more radioactivity than pxa1 after 15 (P ≤ 0.001), 30 (P ≤ 0.001), 45 (P ≤ 0.0001), and 60 (P ≤ 0.0001) min of efflux.
The amount of [3H]-IBA retained in root tips after the 1-h incubation reflects the balance of [3H]-IBA influx and efflux. To distinguish influx from efflux, we incubated root tips of pxa1 and pdr8 pxa1 in [3H]-IBA for 1 h, rinsed the root tips in buffer, and then incubated them in a large buffer volume to minimize reuptake of effluxed [3H]-IBA. After a 1-h efflux period, pxa1 root tips had retained only 61% of the initial [3H]-IBA. The majority of the radioactivity effluxed from pxa1 root tips comigrated with IBA in thin layer chromatography (see Supplemental Figure 2 online), suggesting that the major effluxed radiomolecule assessed in this assay was [3H]-IBA. By contrast, pdr8 pxa1 root tips retained 85% of the initial [3H]-IBA (Figure 8B) and effluxed substantially less [3H]-IBA than pxa1 (see Supplemental Figure 2 online), again suggesting that IBA efflux is reduced in the pdr8 mutant. This PDR8 requirement for efficient IBA efflux is consistent with the possibility that IBA is a substrate of the PDR8 transporter.
Localization of PDR8/ABCG36 in the Root Tip
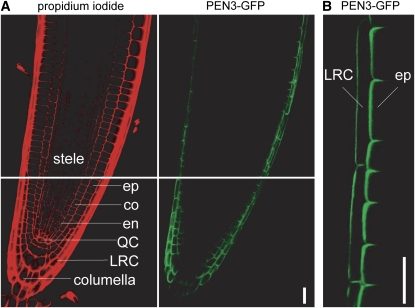
pdr8 mutant phenotypes imply that the PDR8 ABC transporter functions in both root and cotyledon tissues. Moreover, if PDR8 acts directly in IBA efflux, then we would expect that PDR8 would be expressed in root tip cells. A PEN3-GFP (green fluorescent protein) translational fusion reporter driven by the native PEN3/PDR8 promoter (PEN3:PEN3-GFP), which complements the pen3-1 mutant phenotypes, has been used to localize PEN3/PDR8 to the plasma membrane of leaf epidermal cells (Stein et al., 2006). We examined root tips of pen3-1 carrying the PEN3:PEN3-GFP reporter (Stein et al., 2006) and found that the GFP signal emanated from the periphery of lateral root cap and epidermal cells but not from stele or cortex cells (Figure 9). Localization of the GFP signal to the periphery of the cells is consistent with previous reports of PDR8/PEN3 localization to the plasma membrane of leaf epidermal cells (Kobae et al., 2006; Stein et al., 2006). In the region basal to the root tip, PEN3-GFP appeared to be less abundant along epidermal/cortical cell boundaries (Figure 9). The localization of PEN3-GFP around the outer sides of root cells is consistent with the possibility that PDR8/PEN3 functions to efflux a compound or compounds from the root.
Figure 9.
PDR8/PEN3-GFP Is Localized to the Periphery of Lateral Root Cap and Epidermal Cells of the Root Tip.
Confocal images of a root tip of 6-d-old pen3-1 carrying PEN3:PEN3-GFP.
(A) The right panel shows PEN3-GFP; the left panel shows propidium iodide counterstaining of cell walls. The stele, epidermal (ep), cortex (co), endodermal (en), quiescent center (QC), lateral root cap (LRC), and columella cells are labeled on the propidium iodide image.
(B) Magnified view of PEN3-GFP fluorescence, showing PDR8/PEN3-GFP accumulation on the outer edges of the lateral root cap (LRC) and epidermal (ep) cells. Bars = 20 μm.
DISCUSSION
IAA is the predominant active auxin found in plants, and impeding cell-to-cell IAA transport by disrupting influx (aux1 mutants) or efflux (pin and pgp/abcb mutants) results in developmental defects. Plants can store IAA in the form of conjugates or the chain-lengthened precursor IBA (reviewed in Woodward and Bartel, 2005b). However, the roles and mechanisms of auxin storage form transport are largely uncharacterized. We found that the plasma membrane localized (Kobae et al., 2006; Stein et al., 2006) PDR8/PEN3/ABCG36 transporter appears to facilitate IBA efflux, but not IAA efflux. Loss of PDR8 increases sensitivity to the side chain–lengthened IAA precursor IBA and the 2,4-D precursor 2,4-DB, but not to IAA or 2,4-D. The identical IBA and 2,4-DB hypersensitivity of the pen3-4 likely null allele (Stein et al., 2006) and the pdr8-115 allele that we isolated as an ibr5 suppressor (Figures 2A and 2C) suggests that the pdr8-115 Ala-1357 to Val change abolishes PDR8 function. We found that excised pdr8 root tips displayed [3H]-IBA efflux defects but normal [3H]-IAA accumulation (Figure 8) and that a PDR8-GFP fusion protein localized to the periphery of root epidermal and lateral root cap cells (Figure 9), consistent with the possibility that PDR8 specifically promotes IBA efflux. pdr8 mutants also displayed increased lateral root production (Figure 3), lengthened root hairs (Figure 6), and increased cotyledon size (Figure 7). The auxin-related developmental defects of pdr8 mutants suggest that PDR8 and IBA efflux play previously unappreciated roles in plant development.
Roles for IBA Efflux in Auxin-Related Developmental Processes
Plants use postembryonic root organogenesis in the form of lateral branching to alter their subterranean architecture. Auxin promotes lateral root formation, both exogenously (Torrey, 1950; Himanen et al., 2002) and when overproduced due to mutation (Boerjan et al., 1995; Celenza et al., 1995; King et al., 1995). Conversely, auxin-resistant mutants are often deficient in lateral roots (reviewed in De Smet et al., 2006). Furthermore, auxin transport is required for lateral root formation; inhibiting auxin transport with the inhibitor naphthylphthalamic acid decreases lateral rooting (Reed et al., 1998; Casimiro et al., 2001), and the aux1 (Marchant et al., 2002) and lax3 (for like aux1; Swarup et al., 2008) auxin influx mutants are defective in lateral root production or emergence. The proliferation of early lateral root primordia and lateral roots in pdr8 (Figure 3) is consistent with a PDR8 role in auxin efflux in lateral root primordia.
Root hair elongation provides a simple gauge of auxin effects on cell expansion because all growth is from a single cell. Auxin promotes root hair elongation, likely by increasing the duration of tip growth (reviewed in Schiefelbein, 2000; Grierson and Schiefelbein, 2002). For example, decreased auxin efflux in the pgp4 mutant (Santelia et al., 2005; Cho et al., 2007) or increased auxin influx following AUX1 overexpression (Cho et al., 2007) results in longer root hairs. By contrast, decreased auxin influx in the aux1 mutant (Pitts et al., 1998) or increased efflux caused by overexpressing PIN3 (Lee and Cho, 2006; Cho et al., 2007), PID (Lee and Cho, 2006), PGP1 (Cho et al., 2007), PGP4 (Cho et al., 2007), or PGP19 (Cho et al., 2007) results in shorter root hairs. The longer root hairs of pdr8 (Figures 6A and 6C) are consistent with a PDR8 role in auxin efflux.
Cotyledons provide a second cell expansion model because postgerminative Arabidopsis cotyledons grow via cell expansion without cell division (Mansfield and Briarty, 1996). High IAA content is correlated with high rates of cell division during leaf development (Ljung et al., 2001), and mutation of ARF7 and ARF19 results in smaller leaves with more numerous, smaller cells than the wild type (Wilmoth et al., 2005), implicating ARF7 and ARF19 in leaf cell expansion. The pdr8 large cotyledon phenotype (Figures 7A and 7D) suggests that retention of IBA in cotyledons results in increased cell expansion.
All of the pdr8 developmental phenotypes (increased lateral rooting, longer root hairs, and larger cotyledons) can be rationalized by evoking inappropriate auxin accumulation; we did not observe any pdr8 phenotypes that suggest auxin deficiency. This observation suggests that PDR8 functions to prevent excess IBA accumulation and that endogenous IBA may be used primarily in the cells in which it is made rather than being transported elsewhere. Alternatively, increased IAA transport or synthesis may compensate for any reduced IBA transport in pdr8. Furthermore, IBA transport, similar to IAA transport (reviewed in Blakeslee et al., 2005), is likely to involve multiple mechanisms, and redundancy could be masking potential auxin deficiency phenotypes in pdr8. We expect that additional study of pdr8 and IBA transport in general will further illuminate the roles of IBA transport in growth and development.
pdr8 Phenotypes Are Suppressed by Blocking IBA-to-IAA Conversion
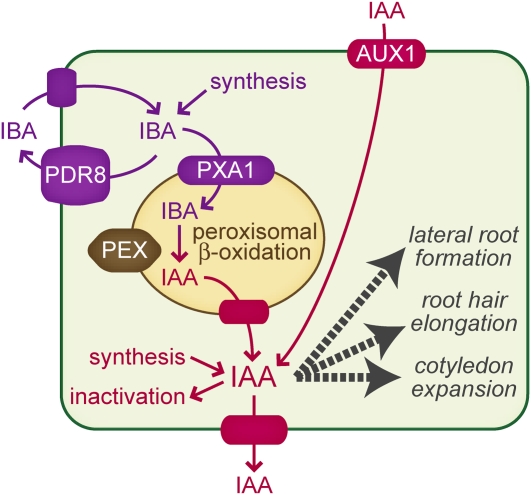
Genetic evidence indicates that peroxisomal carboxyl side chain shortening of IBA to IAA is required for the auxin activity of IBA in Arabidopsis bioassays (Zolman et al., 2000, 2007). IBA-resistant and IAA-sensitive mutants include peroxisome biogenesis mutants, such as pex5 (Zolman et al., 2000) and pex7 (Woodward and Bartel, 2005a), mutants defective in certain peroxisomal β-oxidation enzymes, such as IBR1 (Zolman et al., 2008), IBR3 (Zolman et al., 2007), and IBR10 (Zolman et al., 2008), and mutants defective in the PXA1 transporter that brings β-oxidation substrates into the peroxisome (Zolman et al., 2001). Many of these mutants exhibit decreased lateral root production in the absence of exogenous hormone (Zolman et al., 2001; Woodward and Bartel, 2005a), suggesting that IBA conversion to IAA is important for proper lateral root formation. The pdr8 phenotypes of increased lateral root production, increased root hair growth, and increased cotyledon expansion suggest that certain pdr8 cells hyperaccumulate auxin, and the observation that pdr8 root tips hyperaccumulate [3H]-IBA but not [3H]-IAA suggests that the auxin that accumulates is IBA. Moreover, the pdr8 developmental phenotypes were all suppressed by blocking IBA-to-IAA conversion in the pdr8 pex5 and pdr8 pxa1 double mutants (Figures 5A to 5C, 6G, and 7B). The suppression of pdr8 developmental phenotypes by mutations expected to block IBA-to-IAA conversion suggests a model in which IBA accumulates to inappropriate levels in pdr8 and is converted to IAA, which causes increased growth (Figure 10).
Figure 10.
Model for PDR8/ABCG36 Function in Auxin Homeostasis.
IBA contributes to IAA levels through its peroxisomal β-oxidation to IAA, and depending on the cell type, this IAA promotes various responses (e.g., lateral root formation, root hair elongation, cotyledon expansion, etc.). IAA levels also can be increased through de novo synthesis or by AUX1-mediated import of IAA produced in other cells. IAA levels are decreased by inactivation and efflux. β-Oxidation of IBA to IAA requires the PXA1 peroxisomal ABC transporter and various peroxin (PEX) proteins. The plasma membrane–localized PDR8/ABCG36 transporter appears to limit IBA accumulation in cells by promoting IBA efflux. It is not known whether local IBA synthesis or influx of IBA made elsewhere is more important for the observed auxin responses.
pdr8 Suppresses a Distinct Subset of Auxin Signaling Mutant Phenotypes
We isolated pdr8 as a suppressor of the IBA resistance of ibr5 (MS115; Strader et al., 2008b). Loss of PDR8 did not suppress all ibr5 defects. Although pdr8-115 restored ibr5 IBA and 2,4-DB responsiveness, responses to IAA, 2,4-D, TIBA, ABA, and ACC appear largely unaffected (Figure 1; see Supplemental Figure 1 online; Strader et al., 2008b). Similarly, pdr8 partially restored IBA and 2,4-DB, but not IAA or 2,4-D, responsiveness to tir1-1 and axr1-3 (Figure 4). Although resistant to IBA, ibr5 is not completely insensitive and responds to high IBA concentrations (Monroe-Augustus et al., 2003; Strader et al., 2008a). The IBA resistance of auxin signaling mutants may be offset in pdr8 because applied IBA and 2,4-DB are less efficiently removed from cells in the absence of PDR8, and their consequent hyperaccumulation and conversion to IAA and 2,4-D allows the double mutant to respond to IBA and 2,4-DB similarly to the wild type (Figures 1E to 1G and 4A to 4C).
In contrast with the signaling mutants, pdr8 failed to suppress the auxin-resistant root elongation phenotype of the transport mutant aux1-7 (Figures 4A and 4B). The AUX1 transporter imports 2,4-D and IAA into cells (Maher and Martindale, 1980; Yamamoto and Yamamoto, 1998; Marchant et al., 1999; Yang et al., 2006). IBA does not appear to be an AUX1 substrate; aux1 appears to transport [3H]-IBA normally (Rashotte et al., 2003), excess IBA does not compete with [3H]-IAA uptake by Xenopus laevis oocytes expressing AUX1 (Yang et al., 2006), and excised aux1 root tips do not hypoaccumulate [3H]-IBA (Strader et al., 2008b). However, aux1 is moderately IBA resistant (Zolman et al., 2000), perhaps because the bioassays employed require AUX1-mediated transport of IAA derived from IBA to the responding cells. The aux1 suppression of pdr8 IBA and 2,4-DB hypersensitivity (Figures 4A and 4B) suggests that the auxin that inhibits root elongation is not solely auxin that enters elongating cells directly from the medium, but in addition, cells in other root zones take up IBA (or 2,4-DB) and convert it to IAA (or 2,4-D), which is then transported in an AUX1-dependent manner to the elongation zone where it inhibits cell expansion. The partial restoration of IBA-responsive lateral root production in pdr8 aux1 (Figure 4C) implies that transport of the IAA made from supplied IBA is less important in this bioassay. Although AUX1 is found in atrichoblasts, but not trichoblasts, AUX1-mediated basipetal auxin transport is important for root hair growth (Jones et al., 2009). The ability of pdr8 to fully suppress the short root hairs of aux1 (Figure 6E) implies that excess IAA made from the IBA accumulating in pdr8 is sufficient to compensate for the IAA normally supplied to those trichoblasts by AUX1.
Roles of PDR/ABCG Proteins
Although the pdr8 developmental defects had not been previously noted, PDR8/PEN3/ABCG36 has been implicated in pathogen response; pdr8/pen3 mutants exhibit increased sensitivity to the nonhost pathogens Blumeria graminis f. sp hordei (barley powdery mildew) (Stein et al., 2006), Plectospaerella cucumerina, Erysipha pisa (pea powdery mildew) (Stein et al., 2006), and Phytopthera infestans (potato late blight) (Kobae et al., 2006; Stein et al., 2006), as well as decreased sensitivity to the Arabidopsis pathogens Erysiphe cichoracearum (powdery mildew) (Stein et al., 2006) and Pseudomonas syringae (Kobae et al., 2006). Furthermore, pdr8/pen3 mutants accumulate less extracellular callose in response to the microbe-associated molecular pattern molecule Flg22 (Clay et al., 2009), hyperaccumulate Flg22- or pathogen-elicited 4-methoxyindol-3-ylmethylglucosinolate (Bednarek et al., 2009; Clay et al., 2009), and hyperaccumulate pathogen-elicited camalexin (Bednarek et al., 2009). PDR8 also has been implicated in cadmium efflux (Kim et al., 2007). Whether the developmental roles of PDR8 uncovered here and the roles of PDR8 pathogen response, callose, camalexin, and glucosinolate production, or cadmium extrusion are related or separate functions remains to be explored.
Similar to previously identified auxin effluxers in the ABCB/MDR/PGP subfamily, the 15-member PDR group of the ABCG subfamily of ABC transporters are full-sized ABC transporters with two transmembrane domains each consisting of six membrane-spanning sequences and two apparent nucleotide binding domains (reviewed in van den Brule and Smart, 2002; Crouzet et al., 2006; Verrier et al., 2008). However, few of these transporters have been functionally characterized. PDR9/ABCG37 is a likely 2,4-D (Ito and Gray, 2006) and IBA (Strader et al., 2008b) efflux facilitator. PDR8, which is only 53% identical to PDR9, is the second PDR/ABCG family member implicated in IBA efflux, raising the possibility that multiple members of this family may efflux IBA. PDR family members have diverse expression patterns (Crouzet et al., 2006); PDR8 and PDR9 are more highly expressed in roots than most PDR genes (van den Brule and Smart, 2002). Future studies may elucidate whether ABCG subfamily members participate in long-distance IBA transport, similar to the roles of ABCB subfamily members in IAA transport, or are more important for short-distance IBA transport. The latter possibility is supported by the enrichment of PDR8-GFP on the outward face of root epidermal cells, suggesting that PDR8 may remove IBA from the root tip. Analyses of additional PDR/ABCG family members will be required to fully understand the roles that these proteins play in IBA transport and auxin homeostasis.
METHODS
Plant Materials and Growth Conditions
The Colombia (Col-0) accession of Arabidopsis thaliana was used as the wild type for all experiments. Seeds were surface sterilized (Last and Fink, 1988) and plated on PN (plant nutrient medium; Haughn and Somerville, 1986) supplemented with 0.5% (w/v) sucrose (PNS) and solidified with 0.6% (w/v) agar. All hormones were dissolved in ethanol, and ethanol-supplemented media were used as controls. Hormone treatments were normalized to the same ethanol content (<0.2 μL ethanol/mL medium). Seedlings were grown under continuous illumination through yellow long-pass filters to slow indolic compound breakdown (Stasinopoulos and Hangarter, 1990) at 22°C unless indicated otherwise.
Identification of the pdr8-115 Mutation
MS115 (in the Col-0 background) was outcrossed to Wassilewskija-introgressed ibr5-1 for mapping (Strader et al., 2008b). The resulting F2 seeds were plated on 10 μM IBA or 2 μM 2,4-DB, and DNA from sensitive individuals was isolated (Celenza et al., 1995) for mapping using published genetic markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994) and additional newly developed PCR-based markers (see Supplemental Table 1 online). A candidate gene (PDR8/ABCG36/At1g59870) within the MS115 mapping interval was examined for defects in the mutant. MS115 mutant genomic DNA was amplified using six pairs of oligonucleotides (PDR8-1 and PDR8-2, PDR8-3 and PDR8-4, PDR8-5 and PDR8-6, PDR8-7 and PDR8-8, PDR8-9 and PDR8-10, and PDR8-11 and PDR8-12; see Supplemental Table 3 online). These overlapping PCR fragments, covering the gene from 167 bp upstream of the putative translation start site to 839 bp downstream of the stop codon, were sequenced (Lone Star Labs) with the primers used for amplification.
Double Mutant Isolation
All mutants were in the Col-0 accession and are listed in Supplemental Table 2 online. The pdr8-2 (SALK_142256) mutant (Kobae et al., 2006) was crossed to ibr5-1 (Monroe-Augustus et al., 2003). The pdr8-115 mutant was crossed to tir1-1 (Ruegger et al., 1998), axr1-3 (Lincoln et al., 1990), aux1-7 (Maher and Martindale, 1980), pex5-1 (Zolman et al., 2000), and pxa1-1 (Zolman et al., 2001). PCR analysis of F2 plants was used to identify double mutants. Amplification of PDR8 with PDR8-9 and PDR8-13 yields an ∼1160-bp product in the wild type and no product in pdr8-2. PDR8 amplification with PDR8-9 and LB1-SALK yields an ∼700-bp product in pdr8-2 and no product in the wild type. PDR8 amplification with PDR8-DdeI and PDR8-15 yields a 120-bp product with one DdeI restriction site in the wild type and none in pdr8-115. Oligonucleotide sequences are listed in Supplemental Table 3 online. PCR-based identification of ibr5-1 (Monroe-Augustus et al., 2003), tir1-1 (Strader et al., 2008a), axr1-3 (Strader et al., 2008a), aux1-7 (Strader et al., 2008a), pex5-1 (Zolman et al., 2000), and pxa1-1 (Zolman et al., 2001) alleles were as described previously.
Phenotypic Assays
For auxin-responsive root elongation assays, primary root lengths of seedlings were grown for 8 d on PNS with the indicated auxin or auxin transport inhibitor concentrations, and the lengths of primary roots were measured. For ACC-responsive root elongation assays, primary root lengths of seedlings grown for 10 d on medium supplemented with either ethanol or 100 nM ACC under white light were measured. For ABA-responsive root elongation assays, 4-d-old seedlings grown on unsupplemented medium in the light at 22°C were transferred to medium supplemented with either ethanol or 10 μM ABA, and primary root lengths were measured after an additional 4 d of growth in the light.
For lateral root assays, seedlings were grown for 4 d on unsupplemented medium before transfer to medium supplemented with either ethanol or 10 μM IBA and grown for an additional 4 d. Primordia emerging from the primary root, as seen under a dissecting microscope, were counted as lateral roots. pdr8-115 was crossed to the wild type carrying DR5-GUS (Ulmasov et al., 1997) to generate pdr8-115 DR5-GUS. Seedlings were treated as above and then stained for GUS activity overnight as previously described (Bartel and Fink, 1994). Seedlings were then mounted and imaged through a dissecting microscope.
To quantify LRP, seedlings were treated as above and then cleared by incubating for 1 week at room temperature in a chloral hydrate solution (80 g chloral hydrate, 20 mL glycerol, and 10 mL water). Cleared seedlings were mounted and examined using a Zeiss Axioplan 2 microscope. LRP were classified into three stages. Stage A includes the first anticlinal division of the pericycle cell to a lateral root primordium with three cell layers and corresponds to Stages I to III described by Malamy and Benfey (1997). Stage B consists of preemergent lateral roots with greater than three cells layers, corresponding to Stages IV to VIb described by Malamy and Benfey (1997).
For sucrose dependence assays, seeds were plated on either PN or PNS, incubated under white light at 22°C for 1 d, and then covered in foil and incubated for an additional 4 d. Hypocotyls of dark-grown seedlings were measured.
For cotyledon expansion assays, cotyledons of 7-d-old seedlings grown under continuous white light at 22°C were removed and mounted. Cotyledons were imaged using a dissecting microscope, and cotyledon area was measured using NIH Image software.
For root hair assays, roots of 5-d-old seedlings grown on the surface of vertically oriented plates were imaged using a dissecting microscope and root hair lengths from 4 mm of root adjacent to the root-shoot junction were measured using NIH Image software.
GUS Assays
Seedlings were grown for 4 d on unsupplemented medium before transfer to medium supplemented with either ethanol or various concentrations of IBA and grown for an additional 4 d under continuous yellow-filtered light at 22°C. For each sample, three seedlings were collected into a microcentrifuge tube and frozen. Seedlings were then ground with a pestle in 100 μL GUS extraction buffer (150 mM NaPO4, pH 7.0, 2.5 mM ethylenediaminetetraacetic acid, 10 mM β-mercaptoethanol, 0.1% Triton X-100, and 0.1% sodium dodecyl sulfate). Debris was pelleted by centrifugation for 5 min. A 20-μL aliquot of each supernatant was added to 130 μL prewarmed GUS extraction buffer supplemented with 1 mM 4-methylumbelliferyl β-d-glucuronide hydrate (Sigma-Aldrich). After 4 h at 37°C, 10 μL of the reaction was combined with 190 μL of 200 mM Na2CO3 in an opaque 96-well plate (Nunc). Fluorescence was measured using an Infinite M200 fluorometer (Tecan US) with an excitation wavelength of 355 nm and an emission wavelength of 460 nm.
Confocal Fluorescence Microscopy Analysis
Six-day-old pen3-1 seedlings carrying PEN3:PEN3-GFP (Stein et al., 2006) were counterstained for 10 min in an aqueous solution of 10 μg/mL propidium iodide (Sigma-Aldrich) and then mounted in water under a cover glass. Confocal microscopy was performed on a Zeiss LSM 510 laser scanning confocal microscope equipped with a Meta detector. Samples were imaged through a ×63 oil immersion lens and excited with the 488-nm laser line from an argon laser. The fluorescence was split through a 545-nm secondary dichroic beam splitter. Fluorescence <545 nm was further filtered through a 500- to 530-nm band-pass filter, with pixels resulting from fluorescence detection false-colored green. Fluorescence >545 nm was further filtered through a 560-nm high-pass filter, with pixels resulting from fluorescence detection false-colored red. Images were converted using NIH Image software.
Auxin Accumulation Assays
Excised 5-mm tips of primary roots from 8-d-old light-grown Col-0, pdr8-115, pen3-4, pxa1-1, and pdr8-115 pxa1-1 seedlings were incubated in 40 μL uptake buffer (20 mM MES, 10 mM sucrose, and 0.5 mM CaSO4, pH 5.6) for 10 min at room temperature. Additional uptake buffer containing radiolabeled auxins was added for a final volume of 80 μL containing either 25 nM [3H]-IAA (20 Ci/mmol; American Radiolabeled Chemicals, lot 071130) or 25 nM [3H]-IBA (25 Ci/mmol; American Radiolabeled Chemicals, lot 080801). After 1 h at room temperature, root tips were rinsed with three changes of uptake buffer and placed in 80 μL of fresh uptake buffer. After 20 min incubation at room temperature, root tips were analyzed by scintillation counting in 3 mL of Cytoscint scintillation cocktail (Fisher Scientific). To examine [3H]-IBA efflux, excised 5-mm tips of primary roots from 8-d-old light-grown pxa1-1 and pdr8-115 pxa1-1 seedlings were incubated as above for 1 h, rinsed with three changes of uptake buffer, and placed in 400 μL of fresh uptake buffer. After 0 to 60 min incubation at room temperature, root tips were removed from the efflux buffer and analyzed by scintillation counting in 3 mL of Cytoscint scintillation cocktail.
To monitor the identity of the effluxed compounds, efflux buffers from the 60-min efflux experiments were acidified with 0.1 volume citrate phosphate buffer, pH 2.5, and extracted twice with ethyl acetate. Extracted compounds were cospotted with unlabeled IAA and IBA on a silica thin layer chromatography plate and separated using a solvent of 85% chloroform, 14% methanol, and 1% water. After visualizing the migration of unlabeled IAA and IBA using fluorescence, silica from 3-mm sections of the thin layer chromatography plate were scraped from the glass and analyzed by scintillation counting.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number At1g59870 (PDR8/PEN3/ABCG36). Additional accession numbers are provided in Supplemental Table 2 and Supplemental Table 4 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. pdr8 Fails to Suppress All ibr5 Phenotypes.
Supplemental Figure 2. Effluxed Radioactivity from pxa1 Root Tips Incubated with [3H]-IBA Predominantly Comigrates with IBA.
Supplemental Table 1. New Markers Used in MS115 Mapping.
Supplemental Table 2. Mutant Alleles Used in This Study.
Supplemental Table 3. Oligonucleotides Used in This Study.
Supplemental Table 4. Accession Numbers of PDR/ABCG Family Members.
Supplementary Material
Acknowledgments
We thank Shauna Somerville for pen3-1 carrying PEN3:PEN3-GFP, Mark Estelle for axr1-3 and aux1-7, the ABRC at Ohio State University for pdr8-2 (SALK_142256), pen3-3 (SALK_110926), pen3-4 (SALK_000578), and tir1-1, Grace Lin and Erin Beisner for technical assistance, James McNew for fluorometer use, and Matthew Lingard and Naxhiely Martinez for critical comments on the manuscript. This research was supported by the National Science Foundation (MCB-0745122 to B.B.), the Robert A. Welch Foundation (C-1309 to B.B.), and the National Institutes of Health (F32-GM075689 to L.C.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Bonnie Bartel (bartel@rice.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Bailly, A., Sovero, V., Vincenzetti, V., Santelia, D., Bartnik, D., Koenig, B.W., Mancuso, S., Martinoia, E., and Geisler, M. (2008). Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J. Biol. Chem. 283 21817–21826. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay, A., et al. (2007). Interactions of PIN and PGP auxin transport mechanisms. Biochem. Soc. Trans. 35 137–141. [DOI] [PubMed] [Google Scholar]
- Bartel, B., and Fink, G.R. (1994). Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 91 6649–6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek, P., Pislewska-Bednarek, M., Svatos, A., Schneider, B., Doubsky, J., Mansurova, M., Humphry, M., Consonni, C., Panstruga, R., Sanchez-Vallet, A., Molina, A., and Schulze-Lefert, P. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323 101–106. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Blakeslee, J.J., et al. (2007). Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee, J.J., Peer, W.A., and Murphy, A.S. (2005). Auxin transport. Curr. Opin. Plant Biol. 8 494–500. [DOI] [PubMed] [Google Scholar]
- Blommaert, K. (1954). Growth- and inhibiting-substances in relation to the rest period of the potato tuber. Nature 174 970–972.13214055 [Google Scholar]
- Boerjan, W., Cervera, M.T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., Caboche, M., Van Onckelen, H., Van Montagu, M., and Inzé, D. (1995). superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro, I., Marchant, A., Bhalerao, R.P., Beeckman, T., Dhooge, S., Swarup, R., Graham, N., Inzé, D., Sandberg, G., Casero, P.J., and Bennett, M. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza, J.L., Grisafi, P.L., and Fink, G.R. (1995). A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 9 2131–2142. [DOI] [PubMed] [Google Scholar]
- Cho, M., Lee, S.H., and Cho, H.-T. (2007). P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19 3930–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay, N.K., Adio, A.M., Denoux, C., Jander, G., and Ausubel, F.M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, W.C. (1935). Hormones in relation to root formation on stem cuttings. Plant Physiol. 10 789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzet, J., Trombik, T., Fraysse, A.S., and Boutry, M. (2006). Organization and function of the plant pleiotropic drug resistance ABC transporter family. FEBS Lett. 580 1123–1130. [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (2004). Introduction. In Plant Hormones: Biosynthesis, Signal Transduction, Action! P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–35.
- De Smet, I., Vanneste, S., Inzé, D., and Beeckman, T. (2006). Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 60 871–887. [DOI] [PubMed] [Google Scholar]
- Friml, J., Benkova, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K., Woody, S., Sandberg, G., Scheres, B., Jurgens, G., and Palme, K. (2002. b). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108 661–673. [DOI] [PubMed] [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jürgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 147–153. [DOI] [PubMed] [Google Scholar]
- Friml, J., Wísniewska, J., Benková, E., Mendgen, K., and Palme, K. (2002. a). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415 806–809. [DOI] [PubMed] [Google Scholar]
- Grierson, C., and Schiefelbein, J. (2002). Root hairs. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/, http://www.aspb.org/publications/arabidopsis/.
- Haughn, G.W., and Somerville, C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204 430–434. [Google Scholar]
- Hayashi, M., Toriyama, K., Kondo, M., and Nishimura, M. (1998). 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen, K., Boucheron, E., Vanneste, S., de Almeida Engler, J., Inze, D., and Beeckman, T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14 2339–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie, L., and Estelle, M. (1995). The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J. 7 211–220. [DOI] [PubMed] [Google Scholar]
- Ito, H., and Gray, W.M. (2006). A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol. 142 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A.R., Kramer, E.M., Knox, K., Swarup, R., Bennett, M.J., Lazarus, C.M., Leyser, H.M., and Grierson, C.S. (2009). Auxin transport through non-hair cells sustains root-hair development. Nat. Cell Biol. 11 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.Y., Bovet, L., Maeshima, M., Martinoia, E., and Lee, Y. (2007). The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 50 207–218. [DOI] [PubMed] [Google Scholar]
- King, J.J., Stimart, D.P., Fisher, R.H., and Bleecker, A.B. (1995). A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell 7 2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobae, Y., Sekino, T., Yoshioka, H., Nakagawa, T., Martinoia, E., and Maeshima, M. (2006). Loss of AtPDR8, a plasma membrane ABC transporter of Arabidopsis thaliana, causes hypersensitive cell death upon pathogen infection. Plant Cell Physiol. 47 309–318. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Last, R.L., and Fink, G.R. (1988). Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240 305–310. [DOI] [PubMed] [Google Scholar]
- Lee, J.S., Wang, S., Sritubtim, S., Chen, J.G., and Ellis, B.E. (2009). Arabidopsis mitogen-activated protein kinase MPK12 interacts with the MAPK phosphatase IBR5 and regulates auxin signaling. Plant J. 57: 975–985. [DOI] [PubMed]
- Lee, S.H., and Cho, H.-T. (2006). PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 18 1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung, K., Bhalerao, R.P., and Sandberg, G. (2001). Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 28 465–474. [DOI] [PubMed] [Google Scholar]
- Luschnig, C., Gaxiola, R.A., Grisafi, P., and Fink, G.R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher, E.P., and Martindale, S.J.B. (1980). Mutants of Arabidopsis thaliana with altered responses to auxins and gravity. Biochem. Genet. 18 1041–1053. [DOI] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124 33–44. [DOI] [PubMed] [Google Scholar]
- Mansfield, S.G., and Briarty, L.G. (1996). The dynamics of seedling and cotyledon cell development in Arabidopsis thaliana during reserve mobilization. Int. J. Plant Sci. 157 280–295. [Google Scholar]
- Marchant, A., Bhalerao, R., Casimiro, I., Eklof, J., Casero, P.J., Bennett, M., and Sandberg, G. (2002). AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant, A., Kargul, J., May, S.T., Muller, P., Delbarre, A., Perrot-Rechenmann, C., and Bennett, M.J. (1999). AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 18 2066–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe-Augustus, M., Zolman, B.K., and Bartel, B. (2003). IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 15 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec, J., Kubes, M., Bielach, A., Gaykova, V., Petrasek, J., Skupa, P., Chand, S., Benkova, E., Zazimalova, E., and Friml, J. (2008). Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development 135 3345–3354. [DOI] [PubMed] [Google Scholar]
- Parry, G., and Estelle, M. (2006). Auxin receptors: a new role for F-box proteins. Curr. Opin. Cell Biol. 18 152–156. [DOI] [PubMed] [Google Scholar]
- Parry, G., Ward, S., Cernac, A., Dharmasiri, S., and Estelle, M. (2006). The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18 1590–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts, R.J., Cernac, A., and Estelle, M. (1998). Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16 553–560. [DOI] [PubMed] [Google Scholar]
- Rashotte, A.M., Poupart, J., Waddell, C.S., and Muday, G.K. (2003). Transport of the two natural auxins, indole-3-butyric acid and indole-3-acetic acid, in Arabidopsis. Plant Physiol. 133 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R.C., Brady, S.R., and Muday, G.K. (1998). Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol. 118 1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelia, D., Vincenzetti, V., Azzarello, E., Bovet, L., Fukao, Y., Duchtig, P., Mancuso, S., Martinoia, E., and Geisler, M. (2005). MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Lett. 579 5399–5406. [DOI] [PubMed] [Google Scholar]
- Schiefelbein, J.W. (2000). Constructing a plant cell. The genetic control of root hair development. Plant Physiol. 124 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J., Milpetz, F., Bork, P., and Ponting, C.P. (1998). SMART, a simple modular architecture research tool: Identification of signalling domains. Proc. Natl. Acad. Sci. USA 95 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos, T.C., and Hangarter, R.P. (1990). Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 93 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, M., Dittgen, J., Sanchez-Rodriguez, C., Hou, B.H., Molina, A., Schulze-Lefert, P., Lipka, V., and Somerville, S. (2006). Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18 731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader, L.C., Monroe-Augustus, M., and Bartel, B. (2008. a). The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol. 8 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader, L.C., Monroe-Augustus, M., Rogers, K.C., Lin, G.L., and Bartel, B. (2008. b). Arabidopsis iba response5 (ibr5) suppressors separate responses to various hormones. Genetics 180 2019–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup, K., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10 946–954. [DOI] [PubMed] [Google Scholar]
- Torrey, J.G. (1950). The induction of lateral roots by indoleacetic acid and root decapitation. Am. J. Bot. 37 257–264. [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brule, S., and Smart, C.C. (2002). The plant PDR family of ABC transporters. Planta 216 95–106. [DOI] [PubMed] [Google Scholar]
- Verrier, P.J., et al. (2008). Plant ABC proteins–A unified nomenclature and updated inventory. Trends Plant Sci. 13 151–159. [DOI] [PubMed] [Google Scholar]
- Vieten, A., Sauer, M., Brewer, P.B., and Friml, J. (2007). Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 12 160–168. [DOI] [PubMed] [Google Scholar]
- Wilmoth, J.C., Wang, S., Tiwari, S.B., Joshi, A.D., Hagen, G., Guilfoyle, T.J., Alonso, J.M., Ecker, J.R., and Reed, J.W. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43 118–130. [DOI] [PubMed] [Google Scholar]
- Woodward, A.W., and Bartel, B. (2005. a). The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol. Biol. Cell 16 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, A.W., and Bartel, B. (2005. b). Auxin: Regulation, action, and interaction. Ann. Bot. (Lond.) 95 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, M., and Yamamoto, K.T. (1998). Differential effects of 1-naphthaleneacetic acid, indole-3-acetic acid and 2,4-dichlorophenoxyacetic acid on the gravitropic response of roots in an auxin-resistant mutant of Arabidopsis, aux1. Plant Cell Physiol. 39 660–664. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Hammes, U.Z., Taylor, C.G., Schachtman, D.P., and Nielsen, E. (2006). High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 16 1123–1127. [DOI] [PubMed] [Google Scholar]
- Zolman, B.K., Martinez, N., Millius, A., Adham, A.R., and Bartel, B. (2008). Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman, B.K., Nyberg, M., and Bartel, B. (2007). IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol. Biol. 64 59–72. [DOI] [PubMed] [Google Scholar]
- Zolman, B.K., Silva, I.D., and Bartel, B. (2001). The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 127 1266–1278. [PMC free article] [PubMed] [Google Scholar]
- Zolman, B.K., Yoder, A., and Bartel, B. (2000). Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156 1323–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.