Abstract
Background and Aims
Lychnophora ericoides (Asteraceae) presents disjunct geographical distribution in cerrado rupestre in the south-east and central Brazil. The phylogeography of the species was investigated to understand the origin of the disjunct geographical distribution.
Methods
Populations in the south and centre of Serra do Espinhaço, south-east Brazil and on ten other localities in Federal District and Goiás in central Brazil were sampled. Analyses were based on the polymorphisms at chloroplast (trnL intron and psbA-trnH intergenic spacer) and nuclear (ITS nrDNA) genomes. From 12 populations, 192 individuals were sequenced. Network analysis, AMOVA and the Mantel test were performed to understand the relationships among haplotypes and population genetic structure. To understand better the origin of disjunct distribution, demographic parameters and time to most recent common ancestor (TMRCA) were estimated using coalescent analyses.
Key Results
A remarkable differentiation between populations from the south-east and central Brazil was found and no haplotype was shared between these two regions. No significant effect of isolation by distance was detected. Coalescent analyses showed that some populations are shrinking and others are expanding and that gene flow between populations from the south-east and central Brazil was probably negligible.
Conclusions
The results strongly support that the disjunct distribution of L. ericoides may represent a climatic relict and that long-distance gene flow is unlikely. With an estimated time to most recent common ancestor (TMRCA) dated from approx. 790 655 ± 36 551 years bp (chloroplast) and approx. 623 555 ± 55 769 years bp (ITS), it was hypothesized that the disjunct distribution may be a consequence of an expansion of the geographical distribution favoured by the drier and colder conditions that prevailed in much of Brazil during the Kansan glaciation, followed by the retraction of the distribution due to the extinction of populations in some areas as climate became warmer and moister.
Key words: Disjunct geographical distribution, climatic relict, vicariance, coalescent analysis, phylogeography, endemism, Asteraceae, Lychnophora ericoides, Cerrado, cerrado rupestre, ITS, cpDNA
INTRODUCTION
The origin of the disjunct distributions of plant species has been a major concern in biogeography and several papers tried to disentangle the mechanisms involved in the origin of such distributions (e.g. Azuma et al., 2001; Karanth, 2003; Gaudeul, 2006). Disjunct distributions may be caused by range contraction in an ancient more widely distributed species due to changes in climatic conditions that affected suitable habitat distribution (Cox and Moore, 2005). However, long-distance dispersal to new suitable habitats may also be responsible for disjunct distributions (Davis and Shaw, 2001; Cox and Moore, 2005). Phylogeographical analyses may contribute to distinguish between the two models because each one has explicit phylogenetic predictions, which could be used to determine the model that best explains the disjunct distribution (Avise, 2000; Karanth, 2003).
For instance, in the European Alps many taxa exhibit disjunct distributions that have been explained by the hypothesis of climatic relicts of the ice ages (e.g. Stehlik et al., 2000), or by the hypothesis of glacial refuge with subsequent expansion. For instance, the disjunction in Hypericum nummularium, from the Pyrenees and Alps, is due to bidirectional colonization from a single glacial refuge (Gaudeul, 2006). Long-distance dispersal has been invoked to explain amphi-Atlantic disjunctions in some tropical rain forest tree species such as Symphonia globulifera (Dick et al., 2003) and Ceiba pentandra (Dick et al., 2007). The hypothesis of climatic relicts of interglacial ages was invoked to explain the discontinuous distribution observed for some tree species from seasonal tropical dry forests in south and central America (Prado and Gibbs, 1993; Pennington et al., 2000). However, discontinuity in the distribution of neotropical plant species is still poorly understood.
Climatic oscillations during the Tertiary and Quaternary may have affected species distribution, leading to extinction of some local populations and concomitantly stimulating the evolution and speciation of some groups (Comes and Kadereit, 1998). In such a scenario, traces of bottleneck events may be found in extant populations, since only some genomes may survive and expand in each new cycle (Hewitt, 1996). Moreover, a decrease in genetic variation and an increase in homozygosity might be expected, which in turn may modify gene interactions and cause a change in species response to environmental conditions and adaptability (Hewitt, 1996; Davis and Shaw, 2001). Evidence for the effects of Tertiary and Quaternary climatic oscillations on species distribution in Europe is ubiquitous due to intensive surveys of the fossil record and numerous phylogeographical studies (e.g. Bennett et al., 1991; Taberlet et al., 1998; Hewitt, 1999; Petit et al., 2002).
In neotropical South America, a number of studies based on the fossil pollen record are now available (e.g. Behling, 1998, 2003; Behling and Hooghiemstra, 2000; Marchant et al., 2006). For the cerrado biome in central and south-east Brazil, such studies indicate a drier period in the last glacial, until 6000–5000 years bp (Salgado-Laboriau et al., 1998; Behling, 2003). As a consequence, the distribution of savanna-like vegetation in central Brazil was more extensive in early compared with the late Holocene (Behling and Hooghiemstra, 2000), and this probably favoured the expansion of species adapted to drier climates. Only a few studies on plant phylogeography are available to clarify how these climatic oscillations may have affected species distribution in the cerrado biome. Nevertheless, these studies focus on widely distributed species from cerrado sensu stricto (Collevatti et al., 2003; Ramos et al., 2007) and seasonally dry forest (Pennington et al., 2004; Caetano et al., 2008). The present study is the first attempt to study the disjunct geographical distribution of an essentially rocky savanna (cerrado rupestre) species of the Brazilian cerrado biome.
Cerrado rupestre is present at higher altitudes, on outcrops of sandstone and quartzite soils of the cerrado biome in central and south-east Brazil (Ribeiro and Walter, 1998). It is characterized by the high number of endemic species and the occurrence of fire during the dry season. The cerrado biome covers nearly 22 % of the Brazilian territory (2·5 million km2) and comprises very heterogeneous vegetation, with seasonal savannas (cerrado), hyperseasonal savannas (vereda) on poorly drained soils, rocky savannas (cerrado rupestre), mesophytic evergreen forests (gallery forest) and deciduous and semi-deciduous seasonally dry forests (for detailed description of the cerrado biome, see Furley and Ratter, 1988).
Lychnophora ericoides (Asteraceae) is a diploid shrub species with 34 chromosomes (2n = 34, x = 17; Mansanares et al., 2002). Flowers are purple and pollinated by butterflies and the small seeds are wind dispersed. The species is endemic to outcrop quartzite and sandstone soils of cerrado rupestre of the cerrado biome in altitudes above 950 m, but it can also be found in ironstone and in seasonal savannas (cerrado) in highlands, on rocky and litholic fields that resemble the cerrado rupestre habitat (Semir, 1999). It is considered an endangered species by the Environment Brazilian Ministry (see http://www.mma.gov.br/estruturas/179/_arquivos/179_05122008033615.pdf) due to its natural endemism and habitat vulnerability, in addition to the over-harvesting of buds, leaves and stems that are used by local inhabitants for home-made medicinal recipes, which may affect population dynamics.
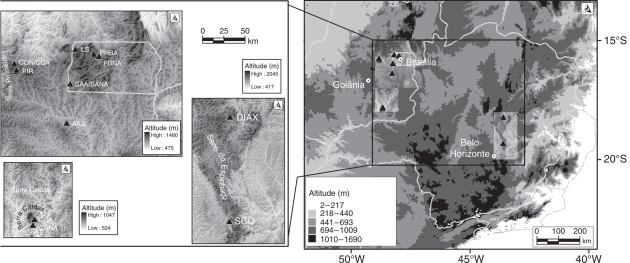
Lychnophora ericoides presents disjunct geographical distribution (Fig. 1) in Serra do Espinhaço, Minas Gerais (MG), south-east Brazil, and on similar habitats in the Federal District (DF) and Goiás (GO) in the Brazilian Shield, central Brazil, such as in Serra dos Pirineus, Serra Dourada and Serra dos Cristais (Coile and Jones, 1981; Semir, 1999). Locally, it is distributed in well-delimited patches, because of the patchily distribution of the suitable habitat.
Fig. 1.
The sample sites of L. ericoides populations. Populations from central Brazil: AAS, Abadiânia, GO; CNA and CNNA, State Park of Serra de Caldas, Caldas Novas, GO; COA and CON, Cocalzinho, GO; PBNA and PNBA, National Park of Brasilia, DF; PIR, Serra dos Pirineus, Pirenópolis, GO; SAA and SANA, Santo Antônio do Descoberto, GO. Populations from south-east Brazil: DIAX, centre of Serra do Espinhaço, Diamantina, MG; SCO, south of Serra do Espinhaço, Serra do Cipó, MG.
The disjunct distribution of L. ericoides may have resulted from one of two different scenarios: long-distance dispersal to suitable habitats or vicariance due to range contraction of a previously more widely distributed species due to changes in climatic conditions that affected suitable habitat distribution. The first scenario should result in isolation by distance, low differentiation among populations, high levels of gene flow and a more recent TMRCA (time to most recent common ancestor) between populations from the south-east and central Brazil (eastern and western localities of the geographical distribution). The latter scenario should result in higher differentiation, no gene flow and a more ancient TMRCA between populations from the south-east and central Brazil.
In the present work, phylogeographical analyses were used to understand the origin of the disjunct geographical distribution of L. ericoides and to try decoupling the mechanisms described above. The working hypothesis was that the disjunct geographical distribution is a climatic relict of the ice ages of the Quaternary. For this, the polymorphisms were analysed at two regions of maternally inherited chloroplast DNA, the non-coding region between the genes psbA and trnH and the trnL gene intron, and one region of the nuclear genome, the internal transcribed spacer (ITS region) of the nrDNA.
MATERIALS AND METHODS
Populations, sampling and DNA extraction
Twelve populations of Lychnophora ericoides Mart. (Asteraceae) were sampled throughout its geographical distribution (Fig. 1). Distance between pairs of populations ranged from 4 km to approx. 727 km. In each of these populations 16 adult individuals were randomly chosen and expanded leaves were sampled and stored at −80 °C. Genomic DNA extraction followed the standard CTAB procedure (Doyle and Doyle, 1987).
Sequencing analysis
Two fragments of chloroplast DNA (cpDNA) and one from ITS nuclear ribosomal gene (nrDNA) were analysed. The intron of trnL gene from cpDNA was amplified using the ‘c–d’ pair of primers as described by Taberlet et al. (1991) and the non-coding region between the psbA and trnH genes was amplified using primers described by Azuma et al. (2001). Primers developed by Desfeux and Lejeune (1996) were used to amplify the region ITS1 + 5·8S + ITS2 (ITS) from nrDNA. Fragments were amplified by PCR in a 20 µL volume containing 1·0 µm of each primer, 1 unit Taq DNA polymerase (Phoneutria, BR), 250 µm of each dNTP, 1× reaction buffer (10 mm Tris–HCl, pH 8·3, 50 mm KCl, 1·5 mm MgCl2), 250 µg of BSA and 10·0 ng of template DNA. Amplifications were performed using a GeneAmp PCR System 9700 (Applied Biosystems, CA, USA) with the following conditions: 96 °C for 2 min (one cycle); 94°C for 1 min, 56 °C for 1 min, 72 °C for 1 min (30 cycles); and 72 °C for 10 min (one cycle). PCR products were sequenced on an ABI Prism 377 automated DNA sequencer (Applied Biosystems) using DYEnamicTM ET terminator cycle sequencing kit (GE HealthCare, Sweden), according to manufacturer's instructions. All fragments were sequenced in forward and reverse directions.
Data analysis
Sequences were aligned using CLUSTALX (Thompson et al., 1997), and characters (each base pair) were equally weighted before analysis. Nucleotide (π) and haplotype (h) diversity (Nei, 1987) were estimated for each population and overall populations using the software Arlequin ver. 2001 (Schneider et al., 2000). To understand the geographical distribution and phylogenetic relationships among haplotypes, intraspecific phylogenies for sequencing data were inferred using median-joining network analysis based on parsimony criteria (Bandelt et al., 1999). The analysis was performed for each region separately with the software Network 4·2·0·1 (Forster et al., 2004).
To understand the origin of disjunct distribution the following statistical analyses were performed. First, an analysis of molecular variance (AMOVA; Excoffier et al., 1992) was performed using Arlequin to test the hypothesis of genetic differentiation among populations. The parameter θ (Weir and Cockerham, 1984) was estimated from AMOVA. Pairwise θ between all pairs of population were also estimated. Significance of θ and pairwise θ were tested by a non-parametric permutation test (Excoffier et al., 1992) implemented in the Arlequin software. The Mantel test was performed between pairwise θ and pairwise geographical distance matrices to test the hypothesis of isolation by distance (Wright, 1943) also using Arlequin software.
Then, a coalescent model (Kingman, 1982) was used to understand the demographic history of the species better and estimate time to most recent common ancestor (TMRCA). The demographic parameters θ = 2 µNe (coalescence force or mutation parameter, 4 µNe for nuclear genome), g (growth force, or exponential growth rate), where θt = θnow exp(−gtμ) and t is time in mutational unit, and M = 2Nem/θ (migration force or immigration rate, 4Nem/θ, for nuclear genome) were estimated based on a maximum likelihood estimation using Markov Chain Monte Carlo approach implemented in LAMARC 2·0·2 software (Kuhner et al., 2006). Four independent analyses were run with ten initial chains and two final chains. TMRCA (time to most recent common ancestor in mutational unit) was estimated based on Bayesian phylogenetic analysis implemented in BEAST 1·4·7 software (Drummond and Rambaut, 2007). For both, nuclear and chloroplast genomes, a relaxed molecular clock was assumed (uncorrelated lognormal), and exponential population growth. Prior Ne was set to assume a lower boundary of zero and infinity the upper boundary with exponential distribution. Four independent analyses were run for 108 generations under HKY + G substitution model. The model of sequence evolution was chosen by hierarchical likelihood ratio tests implemented in Modeltest 3·7 (Posada and Crandall, 1998). Convergence and stationarity of Bayesian phylogenetic analyses were checked using the software Tracer 1·4 (Rambaut and Drummond, 2007). To estimate TMRCA in years, mutation rates previously estimated for chloroplast non-coding regions (Yamane et al., 2006) and for ITS1 + ITS2 (Kay et al., 2006) were used. For chloroplast non-coding regions, Yamane et al. (2006) estimated an indel mutation rate of 0·8 × 10−9 per nucleotide per year and 1·52 × 10−9 for the substitution mutation rate. As the chloroplast regions used in the present work may have both indels and substitutions, a statistical distribution prior in BEAST was used with both the mutation rates as prior distribution, incorporating the uncertainty of mutation rate into the analysis. For ITS, Kay et al. (2006) estimated the mutation rates for different Angiospermae families, including four Asteraceae genera. Mutation rates for Asteraceae ranged from 2·51 × 10−9 (for Eupatorium, an herbaceous genus), 3·00 × 10−9 (for Hawaiian silverswords, woody plants) and 5·00 × 10−9 (for Dendroseris, a woody genus) to 7·83 × 10−9 (for Robinsonian, a woody genus). The full range of variation in mutation rates within Asteraceae was used to estimate divergence times as prior distribution. First preliminary analyses were performed with initial values of Ne ranging from 0·001 to 0·1 to verify the effect on TMRCA but no variation in TMRCA was observed.
RESULTS
Sequence characterization
Amplification of the non-coding regions trnL intron and psbA-trnH intergenic spacer generated fragments of 431 bp and 382 bp, respectively. All sequences were submitted to GenBank database (accession numbers: for trnL, FJ031242 to FJ031400, FJ031413 to FJ031428 and FJ031439 to FJ031454; for psbA-trnH, FJ031669 to FJ031844 and FJ031867 to FJ031882,).
The trnL intron presented eight different haplotypes and 13 polymorphic sites for 192 sequenced individuals. Gene diversity (h) overall population was 0·391 ± 0·044 and nucleotide diversity (π) was 0·003 ± 0·002. For the intergenic spacer psbA-trnH, eight different haplotypes were found, with 19 polymorphic sites for 192 individuals of L. ericoides. Gene diversity for psbA-trnH (h = 0·399 ± 0·044) was very similar to the value found for trnL and nucleotide diversity (π = 0·006 ± 0·004) was higher.
Although higher nucleotide diversity was found for psbA-trnH, chloroplast regions presented similar relationships among haplotypes and all phylogeographic and demographic analyses were performed for combined data. The combined data presented 813 bp with 34 polymorphic sites and 20 haplotypes for the 192 individuals of L. ericoides (see Supplementary data, available online).
For ITS1 + 5·8S + ITS2 (ITS) a fragment of 533 bp was obtained. All sequences were submitted to GenBank database (accession numbers FJ031455 to FJ03163 and FJ031653 to FJ031668). Eleven different haplotypes (see Supplementary data) and 14 polymorphic sites were found. Gene diversity overall population (h) was 0·726 ± 0·029 and nucleotide diversity (π) was 0·003 ± 0·002.
Phylogeography and demography
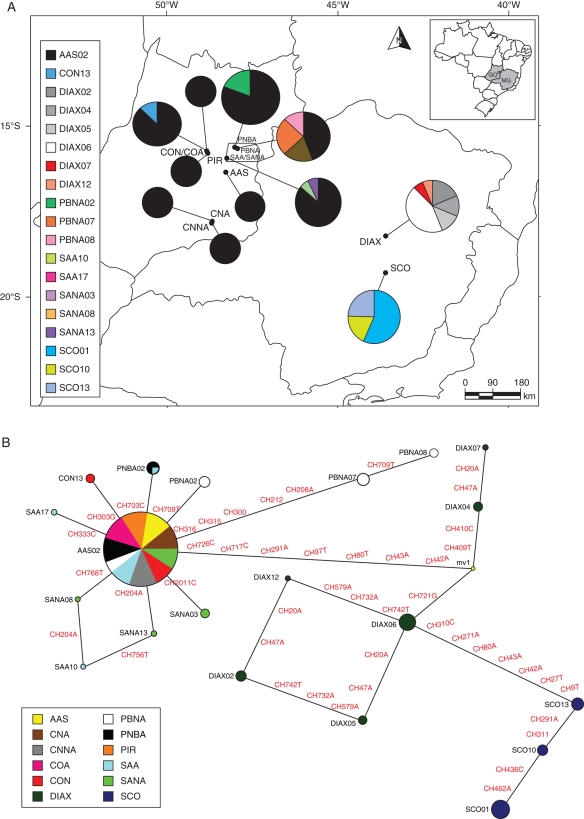
For the combined chloroplast regions, the most frequent haplotype (AAS02; Fig. 2A) was present in all populations from central Brazil, but not in populations from the south-east. Populations from central and south-east Brazil did not share any haplotype (Fig. 2A). Populations of L. ericoides from the eastern localities (DIAX and SCO; Fig. 2B) differed by many mutations from the AAS02 haplotype, and presented a remarkable relationship with haplotypes from the western localities (Fig. 2B).
Fig. 2.
(A) Geographical distribution of the haplotypes based on the combined data of chloroplast regions, for 192 individuals of L. ericoides. Different patterns were assigned for each haplotype accordingly to the legend at the left side of the figure. Circumference size is proportional to the haplotype frequency. (B) Median-joining network for the same data. Circumference size is proportional to the haplotype frequency. All mutations are shown in the network, mv1, median vector. Different patterns were assigned for each population: AAS, yellow; CNA, brown; CNNA, dark gray; COA, magenta; CON, red; DIAX, dark green; PBNA, white; PNBA, black; PIR, orange; SAA, water blue; SANA, light green; SCO, dark blue.
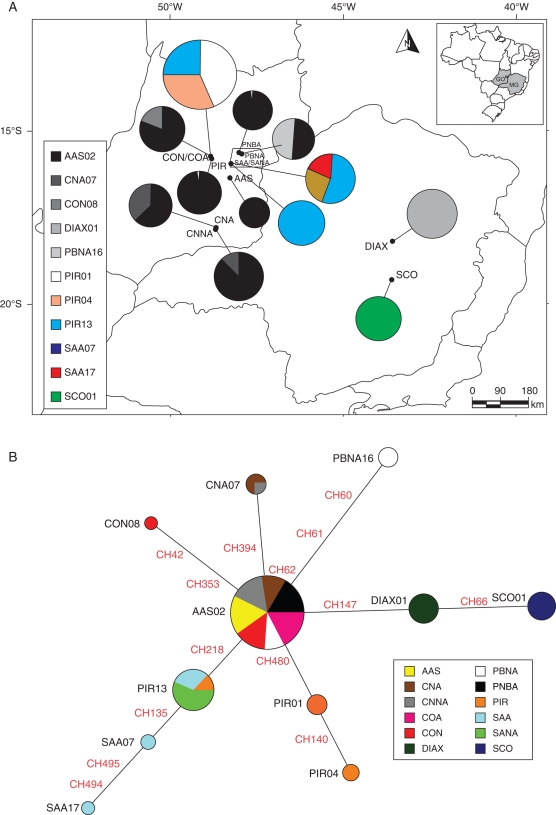
Relationships among ITS haplotypes were very similar to those based on the chloroplast genome (Figs 2B and 3B). Populations from the eastern localities did not share any haplotype with populations from the western (Fig. 3A), but divergences among haplotypes were lower than for the chloroplast genome (Fig. 3B). The most frequent haplotype, AAS02, was present in all populations from central Brazil, except PIR, SAA and SANA (Fig. 3A, B).
Fig. 3.
(A) Geographical distribution of the haplotypes based on the sequence of ITS nrDNA, for 192 individuals of L. ericoides. Different patterns were assigned for each haplotype accordingly to the legend at the left side of the figure. Circumference size is proportional to the haplotype frequency. (B) Median-joining network for the same data. Circumference size is proportional to the haplotype frequency. All mutations are shown in the network. See Fig 2b for legend.
Genetic diversity was slightly higher for the nuclear genome, but nucleotide diversity was higher for chloroplast combined data (Table 1). Some analysed populations presented no polymorphism at all for the two chloroplast regions and for ITS (Table 1). Population PBNA presented the highest gene diversity for chloroplast combined data, and population PIR had the highest gene diversity for ITS, considering populations from the western localities (Table 1). But considering all populations, DIAX presented the highest gene diversity for the chloroplast genome, but no polymorphism for ITS (Table 1).
Table 1.
Genetic diversity and demographic parameters for L. ericoides populations based on maximum likelihood estimations performed with LAMARC software, for combined cpDNA data from trnL and psbA-trnH, and for ITS nrDNA
| Combined cpDNA |
ITS |
|||||||
|---|---|---|---|---|---|---|---|---|
| Pop. | h | π | θ | g | h | π | θ | g |
| AAS | 0·000 | 0·0000 | 0·0051 | 328·52 | 0·000 | 0·0000 | 0·0023 | 335·99 |
| CNA | 0·000 | 0·0000 | 0·0021 | –253·31 | 0·050 | 0·0009 | 0·0019 | –492·52 |
| CNNA | 0·000 | 0·0000 | 0·0032 | –23·61 | 0·233 | 0·0005 | 0·0033 | 1202·40 |
| COA | 0·000 | 0·0000 | 0·0027 | –121·74 | 0·000 | 0·0000 | 0·0006 | –2376·52 |
| CON | 0·233 | 0·0003 | 0·0025 | –548·67 | 0·325 | 0·0013 | 0·0035 | 636·87 |
| DIAX | 0·783 | 0·0042 | 0·0009 | –2236·87 | 0·000 | 0·0000 | 0·0013 | –422·60 |
| PBNA | 0·742 | 0·0039 | 0·0033 | 97·62 | 0·533 | 0·0031 | 0·0038 | 3656·26 |
| PNBA | 0·325 | 0·0004 | 0·0037 | 150·38 | 0·000 | 0·0000 | 0·0047 | 1297·68 |
| PIR | 0·000 | 0·0000 | 0·0023 | –866·66 | 0·692 | 0·0025 | 0·0019 | –212·72 |
| SAA | 0·350 | 0·0007 | 0·0069 | 258·01 | 0·625 | 0·0023 | 0·0022 | 1613·72 |
| SANA | 0·447 | 0·0006 | 0·0015 | –344·69 | 0·000 | 0·0000 | 0·0022 | –945·93 |
| SCO | 0·625 | 0·0024 | 0·0014 | –1878·59 | 0·000 | 0·0000 | 0·0001 | –5000·00 |
n = 16 for all populations (Pop.).
h, haplotype diversity; π, nucleotide diversity; θ, coalescent parameter; g, exponential growth parameter.
AMOVA showed a high differentiation among populations, for both chloroplast (θ = 0·837, P < 0·001) and for nuclear ITS regions (θ = 0·700, P < 0·001). Differentiation was mainly due to differences between populations from south-east and central Brazil that presented high and significant pairwise θ (ranging from 0·713 to 0·973, P < 0·001, for the chloroplast genome). Pairwise θ between populations DIAX and SCO was also significant (pairwise θ = 0·842, P < 0·001). In central Brazil, only population PBNA presented significant differentiation from the other populations (pairwise θ ranging from 0·229 to 0·264, P = 0·004 ± 0·001 to P = 0·025 ± 0·006). For ITS, besides the high differentiation between populations from the south-east and central Brazil (pairwise θ ranging from 0·728 to 1·000, P < 0·001) and between populations DIAX and SCO (pairwise θ = 1·000, P < 0·001), PBNA and other populations from the western localities (PIR, SAA and SANA) also significantly contributed to the high differentiation among populations (pairwise θ ranging from 0·277 to 1·000, P = 0·007 ± 0·002 to P < 0·001).
Differentiation was positively correlated with geographical distance for both chloroplast DNA (r2 = 0·909, P < 0·001) and nrDNA ITS (r2 = 0·354, P = 0·009). However, when analyses were performed without populations from the south-east (DIAX and SCO), correlations were no longer significant (r2 = 0·215, P = 0·084, for chloroplast; r2 = 0·002, P = 0·466, for ITS).
Coalescent analyses performed with LAMARC software showed that most populations are shrinking (Table 1), although some incongruent results were obtained for chloroplast and ITS regions. For the chloroplast genome, gene flow among populations from south-east and central Brazil was negligible: <1·50 × 10−6 migrants per generation for all pairwise comparisons. The same result was found between populations DIAX and SCO. Nevertheless, populations from the western localities showed variable and asymmetrical migration rates. For instance, the highest migration rate was from population CON to SAA (4·03 migrants per generation). Migration from population COA to PIR was also high (2·97 migrants per generation), but from population PIR to COA it was lower with 1·35 migrants per generation. For the nearest populations (Fig. 1), migration rates were highly asymmetrical, such as migration from population SANA to SAA (2·53), and from population SAA to SANA (0·36). The same asymmetry was found from population PNBA to PBNA (1·36 migrants per generation) and from population PBNA to PNBA (8·70 × 10−7). For all other pairwise comparisons, the number of migrants per generation was <0·60.
When the ITS region was analysed, populations from south-east and central Brazil presented higher migration rates than for chloroplast genome, ranging from 7·14 × 10−7 (DIAX to PBNA) to 1·38 (SCO to CNA). Populations DIAX and SCO also presented a higher migration rate –1·30 migrants per generation from DIAX to SCO, and 0·76 from SCO to DIAX. Migration between populations CNA and CNNA was negligible (<3·60 × 10−7). For all other pairwise comparisons, the number of migrants per generation was <1·00.
Coalescent analyses performed with BEAST software indicated an ancient time to most recent common ancestor (TMRCA) for populations of L. ericoides from south-east and central Brazil (Table 2): approx. 709 655 ± 36 551 years bp (years before present) for the chloroplast genome and approx. 623 555 ± 55 769 years bp for ITS, and also for the two populations from Serra do Espinhaço (DIAX and SCO; Table 2). The TMRCA for populations from central Brazil was more recent (approx. 374 138 ± 32 035 to 278 735 ± 22 028 years bp; Table 2).
Table 2.
Time to most recent common ancestor (TMRCA) in years before present estimated with BEAST software for populations of L. ericoides from the west and east and for populations from each region, based on the combined cpDNA data from trnL and psbA-trnH and on ITS nrDNA
| Combined cpDNA |
ITS |
|||
|---|---|---|---|---|
| Population | θ (± s.d.) | TMRCA (± s.d.) | θ (± s.d.) | TMRCA (± s.d.) |
| L. ericoides | 0·00013 ± 0·00005) | 709 655 (± 36 551) | 0·0069 (0·0007) | 623 555 (± 55 769) |
| L. ericoides west | 0·0007 (± 0·0003) | 374 138 (± 32 035) | 0·0016 (0·0001) | 278 735 (± 22 028) |
| L. ericoides east | 0·0012 (± 0·0006) | 545 086 (± 30 491) | 0·0002 (0·00007) | 403 489 (± 26 565) |
DISCUSSION
Lychnophora ericoides presented high gene diversity and genetic differentiation among populations. Polymorphism patterns for the two chloroplast regions were very similar but nuclear ITS presented different patterns of haplotype and nucleotide diversity. Coalescent analyses showed that some populations are in expansion but most are shrinking, even though some incongruence could be found for chloroplast and ITS regions. Populations of L. ericoides usually present a high variation in size and in proportion of adults and juveniles. Most populations had no juveniles at all and were very small, such as PIR (20 adult individuals and no juveniles). The largest populations were PBNA and PNBA, with approx. 25 % of juveniles and nearly 200 individuals. In addition, L. ericoides has low recruitment and growth rates (J. D. Hay, Universidade de Brasília, Brazil, unpubl. res.), which may constrain population growth and resilience. While populations CNA and CNNA had high numbers of adults and juveniles, coalescent analyses showed that these populations are shrinking. On the other hand, for population SAA, a highly disturbed population with no juveniles and small population size (nearly 50 individuals), the result on coalescent analysis showed expansion. These apparently paradoxical results may be caused by population dynamics. The sandstone and quartzite habitat of L. ericoides is highly unstable with high levels of disturbance caused by fire during the dry season and sandstone and quartzite disruption, mainly during the rainy season. These disturbances may be highly variable among habitats and may cause sudden modifications in population size. The effect of population size reduction on haplotype diversity and distribution may be higher in the chloroplast genome than in the nuclear genome, since effective population size for haploid genome is half of the diploid genome due to genetic drift and bottleneck (Birky et al., 1983). This may lead to incongruent results when data from chloroplast and nuclear regions are compared. Notwithstanding, there is evidence of hybridization in the Lychnophora genus (Coile and Jones, 1981). Although there are no reports on hybridization in L. ericoides, differences in phylogenetic relationships between chloroplast and nuclear genomes may also be caused by introgression in the chloroplast genome. Nevertheless, a more in-depth phylogenetic analysis with all Lychnophora species is necessary to elucidate this relationship better. In addition, historical events may also be responsible for the patterns of haplotype diversity and distribution.
The results indicated an ancient divergence between populations from Serra do Espinhaço, south-east Brazil, and populations from the highlands of central Brazil (Table 2). Although estimations presented high variation, the TMRCA for populations from the western and eastern localities dates from the Middle Pleistocene and coincides with the Cromerian Interglacial that occurred between 620 000 and 455 000 years bp. This interglacial was followed by the Anglian Glaciation, also known as the Kansan Glaciation, the most severe glaciation of the Pleistocene that lasted from 455 000 to 300 000 years bp (Gibbard and van Kolfschoten, 2005). The Kansan Glaciation probably had greater effects on species distribution than the more recent glacial period, the Wisconsin Glaciation (approx. 110 000 to 12 000 years bp). In the southern hemisphere, forests shrank due to drier and colder conditions modifying species geographical distribution. The expansion and changing of geographical distribution in the last glaciation is observed in the fossil records for some species at high altitudes, such as Podocarpus (e.g. Colinvaux et al., 1996), Aracauria and other conifer taxa (Ledru, 1993). We hypothesize that L. ericoides expanded its distribution in the Kansan Glacial period, favoured by the colder and drier conditions that created suitable habitats. With the warmer and moisture conditions of the following interglacial (approx. 300 000 to 200 000 years bp), many populations were extinguished and L. ericoides has been restricted to drier quartizite and sandstone soils at high altitudes in Serra do Espinhaço and in similar habitats in central Brazil. Thus, the major phylogeographical break in L. ericoides – the differentiation between populations from the western and eastern localities of the geographical distribution – occurred due to the gene flow ceasing after the retraction of populations. Thus, the present disjunct distribution may represent a climatic relict of an ancient widely distributed population. This same pattern was also suggested for species from seasonally dry tropical forest of South and Central America (Prado and Gibbs, 1993; Pennington et al., 2000, 2004). The high differentiation and low gene flow between populations from the eastern and western localities are also evidence that vicariance is responsible for the disjunct distribution.
The TMRCA for populations from central Brazil is more recent and divergence may have occurred during the Illinoian (Wostonian) Interglacial, when populations where retracting causing a reduction in distribution range. Haplotype sharing among populations from Serra de Caldas, Serra dos Pirineus and Federal District is also evidence of a common origin of these populations. Furthermore, the sequence of several glaciations led to an advance of the savanna-like vegetation in cerrado and retreat of the tree species and forest vegetation during glaciations and the opposite in interglacial periods (Salgado-Laboriau et al., 1998; Burnham and Graham, 1999). This pattern of change may have caused several cycles of spreading and extinction of L. ericoides populations, leading to the extinction of haplotypes with low frequency during interglacial periods. Although the published cpDNA mutation rates are based on herbs, which may present 2-fold faster rates of substitution than woody plants (e.g. Smith and Donoghue, 2008) leading to an underestimation of divergence times, the TMRCA range was very similar to values obtained for ITS in which the substitution rate is based mainly on woody Asteraceae genus.
Although differentiation was correlated with geographical distance, this effect was due to the high differentiation between populations from the south-east and central Brazil. In fact, coalescent analyses showed no migration between populations from those localities. Migration was highly asymmetrical and lower than 1·0 migrant per generation for most pairs of populations from central Brazil. This may be caused by pollination and dispersal systems of L. ericoides. This species is pollinated by butterflies, and seeds (small achenes) are wind dispersed. Although butterfly pollinators may present variable flight distance and may fly long distances (Proctor et al., 1996), the patchy distribution of L. ericoides may favour the isolation of pollinators on patches of plants, restricting pollen flow due to the long distance between suitable habitats. Wind may promote long-distance seed dispersal (Horn et al., 2001), but habitat distribution may pose limitations on the expansion of populations by diffuse or jump dispersal. Dispersal may be constrained because the species is unable to cross barriers or because the species is a habitat specialist and may not succeed in habitats available in the barrier (Cox and Moore, 2005). The life history of Lychnophora ericoides suggests that both factors may constrain species expansion. Hence, a modification in habitat availability would be necessary for an expansion in the geographical distribution of the species.
In conclusion, our results strongly support that the disjunct distribution of L. ericoides may represent a climatic relict and that long-distance gene flow is unlikely. Moreover, the present work showed that phylogeographical analyses allowed the adequate hypothesis testing to decouple the mechanisms involved in the origin of the disjunct distribution of an endangered cerrado rupestre species. Finally, this report shows that besides the small population size, highly fragmented suitable habitat and harvesting of wild populations, L. ericoides is also threatened due to the lack of polymorphism in many populations, low migration among populations and because most populations are shrinking. Thus, the probability of local extinction due to demographic and genetic stocasticity could be very high. This information is highly important for planning and executing scientifically sound conservation programmes for the species.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and gives a list of haplotypes of L. ericoides based on the combined cpDNA data from trnL and psbA-trnH, and on ITS nrDNA.
ACKNOWLEDGEMENTS
We acknowledge Universidade Católica de Brasília and FNMA/MMA (National Fund for the Environment, Environment Brazilian Ministry) for financial support, Sergio Noronha for designing the maps of sample sites and haplotype distribution, Aécio A. dos Santos for kindly helping with the field work, officials at Ibama (Brazilian Institute for the Environment) for authorizing sampling in the National Park of Brasilia and SEMARH-GO (State Environmental Institute of Goiás) for sampling in the State Park of Serra de Caldas. We also acknowledge two anonymous reviewers for helpful comments that significantly improved this paper.
LITERATURE CITED
- Avise JC. Phylogeography: the history and formation of species. London: Harvard University Press; 2000. [Google Scholar]
- Azuma H, García-Franco JG, Rico-Gray V, Thien LB. Molecular phylogeny of the Magnoliaceae: the biogeography of tropical and temperate disjunctions. American Journal of Botany. 2001;88:2275–2285. [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Behling H. Late quaternary vegetational and climatic changes in Brazil. Review of Paleobotany and Palynology. 1998;99:143–156. [Google Scholar]
- Behling H. Late glacial and Holocene vegetation, climate and fire history inferred from Lagoa Nova in the southeastern Brazilian lowland. Vegetation History Archaeobotany. 2003;12:263–270. [Google Scholar]
- Behling H, Hooghiemstra H. Neotropical savanna environments in space and time: Late Quaternary. In: Markgraf V, editor. Interhemispheric climate linkages. Oxford: Academic Press; 2000. pp. 307–323. [Google Scholar]
- Bennett KD, Tzedakis PC, Willis KJ. Quaternary refugia of North European trees. Journal of Biogeography. 1991;18:103–115. [Google Scholar]
- Birky CW, Maruyama T, Fuerst P. An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplast, and some results. Genetics. 1983;103:513–527. doi: 10.1093/genetics/103.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham RJ, Graham A. The history of Neotropical vegetation: new developments and status. Annals of the Missouri Botanical Garden. 1999;86:546–589. [Google Scholar]
- Caetano S, Prado D, Pennington RT, et al. The history of seasonally dry tropical forests in eastern south America: inferences from the genetic structure of the tree Astronium urundeuva (Anacardiaceae) Molecular Ecology. 2008;17:3147–3159. doi: 10.1111/j.1365-294X.2008.03817.x. [DOI] [PubMed] [Google Scholar]
- Coile NC, Jones SB. Lychnophora (Compositae: Vernoniae), a genus endemic to the Brazilian Planalto. Brittonia. 1981;33:528–542. [Google Scholar]
- Colinvaux PA, de Oliveira PE, Moreno JE, Miller MC, Bush MB. A long pollen record from lowland Amazonia: forest and cooling in glacial times. Science. 1996;274:85–88. [Google Scholar]
- Collevatti RG, Grattapaglia D, Hay JD. Evidences for multiple maternal lineages of Caryocar brasiliense populations in the Brazilian Cerrado based on the analysis of chloroplast DNA sequences and microsatellite haplotype variation. Molecular Ecology. 2003;12:105–115. doi: 10.1046/j.1365-294x.2003.01701.x. [DOI] [PubMed] [Google Scholar]
- Comes HP, Kadereit JW. The effects of Quaternary climatic changes on plant distribution and evolution. Trends in Plant Science. 1998;3:432–438. [Google Scholar]
- Cox CB, Moore PD. Biogeography: an ecological and evolutionary approach. 7th edn. Oxford: Blackwell Publishing; 2005. [Google Scholar]
- Davis MB, Shaw RG. Range shifts and adaptive responses to Quaternary climate change. Science. 2001;292:673–679. doi: 10.1126/science.292.5517.673. [DOI] [PubMed] [Google Scholar]
- Desfeux C, Lejeune B. Systematics of euromediterranean Silene (Caryophylaceae): evidence from a phylogenetic analysis using ITS sequence. Les Comptes rendus de l'Académie des sciences, Série III. 1996;319:351–358. [PubMed] [Google Scholar]
- Dick CW, Abdul-Salim K, Bermingham E. Molecular systematics reveals cryptic Tertiary diversification of a widespread tropical rainforest tree. American Naturalist. 2003;162:691–703. doi: 10.1086/379795. [DOI] [PubMed] [Google Scholar]
- Dick CW, Bermingham E, Lemes MR, Gribel R. Extreme long-distance dispersal of the lowland tropical rainforest tree Ceiba pentandra L. (Malvaceae) in Africa and the Neotropics. Molecular Ecology. 2007;16:3039–3049. doi: 10.1111/j.1365-294X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1987;12:13–15. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian Evolutionary Analysis by Sampling Trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse P, Quattro J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitocondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Bandelt HJ, Röhl A, et al. Network 4·2·0·1. 2004 Software available free at www.fluxus-engineering.com . Fluxus Technology Ltda. [Google Scholar]
- Furley PA, Ratter JA. Soil resource and plant communities of the central Brazilian Cerrado and their development. Journal of Biogeography. 1988;15:97–108. [Google Scholar]
- Gaudeul M. Disjunct distribution of Hypericum nummularium L. (Hypericaceae): molecular data suggest bidirectional colonization from a single refugium rather than survival in distinct refugia. Biological Journal of the Linnean Society. 2006;87:437–447. [Google Scholar]
- Gibbard P, van Kolfschoten T. The Pleistocene and Holocene epochs. In: Gradstein FM, Ogg JG, Smith AG, editors. A geologic time scale 2004. Cambridge: Cambridge University Press; 2005. pp. 441–452. [Google Scholar]
- Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society. 1996;58:247–276. [Google Scholar]
- Hewitt GM. Post-glacial recolonization of European biota. Biological Journal of the Linnean Society. 1999;68:87–112. [Google Scholar]
- Horn HS, Nathan R, Kaplan SR. Long-distance dispersal of tree seeds by wind. Ecological Research. 2001;16:877–885. [Google Scholar]
- Karanth KP. Evolution of disjunct distributions among wet-zone species of the Indian subcontinent: testing various hypotheses using a phylogenetic approach. Current Science. 2003;85:1276–1283. [Google Scholar]
- Kay KM, Whittall JB, Hodges SA. A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: an approximate molecular clock with life history effects. BMC Evolutionary Biology. 2006;6:36–45. doi: 10.1186/1471-2148-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman JFC. The coalescent. Stochastic Processes and their Applications. 1982;13:235–248. [Google Scholar]
- Kuhner MK. LAMARC 2·0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics. 2006;22:768–770. doi: 10.1093/bioinformatics/btk051. [DOI] [PubMed] [Google Scholar]
- Ledru MP. Late Quaternary environmental and climatic change in central Brazil. Quaternary Research. 1993;39:90–98. [Google Scholar]
- Mansanares ME, Forni-Martins ER, Semir J. Chromosome numbers in the genus Lychnophora Mart. (Lychnophorinae: Vernonieae: Asteraceae) Caryologia. 2002;55:367–374. [Google Scholar]
- Marchant R, Berrıó JC, Behling H, Boom A, Hooghiemstra H. Colombian dry moist forest transitions in the Llanos Orientales: a comparison of model and pollen-based biome reconstructions. Palaeogeography, Palaeoclimatology, Palaeoecology. 2006;234:28–44. [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York, NY: Columbia University Press; 1987. [Google Scholar]
- Pennington RT, Lavin M, Prado DE, Pendry CA, Pell SK, Butterworth CA. Historical climate change and speciation: Neotropical seasonally dry forest plants show patterns of both Tertiary and Quaternary diversification. Philosophical Transactions of the Royal Society of London B. 2004;359:515–538. doi: 10.1098/rstb.2003.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington RT, Prado DA, Pendry C. Neotropical seasonally dry forests and Pleistocene vegetation changes. Journal of Biogeography. 2000;27:261–273. [Google Scholar]
- Petit RJ, Brewer S, Bordács S, et al. Identification of refugia and post-glacial colonisation routes of European white oaks based on chloroplast DNA and fossil pollen evidence. Forest Ecology Management. 2002;156:49–74. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Prado DE, Gibbs PE. Patterns of species distributions in the dry seasonal forests of South America. Annals of the Missouri Botanical Garden. 1993;80:902–927. [Google Scholar]
- Proctor M, Yeo P, Lack A. The natural history of pollination. Portland, OR: Timber Press; 1996. [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1·4. 2007. Available free from http://beast.bio.ed.ac.uk/Tracer .
- Ramos ACS, Lemos-Filho JP, Ribeiro RA, Santos FR, Lovato MB. Phylogeography of the tree Hymenaea stigonocarpa (Fabaceae: Caesalpinioideae) and the influence of Quaternary climate changes in the Brazilian Cerrado. Annals of Botany. 2007;100:1219–1228. doi: 10.1093/aob/mcm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JF, Walter BMT. Fitofisionomias do Bioma Cerrado. In: Sano SM, Almeida SP, editors. Cerrado: ambiente e flora. Brasília: Embrapa; 1998. pp. 89–168. [Google Scholar]
- Salgado-Labouriau ML, Barberi M, Ferraz-Vicentini KR, Parizzi MG. A dry climatic event during the late Quaternary of tropical Brazil. Review of Paleobotany and Palynology. 1998;99:115–129. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Switzerland: Genetics and Biometry Laboratory, University of Geneva; 2000. Arlequin Ver. 2000: software for population genetic data analysis. [Google Scholar]
- Semir J. Universidade Estadual de Campinas; 1999. Revisão taxonômica de Lychnophora Mart. (Vernonieae: Compositae) PhD thesis. [Google Scholar]
- Smith SA, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322:86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- Stehlik I, Holderegger R, Schneller JJ, Abbott RJ, Bachmann K. Molecular biogeography and population genetics of alpine plant species. Bulletin of the Geobotanical Institute ETH. 2000;66:47–59. [Google Scholar]
- Taberlet P, Fumagalli L, Wust-Saucy AG, Cossons JF. Comparative phylogeography and postglacial colonization routes in Europe. Molecular Ecology. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Yano K, Kawahara T. Pattern and rate of indel evolution inferred from whole chloroplast intergenic regions in sugarcane, maize and rice. DNA Research. 2006;13:197–204. doi: 10.1093/dnares/dsl012. [DOI] [PubMed] [Google Scholar]