Abstract
Background and Aims
Incongruence between chloroplast and nuclear DNA phylogenies, and single additive nucleotide positions in internal transcribed spacer (ITS) sequences of polyploid Australian/New Zealand (NZ) Lepidium species have been used to suggest a bicontinental hybrid origin. This pattern was explained by two trans-oceanic dispersals of Lepidium species from California and Africa and subsequent hybridization followed by homogenization of the ribosomal DNA sequence either to the Californian (C-clade) or to the African ITS-type (A-clade) in two different ITS-lineages of Australian/NZ Lepidium polyploids.
Methods
Genomic in situ hybridization (GISH) was used to unravel the genomic origin of polyploid Australian/NZ Lepidium species. Fluorescence in situ hybridization (FISH) with ribosomal DNA (rDNA) probes was applied to test the purported ITS evolution, and to facilitate chromosome counting in high-numbered polyploids.
Key Results
In Australian/NZ A-clade Lepidium polyploids, GISH identified African and Australian/NZ C-clade species as putative ancestral genomes. Neither the African nor the Californian genome were detected in Australian/NZ C-clade species and the Californian genome was not detected in Australian/NZ A-clade species. Five of the eight polyploid species (from 7x to 11x) displayed a diploid-like set of rDNA loci. Even the undecaploid species Lepidium muelleriferdinandi (2n = 11x = 88) showed only one pair of each rDNA repeat. In A-clade allopolyploids, in situ rDNA localization combined with GISH corroborated the presence of the African ITS-type.
Conclusions
The nuclear genomes of African and Australian/NZ C-clade species were detected by GISH in allopolyploid Australian/NZ Lepidium species of the A-clade, supporting their hybrid origin. The presumed hybrid origin of Australian/NZ C-clade taxa could not be confirmed. Hence, it is assumed that Californian ancestral taxa experienced rapid radiation in Australia/NZ into extant C-clade polyploid taxa followed by hybridization with African species. As a result, A-clade allopolyploid Lepidium species share the Californian chloroplast type and the African ITS-type with the C-clade Australian/NZ polyploid and African diploid species, respectively.
Key words: Lepidium, Brassicaceae, FISH, GISH, hybridization, polyploidy, long-distance dispersal, ITS, rDNA, Australia, New Zealand
INTRODUCTION
Lepidium is one of the largest genera in the Brassicaceae, consisting of approximately 230 species distributed worldwide (Al-Shehbaz et al., 2006). This genus is characterized by reduced floral organs, an autogamous mating system and mucilaginous seeds (Al-Shehbaz, 1986). Recent molecular phylogenetic studies using the nuclear ribosomal DNA (rDNA) internal transcribed spacer (ITS), non-coding chloroplast DNA (cpDNA) and single-copy nuclear DNA sequences (an intron of the PISTILLATA gene, PI), respectively, clarified only some relationships within the genus (Bowman et al., 1999; Mummenhoff et al., 2001; Lee et al., 2002). All molecular phylogenies support three main infrageneric lineages, corresponding to (1) section Monoploca sensu stricto (s.s.) (Australia), (2) section Lepia with Cardaria included (Eurasia) and (3) Lepidium s.s. (referred to hereafter as Lepidium) representing by far the bulk of Lepidium species on every continent. The fossil data, easily dispersible mucilaginous seeds, common autogamous breeding systems and low levels of sequence divergence between Lepidium species from different continents or islands suggest a rapid radiation of Lepidium by long-distance dispersal in the Pliocene/Pleistocene (Mummenhoff et al., 2001). The large number of polyploid species (Warwick and Al-Shehbaz, 2006) suggests reticulate evolution in Lepidium and this was indicated also in the analysis of PISTILLATA intron sequences, in which many polyploid taxa harbour two or more phylogenetically distinct sequences (Lee et al., 2002). Chromosome numbers have been reported for about 70 Lepidium species and subspecies (Warwick and Al-Shehbaz, 2006), of which all except five have x = 8 chromosomes (Al-Shehbaz 1986). Vaarama (1951) speculated that species with 2n = 24 (e.g. L. sativum) are hexaploids based on x = 4, but this interpretation was not supported by any further research. More than half of Lepidium species studied are polyploids, and chromosome numbers vary widely (2n = 16, 24, 28, 32, 40, 48, 64 and 80; Warwick and Al-Shehbaz, 2006).
In Australia and New Zealand (NZ), Lepidium is represented by 19 and seven native species, respectively. Incongruence between the chloroplast and nuclear DNA phylogenetic analyses indicated a hybridogenous genomic constitution of Australian/NZ Lepidium species: all 18 studied species shared a Californian cpDNA-type, while 11 Australian/NZ species appeared to harbour a Californian ITS-type (C-clade) and a group of the remaining seven species shared an African ITS-type (A-clade) (Mummenhoff et al., 2004; Table 1, Fig. 1). This pattern was explained by two trans-oceanic dispersals of ancestral taxa from California and Africa to Australia/NZ and subsequent hybridization followed by sequence homogenization of the rDNA either to the Californian or to the African ITS-type in the two different lineages, with single additive nucleotide positions remaining, representing the ancestral parental input (Table 1, Fig. 1). However, Mummenhoff et al. (2004) did not assess the ploidy level of Australian/NZ species and the conclusion of a bicontinental hybridogenous genomic constitution was based only on two markers representing nuclear (ITS) and chloroplast genomes (three non-coding cpDNA regions).
Table 1.
Composition of the ITS sequences of Californian, Australian/New Zealand and African Lepidium species
|
ITS1 ITS2 |
|
|---|---|
| 1111111112233344 | |
| 67880013446775936933 | |
| 83478965680241795402 | |
| Californian C-clade species | |
| L. nitidum, L. oxycarpum, L. dictyotum | CATTAGCCCGTCATCGG–TA |
| Australian/NZ C-clade polyploids | |
| L. aschersonii | CATTATCCCGTCATCGG–TW |
| L. muelleriferdinandi | CATTRCCCCGTCATCGG–TA |
| Australian/NZ A-clade polyploids | |
| L. hyssopifolium (KM 1670) | YWYKRSSYSSWACYTA-TCT |
| L. hyssopifolium (KM 1669) | TTCGGCGTGCAACCTA–TCT |
| L. pseudohyssopifolium | TTCGGCGTGCAACCYA–TCT |
| African A-clade species | |
| L. africanum, L. transvaalense | TTCGGCGTGCAACCTA–TCT |
This data matrix contains only those positions of a complete alignment (not shown) that distinguish Californian from African Lepidium taxa. Californian and African species are extant members of two different lineages suggested to have been involved in the hybridogenous origin of Australian/NZ Lepidium species. R = A and G; W = A and T; Y = C and T; K = G and T; S = G and C. Site numbers are those of the complete alignment. For details see Mummenhoff et al. (2004).
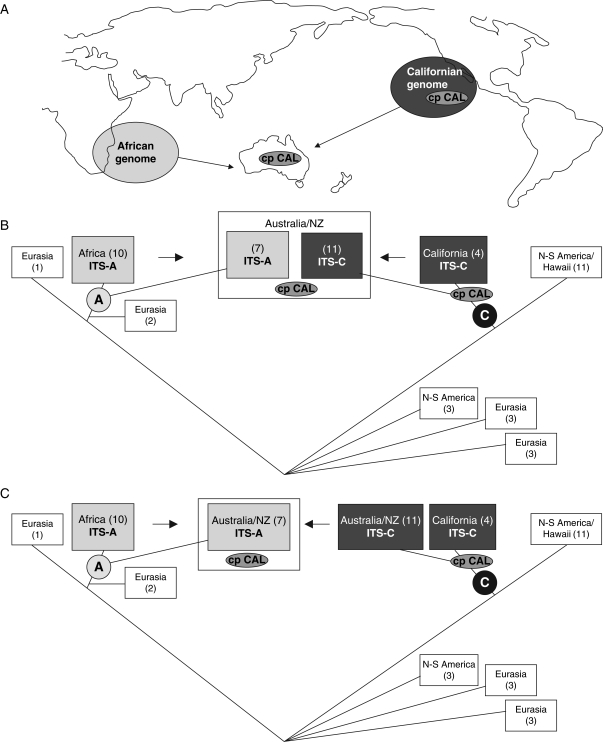
Fig. 1.
Organismal and geographical context of the genome evolution of polyploid Australian/NZ A- and C-clade Lepidium species. (A) Geographical context of the evolution of polyploid Australian/NZ Lepidium species further outlined in (B) and (C). (B) Evolution of polyploid Australian/NZ A- and C-clade Lepidium species as inferred from ITS- and cpDNA sequence data (Mummenhoff et al., 2004). All Australian/NZ species harbour a Californian chloroplast type (cp CAL), while bidirectional concerted evolution of ribosomal DNA (ITS) subsequent to hybridization between African and Californian parental species resulted in the presence of either of the parental ITS-types, i.e. African (ITS-A) and Californian (ITS-C), in the two lineages (Australian/NZ A- and C-clade species, respectively). Arrows indicate hybridization. Numbers in boxes represent number of analysed species of given geographical origin. Encircled letters A and C refer to A- and C-clade species, respectively. See text for more details. (C) Evolution of polyploid Australian/NZ A- and C-clade Lepidium species as inferred from ITS and cpDNA sequence data (Mummenhoff et al., 2004) and current GISH experiments. In this alternative scenario polyploid Australian/NZ C-clade species are the outcome of rapid radiation of an ancient Californian ancestor in Australia/New Zealand. Australian/NZ A-clade species originated from hybridization between African and Australian/NZ C-clade species, still harbouring the ancestral Californian chloroplast type. In this scenario concerted evolution operated unidirectionally towards the African ITS-type in Australian/NZ A-clade hybrids. Arrows indicate hybridization. Numbers in boxes represent number of analysed species of given geographical origin. Encircled letters A and C refer to A- and C-clade species, respectively. See text for more details.
The present study aimed to (1) assess ploidy level and chromosome number variation among Australian/NZ Lepidium species, (2) test the purported allopolyploid bicontinental origin of Australian/NZ species of the A- and C-clades using genomic in situ hybridization (GISH) and (3) gain insight into the physical organization of rDNA loci in the presumed parental species and their Australian/NZ hybrid derivatives.
MATERIALS AND METHODS
Plant material
Collection data of selected species of Lepidium L. used in the current study are given in Table 2. The presumed parental and hybrid species were used exemplarily, based on results of previous studies (Mummenhoff et al., 2001, 2004, 2009).
Table 2.
Origin, chromosome number, ploidy level and number of rDNA loci for Lepidium species studied
| Species | Accession no. | Origin/Collector | Chromosome no. | No. of 45S rDNA loci | No. of 5S rDNA loci |
|---|---|---|---|---|---|
| L.africanum (Burm.f.) DC.* | KM 1702 | Australia, Melbourne, Royal Park, South of Park Street/Scarlett, N. | 2n = 2x = 16 | 2 | 2 |
| KM 1793 | South Africa, Eastern Cape/Clark, V. R. | 2n = 2x = 16 | 2 | 2 | |
| KM 1794 | South Africa, Eastern Cape/Clark, V. R. | 2n = 2x = 16 | 2 | 2 | |
| L. transvaalense Marais | KM 1792 | South Africa, Eastern Cape/Clark, V. R. | 2n = 2x = 16 | 2 | 2 |
| L. nitidum Nutt. | KM 1711 | USA, California, Tucker Herbarium at UC Davis/Bowman, J. | 2n = 7x = approx. 56 | 2 | 2 |
| L. oxycarpum Torr. & Gray | KM 1713 | USA, California, Central Valley, Los Banos/Bowman, J. | 2n = 7x = approx. 56 | 2 | 2 |
| L. dictyotum Gray | KM 1714 | USA, California, Carrizo plain/Bowman, J. | 2n = 7x = approx. 56 | 2 | 4 |
| L. hyssopifolium Desv. | KM 1669 | Australia, New South Wales, Bathurst/Scarlett, N. | 2n = 9x = 72 | 2 | 4 |
| KM 1670 | Australia, Victoria, Belfast Lough, Port Fairy end of the Lough/Scarlett, N. | 2n = 9x = 72 | 2 | 4 | |
| L. pseudohyssopifolium Hewson | KM 1666 | Australia, Victoria, Melbourne, Fairfield, Banks of the Yarra River/Parsons, R. F. | 2n = 9x = 72 | 2 | 2 |
| L. ginninderrense Scarlett | KM 1673 | Australia, Australian Capital Territory, Belconnen Naval Station, floodplain of Ginninderra Creek/Scarlett, N. | 2n = 14x = 112 | 4 | 2 |
| L. muelleriferdinandi Thell. | KM 1710 | Australia, Northern Territory, Alice Springs Desert Park, outside herbarium/Albrecht, D. E. | 2n = 11x = 88 | 2 | 2 |
| L. aschersonii Thell. | KM 1668 | Australia, Victoria, Lake Beeac/Scarlett, N. | 2n = 7x = 56 | 2 | 2 |
* L. africanum is native to South Africa, and an introduced weed in Australia.
Chromosome preparations
Plants were grown from seeds in plastic Petri dishes on sieved potting soil in a phytotron with long day illumination (16 h light at 20 °C, 8 h dark at 15 °C). Young inflorescences were fixed in ethanol/acetic acid (3:1, v/v) fixative for 24 h at 4 °C. Fixative was replaced by 70 % ethanol and the material stored at –20 °C until further use. Chromosome spreads were prepared as described by Lysak et al. (2006). Slides were examined under phase contrast for the presence of suitable mitotic metaphase spreads. Selected slides were post-fixed in 4 % formaldehyde in 2× sodium saline citrate (SSC, 5 min), washed in 2× SSC (twice for 5 min), and dehydrated in an ethanol series (70, 90 and 100 %, 2 min each). The preparations were stained with 2 µg mL−1 4,6-diamino-2-phenylindole (DAPI) in Vectashield antifade (Vector Laboratories, Burlington, Ontario, Canada) and screened for the quality of metaphase and the presence of cytoplasm under an epifluorescence microscope (BX-61, Olympus, Tokyo, Japan). If appropriate, the slides were treated with pepsin (0·1 g L−1) in 0·01 m HCl at 39 °C for 15–45 min, and post-fixed and dehydrated as described above.
DNA probes
The Arabidopsis thaliana BAC clone T15P10 (AF167571) bearing 45S rRNA gene repeats was used for in situ localization of 45S rDNA, and A. thaliana clone pCT 4·2 (M65137), corresponding to a 500-bp 5S rRNA repeat, was used for localization of 5S rDNA loci. For GISH, total genomic DNA (gDNA) was extracted from healthy young leaves according to Dellaporta et al. (1983) followed by RNase treatment (50 µg mL−1). Extracted gDNA was checked for protein, starch or RNA contamination using a Beckmann photospectrometer and run on a 1 % (w/v) agarose gel in 1× Tris-acetate-EDTA (TAE) buffer. DNA probes were labelled either by biotin- or by digoxigenin-dUTP using the Nick Translation Mix (Roche, Mannheim, Germany) according to the manufacturer's instructions.
In situ hybridization
Labelled 5S and 45S rDNA probes were precipitated and the pellet was resuspended in 20 µL hybridization mixture containing 50 % formamide and 10 % dextrane sulfate in 2× SSC. The probe was applied to the slide, covered with a cover slip and denaturated on a hot plate at 80 °C for 2 min. Slides were hybridized at 37 °C for approx. 12 h. To prevent drying, cover slips were framed by rubber cement. Labelled gDNA probes were prepared and denatured as described above. Hybridization time for GISH was between 12 and 48 h.
Following hybridization, stringent washing was carried out in 50 % formamide in 2× SSC (v/v) at 42 °C three times for 5 min. Detection of hybridization signals was performed according to Lysak et al. (2006). The biotin-labelled probes were detected by avidin–Texas Red (Vector Laboratories), and signals amplified by biotinylated goat anti-avidin (Vector Laboratories) and avidin–Texas Red. Digoxigenin-labelled probes were detected by mouse anti-digoxigenin (Roche) and goat anti-mouse–Alexa Fluor 488 antibodies (Molecular Probes, New Haven, CT, USA). Chromosomes were counterstained with DAPI (2 µg mL−1) in Vectashield. Fluorescence signals were analysed with an Olympus BX-61 epifluorescence microscope and AxioCam CCD camera (Carl Zeiss, Jena, Germany). Individual images were merged and processed by using Photoshop CS software (Adobe Systems).
RESULTS
Chromosome counts
In the analysed Lepidium species, chromosome numbers varied from 2n = 16 to 2n = 112 (see Table 2 for all chromosome counts). The two African species (L. africanum, L. transvaalense) have a diploid chromosome number (2n = 2x = 16), whereas all Australian/NZ and Californian species are polyploid (Fig. 2). Four different ploidy levels (7x, 9x, 11x and 14x) were discerned in Australian/NZ species. In the three Californian species, we could not obtain suitable chromosome spreads to determine exact chromosome numbers. Observed chromosome numbers were tentatively interpreted as heptapolyploid (2n = 7x = 56).
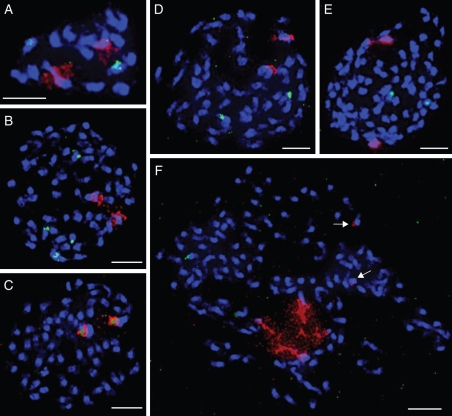
Fig. 2.
DAPI-stained mitotic chromosome spreads from flower bud tissue of Lepidium species. Chromosomes were hybridized with 5S (green) and 45S (red) rDNA probes and counterstained with DAPI. (A) L. africanum (2n = 2x = 16), (B) L. dictyotum (2n = 7x = 56), (C) L. oxycarpum (2n = 7x = 56), (D) L. aschersonii (2n = 7x = 56), (E) L. muelleriferdinandi (2n = 11x = 88), (F) L. ginninderrense (2n = 14x = 112). In (F) one pair of major 45S rDNA loci is decondensed around the nucleolus; two minor 45S rDNA loci are indicated by arrows. Scale bars = 5 µm.
Mitotic chromosomes of Lepidium species are small (approx. 2–5 µm) with the bulk of heterochromatin concentrated within pericentromeres, and often fuzzy euchromatic chromosome ends (Figs 2 and 3). In the two African species, chromosomes are larger and possess a less distinct eu-/heterochromatin profile than chromosomes in the other two geographical groups. The presumably different chromosome structure of African species is particularly apparent upon GISH in the A-clade allopolyploids, whereby chromosomes of African origin show a more even labelling pattern and appear to be larger than the remaining chromosomes (Fig. 3).
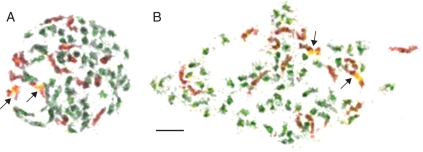
Fig. 3.
GISH in two nonaploid (2n = 9x = 72) Australian/NZ Lepidium A-clade species. Chromosomes of L. hyssopifolium (A) and L. pseudohyssopifolium (B) hybridized with gDNA of L. africanum (16 red chromosomes) and gDNA of an Australian/NZ C-clade species, i.e. L. muelleriferdinandi (56 green chromosomes). Arrows mark 45S rDNA-bearing chromosomes (yellow/red). Scale bar = 5 µm.
Localization of rDNA loci
Most Lepidium species exhibit one pair of 5S and 45S rDNA loci (Table 2 and Fig. 2). This pattern was also observed in high-numbered polyploid species such as L. nitidum, L. oxycarpum, L. pseudohyssopifolium, L. muelleriferdinandi and L. aschersonii. Two species (L. dictyotum and L. hyssopifolium) possess two pairs of 5S rDNA, and one species (L. ginninderrense) possesses two pairs of 45S rDNA loci (Table 2 and Fig. 2). In L. oxycarpum, 5S and 45S loci were found to be partly co-localized (Fig. 2C). 5S rDNA loci are positioned on chromosomes interstitially, whereas 45S rDNA loci showed terminal localization (Fig. 2).
GISH in polyploid Australian/NZ Lepidium species
With the aim to discern the origin of Australian/NZ Lepidium polyploids, labelled gDNA of putative African and Californian parental species was hybridized to mitotic chromosomes of Australian/NZ polyploid A- and C-clade taxa. In two A-clade species (L. hyssopifolium, L. pseudohyssopifolium), gDNA of presumed African parents (L. africanum, L. transvaalense) labelled 16 of the 72 chromosomes, whereas no chromosomes were identified using gDNA of presumed Californian parental species (L. dictyotum, L. oxycarpum) (data not shown). No chromosome-specific hybridization signals were observed in C-clade species (L. aschersonii, L. muelleriferdinandi) using the same probes as used in the A-clade species (data not shown).
In follow-up GISH experiments, 56 of 72 chromosomes in the Australian/NZ A-clade species L. hyssopifolium and L. pseudohyssopifolium were detected using gDNA of two Australian/NZ C-clade species, i.e. L. muelleriferdinandi (Fig. 3) and L. aschersonii (data not shown). In the two A-clade allopolyploids (2n = 72), 16 chromosomes correspond to L. africanum and 56 chromosomes were labelled by gDNA of L. aschersonii/L. muelleriferdinandi (Fig. 3). This experiment also showed that 45S rDNA in L. hyssopifolium and L. pseudohyssopifolium has probably been contributed by the African parental genome(s).
DISCUSSION
rDNA patterns in polyploid Lepidium species
The present study followed three principal goals as far as in situ localization of rDNA repeats is concerned. First, we were interested to see if the three geographical species groups (Africa, Australia/NZ and California) are characterized by specific rDNA patterns. Secondly, the number of rDNA loci in Australian/NZ polyploids was compared with the number of the same loci in the presumed progenitor species, and finally, the origin of 45S rDNA loci in Australian/NZ polyploid species was inferred from GISH analysis and compared with conclusions based on ITS sequence data (Mummenhoff et al., 2004).
From the limited data set (only ten of 230 Lepidium species were analysed) no apparent geographical pattern of rDNA variation emerges. In all but one polyploid species analysed, only a diploid-like number of 45S rDNA loci were found, with two pairs of 45S identified in the tetradecaploid L. ginninderrense (2n = 112). This finding is somewhat surprising given the high ploidy levels (from 7x to 14x) of the analysed taxa. Regardless of prevailing modes of polyploid evolution (auto- vs. allopolyploidy) in the Californian and Australian/NZ groups, respectively, the present data indicate that there is a strong tendency toward diploidization of rDNA loci in both geographical species groups. Although less likely, preferential loss of rDNA-bearing chromosomes in odd-numbered (7, 9 and 11x) Lepidium polyploids cannot be ruled out.
Ali et al. (2005) analysed three different crucifer species with the same polyploid chromosome number (2n = 48). In two species, the number of 45S was high (approx. 14 in Camelina microcarpa, and 10 + 6 minor loci in Olimarabidopsis cabulica), whereas only one pair has been recorded in Aethionema schistosum (Ali et al., 2005). It could be speculated that this limited comparison corroborates the generally accepted correlation between the number of 45S rDNA loci and the age of polyploid species as the genus Aethionema is sister to and older than the rest of Brassicaceae (Al-Shehbaz et al., 2006). However, the Aethionema polyploid species may be of a similar age to two Camelineae species analysed. Hence, it appears to be difficult to establish a direct link between the age of a phylogenetic split and the diploidization rate in crucifer taxa.
With each polyploidization event the number of rDNA loci increases and some rDNA sequences become redundant. These ITS sequences can be changed by bidirectional interlocus concerted evolution, resulting in complete homogenization to either of the parental types or to mosaic-like sequences with additive nucleotides from both parents (Wendel, 2000). In the long term, genome diploidization events as discovered for ancient allopolyploid Nicotiana species of the section Repandae (4·5 Myr old) can occur, resulting in the loss of redundant rDNA loci and a diploid-like number of rDNA loci, which can be even lower than the number of 45S loci found in the progenitor species (Clarkson et al., 2005). However, more recently formed Nicotiana allopolyploids (e.g. N. tabacum, 0·2 Myr old) display the sum of the rDNA loci of their progenitors (Clarkson et al., 2005). As a maximum estimated age for the origin of polyploid Australian/NZ Lepidium species is approx. 1·3 Ma (Mummenhoff et al., 2004), this or a shorter period of time has been sufficient for (nearly) complete diploidization of rDNA loci (Table 2).
Within the set of the 72 chromosomes of Australian/NZ A-clade species (L. hyssopifolium, L. pseudohyssopifolium) combined GISH and rDNA fluorescence in situ hybridization (FISH) localization detected two 45S rDNA loci on two chromosomes belonging to the 16 chromosomes of African origin. The 56 remaining chromosomes displayed no 45S rDNA loci (Fig. 3). This is in agreement with the ITS phylogenetic data showing that all Australian/NZ A-clade species harbour the African ITS-type (Mummenhoff et al., 2004; Fig. 1C, Table 1). For these species, physical loss of C-clade rDNA loci must be assumed subsequent to hybridization. Furthermore, the African ITS-type in polyploid Australian/NZ A-clade species can be explained by unidirectional concerted evolution to the African ITS-type with single additive nucleotides remaining (Table 1, L. hyssopifolium KM 1670).
Previous hypothesis on the origin of polyploid Australian/NZ Lepidium species
The rDNA ITS regions are the most widely used nuclear-encoded phylogenetic markers (Álvarez and Wendel, 2003). Their high evolutionary rate permits discrimination of closely related putative parental species and the identification of additive patterns in hypothesized allopolyploids (Marhold and Lihová, 2006). Although concerted evolution can erase nucleotide additivity, bidirectional, nearly complete homogenization or intergenomic recombination may result in a mosaic-like structure of ITS sequences, which has been described in the literature (Marhold and Lihová, 2006). In the Brassicaceae, such analyses unravelled hybridization events in Thlaspi (Mummenhoff et al., 1997), Cardamine (Franzke and Mummenhoff, 1999; Lihová et al., 2006) and Boechera (Koch et al., 2003). Support for the bicontinental hybrid origin of polyploid Australian/NZ Lepidium species was provided by bidirectional, nearly complete concerted evolution of the ITS to either of the two presumed parental ITS-types (African and Californian) with single additive (parental) nucleotides remaining (Mummenhoff et al., 2004; Table 1). It appears that all Australian/NZ polyploids share a Californian cpDNA-type (Mummenhoff et al., 2004), but this cpDNA evidence was detected only in 60 % of the maximally parsimonious trees (Mummenhoff et al., 2004). Furthermore, this scenario of a bicontinental hybridogenous genomic constitution was based on only two markers representing the nuclear (ITS) and chloroplast genome (Mummenhoff et al., 2004). Thus, GISH experiments were performed to gain deeper insight into the origin and evolution of polyploid Australian/NZ Lepidium species.
Origin of Australian/NZ C-clade species: neither African nor Californian progenitors detected by GISH
In previous studies based on sequence analysis of nuclear ITS and non-coding cpDNA (Mummenhoff et al., 2004), a bicontinental hybrid origin of polyploid Australian/NZ Lepidium species has been suggested. In GISH experiments, gDNA probes of presumed African (L. africanum, L. transvaalense) and Californian parental species (L. dictyotum, L. oxycarpum) did not label specific chromosomes in the Australian C-clade species. This result is in contrast to the ITS and cpDNA analysis (Mummenhoff et al., 2004). The evidence of a bicontinental hybridogenous genomic constitution of C-clade species is based on weak cpDNA evidence (see above), ITS sequence homology of Australian/NZ species with Californian taxa, and single additive nucleotide positions representing both parental ITS-types. If this additivity in single ITS nucleotide positions is not random and indicates an African input, present-day African species used for GISH experiments do not represent the original African progenitors. This means that the genome of the original African species contributing to the ITS additivity in the polyploid Australian/NZ C-clade species is too distinct from the genome of extant African taxa used in the present study (L. africanum, L. transvaalense), and thus gDNA probes of L. africanum/L. transvaalense could not be used to detect the original African parental genome in C-clade species.
Ancestral Californian species (migrating to Australia) may have transmitted the Californian ITS-type to the allopolyploid Australian/NZ C-clade species via hybridization with an unknown taxon, potentially of African origin. At the same time the ancestral Californian taxa might have evolved into the present-day polyploid Californian species, probably by several rounds of hybridization and polyploidization events, accompanied by the dynamic evolution of genome-specific dispersed repeats. As a consequence, the extant Californian species differ significantly in genome constitution from their presumed polyploid Australian/NZ descendants, and thus their gDNAs do not reveal the ancestral Californian genome in the polyploid Australian/NZ C-clade species. This can be analogous to the decreased efficiency of GISH and its failure in approx. 1- and 5-Myr-old Nicotiana allopolyploids, respectively (Lim et al., 2007).
Australian/NZ C-clade species: simply descendants of Californian ancestors?
Negative results using gDNA of Californian and African species in C-clade Australian/NZ polyploids suggest long-distance dispersal of ancestral Californian species to Australia followed by radiation into extant C-clade polyploid species without hybridization with African species being involved. All Australian/NZ C-clade species appear to harbour a Californian ITS- and cpDNA-type (Mummenhoff et al., 2004) and only one additive nucleotide position each (representing African and Californian ancestral nucleotides, respectively) in the ITS sequences of two C-clade species studied (L. aschersonii, L. muelleriferdinandi). This single African nucleotide in these two C-clade species could also be explained by random mutations or could represent an input by hybridization with an unknown species. The remaining 32 chromosomes of the genome of present-day L. muelleriferdinandi (2n = 88) not detected in Australian A-clade polyploids (2n = 72) could indeed indicate some sort of hybridization/polyploidization with unknown taxa in the evolution of L. muelleriferdinandi subsequent to the hybridization of an ancestor of L. muelleriferdinandi (presumably with 2n = 56) with African species (2n = 16). GISH signals of an African genome input could not be detected, suggesting that Australian/NZ C-clade species are not the outcome of a bicontinental allopolyploidization scenario as originally described (Mummenhoff et al., 2004). Calibration of molecular trees yielded ages of approx. 0·7–1·3 and 0·3–0·55 Ma for the Australian/NZ species of the C- and A-clades, respectively (Mummenhoff et al., 2004). These age differences might indicate that the radiation of Californian ancestors into Australian/NZ C-clade species pre-dates the later arrival of African ancestors and the origin of A-clade allopolyploids.
Origin of Australian/NZ A-clade species: allopolyploid hybrids between African and Australian/NZ C-clade species
GISH experiments clearly detected 16 chromosomes of African origin (L. africanum, L. transvaalense) in the two polyploid Australian/NZ A-clade species analysed (L. hyssopifolium, L. pseudohyssopifolium). The remaining 56 chromosomes in the A-clade allopolyploid genomes were not labelled by any of the Californian presumed parental gDNAs used. Thus, the presumed direct Californian genomic input into the Australian/NZ A-clade Lepidium species could not be confirmed by GISH.
One might thus speculate that Australian/NZ C-clade taxa have contributed the Californian cpDNA-type to the Australian/NZ A-clade taxa by hybridization with African species in Australia. To test this scenario, GISH experiments were conducted using gDNAs of Australian/NZ C-clade species (L. aschersonii 2n = 56, L. muelleriferdinandi 2n = 88) as probes onto the Australian/NZ A-clade polyploid species L. hyssopifolium and L. pseudohyssopifolium (2n = 72). Of the 72 chromosomes, 56 were labelled by gDNA of Australian/NZ C-clade species and, as outlined above, the remaining 16 chromosomes were labelled by gDNA of L. africanum/L. transvaalense. The remaining 32 chromosomes of the genome of present-day L. muelleriferdinandi (2n = 88) not detected in Australian A-clade polyploids (2n = 72) could indicate some sort of hybridization/polyploidization with unknown taxa in the evolution of L. muelleriferdinandi subsequent to the hybridization of an ancestor of L. muelleriferdinandi (presumably with 2n = 56) with African species (2n = 16).
This general scenario is in agreement with the cpDNA phylogeny in which the Australian/NZ A-clade hybrids harbour a Californian chloroplast type (Mummenhoff et al., 2004). There is some additional evidence for this alternative scenario: one accession of an Australian/NZ A-clade polyploid (L. hyssopifolium, KM 1670) shows additivity in almost every diagnostic nucleotide position that differs between the C-clade and A-clade species group (Table 1). Regardless, all GISH and rDNA FISH experiments using two different accessions of L. hyssopifolium (KM 1670, KM 1669) show the same results. As ITS homogenization can occur very rapidly, as also observed in experiments with recent and synthetic allopolyploids where 45S rDNA homogenization was apparent within a few generations (Franzke and Mummenhoff, 1999; Skalická et al., 2003; Kovarik et al., 2005; Shcherban et al., 2008), it is possible that additivity in almost all diagnostic nucleotide position in L. hyssopifolium (KM 1670) indicates recent and/or recurrent hybridization between African and Australian/NZ C-clade species in Australia.
Thus, the main conclusions from GISH analysis are as follows. The nuclear genomes of African and Australian/NZ C-clade species were detected in allopolyploid Australian/NZ Lepidium A-clade species. The presumed hybrid origin of Australian/NZ C-clade taxa with African and Californian parents involved (Mummenhoff et al., 2004) could not be confirmed. This does not mean that hybridization processes did not play a role in the evolution of polyploid Australian C-clade species, for example L. muelleriferdinandi with 2n = 88 chromosomes. Hence, it is assumed that ancestral Californian taxa subsequent to their dispersal to Australia experienced a rapid radiation in Australia and New Zealand into extant C-clade taxa that hybridized with African species. As a result, A-clade allopolyploid Lepidium species in Australia/NZ share the Californian chloroplast type and the African ITS-type with polyploid Australian/NZ C-clade and diploid African species, respectively.
ACKNOWLEDGEMENTS
We thank all collectors and institutions for providing plant material, Ulrike Coja for technical assistance and Andreas Franzke and two anonymous reviewers for valuable comments. T.D. was supported by the DAAD (German Academic Exchange Service), and T.M. and M.A.L. were supported by research grants nos KJB601630606 and IAA601630902 from the Grant Agency of the Czech Academy of Science, and a grant from the Czech Ministry of Education (no. MSM0021622415).
LITERATURE CITED
- Al-Shehbaz IA. The genera of Lepidieae (Cruciferae; Brassicaceae) in the southeastern United States. Journal of the Arnold Arboretum. 1986;67:265–311. [Google Scholar]
- Al-Shehbaz IA, Beilstein MA, Kellogg EA. Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Systematics and Evolution. 2006;259:89–120. [Google Scholar]
- Ali HBM, Lysak MA, Schubert I. Chromosomal localization of rDNA in the Brassicaceae. Genome. 2005;48:341–346. doi: 10.1139/g04-116. [DOI] [PubMed] [Google Scholar]
- Álvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution. 2003;29:417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Brüggemann H, Lee J-Y, Mummenhoff K. Evolutionary changes in floral structure within Lepidium L. (Brassicaceae) International Journal of Plant Sciences. 1999;160:917–929. doi: 10.1086/314194. [DOI] [PubMed] [Google Scholar]
- Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S, Leitch AR. Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae) New Phytologist. 2005;168:241–252. doi: 10.1111/j.1469-8137.2005.01480.x. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Molecular Biology Reporter. 1983;1:19–21. [Google Scholar]
- Franzke A, Mummenhoff K. Recent hybrid speciation in Cardamine (Brassicaceae) conversion of nuclear ribosomal ITS sequences in statu nascendi. Theoretical and Applied Genetics. 1999;98:831–834. [Google Scholar]
- Kovarik A, Pires JC, Leitch AR, et al. Rapid concerted evolution of nuclear ribosomal DNA in two Tragopogon allopolyploids of recent and recurrent origin. Genetics. 2005;169:931–944. doi: 10.1534/genetics.104.032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M, Al-Shehbaz IA, Mummenhoff K. Molecular systematics, evolution, and population biology in the mustard family (Brassicaceae) Annals of the Missouri Botanical Garden. 2003;90:151–171. [Google Scholar]
- Lee J-Y, Mummenhoff K, Bowman JL. Alloploidization and evolution of species with reduced floral structures in Lepidium L. (Brassicaceae) Proceedings of the National Academy of Sciences. 2002;16:835–840. doi: 10.1073/pnas.242415399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lihová J, Shimizu KK, Marhold K. Allopolyploid origin of Cardamine asarifolia (Brassicaceae): incongruence between plastid and nuclear ribosomal DNA sequences solved by a single-copy nuclear gene. Molecular Phylogenetics and Evolution. 2006;39:759–786. doi: 10.1016/j.ympev.2006.01.027. [DOI] [PubMed] [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, et al. Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytologist. 2007;175:756–763. doi: 10.1111/j.1469-8137.2007.02121.x. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proceedings of the National Academy of Sciences of the USA. 2006;103:5224–5229. doi: 10.1073/pnas.0510791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhold K, Lihová J. Polyploidy, hybridization and reticulate evolution: lessons from the Brassicaceae. Plant Systematics and Evolution. 2006;259:143–174. [Google Scholar]
- Mummenhoff K, Franzke A, Koch M. Molecular phylogenetics of Thlaspi s.l. (Brassicaceae) based on chloroplast DNA restriction site variation and sequences of the internal transcribed spacers of nuclear ribosomal DNA. Canadian Journal of Botany. 1997;75:469–482. [Google Scholar]
- Mummenhoff K, Brüggemann H, Bowman JL. Chloroplast DNA phylogeny and biogeography of Lepidium (Brassicaceae) American Journal of Botany. 2001;88:2051–2063. [PubMed] [Google Scholar]
- Mummenhoff K, Linder P, Friesen N, Bowman JL, Lee J-Y, Franzke A. Molecular evidence for bicontinental hybridogenous genomic constitution in Lepidium sensu stricto (Brassicaceae) species from Australia and New Zealand. American Journal of Botany. 2004;91:254–261. doi: 10.3732/ajb.91.2.254. [DOI] [PubMed] [Google Scholar]
- Mummenhoff K, Polster A, Mühlhausen A, Theißen G. Lepidium as a model system for studying the evolution of fruit development in Brassicaceae. Journal of Experimental Botany. 2009;60:1503–1513. doi: 10.1093/jxb/ern304. [DOI] [PubMed] [Google Scholar]
- Shcherban AB, Sergeeva EM, Badaeva ED, Salina EA. Analysis of 5S rDNA changes in synthetic allopolyploids Triticum × Aegilops. Molecular Biology. 2008;42:536–542. [PubMed] [Google Scholar]
- Skalická K, Lim KY, Matyasek R, Koukalová B, Leitch A, Kovarik A. Rapid evolution of parental rDNA in a synthetic tobacco allotetraploid line. American Journal of Botany. 2003;90:988–996. doi: 10.3732/ajb.90.7.988. [DOI] [PubMed] [Google Scholar]
- Vaarama A. Chromosome number and cryptic polyploidy in Lepidium sativum. Hereditas. 1952;37:290–292. [Google Scholar]
- Warwick SI, Al-Shehbaz IA. Brassicaceae: chromosome number index and database on CD-Rom. Plant Systematics and Evolution. 2006;259:237–248. [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Molecular Biology. 2000;42:225–249. [PubMed] [Google Scholar]