Abstract
Background and Aims
Quercus suber and Q. ilex are distantly related and their distributions partially overlap. They hybridize occasionally, but the complete replacement of Q. suber chloroplast DNA (cpDNA) by that of Q. ilex was identified in two specific geographical areas. The objective of this study was to determine whether the contrasting situation reflected current or recent geographical interspecies gene flow variation or was the result of ancient introgression.
Methods
cpDNA PCR-RFLPs (restriction fragment length polymorphisms) and variation at ten nuclear microsatellite loci were analysed in populations of each species, in 16 morphologically intermediate individuals and the progeny of several of them. Interspecies nuclear introgression was based on individual admixture rates using a Bayesian approach with no a priori species assignment, and on a maximum-likelihood (ML) method, using allele frequencies in the allopatric populations of each species as controls. Gene flow was compared specifically between populations located within and outside the specific areas.
Key Results
High interspecies nuclear genetic differentiation was observed, with twice the number of alleles in Q. ilex than in Q. suber. According to Bayesian assignment, approx. 1 % of individuals had a high probability of being F1 hybrids, and bidirectional nuclear introgression affected approx. 4 % of individuals in each species. Hybrid and introgressed individuals were identified predominantly in mixed stands and may have a recent origin. Higher proportions including allospecific genes recovered from past hybridization were obtained using the ML method. Similar rates of hybridization and of nuclear introgression, partially independent of cpDNA interspecies transfer suggestive of gene filtering, were obtained in the populations located within and outside the areas of complete cpDNA replacement.
Conclusions
The results did not provide evidence for geographical variation in interspecies gene flow. In contrast, historical introgression is supported by palynological records and constitutes the more reliable origin of cpDNA replacement in specific regions.
Key words: cpDNA PCR-RFLPs, nuclear microsatellite (nSSR) variation, hybridization, interspecies genetic introgression, Quercus suber, Quercus ilex
INTRODUCTION
The integration of foreign genes into a recipient species' gene pool as the result of the backcrossing of hybrid offspring with a parental species (genetic introgression) is an evolutionarily important process contributing to adaptation. This may occur either through the transfer of adaptive traits of one species to another or by producing novel adaptive variation (Cronn and Wendel, 2004). In addition, the movement of genes across species boundaries can promote colonizing abilities (Petit et al., 2004). In plants, genetic introgression is often more easily observed for maternally inherited genes [chloroplast DNA (cpDNA) and mitochondrial DNA] than for biparentally inherited ones (Avise, 2004). This is presumably due to the mode of inheritance and absence of recombination in cytoplasmic genomes, possibly combined with a more limited influence of selection as a result of lower effective population size (Martinsen et al., 2001; Avise, 2004). Consequently, a species can retain the cytoplasmic genome of another one over very long periods, if not indefinitely, and cytoplasmic DNA has often been used to describe ancient episodes of gene flow between species (e.g. Magri et al., 2007). Conversely, nuclear introgression disappears with successive backcrosses, more particularly if these involve the same parental species. Therefore, evidence for contemporary interspecies gene flow has been mainly provided by using nuclear loci which are useful to identify hybrids and early-generation introgressed individuals (e.g. Lexer et al., 2005). In most plant studies, e.g. in European white oak species (e.g. Valbuena-Carabaña et al., 2007) or in European ashes (Heuertz et al., 2006), hybridization and genetic introgression referred to closely related species which usually show high levels of interspecies gene exchange and for which specific diagnostic markers are not easily identified. Lower interspecies gene flow is expected to occur between more distantly related species. However, in a few case studies, introgression over large geographical areas was reported to occur between species that are not closely related. These situations reflect either local environmental and/or genetic conditions favouring interspecies genetic exchanges, or the historical dispersal of introgressed genotypes, especially of cytoplasmic DNA. For instance, Martinsen et al. (2001) analysed cytoplasmic and nuclear introgression between two distantly related Populus species and concluded that their hybrid area was ancient.
Here the focus is on gene exchange between two distantly related Mediterranean oaks, Quercus suber (cork oak) and Q. ilex (holm oak). Within section Cerris, Q. suber and Q. ilex belong to distinct clades, Cerris and Ilex, respectively (Manos et al., 1999). According to fossil records, these groups were already distinct at the end of the Tertiary period (Kvacek and Walther, 1989). Phylogenetic divergence between the two oak species is clearly supported by cpDNA, allozymes, internal transcribed spacer (ITS) and amplified fragment length polymorphism (AFLP) variation (Toumi and Lumaret, 2001; Jiménez et al., 2004; Lumaret et al., 2005; López de Heredia et al., 2007a, b). The two evergreen species possess the same chromosome number (2n = 2x = 24), are both widespread in the western Mediterranean Basin and along the Atlantic coast of southern Europe, and their geographical distributions partly overlap (Toumi and Lumaret, 2001). Quercus suber and Q. ilex individuals are easily discriminated by a few morphological traits, including bark (cork is exclusive to Q. suber), leaf and fruit features (Tutin et al., 1993). Rare morphologically intermediate individuals, possessing an ‘ilex’ chlorotype (Lumaret et al., 2002, 2005) and for which the hybrid origin was inferred using nuclear molecular markers, have been identified over the entire sympatric range (Elena-Rossello et al., 1992; Toumi and Lumaret, 1998; Oliveira et al., 2003; Lumaret et al., 2005; Mir et al., 2009). Asymmetric hybridization between maternal Q. ilex individuals and Q. suber was confirmed by experimental crosses (Boavida et al., 2001).
According to previous studies totalling approx. 400 populations sampled over the whole cork oak range, the complete replacement of Q. suber cpDNA by that of Q. ilex was observed exclusively in the populations of eastern Iberia and adjacent French Catalonia and south-eastern Morocco (Belahbib et al., 2001; Lumaret et al., 2005; López de Heredia et al., 2007b; Magri et al., 2007; Mir et al., 2009). In these two areas, that constitute approx. 25 % of the whole sympatric range, up to 14 distinct chlorotypes occur in Q. suber. All belong to the ‘ilex’ lineage, and show a structured geographical distribution, suggesting the occurrence of multiple events of local hybridization and of subsequent genetic introgression between the two species (Behabib et al., 2001; López de Heredia et al., 2007b). In the remainder of the sympatric geographical distribution, isolated Q. suber individuals possessing an ‘ilex’ chlorotype appear very occasionally and, exceptionally, Q. suber chlorotypes were observed in Q. ilex (Lumaret et al., 2005; López de Heredia et al., 2007b). Geographical variation in cytoplasmic DNA transfer may be responsible for differences in the extent and direction of interspecific gene flow between regions. Alternatively, it may reflect past hybridization with subsequent dispersal of the chlorotypes recovered from the opposite species. This contrasting geographical situation has been already noted in previous studies regarding mainly range-wide cpDNA variation in Q. suber and allozyme and cpDNA variation in both species at a regional scale (e.g. López de Heredia et al., 2007b; Mir et al., 2009; Lumaret et al., 2009). However, this issue has not yet been clarified.
In the present study, cpDNA, maternally inherited in oaks (Dumolin et al., 1995), and nuclear microsatellite (nSSR) polymorphisms analysed in the same individuals, were used to evaluate the range-wide geographical variation of gene flow between Q. suber and Q. ilex. In particular, we compared hybridization, and cytoplasmic and nuclear interspecies gene flow levels between populations located within and outside the areas of total replacement of Q. suber cpDNA by that of Q. ilex. In addition, individuals morphologically intermediate between the two species, and their open-pollinated progeny, were scored for cpDNA and nSSRs to determine gene flow direction in initial hybridization and backcrosses. The hybrid pedigree of the morphologically intermediate individuals had been inferred on the basis of allozyme markers (Staudt et al., 2004). The objectives of the study were: (a) to determine if the two oak species are highly differentiated for nSSR markers and if diagnostic alleles could be used to identify hybrids and recently introgressed individuals; (b) to assess cross direction in hybridization and in backcrosses by identifying chlorotype lineage in hybrids, in introgressed individuals and in their progeny; and (c) to determine if interspecies gene flow varies geographically and, more particularly, between the populations located inside and outside the areas where Q. suber populations possess an ‘ilex’ cpDNA. If no geographical variation in interspecies gene flow were observed, ancient introgression, as supported by palynological records, would constitute the more plausible origin of cpDNA replacement in specific regions.
MATERIALS AND METHODS
Sampling design and DNA extraction
Quercus suber L. is thermophilous and avoids limestone substrates. Its distribution is restricted to several discontinuous areas in the western Mediterranean Basin, whereas Q. ilex L., which shows wider ecological amplitude, has a wider range with an eastward extension to Greece (Toumi and Lumaret, 2001). In the geographic areas where a single species was present (allopatric area), analyses in several distant populations were used as a control to infer species genetic identity. In the sympatric area, the populations located within and outside the areas characterized by the complete replacement of Q. suber cpDNA by that of Q. ilex were considered separately. Mixed and pure stand conditions were also distinguished.
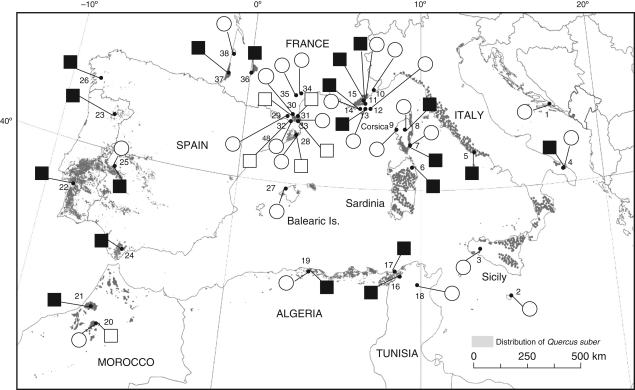
Leaves were collected on individual trees in eight Q. suber and nine Q. ilex allopatric populations, and in five Q. suber, three Q. ilex and 13 mixed populations of the sympatric area (Fig. 1). The list of populations, geographical coordinates, sample sizes and population status is available as Supplementary Data, online. A total of 580 Q. suber and of 592 Q. ilex individuals were scored for both nSSR and cpDNA variation. Species identification was based on morphological characters as described in Flora Europaea (Tutin et al., 1993). In addition, by using several leaf, acorn and trunk characters (details in Flora Europaea), 16 trees from eight populations (one individual from sites 7, 19 and 32, two from sites 11, 14, 15 and 30, and five from site 13) were identified as being morphologically intermediate between Q. ilex and Q. suber, and were also scored for both cpDNA and nSSR variation. Finally, in site 13, four mother trees and their open pollinated progeny were scored for cpDNA and nSSRs. These trees were identified morphologically as two intermediate and two Q. ilex individuals whose pedigree, i.e. F1 hybrid and Q. ilex introgressed by Q. suber, respectively, had been inferred on the basis of allozyme diagnostic markers (Staudt et al., 2004). The objective was to determine the relative proportion of each parental genome and to identify the pollen-bearing species. Mature acorns were collected on each mother tree and were sown individually in large pots at the CEFE-CNRS experimental garden in Montpellier. They were grown for 7 years in homogeneous conditions and then characterized morphologically and analysed for molecular markers. Several diagnostic characters, e.g. the occurrence of cork on the trunk which is specific to Q. suber, are not expressed before the saplings are 5–7 years old.
Fig. 1.
Geographic distribution of chlorotype lineages ‘suber’ (black) and ‘ilex’ (white), in 26 Q. suber populations (squares) and 25 Q. ilex populations (circles) scored for PCR-RFLP variation at five cpDNA fragments. The dark grey shading on the map indicates the natural range of Q. suber.
Total DNA was extracted either from freeze-dried powder or silica-gel-dried leaf material using a DNeasy Plant Minikit (QIAGEN).
CpDNA analyses and data treatment
CpDNA was amplified using five pairs of universal chloroplast primers corresponding to fragments TF, AS, CD, DT and SR (Grivet et al., 2001). The two CD and DT amplified fragments and the three AS, TF and SR fragments were cut using endonucleases TaqI and HinfI, respectively. Experimental conditions for PCR, amplified fragment digestion and gel migration are described in Lumaret et al. (2005). Fragment variants and chlorotypes were identified according to the detailed description of Jiménez et al. (2004).
Chlorotype lineage was assessed in each oak species and was mapped for the 38 sampled sites (Fig. 1). Chloroplast DNA haplotypes were identified in each population, in each species and in morphologically intermediates individuals. The rate of cpDNA exchange was directly estimated per population and species as the frequency of chlorotypes belonging to the lineage of the opposite species. In each species, populations were separated into three groups, allopatric populations (control populations) and the sympatric populations located inside and out of the areas where the complete replacement of Q. suber cpDNA by that of Q. ilex was observed, respectively. Haplotype diversity within a population (hs), total species diversity (ht) and Gst values were computed according to the method of Nei (1987) which is based on haplotype frequencies (unordered alleles). All calculations were performed with the program HAPLODIV (available at http://www.pierroton.inra.fr).
Nuclear DNA variation analysis and data treatment
Individuals were genotyped at ten polymorphic microsatellite loci, MSQ4, MSQ13 (Dow et al., 1995), QpZAG9, QpZAG15, QpZAG36 and QpZAG46 (Steinkellner et al., 1997), and QrZAG7, QrZAG11, QrZAG20 and QrZAG112 (Kampfer et al., 1998). In previous studies, the genetic variation at these loci was used to analyse local genetic diversity in Q. suber and Q. ilex (Soto et al., 2007).
The PCRs were multiplexed for three subsets of loci (QpZAG9, QpZAG15, MSQ4; MSQ13 QpZAG36, QpZAG46; and QrZAG7, QrZAG11, QrZAG20 and QrZAG112) in 10 µL final volume including 0·2 µm of each primer and 5 ng of genomic DNA, using the QIAGEN multiplex PCR kit. PCRs were conducted using a Peltier Thermal Cycler (PTC100; MJ Research, Waltham, MA, USA) under the following conditions: 15 min activation of Hotstar Taq DNA polymerase at 95 °C, 30 cycles including 30 s initial denaturation at 94 °C, 1 min and 30 s annealing at 55 °C, and 60 s extension at 72 °C, and then 30 min final extension at 60 °C. A 1-μL aliquot of diluted PCR product was pooled in 15 µL of deionized formamide and 0·2 µL of GeneScan-500XL ROX Size Standard (Applied BioSystems, Streetville, Ontario, Canada), and analysed on an ABI PRISM 310 Genetic Analyser (Applied BioSystems, Streetville, Ontario, Canada).
At each locus, alleles were characterized exclusively by the length of the DNA fragments generated by the amplification process. In each species, allelic frequencies, total and private (species-specific) allele numbers, Nei's expected heterozygosity and fixation index (Fis) were estimated per nSSR locus and overall loci, and were averaged over allopatric populations (control populations) and over all populations, using GENETIX 4·04 (Belkhir et al. 2001). In addition, following Shriver et al. (1997), the allele frequency differential between the two species (δ) was estimated to assess the discriminating power of each marker in allopatric populations and in all sampled populations. At each locus, δ is calculated as half the sum of the absolute value of allele frequency differences between species.
Differences for allele numbers between control populations and all populations were tested using Mann–Whitney U-tests. In multiple comparisons, the Bonferroni correction was used. Differences for heterozygote frequencies between control populations and all populations were tested using a two-tailed Fisher exact test. Using GENETIX 4·04, in each species, mean allele number per locus and per population (Am) with standard error, total genetic diversity (Ht) and the proportion of diversity resulting from gene differentiation among populations (Gst; Nei, 1987) were estimated in allopatric (control) and sympatric populations, and overall. In addition, Fst values and Rst values, the latter of which take into account allele sizes, were estimated within each species. The test developed by Hardy et al. (2003) was used to indicate whether Rst performs better than Fst, i.e. whether stepwise-like mutations contributed to genetic differentiation. The analyses were carried out using the program SPAGeDi 1·1 (Hardy and Vekemans, 2002).
In addition, the genetic structure in each species was analysed using a Bayesian clustering approach with software STRUCTURE version 2·0 (Pritchard et al., 2000) and admixture option. The independent allele frequency option was chosen owing to the substantial interspecies genetic differentiation (see below). Several preliminary runs with K values ranging from one to five were performed to find the most likely number of clusters. The final run was performed using a burn-in period of 105, and a running length period of 106. Multilocus allele frequencies were used to compute the likelihood (Pi) that a given genotype originated from a given cluster.
To confirm species identity of each individual and to analyse the patterns of introgression between species, two independent approaches were used. First, admixture proportions were estimated for all individuals, using a Bayesian clustering approach (STRUCTURE version 2·0). Secondly, hybrid indices (HIs) were calculate using a maximum-likelihood (ML) approach to compare allele frequencies for each individual with average allele frequencies in the parental taxa (Rieseberg et al., 1998; Watano et al., 2004). In the present study, the individual HIs were calculated using Mathematica 4·0 (Wolfram, 1996) as the sum, over each individual's alleles, of the probability that the allele was derived from Q. suber (species arbitrarily chosen as reference) rather than from Q. ilex. Average allele frequencies of the Q. suber and Q. ilex allopatric (control) populations were used as parental references for HI calculation. For both methodological approaches, data were compared between the populations within and outside the area where Q. suber populations possess an ‘ilex’ cpDNA molecule, and between species. The analysis of variance (ANOVA) was performed with SAS (SAS Institute Inc., 2004) after normalizing individual Pi and HI values. Pi and HI were also estimated for morphological intermediate individuals and their open pollinated progeny. The frequency of populations including intermediate or introgressed individuals, and the frequencies of intermediate and introgressed individuals were compared between the populations located inside and outside of the area where Q. suber populations possess an ‘ilex’ cpDNA molecule, and between species, using a two-tailed Fisher exact test.
RESULTS
Chloroplast DNA variation
Eleven distinct chlorotypes belonging to lineage ‘ilex’ were identified in Q. ilex (Fig. 1, and Supplementary Data, available online). Chlorotype A, closely related to chlorotype 78, has not been identified in previous studies and was found exclusively in Tunisia and in Algeria. In addition, a chlorotype belonging to lineage ‘suber’ was found in a single Q. ilex individual sampled in site 13. Three and four chlorotypes belonging to lineages ‘suber’ and ‘ilex’, respectively, were identified in Q. suber. Three ‘ilex’ chlorotypes were observed in all Q. suber individuals sampled in Moroccan site 20 and in Spain and French Catalonia, whereas the fourth ‘ilex’ chlorotype was identified in a single Q. suber individual sampled in site 13. Overall, 23·3 % of Q. suber and 0·1 % of Q. ilex individuals possessed a chlorotype belonging to the lineage of the opposite species. Cytoplasmic introgression per population ranged from 0 to 100 % in Q. suber and from 0 to 3·3 % in Q. ilex. Average genetic diversity per population, total genetic diversity and the proportion of diversity attributable to between-population differentiation were equal to 0·043 (±0·008), 0·827 (±0·023) and 0·948 (±0·009), respectively, in Q. suber and to 0·034 (±0·009), 0·926 (±0·002), and 0·963 (±0·006), respectively, in Q. ilex.
Four chlorotypes of lineage ‘ilex’ were identified in the 16 individuals morphologically intermediate between the two oak species (Supplementary Data). Most of these individuals grew in mixed stands. Usually, the same chlorotype was identified in the morphologically intermediate individuals and in the Q. ilex trees of the same site.
Nuclear microsatellite variation
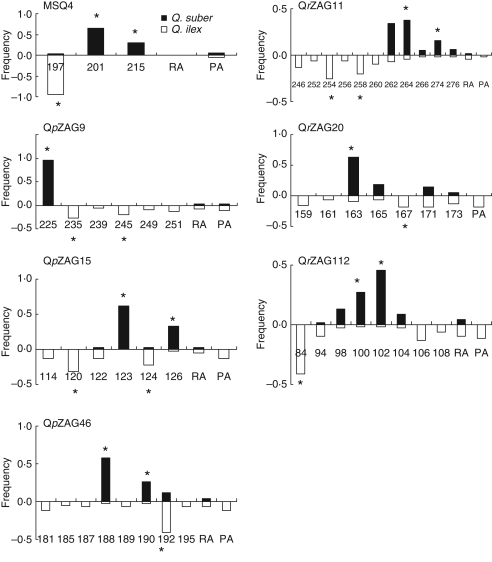
In the present study, 95 and 94 % of the nSSR fragments amplified at each locus in Q. ilex and in Q. suber, respectively, corresponded to a continuous range of successive fragment sizes differing by 2 bp intervals. Substantial polymorphism was observed at the ten nSSR loci (Table 1). Overall, Q. ilex showed at least twice the number of alleles (P < 001) and a significantly higher proportion of species-private alleles (P < 0·01) than did Q. suber. However, many alleles observed in a single species were found at low frequency (<0·05). As compared with allopatric populations, a higher number of alleles were shared by the two species in the sympatric area. In Q. suber, this result was reflected by a significant increase in total allele number (P < 0·01) and a decrease in private allele number (P < 0·05), when all the populations were considered. In Q. ilex, both total and species-private allele numbers increased with population number, due to additional rare alleles, but the proportion of private alleles was significantly higher in the control populations (P < 0·05). Moreover, 23·6, 37·3 and 37·6 % of shared fragments were identified in allopatric situations, in the sympatric range within the area where Q. suber populations possessed exclusively ‘ilex’ chlorotypes, and in the sympatric populations outside this area, respectively. Based on private allele numbers, the highest interspecies differentiation was observed at MSQ13, which is completely diagnostic, at MSQ4, QrZag7 and QpZAG9. However, in Q. ilex exclusively, complex amplification patterns were observed in approx. 33 and 9 % of individuals, at MSQ13 and QrZAG7, respectively, with multiple peaks corresponding to three and four alleles per individual, suggesting gene duplication. In addition, in the same species, a high frequency of null alleles was detected at locus QpZAG36. Therefore, in that species, and in between-species comparisons, the three loci were not included to estimate genetic parameters. At the seven other loci analysed in all populations, allele frequencies differed substantially between species (Fig. 2), as also shown by the allele frequency differential values (δ) ranging from 0·663 and 0·640 at locus QrZAG20 to 0·979 and 0·971 at MSQ4 in allopatric populations and in all populations, respectively (Table 1). Moreover, the average frequency differential value over seven loci slightly decreased from 0·863 in allopatric populations to 0·847 when all populations were considered. Twelve and ten diagnostic alleles (with frequency >20 % in one species and <5 % in the other species) were identified in Q. suber and in Q. ilex, respectively (Fig. 2).
Table 1.
Total allele number, species-private allele number (in parentheses), allele size range, expected heterozygosity (He), fixation index (Fis) and interspecies allele frequency differential (δ), at ten nSSR loci in (A) allopatric and (B) all Q. suber and Q. ilex populations
| Population no. | Sample size | QrZAG7 | QrZAG11 | QrZAG20 | QrZAG112 | QpZAG9 | QpZAG15 | QpZAG36 | QpZAG46 | MSQ4 | MSQ13 | 10 loci | 7 loci | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (A) Allopatric populations | ||||||||||||||
| Q. suber | 8 | 171 | 8 (7) | 6 (1) | 4 (0) | 6 (0) | 7 (2) | 6 (2) | 10 (3) | 4 (1) | 7 (6) | 3 (3) | 61 (25) | 40 (12) |
| SSR size | 118–134 | 262–276 | 163–173 | 90–104 | 225–251 | 122–140 | 210–228 | 180–192 | 197–221 | 219–237 | ||||
| He | 0·714 | 0·618 | 0·457 | 0·695 | 0·151 | 0·492 | 0·700 | 0·500 | 0·485 | 0·158 | 0·505 | 0·485 | ||
| Fis | 0·01 | −0·05 | 0·20* | –0·13 | –0·04 | 0·06 | 0·04 | 0·05 | 0·15 | 0·10 | 0·04 | 0·03 | ||
| Q. ilex | 9 | 220 | 6 (5) | 16 (11) | 17 (13) | 16 (10) | 16 (11) | 15 (11) | 10 (3) | 15 (12) | 3 (2) | 13 (13) | 127 (91) | 98 (70) |
| SSR size | 114–136 | 244–278 | 159–199 | 82–112 | 223–261 | 112–142 | 204–224 | 168–201 | 195–199 | 194–220 | ||||
| He | – | 0·803 | 0·768 | 0·656 | 0·800 | 0·677 | – | 0·694 | 0·109 | – | – | 0·644 | ||
| Fis | – | 0·03 | 0·11 | 0·18* | −0·01 | 0·06 | – | 0·16* | 0·15 | – | – | 0·09 | ||
| δ | – | 0·876 | 0·663 | 0·833 | 0·936 | 0·945 | – | 0·806 | 0·979 | 1·000 | – | 0·863 | ||
| (B) All populations | ||||||||||||||
| Q. suber | 26 | 580 | 11 (3) | 11 (0) | 4 (0) | 8 (0) | 8 (1) | 9 (1) | 10 (1) | 6 (0) | 10(6) | 3 (2) | 80 (14) | 56 (8) |
| SSR size | 116–136 | 254–284 | 163–173 | 84–104 | 225–251 | 120–140 | 210–228 | 180–194 | 197–221 | 219–239 | ||||
| He | 0·739 | 0·656 | 0·461 | 0·612 | 0·102 | 0·462 | 0·675 | 0·510 | 0·460 | 0·203 | 0·482 | 0·475 | ||
| Fis | 0·01 | –0·06 | 0·31* | –0·08 | 0·07 | 0·06 | –0·06 | 0·05 | 0·03 | 0·02 | 0·03 | 0·02 | ||
| Q. ilex | 25 | 592 | 11 (3) | 20 (9) | 21 (17) | 17 (9) | 18 (11) | 17 (9) | 15 (6) | 25 (19) | 6 (2) | 16 (15) | 166 (100) | 124 (76) |
| SSR size | 114–147 | 244–288 | 157–199 | 80–112 | 219–261 | 110–140 | 198–226 | 168–202 | 193–215 | 190–220 | ||||
| He | – | 0·795 | 0·784 | 0·729 | 0·808 | 0·738 | – | 0·709 | 0118 | – | – | 0·672 | ||
| Fis | – | 0·07 | 0·06 | 0·20* | 0·04 | 0·06 | – | 0·17* | 0·21* | – | – | 0·11* | ||
| δ | – | 0·842 | 0·640 | 0·815 | 0·917 | 0·938 | – | 0·806 | 0·971 | 0·847 | ||||
*Significant heterozygote deficiency (P < 0·05).
– indicates that there were no available data for Q. ilex (see text).
Fig. 2.
Allelic frequency for shared and species-private alleles identified at seven nSSR loci in 580 individuals of Q. suber and 592 individuals of Q. ilex, as indicated, sampled over the western Mediterranean Basin. Allele sizes are given in base pairs. Shared and private low-frequency alleles (<0·05) were pooled in single categories named RA and PA, respectively. Asterisks indicate diagnostic alleles that may be suitable for species discrimination.
When nSSR variation was analysed in Q. suber using STRUCTURE, the model-based clustering revealed an optimal number of two groups of populations located in the eastern part (sites 1–19 in Fig. 1) and the western part (sites 20–38) of the species range, respectively, indicating that nuclear microsatellite variation is structured geographically. Using the same treatment in Q. ilex, however, no clear geographical pattern was observed. Only two small allopatric populations (two from Malta and 18 from Tunisia), both isolated from the main distribution area, were highly differentiated from each other and from all other populations.
Expected heterozygosity and fixation indices were calculated at the seven nSSR loci in both species (Table 1). Significant excess of homozygotes (P < 0·05) was observed at QrZAG20 in Q. suber and at QrZAG112, QpZAG46 and MSQ4, and over all loci, in Q. ilex. Over seven nSSR loci, mean allele number per locus and per population, and total genetic diversity were higher in Q. ilex than in Q. suber (P < 0·01). In each species, genetic differentiation was not significantly different between allopatric populations and all populations studied (Table 2). A permutation test (10 000 iterations) of the allele sizes indicated that differentiation estimated by Rst was significantly higher than that estimated by Fst in Q. suber, but not in Q. ilex.
Table 2.
Mean number of alleles per locus and per population (Am), total genetic diversity (Ht), proportion of diversity among populations (Gst), genetic differentiation between populations (Fst) and Rst statistic over seven nuclear microsatellite loci analysed in allopatric populations and in all the populations of Quercus suber and Q. ilex
| Species | Distribution area | Population no. | Am | Ht | Gst | Fst | Rst |
|---|---|---|---|---|---|---|---|
| Q. suber | Allopatric | 8 | 3·67 (0·279) | 0·546 | 0·060 | 0·071 (0·013) | 0·092 (0·010) |
| Overall | 26 | 3·68 (0·264) | 0·537 | 0·075 | 0·085 (0·006) | 0·093 (0·008) | |
| Q. ilex | Allopatric | 9 | 6·34 (0·963) | 0·700 | 0·105 | 0·097 (0·014) | 0·099 (0·017) |
| Overall | 25 | 6·72 (0·982) | 0·717 | 0·078 | 0·064 (0·008) | 0·069 (0·008) |
Standard deviations are indicated in parentheses.
Fourteen of the 16 morphologically intermediate individuals combined alleles of both species or, less often, alleles shared by both species, at every locus. These individuals therefore had a very high likelihood of being F1 hybrids.
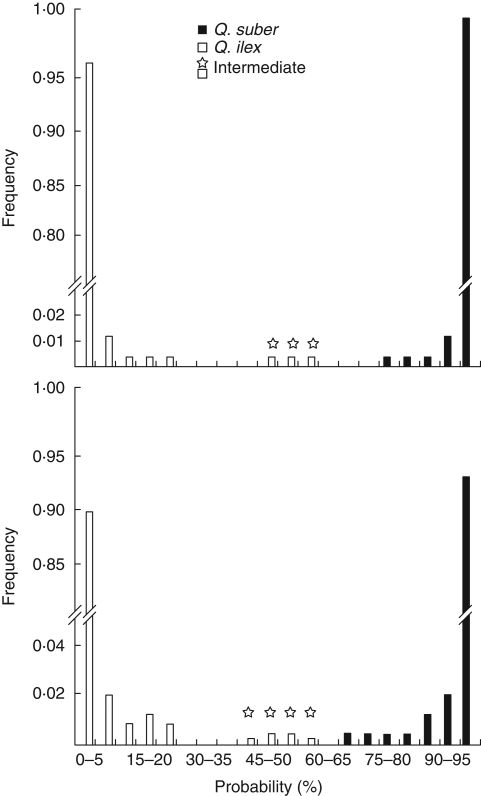
The model-based clustering (STRUCTURE) was used with genotype data from all the individuals analysed at eight SSR loci. These included MSQ13, a highly informative locus for which allele sizes do not overlap between the two species (Soto et al., 2003) but which may show more than two distinct alleles in Q. ilex (see above). To include this locus in the Bayesian analyses, the alleles specific to each species were pooled. Disregarding species assignment of individual trees on the basis on morphology, an optimal number of populations (K) of two was obtained. The two genetically distinct clusters found in the analysis corresponded well to our assignment of individuals to Q. suber and Q. ilex. Overall, we found approx. 5·0 % of introgressed and potentially first-generation hybrid trees, i.e. individuals having <95 % probability of belonging to its own species (Fig. 3). All these were found to possess at least one allele (either diagnostic or private) of the opposite species in their nSSR multilocus genome. Introgression was bidirectional and not significantly different between species (4·14 and 3·90 % of introgressed individuals in Q. suber and Q. ilex, respectively). Approximately 1·2 % of trees had a high probability of being F1 hybrids (i.e. those having a probability of 45–60 %; Fig. 3) which is consistent with the number of morphologically intermediate individuals observed in the survey. HIs were also computed for the same individuals using the ML approach based on average allele frequencies in parental taxa (Fig. 3). The number of potential F1 hybrids was similar to that found using the Bayesian clustering method, but the range of probability was wider (40–60 %; Fig. 3). As compared with the assignments obtained using Bayesian clustering, the fraction of potentially introgressed individuals identified using the ML approach increased significantly, i.e. from 6·0 to 7·6 % and from 4·0 to 5·1 % over the individuals of the sympatric area and over all individuals, respectively (P < 0·05). Most of the additional introgressed individuals still had a high probability (90–95 %) of belonging to their own species. Frequencies of introgressed individuals and the mean probability of belonging to Q. suber were calculated over the introgressed and over all the individuals of the sympatric area (Table 3). The values were not significantly different between or within species, between the populations located inside and out of the areas where Q. suber possesses ‘ilex’ chlorotypes.
Fig. 3.
Frequency distribution of individual probabilities of belonging to Q. suber in individuals identified morphologically as Q. suber, as Q. ilex and as intermediate between the two species, as indicated. The 1188 individuals were scored at eight nSSR loci and analysed by using either a Bayesian clustering method (STRUCTURE; top) or a maximum-likelihood approach based on parental species nSSR frequencies in allopatric populations used as controls (below).
Table 3.
Number of populations sampled in the sympatric area, number of those including individuals introgressed for the nuclear genome, sample size, percentage of introgressed individuals and mean probability of belonging to Q. suber according to Bayesian clustering (M1) or to the maximum-likelihood method (M2), in Q. suber and Q. ilex populations located inside the area characterized by the complete replacement of Q. suber cpDNA by Q. ilex cpDNA (A) and outside this area (B)
| A |
B |
||||
|---|---|---|---|---|---|
| Q. suber | Q. ilex | Q. suber | Q. ilex | ||
| Sampled populations | 6 | 5 | 12 | 11 | |
| Populations including introgressed individuals | 5 | 4 | 7 | 7 | |
| Sample size | 212 | 174 | 197 | 198 | |
| Introgressed individuals ( %) | M1 | 5·70 | 6·32 | 6·10 | 6·06 |
| M2 | 6·60 | 8·04 | 7·10 | 9·09 | |
| Mean probability of belonging to Q. suber over introgressed individuals | M1 | 0·884 | 0·129 | 0·892 | 0·135 |
| M2 | 0·838 | 0·132 | 0·871 | 0·142 | |
| Mean probability of belonging to Q. suber over all individuals | M1 | 0·994 | 0·011 | 0·988 | 0·014 |
| M2 | 0·986 | 0·018 | 0·980 | 0·027 | |
| Intermediate individuals | 3 (2) | 13 (6) | |||
Numbers of morphologically intermediate individuals and of sites where they were found (in parentheses) are also indicated.
In each species and at each locus, the percentage of private and diagnostic alleles of the opposite species was estimated over the individuals classified as introgressed according to the model-based clustering method (see above). In both species, this percentage was very low (range from 0 to 10 %) at loci MSQ4, MSQ13 and QpZAG9, which also showed the highest discriminating power between species (see above). The frequency of alleles derived from interspecies introgression was significantly higher (P < 0·01) in Q. suber (43 %) than in Q. ilex (8 %) at QpZAG15, and significantly lower (P < 0·01) in Q. suber (4–9 %) than in Q. ilex (39–54 %) at QpZAG46, QrZAG11, QrZAG20 and QrZAG112.
Genotype distribution at nuclear microsatellite loci in open-pollinated progeny of F1 hybrids and of Q. ilex trees slightly introgressed by Q. suber
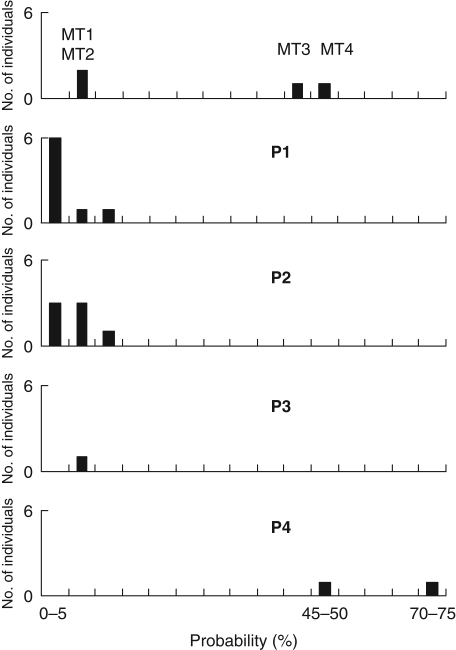
In site 13, 10 and 11, acorns (the whole individual acorn annual production under dry climate conditions) were collected from two Q. ilex mother trees, MT1 and MT2, with a 0·072 and 0·086 probability of belonging to Q. suber, respectively, using the model-based clustering (STRUCTURE). In the same site, 12 and one acorns were collected from two morphologically intermediate mother trees, MT3 and MT4, with a 0·528 and 0·472 probability of belonging to Q. suber, respectively, and which were probably first-generation hybrids. The four mother trees possessed an ‘ilex’ chlorotype. After 7 years growth under similar conditions in the experimental field, mean mortality was 33·3 and 76·9 % in Q. ilex and hybrid mother tree progeny, respectively. As shown by their genotype analysed at the ten SSR loci, all offspring derived from outcrossing, as alleles absent in the mother tree were identified at least at one locus in each individual.
In P1, P2 and P3, the model-based clustering (STRUCTURE) probability of belonging to Q. suber ranged from 0 to 15 % (Fig. 4), suggesting that these mother trees were pollinated exclusively by Q. ilex. All these individuals were identified morphologically as Q. ilex and showed normal growth. In P4, the two saplings had a probability of 54 and 70 %, respectively, of belonging to Q. suber. Both derived probably from MT4 pollination by Q. suber, as they were homozygotes for alleles diagnostic of that species at several loci. These individuals both possessed leaves characteristic of Q. suber but they had no corky trunk, contrary to what was observed in all 7-year-old saplings from Q. suber mother trees of the same site and growing in similar conditions. In addition, the sapling with the higher probability of belonging to Q. suber was a dwarf with very low probability of survival under natural conditions.
Fig. 4.
Distribution of individual probabilities of belonging to Q. suber, according to nSSR variation at eight loci and by using a Bayesian clustering method (STRUCTURE; Pritchard et al., 2000), in mother trees MT1 and MT2 identified morphologically as Q. ilex, in mother trees MT3 and MT4 intermediate between Q. suber and Q. ilex, and in their respective progeny (P1–P4) collected in mixed population 13.
DISCUSSION
Direction and geographical distribution of plastid gene flow
Our data are consistent with previous reports showing clear geographical chlorotype patterns in both species and higher chlorotype variation in Q. ilex than in Q. suber (Lumaret et al., 2002, 2005; López de Heredia et al., 2007b). These results may suggest the occurrence of slow cpDNA evolution in Q. suber, as was reported previously in the same species, on the basis of chloroplast microsatellite variation (Magri et al., 2007), and in several other Fagaceae, e.g. Quercus rubra and Fagus sylvatica (Magni et al., 2005), but note that diversity at nuclear genetic markers was greater in Q. ilex as well (Table 2). Moreover, our data are consistent with previous studies (e g. López de Heredia et al., 2007a, b) and confirm the complete replacement of Q. suber cpDNA by a molecule of lineage ‘ilex’ in specific regions, namely eastern Spain with adjacent French Catalonia, and the East and the South of Morocco. In their exhaustive survey over Iberian populations, López de Heredia et al. (2007a) described additional Q. suber populations with both ‘ilex’ and ‘suber’ chlorotypes in central Spain and in southern Portugal. However, in several distinct areas (e.g. Morocco and the French Riviera), rare Q. ilex individuals were observed to possess chlorotypes of lineage ‘suber’ (Belhabib et al., 2001; Mir et al., 2009; this study), and Staudt et al. (2004) reported that approx. 5 % of morphologically intermediate individuals, for which the F1 hybrid origin had been confirmed on the basis of nuclear diagnostic markers, possessed ‘suber’ chlorotypes. These results suggest that both in initial hybridization and in subsequent backcrosses, Q. ilex is very predominantly, but not exclusively, the maternal species. As expected, interspecies cytoplasmic transfer was not observed in the allopatric distribution area of either species.
Nuclear SSR variation and interspecies genetic differentiation
As compared with Q. suber, we observed similar genetic differentiation levels between populations, but higher allele numbers per locus and higher genetic diversity (per population and overall) were observed in Q. ilex. This is consistent with previous studies based on allozyme polymorphism (Michaud et al., 1995; Toumi and Lumaret, 1998). The weak geographical structure of nSSR variation observed in Q. ilex, and the clear separation between eastern and western Q. suber populations, support previous findings based on allozyme polymorphism (Michaud et al., 1995; Toumi and Lumaret, 1998). However, SSR variation provides new insights that should be useful for interpreting the history of the two species across their ranges. For instance, the fact that shifts in average allele sizes contribute significantly to population differentiation in Q. suber but not in Q. ilex may indicate that eastern and western Q. suber populations diverged a very long time ago, and exchanged migrants at a low rate, whereas higher mutation or exchange rates may have occurred in Q. ilex.
As expected in distantly related species, substantial nSSR genetic differentiation was observed between the two oak species, a result that is consistent with previous findings on the basis of allozyme variation (Toumi and Lumaret, 2001; Mir et al., 2009). Large interspecies differences were found in allele sizes, and in allele frequencies, even for alleles shared by both species. Such shifts in allele size are considered to occur over longer divergence times than changes in frequencies (Slatkin, 1995), suggesting historically low levels of nuclear introgression. However, in this study, nuclear homology of the amplified fragments within and between species has not been tested. Thus, the possibility that a part of the interspecies DNA fragment length differences is due to DNA variation in the flanking regions of the microsatellites cannot be ruled out. In several oak species of sections Quercus and Cerris, the occurrence of non-homologous nSSR fragments of the same size (size homoplasy) at both intra- and interspecies levels has been reported (Curtu et al., 2004; Gugerli et al., 2008). However, in the present study, within each species, fragment size changes were predominantly characterized by 2 bp intervals, corresponding to repeat numbers of the SSR motifs; this suggests that the bias in estimating the extent of divergence among populations may be low.
Distribution of hybridization and of nuclear introgression within and outside the areas where Q. suber populations possess ‘ilex’ chlorotypes
Although rare, natural hybridization occurs between the two genetically distant oak species, as shown in previous studies (see above). However, the hybrids were found exclusively in sympatric areas, and, with rare exceptions, in mixed stands. This suggests that hybridization may be more favoured in contact areas, due to physical proximity promoting extensive and genetically diversified pollen clouds. Alternatively, given the different ecological requirements of Q. suber and Q. ilex, contact areas may represent ecotones favouring the establishment of intermediate individuals, as suggested for other pairs of oak species (Williams et al., 2001; Valbuena-Carabaña et al., 2005). In the western part of their sympatric range (Iberia), only very short overlaps were reported between the flowering periods of the two study species (Elena-Rossello et al., 1992), whereas in the eastern part (e.g. eastern France and Italy) a wider overlap was often observed, as in site 13 of the present study. However, due to high interannual microclimate variation, longer overlaps in flowering period may occur, even in the western range.
In the present work, interspecies gene flow estimation was based on fragment length variation whatever the underlying DNA changes among the amplified fragments. Likewise, the occurrence of fragments of the same size that are non-homologous between the species is possible. However, homoplasy rates are not expected to differ considerably between the populations located in allopatric areas and those growing in sympatry, as both population groups are distributed over the whole species range. An increase of shared fragments in the populations of the sympatric area as compared with those being allopatric clearly supports interspecies gene flow. In the sympatric area, introgressed individuals were observed in both species in more than half of the populations sampled, supporting previous reports based on enzyme markers (e.g. Mir et al., 2009). Introgression levels were low and similar in both species for the number of introgressed individuals (approx. 4 %) and for introgressed gene pool proportion (approx. 1 %). These results are globally consistent with the bidirectional nSSR introgression rate (<2 %) estimated by Burgarella et al. (2009) in five large mixed populations of Q. suber and Q. ilex. In the present study, nuclear introgression was observed primarily in mixed stands where first-generation hybrids were also detected. It may be the result of first-generation backcrosses occurring in both directions, as shown by the genotypes of the hybrids' progeny sampled in site 13. As compared with the Bayesian approach, the ML method is based on allele frequencies in allopatric populations and may reflect more accurately the dispersal of alleles derived from past interspecies introgression. The higher proportions of introgressed trees obtained using the ML method suggests that recurrent hybridization events and subsequent introgression may have occurred at different places in the past. Introgressed individuals were subsequently dispersed over larger areas. The symplesiomorphy hypothesis, according to which diagnostic alleles of one species found at low frequency in another species represent shared ancestral characters (Martinsen et al., 2001), is unlikely to be validated in this case because the species are very distantly related.
Two striking points are raised by this study. First, outside the areas where Q. suber populations are characterized by cytoplasmic introgression, most individuals showing an introgressed nuclear genome possessed chlorotypes of their own lineage, indicating that nuclear gene flow may occur in the absence of significant cytoplasm transfer. This result is unusual, as in most other case studies, e.g. in Helianthus (Rieseberg et al., 1991) and in Populus (Martinsen et al., 2001), the rates of cytoplasmic transfer were greater than the rates of nuclear introgression (but see Lexer et al., 2005, for an example of the opposite pattern). In two regions of France, independence between nuclear and cytoplasmic gene flow between Q. ilex and Q. suber was reported by Mir et al. (2009) on the basis of allozyme and cpDNA markers. Moreover, if in initial hybridization Q. ilex is confirmed to be predominantly the mother species, the genetic analysis of the hybrid's progeny indicates that these can be pollinated by both parental species. When the pollen-bearing species is Q. ilex, the predominant species in all the sites where hybrids were identified, progeny individuals are produced and are characterized by ‘ilex’ chlorotypes and by ‘ilex’ nuclear genomes partially introgressed by Q. suber. In addition, the occurrence of Q. suber individuals possessing ‘suber’ chlorotypes and ‘suber’ nuclear genomes introgressed by Q. ilex suggests that the hybrids may also act as pollen donors in backcrosses. In this oak pair, the wide range of modalities in crosses between hybrids and their parental species may play a role in the low geographical extent of interspecies cytoplasmic transfers that is currently observed.
Secondly, nuclear and DNA markers introgress at different rates. In addition, as reported in other plant genera, e.g. Populus (Martinsen et al., 2001), selective filtering of the introgressed genome is expected to be most intense in the earliest backcross generations. Indeed, in our study, there were few individuals showing 60–80 % probability of belonging to their own species according to the two clustering methods, and slow growth was observed in the progeny of an F1 hybrid collected in site 13, which included individuals corresponding to this range of probability. So the type of selection expected under a ‘gene filtering’ scenario (Martinsen et al., 2001) may operate in these species and hybrids.
In the present work, evidence was provided for similar rates of current hybridization and interspecies nuclear introgression within and outside the areas where Q. suber populations possess ‘ilex’ chlorotypes. Moreover, in Catalonia, a relatively large area where Q. suber possesses exclusively ‘ilex’ lineage chlorotypes, López de Heredia et al. (2007a) and Mir et al. (2009) showed that the two species do not always share the same chlorotypes, even in mixed stands, and that, when they do, chlorotype frequencies differ substantially between them. This finding suggests that current interspecies cytoplasmic transfer may be limited, even in Catalonia.
Evidence for ancient interspecies cytoplasmic transfer in specific areas
A first line of evidence for historical replacement of Q. suber cpDNA by that of Q. ilex comes from the observation that two ‘ilex’ chlorotypes identified as Nos 66 and 29 in the present study (Supplementary Data, available online), are predominant or even exclusive in most of the Q. suber populations distributed over very large areas in Catalonia and in a part of Morocco, respectively (Belahbib et al., 2001; Lumaret et al., 2005; López de Heredia et al., 2007a). The wide geographical distribution of chlorotype 66 transmitted exclusively by seeds suggests that the dispersal began a long time ago. In Morocco, the same ‘ilex’ chlorotypes were more often shared by both species, which have a patchy distribution, suggesting the occurrence of multiple independent cytoplasmic transfer events (Belahbib et al., 2001).
A second line of evidence is based on pollen and macrofossil data. López de Heredia et al. (2007a) made an extensive review of palynological data based on pollen and charcoal in evergreen oak species from Iberia, including Q. ilex and Q. suber which possess morphologically distinct pollen grains. The authors concluded that Iberian evergreen oaks occurred in several refugia during the Würmian period, including eastern Spain. In that region characterized by the predominance of limestone, a few Q. suber populations apparently survived during the glaciation periods, in isolated decarbonated sites where they grew in mixtures with predominant Q. ilex, a cold-tolerant species indifferent to soil type. As reported by López de Heredia et al. (2007a), the possibility that some populations of Q. suber withstood the glacial conditions in that Spanish region by hybridizing and introgressing with Q. ilex cannot be ruled out.
On the basis of cpDNA phylogeographical variation, North Africa was also identified as one of the potential glacial refuge areas for Q. suber and Q. ilex (Lumaret et al., 2002, 2005). In Morocco, the overall drought during and after the ice periods led the Mediterranean oak forests to move either to the most humid coasts or to mountains (Quezel and Médail, 2003). This assessment is supported by the presence, since the Boreal, of Q. suber and Q. ilex pollen types at low and high densities, respectively, in fossil sediments of the high Atlas, at 2000 m elevation (Bernard and Reille, 1987). Interestingly, all the Moroccan Q. suber populations possessing exclusively ‘ilex’ chlorotypes grow at elevations ranging from 750 to 2000 m (Belahbib et al., 2001; Lumaret et al., 2005). This indicates that the populations acquired the ability to cope with cold winter conditions, which is very unusual in this thermophilic species. This finding suggests that hybridization and introgression with Q. ilex may have played a significant adaptive role, given that the latter species is more cold tolerant that Q. suber.
In the present study, low current rates of hybridization of asymmetrical cytoplasmic interspecies transfers were estimated between Q. suber and Q. ilex, as expected in distantly related oak species. In addition, bidirectional nuclear introgression, with the possibility of gene flow filtering according to loci and species, was clearly identified. Current interspecies gene flow was not found to vary significantly according to geographical location and cannot explain the occurrence of the two large and distant areas characterized by the total cytoplasmic introgression of Q. suber by Q. ilex. As shown by paleontological records, these areas may have a historical origin and date back to the ice age (or even before), when Q. suber was subjected to high selective pressure. If this scenario is correct, hybridization and introgression between these two genetically distant species may not be merely accidental, but they may play a significant adaptive role, a hypothesis that can be tested more accurately by physiological experiments such as those already reported by Staudt et al. (2004).
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of location, sample size, chlorotype identification and population status (allopatric, sympatric and mixed populations) in the 38 sites sampled in the study.
ACKNOWLEDGEMENTS
We are grateful to M. Acherar, J. Aronson, G. Blanc, L. Bertrand-Battin, J. M. Bertrand-battin, P. D'Onofrio, L. Rocca-Serra, G. Ruiz, D. Ghaioule, E. Sahuquillo-Balbuena, D. Slade, L. Toumi and L. Zanone for their assistance in collecting plant material, to H. Bohbot, V. Sarda, C. Debain, J. Gimenes, C. Lind and S. Meski, for technical assistance, and to J. Aronson and C. Fontaine for making suggestions to improve the manuscript. This work was funded by the French GIP-Ecofor program ‘Biodiversité et Gestion forestière’ No 2001-23 and partly by the EU program CREOAK, QLK5-CT-2002-015.
LITERATURE CITED
- Avise JC. Molecular markers, natural history and evolution. Sunderland, PA: Sinauer Associates, Inc; 2004. [Google Scholar]
- Belahbib N, Pemonge MH, Ouassou A, Sbay H, Kremer A, Petit RJ. Frequent cytoplasmic exchanges between oak species that are not closely related: Quercus suber and Q. ilex in Morocco. Molecular Ecology. 2001;10:2003–2012. doi: 10.1046/j.0962-1083.2001.01330.x. [DOI] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikli L, Raufaste N, Bonhomme F. GENETIX, logiciel sous Windows TM pour la génétique des populations. Montpellier, France: Laboratoire Génome, Populations; 2001. Interactions CNRS UMR 5000, UMII. [Google Scholar]
- Bernard J, Reille M. Nouvelles analyses polliniques dans l'Atlas de Marrakech, Maroc. Pollen et Spores. 1987;39:225–240. [Google Scholar]
- Boavida LC, Silva JP, Feij JA. Sexual reproduction in the cork oak (Quercus suber L.) II. Crossing intra- and inter-specific barriers. Sexual Plant Reproduction. 2001;14:143–152. [Google Scholar]
- Burgarella C, Lorenzo Z, Jabbour-Zahab R, et al. Detection of hybrids in nature: application to oaks (Quercus suber and Q. ilex) Heredity. 2009;102:442–452. doi: 10.1038/hdy.2009.8. [DOI] [PubMed] [Google Scholar]
- Cronn R, Wendel J. Cryptic trysts, genome mergers, and plant speciation. New Phytologist. 2004;161:133–142. [Google Scholar]
- Curtu AL, Finkeldey R, Gailing O. Comparative sequencing of a microsatellite locus reveals size homoplasy within and between European oak species (Quercus spp.) Plant Molecular Biology Reporter. 2004;22:339–346. [Google Scholar]
- Dow B, Ashley M, Howe H. Characterization of highly variable (GA/CT)n microsatellites in the bur oak, Quercus macrocarpa. Theoretical and Applied Genetics. 1995;9:137–141. doi: 10.1007/BF00220870. [DOI] [PubMed] [Google Scholar]
- Dumolin S, Demesure B, Petit RJ. Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theoretical and Applied Genetics. 1995;91:1253–1256. doi: 10.1007/BF00220937. [DOI] [PubMed] [Google Scholar]
- Elena-Rosselló JA, Lumaret R, Cabrera E, Michaud H. Evidence for hybridization between symparic holm-oak and cork oak in Spain based on diagnostic enzyme markers. Vegetatio. 1992;99–100:115–118. [Google Scholar]
- Grivet D, Heinze B, Vendramin GG, Petit RJ. Genome walking with consensus primers: application to the large single copy region of chloroplast DNA. Molecular Ecology Notes. 2001;1:345–349. [Google Scholar]
- Gugerli F, Brodbeck S, Holderegger R. Insertions–deletions in a microsatellite flanking region may be resolved by variation in stuttering patterns. Plant Molecular Biology Reporter. 2008;26:255–262. [Google Scholar]
- Hardy OJ, Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2:618–620. [Google Scholar]
- Hardy OJ, Charbonnel N, Fréville H, Heuertz M. Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics. 2003;163:1467–1482. doi: 10.1093/genetics/163.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuertz M, Carnevale S, Fineschi S, et al. Chloroplast DNA phylogeography of European ashes, Fraxinus sp. (Oleaceae): roles of hybridization and life history traits. Molecular Ecology. 2006;15:2131–2140. doi: 10.1111/j.1365-294X.2006.02897.x. [DOI] [PubMed] [Google Scholar]
- Jimenez P, López de Heredia U, Collada C, Lorenzo Z, Gil L. High variability of chloroplast DNA in three Mediterranean evergreen oaks indicates complex evolutionary history. Heredity. 2004;93:510–515. doi: 10.1038/sj.hdy.6800551. [DOI] [PubMed] [Google Scholar]
- Kampfer S, Lexer C, Glössl J, Steinkellner H. Characterization of (GA)n microsatellite loci from Quercus robur. Hereditas. 1998;129:183–186. [Google Scholar]
- Kvacek Z, Walther H. Paleobotanical studies in Fagaceae of the European Tertiary. Plant Systematics and Evolution. 1989;162:213–229. [Google Scholar]
- Lexer C, Fay MF, Joseph JA, Nica MS, Heinze B. Barrier to gene flow between two ecologically divergent Populus species, P. alba (white poplar) and P. tremula (European aspen); the role of ecology and life history in gene introgression. Molecular Ecology. 2005;14:1045–1057. doi: 10.1111/j.1365-294X.2005.02469.x. [DOI] [PubMed] [Google Scholar]
- López de Heredia U, Carrión JS, Jiménez P, Collada C, Gil L. Journal of Biogeography. 2007a. Molecular and palaeobotanical evidence for multiple glacial refugia for evergreen oaks on the Iberian Peninsula. doi:10.1111/j.1365-2699.2007.01715.x. [Google Scholar]
- López de Heredia U, Jiménez P, Collada C, et al. Multi-marker phylogeny of three evergreen oaks reveal vicariant patterns in the Western Mediterranean. Taxon. 2007b;56:1209–1220. [Google Scholar]
- Lumaret R, Mir C, Michaud H, Raynal V. Phylogeographical variation of chloroplast DNA in holm oak (Quercus ilex L.) Molecular Ecology. 2002;11:2327–2336. doi: 10.1046/j.1365-294x.2002.01611.x. [DOI] [PubMed] [Google Scholar]
- Lumaret R, Tryphon-Dionnet M, Michaud H, et al. Phylogeographical variation of chloroplast DNA in cork oak (Quercus suber) Annals of Botany. 2005;96:853–861. doi: 10.1093/aob/mci237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumaret R, López de Heredia U, Soto A. Origin and genetic variability of Cork-oak. In: Aronson J, Pereira JS, Pausas J, editors. Cork oak woodlands on the edge: ecology, adaptive management and restoration. Washington, DC: Island Press; 2009. pp. 25–32. [Google Scholar]
- Magni CR, Ducousso A, Caron H, Petit RJ, Kremer A. Chloroplast DNA variation of Quercus rubra L. in North America and comparison with other Fagaceae. Molecular Ecology. 2005;14:513–524. doi: 10.1111/j.1365-294X.2005.02400.x. [DOI] [PubMed] [Google Scholar]
- Magri D, Fineschi S, Bellarosa R, et al. The distribution of Quercus suber chloroplast haplotypes matches the palaeogeographical history of the western Mediterranean. Molecular Ecology. 2007;16:5259–5266. doi: 10.1111/j.1365-294X.2007.03587.x. [DOI] [PubMed] [Google Scholar]
- Manos PS, Doyle JJ, Nixon KC. Phylogeny, biogeography, and processes of molecular differentiation in Quercus subgenus Quercus (Fagaceae) Molecular Phylogenetics and Evolution. 1999;12:333–349. doi: 10.1006/mpev.1999.0614. [DOI] [PubMed] [Google Scholar]
- Martinsen GD, Whitham TG, Turek RJ, Keim P. Hybrid populations selectively filter gene introgression between species. Evolution. 2001;55:1325–1335. doi: 10.1111/j.0014-3820.2001.tb00655.x. [DOI] [PubMed] [Google Scholar]
- Michaud H, Toumi L, Lumaret R, Li TX, Romane F, Di Giusto F. Effect of geographical discontinuity on genetic variation in Quercus ilex L. (holm oak). Evidence from enzyme polymorphism. Heredity. 1995;74:590–606. [Google Scholar]
- Mir C, Jarne P, Sarda V, Bonin A, Lumaret R. Contrasting nuclear and cytoplasmic exchanges between phylogenetically distant oak species (Quercus suber L. and Q. ilex L.) in Southern France: inferring crosses and dynamics. Plant Biology. 2009;11:213–226. doi: 10.1111/j.1438-8677.2008.00106.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Oliviera P, Custodio AC, Branco C, et al. Hybrids between cork oak and holm oak: isoenzyme analysis. Forest Genetics. 2003;10:283–298. [Google Scholar]
- Petit RJ, Bodénès C, Ducousso A, Roussel G, Kremer A. Hybridization as a mechanism of invasion in oaks. New Phytologist. 2004;161:151–164. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezel P, Médail F. Ecologie et biogéographie des forêts du Bassin méditerranéen. Paris: Elsevier; 2003. [Google Scholar]
- Rieseberg LH, Choi HC, Ham D. Differential cytoplasmic versus nuclear introgression in Helianthus. Journal of Heredity. 1991;82:489–493. [Google Scholar]
- Rieseberg LH, Bair SJE, Desrochers AM. Patterns of mating in wild sunflower hybrid zones. Evolution. 1998;52:489–493. doi: 10.1111/j.1558-5646.1998.tb03696.x. [DOI] [PubMed] [Google Scholar]
- Shriver MD, Smith MW, Jin L, et al. Ethnic-affiliation estimation by use of population-specific DNA markers. American Journal of Human Genetics. 1997;60:957–964. [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;130:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto A, Lorenzo Z, Gil L. Nuclear microsatellites markers for the identification of Quercus ilex L. and Quercus suber L. hybrids. Silvae Genetica. 2003;52:63–66. [Google Scholar]
- Soto A, Lorenzo Z, Gil L. Differences in fine-scale genetic structure and dispersal in Quercus ilex L. and Q. suber L.: consequences for regeneration of Mediterranean open woods. Heredity. 2007;99:601–607. doi: 10.1038/sj.hdy.6801007. [DOI] [PubMed] [Google Scholar]
- Staudt M, Mir C, Joffre R, et al. Isoprenoid emissions of Quercus spp. (Q. suber and Q. ilex) in mixed stands contrasting in interspecific genetic introgression. New Phytologist. 2004;163:573–584. doi: 10.1111/j.1469-8137.2004.01140.x. [DOI] [PubMed] [Google Scholar]
- Steinkellner H, Fluch S, Turetschek E, et al. Identification and characterization of (GA/CT)n – microsatellite loci from Quercus petraea. Plant Molecular Biology. 1997;3:1093–1096. doi: 10.1023/a:1005736722794. [DOI] [PubMed] [Google Scholar]
- Toumi L, Lumaret R. Allozyme variation in cork oak (Quercus suber L): the role of phylogeography, genetic introgression by other Mediterranean oak species and human activities. Theoretical and Applied Genetics. 1998;97:647–656. [Google Scholar]
- Toumi L, Lumaret R. Allozyme characterisation of four Mediterranean evergreen oak species. Biochemical Systematics and Ecology. 2001;29:799–817. doi: 10.1016/s0305-1978(01)00024-2. [DOI] [PubMed] [Google Scholar]
- Tutin TG, Heywood VH, Burges NA, et al. Flora Europaea. London: Cambridge University Press; 1993. [Google Scholar]
- Valbuena-Carabaña M, González-Martínez SC, Sork VL, et al. Gene flow and hybridization in a mixed oak forest (Quercus pyrenaica Willd. and Quercus petraea (Matts.) Liebl.) in central Spain. Heredity. 2005;95:1–5. doi: 10.1038/sj.hdy.6800752. [DOI] [PubMed] [Google Scholar]
- Valbuena-Carabaña M, Gonzalez-Martinez SC, Hardy OJ, Gil L. Fine-scale spatial genetic structure in mixed oak stands with different levels of hybridization. Molecular Ecology. 2007;16:1207–1219. doi: 10.1111/j.1365-294X.2007.03231.x. [DOI] [PubMed] [Google Scholar]
- Watano Y, Kanai A, Tani N. Genetic structure of hybrid zones between Pinus pumila and P. parviflora var. pentaphylla (Pinaceae) revealed by molecular hybrid index analysis. American Journal of Botany. 2004;91:65–72. doi: 10.3732/ajb.91.1.65. [DOI] [PubMed] [Google Scholar]
- Williams JH, Boecklen WJ, Howard DJ. Reproductive processes in two oak (Quercus) contact zones with different levels of hybridization. Heredity. 2001;87:680–690. doi: 10.1046/j.1365-2540.2001.00968.x. [DOI] [PubMed] [Google Scholar]
- Wolfram S. The mathematica book. Cambridge: Cambridge University Press; 1996. [Google Scholar]