Abstract
The rarity and inaccessibility of the earliest primordial germ cells (PGCs) in the mouse embryo thwarts efforts to investigate molecular mechanisms of germ cell specification. Stella marks the minute founder population of the germ lineage1,2. Here we differentiate mouse embryonic stem cells (ESCs) carrying a Stella transgenic reporter into putative PGCs in vitro. The Stella+ cells possess a transcriptional profile similar to embryo-derived PGCs, and like their counterparts in vivo, lose imprints in a time-dependent manner. Using inhibitory RNAs to screen candidate genes for effects on the development of Stella+ cells in vitro, we discovered that Lin28, a negative regulator of let-7 microRNA processing3-6, is essential for proper PGC development. We further show that Blimp1, a let-7 target and a master regulator of PGC specification7-9, can rescue the effect of Lin28-deficiency during PGC development, thereby establishing a mechanism of action for Lin28 during PGC specification. Over-expression of Lin28 promotes formation of Stella+ cells in vitro and PGCs in chimeric embryos, and is associated with human germ cell tumours. The differentiation of putative PGCs from ESCs in vitro recapitulates the early stages of gamete development in vivo, and provides an accessible system for discovering novel genes involved in germ cell development and malignancy.
The transcriptional program responsible for specifying PGCs has been investigated through molecular analysis of cDNA created from single cells10,11, but because transgenic or knock-out methods required to verify roles in germ cell formation are cumbersome, only a small number of genes have been demonstrated to be essential in germ cell formation, most notably Blimp1 and Prdm148,9,12. Building upon our previous demonstration of germ cell formation from ESCs in vitro13, we have further characterized differentiation into putative PGCs, and now exploit the system to screen candidate genes via RNA interference (RNAi) to discover novel regulators of PGC development. For these studies, we used embryoid body (EB) differentiation of ESCs carrying a transgenic StellaGFP reporter that specifically marks the germ cell lineage in vivo2 (Fig. 1a, Supplementary Fig. 1). The StellaGFP reporter, which is expressed in approximately 20% of cells within each ESC colony, was rapidly down-regulated during EB differentiation (Fig. 1a, Supplementary Fig. 2a). Reporter gene expression in ESCs reflects known Stella expression in the inner cell mass (ICM) and does not appear to represent pre-existing germ cells14,15. Rather, as shown below, a small percentage of Stella+ cells arise in punctate structures within the EB, which represent putative PGCs (Fig. 1a and Supplementary Fig. 2)2,15.
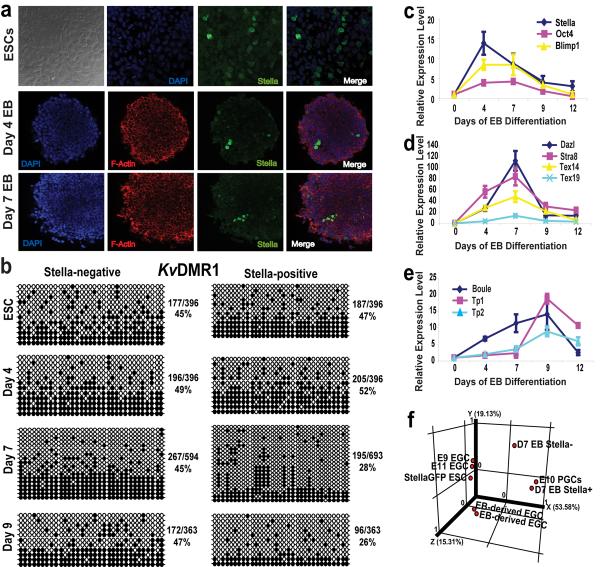
Figure 1. ESC differentiation into putative PGCs in vitro is marked by Stella expression.
a, Immunofluorescent detection of StellaGFP, F-actin and DAPI expression in ESCs and EBs at days 4 and 7 (63× confocal objective). b, Bisulfite sequencing at the KvDMR1 imprinted locus. White circles, unmethylated CpG dinucleotides; black circles, methylated CpGs. The percentage of methylated CpGs is noted next to each panel. c-e, Relative gene expression within Stella+ cells during EB differentiation, including the pre-migratory PGC markers, Stella, Oct4, and Blimp1 (c), migratory and post-migratory markers Dazl, Stra8, Tex14, and Tex19 (d), and Boule, a marker of meiosis, and the transition proteins Tp1 and Tp2 (e), by real time-PCR. Levels are relative to day 0 Stella+ cells and the data are represented as a mean +/- the s.d. n=3. f, PCA of Stella ESCs, EB-derived and embryo-derived EGCs (E9 and E11), day 7 EB cells (both Stella+ and Stella-negative) and E10.5 embryo-derived PGCs.
During mouse development, loss of genomic imprinting occurs solely in the germ line and is a prerequisite for the sex-specific reestablishment of imprints during gametogenesis, thus establishing loss of imprinting as a unique marker of the germ lineage16. Given the fidelity of the Stella reporter in vivo1, we predicted that the Stella+ population derived during ESC differentiation would demonstrate a time-dependent loss of imprints when compared to Stella-negative fractions and the parental ESCs. Indeed, Stella+ cells purified directly from EBs or clonally derived under EGC growth conditions showed extensive loss of methylation at imprinted loci from day 7 of differentiation onwards (Fig. 1b and Supplementary Fig. 3,4). These EB-derived EGCs displayed pluripotency properties similar to their embryo-derived counterparts (Supplementary Fig. 5)17.
Quantitative gene expression analysis revealed that germ cell-specific genes become highly enriched within the EB-derived Stella+ cell fraction in a temporal pattern that recapitulates gene activation during germ cell specification in vivo (Fig. 1c-e). Using microarrays, we observed that Stella+ cells purified from day 7 EBs display a transcriptional profile with significant similarities to embryo-derived PGCs. Unsupervised principal component analysis (PCA) of the microarray data revealed the close clustering of day 7 Stella+ EB-derived cells with embryo-derived E10.5 PGCs (Fig. 1f). Additionally, among a set of 178 genes that displayed at least a 2-fold change in Stella+ cells from day 7 EBs (when compared to Stella+ ESCs, Stella-negative ESCs, and Stella-negative cells from day 7 EBs), germ cell-specific transcripts were highly represented in the EB-derived Stella+ cell population (p=0.0009; Supplementary Fig. 6a,b). Scatter plot representations of the microarray data comparing Stella+ cells purified from day 7 EBs versus either StellaGFP ESCs or embryo-derived E10.5 PGCs were created to highlight individual gene expression similarities and differences (Supplementary Fig. 6c,d). These microarray data reveal that the overall transcriptional profile of EB-derived Stella+ cells is highly correlated with PGCs.
We next sought to use this system of in vitro germ cell specification to characterize the loss-of-function phenotypes for a number of candidate genes (n= 30) identified through our microarray analysis and reports of transcriptional profiling of PGCs10. We assessed the effects of gene knockdown on both tissue-nonspecific alkaline phosphatase-positive (TNAP+) EGC colony formation and on the loss of imprints during differentiation to link candidate gene function to germ lineage specification. TNAP staining is a hallmark of PGCs and EGCs. We knocked down endogenous expression of each gene within StellaGFP ESCs by delivering short hairpin RNAs (shRNAs) via lentiviral transduction (Supplementary Fig. 7). shRNAs directed against Blimp1, an established regulator of PGC development8,9, and LacZ served as controls. We differentiated ESCs carrying shRNAs for nine days and assayed StellaGFP+ cells for their ability to form TNAP+ EGC colonies after 5 days of retinoic acid (RA) treatment. The addition of RA to our selection media after EB differentiation served to promote the self-renewal of PGCs and to differentiate potential contaminating ESCs. Interestingly, transgenic StellaGFP expression during EB differentiation was largely unaffected under all conditions, including Blimp1 knockdown (data not shown), while endogenous Stella is modulated by gene manipulation (see below). This suggests that the endogenous Stella locus is more tightly regulated than the transgene in vitro. Although Blimp1 knockdown would be expected to completely abrogate Stella expression, Blimp1 homozygous knockout mice still form Stella-expressing cells, although these cells are rare and fail to proliferate and develop properly8,10. Thus, we focused on functional aspects of germ cell formation, including TNAP-colony formation and loss of genomic imprints. Among the candidate genes that we assessed, knockdown of Lin28 demonstrated the most quantitative reduction in TNAP-positive colony formation (Fig. 2a and Supplementary Fig. 8a). Further corroborating the loss of germ cells, we verified that knockdown of Lin28 abrogates the ability to derive imprint-erased clones after RA-selection of EB-derived Stella+ cells (Fig. 2b).
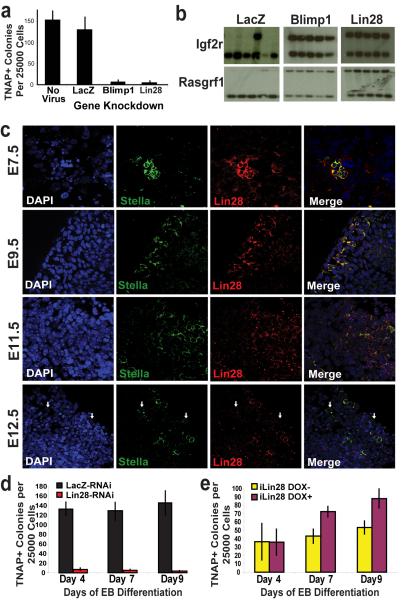
Figure 2. Lin28 regulates PGC development.
a, The effects of candidate gene knockdown on TNAP+ EGC-colony formation from day 9 EB-derived Stella+ cells following in vitro differentiation of ESCs carrying shRNA-mediated gene knockdown, as indicated. n=3 b, Imprint status at the Igf2r and Rasgrf1 loci of individual clones derived from day 9 EB-derived Stella+ cells carrying gene knockdown of either LacZ, Blimp1, or Lin28. c, Expression of Lin28 during embryonic PGC development. By E12.5, multiple Stella+ PGCs within the genital ridge are negative for Lin28 (white arrows). (63× confocal objective) d, Lin28-RNAi prevents TNAP+ EGC-colony formation during EB differentiation. n=3 e, Induced Lin28 expression enhances TNAP+ EGC-colony formation on and after day 7 of EB differentiation compared to the uninduced control. n=3 All error bars depicted represent the S.E.M.
Lin28 selectively blocks the processing of let-7 precursors into the corresponding mature miRNA species3-6. Although not previously suspected as a regulator of PGC formation, we included Lin28 in our screen because it was more highly expressed in day 7 EB-derived Stella+ cells than in ESCs and EB-derived Stella-negative cells (as determined by microarray). Interrogation of a microarray dataset of embryo-derived single cells from the mouse PGC lineage indicated high Lin28 expression in the proximal epiblast, PGC-precursors, and lineage-restricted PGCs, as well as in the posterior mesoderm surrounding these cells10. We evaluated Lin28 protein expression during PGC development in mouse embryos and observed high levels of Lin28 staining within Stella+ PGCs at E7.5, and only diffuse low-level Lin28 staining in surrounding somatic cells. We observed diminishing yet persistent expression of Lin28 within PGCs through E12.5, at which time Lin28-negative PGCs become apparent (Fig. 2c).
To further elucidate the role of Lin28 on the development of germ cells, we characterized the effects of modulating Lin28 expression on germ cell-marker gene expression during ESC differentiation in vitro. Quantitative gene expression analysis revealed that Lin28-RNAi decreased the expression of multiple germ cell markers in StellaGFP+ cells during EB differentiation, including key genes implicated in PGC commitment, such as Blimp1, Prdm14, and endogenous Stella (Supplementary Fig. 9ai). Moreover, we confirmed that Lin28-RNAi resulted in increased levels of mature let-7 miRNA family members by approximately 5- to 6-fold in Stella+ cells (Supplementary Fig. 9z-bb). Lin28-RNAi dramatically reduced TNAP-colony formation throughout EB differentiation of StellaGFP ESCs, consistent with a reduction in the generation of putative PGCs (Fig. 2d). Three individual shRNAs directed against Lin28 resulted in a similar phenotype, and ectopic Lin28 expression resistant to shRNA inhibition was able to stimulate formation of TNAP+ colonies and re-expression of germ cell-specific genes, thereby arguing against off-target effects of Lin28-RNAi (Supplementary Fig. 10).
We next drove ectopic Lin28 expression during EB differentiation from a tetracycline-inducible ESC line (iLin28 ESCs). Addition of doxycycline to the media resulted in a pronounced up-regulation of Lin28 throughout differentiation, and a corresponding suppression of let-7 maturation (Supplementary Fig. 9j,q,cc-ee). Induction of Lin28 during EB differentiation was accompanied by increased expression of multiple germ cell markers, including Stella, Prdm14, and Tex14 within whole EBs as well as within SSEA1+ cells (Supplementary Fig. 9j-y). Interestingly, we found no evidence that ectopic Lin28 expression influenced Blimp1 mRNA levels (Supplementary Fig. 9l,s), which could be a result of the reported post-transcriptional regulation of Blimp1 by let-77. Functionally, ectopic Lin28 expression during EB differentiation resulted in significant increases in TNAP+ EGC-colony formation when compared to uninduced controls, suggesting that enforced expression of Lin28 promotes expansion of PGC pools (Fig. 2e).
To corroborate our in vitro results of Lin28 knockdown within a physiological setting in vivo, we injected knockdown ESCs into blastocysts and monitored the germ cell contribution in the resulting chimeric embryos via StellaGFP transgenic reporter expression. Control embryos chimerized with StellaGFP ESCs expressing a constitutive LacZ-RNAi construct showed extensive contribution of GFP+ germ cells in the genital ridge, whereas Blimp1-RNAi ablated germ cell commitment (Fig. 3a,b). Lin28 knockdown compromised PGC formation to the same extent as Blimp1-knockdown. Failure of Lin28-RNAi ESCs to contribute to the germ line was not due to a generalized deficit in chimerization potential, as Lin28-RNAi ESCs were able to chimerize tissues of the head and limbs in virtually all embryos tested (Supplementary Fig. 11). StellaGFP expression was not observed in somatic areas of chimeric embryos, indicating the specificity of the Stella reporter2. Furthermore, these data suggest a cell autonomous role for Lin28 in PGC formation as chimeric embryos possess mosaic tissues with presumably normal elements derived from the host, which would be expected to complement deficits caused by Lin28 deficiency in non-germ cells. Thus, our data argue for a physiologic role for Lin28 in PGC development in vivo.
Figure 3. Lin28 and Blimp1 regulate PGC development in vivo.
a, Representative images of genital ridges from E12.5 embryos chimerized with StellaGFP ESCs harbouring either LacZ-RNAi, Blimp1-RNAi, ectopic Blimp1 expression, Lin28-RNAi, ectopic Lin28 expression, Lin28-RNAi plus ectopic Blimp1 expression, or ectopic Lin28 plus Blimp1-RNAi. Scale bar = 100 μm. b, Quantification by flow cytometry of the percent of Stella+ PGCs in E12.5 genital ridges dissected from chimeric embryos created with StellaGFP ESCs containing the following: LacZ-RNAi, Blimp1-RNAi, ectopic Blimp1 expression, Lin28-RNAi, ectopic Lin28 expression, Lin28-RNAi plus ectopic Blimp1 expression, or ectopic Lin28 plus Blimp1-RNAi. Black horizontal bars represent the median. Non-significant p-values, n.s. c, Immunoblot for Blimp1 expression in extracts from ESCs and day 5 EBs with or without doxycycline induction of let-7s-21L. Gapdh served as a loading control.
As a complementary approach to examine the impact of Lin28 on PGC numbers in vivo, we transduced StellaGFP ESCs with a retrovirus that drives the ectopic expression of Lin28, injected blastocysts, and quantified ESC contribution to the genital ridge. Ectopic Lin28 expression resulted in a significant enhancement of PGC numbers in chimeric embryos (Fig. 3a,b), thus suggesting that Lin28 can both positively and negatively regulate the pool of germ cells.
As the 3' untranslated region (UTR) of Blimp1 has been identified as a direct target for let-7 miRNAs7, we hypothesized that Lin28 influences PGC development through let-7 mediated effects on Blimp1. Blimp1 is a key regulator of germ cell commitment8,9, and we sought to determine if ectopic expression of Blimp1 lacking its 3'UTR could rescue PGC development in the setting of Lin28 depletion. We engineered StellaGFP ESCs possessing both Lin28-RNAi and ectopic Blimp1 expression and assayed the ability of these cells to contribute to the germ line in chimeric E12.5 embryos. In this context, Blimp1 rescues PGC numbers to control levels (Fig. 3a,b). Additionally, Blimp1-RNAi ablates PGC development from ESCs ectopically expressing Lin28 (Fig. 3a,b). These data suggest Lin28 as an upstream regulator of PGC development relative to Blimp1, potentially acting via repression of the let-7 miRNA family within PGCs or their precursors. To further test this hypothesis, we engineered an doxycycline-inducible ESC line to express a form of let-7s (7s-21L) that is resistant to Lin28 by virtue of a terminal loop substitution with that of miR-21 (a miRNA not regulated by Lin28)18 and determined the effect of induction on Blimp1 expression in EBs. We found that induction of let-7s-21L abrogates Blimp1 protein expression during EB differentiation (Fig. 3c), further supporting Lin28 regulation of Blimp1 via let-7.
Expression of Lin28b, a homologue of Lin28, is largely absent in early PGCs10, but is expressed in ESCs. Thus, we also assessed the impact of Lin28b-RNAi on PGC formation. We observed a compromised ability to form EB-derived PGCs due to Lin28b knockdown and an intermediate phenotype in vivo (Supplementary Fig. 8). Knockdown of Lin28b in ESCs likewise results in up-regulation of let-7 and compromises germ cell formation, further linking let-7 to PGC development but suggesting that Lin28 is the dominant regulator in vivo.
Given our data linking Lin28 and germ cell numbers, we surmised that aberrant over-expression of Lin28 might dysregulate the growth or differentiation of pluripotent cells including germ cells. Furthermore, recent reports have linked Lin28 and its homologue Lin28b to oncogenesis19-21. In teratomas formed from ESCs engineered to express Lin28 in response to doxycycline induction (iLin28 ESC) we observed that primitive neural tissue predominated in regions of transgene induction (Supplementary Fig. 12a). High grade, aggressive human teratocarcinomas contain abundant immature neural tissue while low grade, benign teratomas contain no immature neural tissue. This phenomenon is also observed in teratomas formed from inducible Oct4 ESCs22. In contrast, Lin28-RNAi during teratoma formation resulted in reduced tumour size, and these teratomas were comprised of mature tissues from all three germ layers (Supplementary Fig. 12b,c). Lin28 and Lin28b are more prominently over-expressed in the transformed embryonal carcinoma (EC) cell line P19 relative to ESCs than corresponding Oct4 levels in these cells (Supplementary Fig. 13a). Modulation of Lin28 expression by RNAi in P19 cells reduces both cell proliferation and tumour formation in immunodeficient mice, suggesting that Lin28 contributes to the malignant phenotype of embryonal carcinoma cells (Supplementary Fig. 13b-e). In addition, tumours with ectopic Lin28 displayed local invasion into the host tissues, while teratomas formed from pluripotent cells treated with Lin28-RNAi were differentiated, non-invasive, and well-encapsulated (Supplementary Fig. 13d,e), paralleling recent reports linking Lin28 to metastasis20,21.
We sought to determine whether LIN28 is expressed in human germ cell tumours (GCTs). We interrogated a GCT microarray dataset and profiled multiple primary human tumours for the expression of LIN28, LIN28B, and OCT4, a known marker of GCTs23. We observed that malignant tumours of a germ cell origin, including mixed germ cell tumours, yolk-sac tumours, choriocarcinomas, embryonal carcinomas, and seminomas, demonstrated over-expression of LIN28 and/or LIN28B (Fig. 4a). Importantly, benign teratomas and normal testis do not show high levels of LIN28 and LIN28B expression relative to malignant tumour types, and OCT4 is not expressed in yolk-sac tumours or choriocarcinomas24. The expression of LIN28/28B in all malignant GCT subtypes implicates LIN28 as a consistent marker of GCTs. We confirmed LIN28 protein expression in a primary human embryonal carcinoma by immunohistochemistry (Fig. 4b). Importantly, we noted that LIN28 expression was specifically observed in malignant components of the tumour, but not in benign elements of the tumour or in the adjacent stroma. Interestingly, regions of intratubular germ cell neoplasia (a precursor lesion for most germ cell tumours) stained strongly positive for LIN28 while normal seminiferous tubules displayed little to no staining (Fig. 4b, right panel), suggesting that aberrant expression of LIN28 may be an early lesion in germ cell tumourigenesis. In a separate study, we have documented that LIN28 over-expression facilitates cellular transformation in NIH-3T3 and BaF3 cells, consistent with a functional role for LIN28 in malignancy21.
Figure 4. LIN28 expression in human germ cell tumours.
a, Primary malignant germ cell tumours of multiple histologies show consistent high expression of LIN28 and LIN28B. Benign teratomas and normal testis show no significant expression of LIN28 or LIN28B. Mixed germ cell tumour (GCT); yolk-sac tumour (YST); Choriocarcinoma (C); Embryonal carcinoma (EC); Seminoma (S); Teratoma (Ter); and Normal testis (Tes). b, Immunohistochemical detection of LIN28 in primary embryonal carcinoma. Left panel: Cytoplasmic immunoreactivity of embryonal carcinoma cells for LIN28 (between arrowheads). Endodermal epithelial elements of the teratoma (arrows) and tumoural stroma are not immunoreactive. Middle panel: Higher magnification of immunoreactive embryonal carcinoma. Adjacent seminiferous tubules are negative (between arrows). Right panel: Intratubular germ cell neoplasia within the embryonal carcinoma demonstrates LIN28 immunoreactivity. (40× magnification).
Lin28 selectively blocks the processing of the let-7 family of miRNAs3-6. Let-7 miRNAs are upregulated only during later stages of PGC development25, establishing that let-7 expression is dynamically regulated within the germ lineage. Our data suggest that Lin28 suppression of let-7 maturation is required for proper Blimp1-mediated development of the germ lineage. Consistent with this hypothesis, we have observed that Lin28-RNAi is associated with up-regulation of let-7 miRNAs and the abrogation of Blimp1 expression in Stella+ cells during in vitro differentiation (Supplementary Fig. 9c, z, aa-ee). Additionally, ectopic Blimp1 expression rescues PGC formation from ESCs harbouring Lin28 knockdown. Our Lin28 expression data in PGCs correlate inversely with the expression of let-7 miRNAs25 and are coincident with Blimp1 protein dynamics in PGCs26. Although Blimp1 appears critical, additional let-7 targets may yet prove relevant to PGC specification.
Our functional data indicate that Lin28 is a key lineage-regulator for germ cell numbers (Supplementary Fig. 14). As multiple lineage-specifying factors such as MITF (Microphthalmia-associated transcription factor)27 and SCL (Stem cell leukaemia gene)28 have been implicated as oncogenes, Lin28 could play a dual role in both germ cell development and malignancy. As let-7 miRNAs function as tumour suppressors by directly and indirectly repressing a number of cell proliferation pathways, Lin28 de-repression of let-7 oncogenes has been established as a key oncogenic factor19-21. Multiple tumour types show reduced levels of let-7 miRNAs, and several let-7 targets, including c-Myc, k-Ras, and HmgA2 are oncogenic21,29. In addition, somatic cell reprogramming factors (including presumably Lin28) also play prominent roles in cellular transformation. Oct4 has been identified as an oncogenic driver for certain GCTs, including embryonal carcinomas and seminomas but not yolk-sac tumours and choriocarcinomas22,24. Here we show that Lin28 and its homologue Lin28b are more consistently expressed in malignant GCTs than even Oct4, including within yolk-sac tumours and choricarcinomas. Together, our data implicate Lin28 as a novel regulator of germ cell development and a consistent marker of germ cell malignancy, and provide new insights into germ cell formation, development, and cancer.
METHODS SUMMARY
shRNA constructs in the pLKO.1-Puro vector system for RNAi-mediated gene knockdown were obtained from Sigma and Open Biosystems (MISSION™ TRC-Mm 1.0). TRC numbers for individual hairpins as well as primer information for real-time PCR analysis can be found in Supplementary Tables 1 and 2. EB-derived PGC microarray analysis used Affymetrix mouse 430 2.0 whole genome arrays, and data analysis employed MAS5.0 standard normalization and differential expression analysis techniques using GeneSpring GX 7.3 software. Embryo-derived PGC and PGC-precursor microarray datasets were published previously10 and have the Gene Expression Omnibus accession number GSE11128. Primary human tissue microarray datasets were published previously23 and have the Gene Expression Omnibus (GEO) accession number GSE3218. R. Yu and C. Estrada kindly provided the pCDH-CMV-mPrdm1 construct. Antibodies used in this study include: mouse anti-SSEA1 (MC480; Developmental Studies Hybridoma Bank at the U. of Iowa), rabbit anti-GFP (BioVision), goat anti-Lin28 for immunofluoresence and western blotting (Santa Cruz), anti-LIN28 for human GCT immunohistochemistry (Proteintech Group Inc.), anti-Blimp1 (R&D Systems), and Alexa-594 phalloidin (Molecular Probes). In Fig. 3b, p-values were calculated by the Mann Whitney rank-sum test and are relative to the control, LacZ RNAi.
Methods
ESC Cell Culture, EB Differentiation, PGC selection, and EGC culture
Transgenic StellaGFP and StellaBAC ESCs (B6/CBA) were cultured as described, and the hanging drop method was used for ESC differentiation as EBs2,13,30-32. All data presented used the StellaGFP transgenic ESC line unless indicated2. Inducible Lin28 ESCs (iLin28 ESCs) and inducible let-7s-21L ESCs were created from the KH2 ESC line as described18,33, and 2 ug/ml doxycycline was added for induced expression during differentiation.
For PGC selection, EBs were dissociated and the single cell suspension was subjected to FACS using a BD FACS Aria cell sorting system to purify StellaGFP-positive cells. For the iLin28 ESC line, SSEA1-purification was utilized for germ cell isolation, as described13,32. For EGC culture, FACS-processed cells were plated on MEFs, with mESC media containing 15 ng/ml of bFGF (Invitrogen) and 30 ng/ml of SCF (Peprotech), plus 2 uM trans-retinoic acid (Sigma). Retinoic acid (RA) promotes germ cell self-renewal while serving as a differentiation agent for ESCs34. After 5 to 7 days, replacing the media daily, cultures were fixed and stained for tissue-nonspecific alkaline phosphatase (TNAP), a germ cell specific marker35, or clonal EGC colonies were isolated and cultured as previously described and expanded for analysis32. For TNAP staining, cultured cells were washed once with PBS, fixed in 4% paraformaldyhyde (PFA) for 1 minute, and then washed in PBS three times. AB staining solution [10 mL buffer (100 mM Tris HCl pH 9.5, 50 mM MgCl2, 100 mM NaCl, and 0.1% Tween-20), 22.5 uL of 50 mg/mL nitro-blue tetrazolium in 70% dimethylformamide (NBT; Promega), and 17.5 uL of 50 mg/mL 5-bromo-4-chloro-3-indoyl-1-phosphate in 100% dimethylformamide (BCIP; Promega)] was added to the cells and incubated at room temperature for 5 to 10 minutes, and then the cells were washed in PBS several times and imaged.
Confocal Imaging
Embryos or EBs were fixed in 4% PFA (in PBS) for 1 hour (h) at room temperature (RT), washed 3× in PBS (5 min. per wash) and permeabilized for 1 hour at RT in 1× IF buffer (PBS plus 0.1% BSA, 0.2% Triton X-100, 0.04% Tween-20). They were then incubated with 1:200 primary rabbit anti-GFP (BioVision) or 1:200 primary goat anti-Lin28 (Santa Cruz) in IF buffer at 37°C for 1h, washed 3× with IF buffer 5 min. each at RT with a final wash in IF buffer at 37°C for 10 min. They were then incubated with 1:500 secondary (Alexa488 and/or Alexa594, Molecular Probes) or 1:200 Alexa-594 phalloidin (Molecular Probes) and DAPI (1:100 from 5 mg/ml stock) in IF buffer for 1h at 37°C. Samples were placed in coverslip-bottom dishes (MatTek) in Fluoromount G mounting medium (Southern Biotech) and imaged on a Leica confocal microscope (63× objective with oil).
Quantitative real time RT-PCR
Quantitative real time PCR of gene expression utilized SYBR green. Multiple, independent biological samples were obtained for RNA isolation. Levels of mature miRNAs were measured using Taqman probes (Applied Biosystems). Beta-actin expression was used to normalize all samples except for miRNA expression, which were normalized to sno142 RNA. Relative fold changes were calculated using the ΔΔCt method. See Supplementary Table 1 for primer sequences.
Microarray hybridization and data analysis
Each sample contained two to four biological replicates. RNA was isolated using an RNeasy kit (Qiagen) according to the manufacturer's protocol. Biotin-labelled RNA was hybridized to Affymetrix mouse 430 2.0 whole genome arrays at the Dana-Farber Cancer Institute Microarray Core. GeneSpring GX 7.3 software was used for all data analysis, including scatter plot generation, employing MAS5.0 standard normalization and differential expression analysis techniques. Principal component analysis (PCA) was performed via GeneSpring using mean centring and scaling for all genes. Raw data for EB-derived PGCs and associated cell types are deposited in the public gene expression database GEO (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE7948. Embryo-derived PGC and PGC-precursor microarray datasets were published previously10 and have the Gene Expression Omnibus accession number GSE11128.
Southern blot analysis
Imprint analysis for the Igf2r locus was at the maternally methylated DMR1. Genomic (g) DNA was digested with PvuII/MluI and probed via Southern blot hybridization as described36. For the paternally methylated Rasgrf1 locus, Southern blot hybridization of gDNA digested with PstI/NotI was probed as described37.
Bisulfite Sequencing
Bisulfite treatment of gDNA was carried out using a Chemicon CpGenome DNA Modification Kit according to the manufacturer's protocol. Sample treatment and processing were performed simultaneously for quality control. Nested PCR was then used to amplify KvDMR138 and the Snrpn DMR1 as described39. The KvDMR1 region examined is on distal chromosome 7, contains 33 CpGs within intron 10 of the Kcnq gene, and controls the expression of six paternally repressed genes: Ascl2, Tssc4, Kcnq, Cdkn1c, Slc221l, and Tssc238. The maternally repressed Snrpn DMR1 examined is located on central chromosome 7 and contains 11 CpGs39. PCR products were cloned from two independent PCR reactions. Bisulfite conversion efficiency ranged from 85% to 99%, and 83 to 100% of individual clones for each sample showed unique patterns of conversion.
RNA Interference
shRNA constructs in the pLKO.1-Puro vector system for RNAi-mediated gene knockdown were obtained from Sigma and Open Biosystems (MISSION™ TRC-Mm 1.0). Expression of the shRNA was constitutively driven by the human U6 promoter while the human phosphoglycerate kinase promoter (PGK) independently drove the expression of a puromycin resistance gene to allow selection for cells harbouring the shRNA-mediated gene knockdown. TRC numbers for hairpins used are listed in Supplementary Table 2. For Lin28, only shRNA TRCN0000102579 was used for all experiments, unless otherwise noted. Viral production in 293T cells and ESC infection were performed according to product literature and as described 40. Puromycin-selection was applied continuously during all subsequent cell culture including EB differentiation.
Ectopic Gene Expression Vectors
Ectopic Lin28 was provided by either pBABE-puro-Lin28 or MSCV-neo-Lin28, while ectopic Blimp1 was driven by pCDH-CMV-mPrdm1 (System Biosystems; We are grateful to Richard Yu and Carlos Estrada of Children's Hospital Boston for engineering and sharing this reagent).
Tumour Formation
All tumour-forming assays, included teratoma formation, were performed using NOD-SCID mice (Charles River Laboratories). 1×106 cells in 150μl PBS were injected subcutaneously for all experiments.
Production of chimeric embryos
C57BL6 females were superovulated by PMSG followed 48 hours later by hCG and were mated with C57BL6 males. Blastocysts were collected from the uterus 3.5 days after vaginal plugging. 12-15 ESCs were injected into each blastocyst which was then transferred to the uterus of a 2.5 days postcoitum pseudo-pregnant CD-1 female. Ectopic Lin28 expression was accomplished by retroviral infection of StellaGFP ESCs with pBABE-Lin28. Statistical significance of germ cell chimerism was determined by the Mann-Whitney rank-sum test.
Human primary tumour and cell line information
Primary tissue microarray datasets were published previously23 and have the Gene Expression Omnibus accession number GSE3218. Log2 transformed expression data were normalized to the average expression signal for normal tissue. Cut-off for over-expression was as a normalized expression value of 2 or greater. Immunohistochemistry of paraffin-embedded tissues were performed using standard techniques with LIN28 primary antibody staining (1/200; Proteintech Group Inc., Chicago, IL) followed by streptavidin peroxidase incubation (Lab Vision, Fremont, CA) and DAB kit colour developing (Vector Laboratories, Burlingame, CA) according to the manufacturers' protocol. Sections were counterstained with haematoxylin.
Supplementary Material
Acknowledgments
We wish to thank M.W. Lensch for his helpful comments on this manuscript, D.K. Gifford, G. Gerber, C. Reeder, and J. Baughman for useful comments and input regarding microarray analysis, G. Losyev for flow cytometry expertise, and S. Winkler of the British Consulate for providing support for collaboration between the Daley and Surani laboratories. This study was supported by grants from the NIH, the NIH Director's Pioneer Award of the NIH Roadmap for Medical Research, and by support from the germ cell program of the Harvard Stem Cell Institute. G.Q.D. is a recipient of the Burroughs Wellcome Fund Clinical Scientist Award in Translational Research.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Payer B, Saitou M, Barton SC, et al. Curr Biol. 2003;13(23):2110. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 2.Payer B, Chuva de Sousa Lopes SM, Barton SC, et al. Genesis. 2006;44(2):75. doi: 10.1002/gene.20187. [DOI] [PubMed] [Google Scholar]
- 3.Heo I, Joo C, Cho J, et al. Molecular Cell. 2008;32(2):276. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Newman MA, Thomson JM, Hammond SM. RNA. 2008;14(8):1539. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rybak A, Fuchs H, Smirnova L, et al. Nature Cell Biology. 2008;10(8):987. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 6.Viswanathan SR, Daley GQ, Gregory RI. Science. 2008;320(5872):97. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nie K, Gomez M, Landgraf P, et al. The American Journal of Pathology. 2008;173(1):242. doi: 10.2353/ajpath.2008.080009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohinata Y, Payer B, O'Carroll D, et al. Nature. 2005;436(7048):207. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 9.Vincent SD, Dunn NR, Sciammas R, et al. Development. 2005;132(6):1315. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- 10.Kurimoto K, Yabuta Y, Ohinata Y, et al. Genes & Development. 2008;22(12):1617. doi: 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saitou M. Front Biosci. 2009;14:1068. doi: 10.2741/3294. [DOI] [PubMed] [Google Scholar]
- 12.Yamaji M, Seki Y, Kurimoto K, et al. Nature Genetics. 2008;40(8):1016. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- 13.Geijsen N, Horoschak M, Kim K, et al. Nature. 2004;427(6970):148. doi: 10.1038/nature02247. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi K, Lopes SM, Tang F, et al. Cell stem cell. 2008;3(4):391. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei W, Qing T, Ye X, et al. PLoS ONE. 2008;3(12):e4013. doi: 10.1371/journal.pone.0004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reik W, Walter J. Nat Rev Genet. 2001;2(1):21. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 17.Donovan PJ, de Miguel MP. Curr Opin Genet Dev. 2003;13(5):463. doi: 10.1016/j.gde.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Piskounova E, Viswanathan SR, Janas M, et al. The Journal of Biological Chemistry. 2008;283(31):21310. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 19.Chang TC, Zeitels LR, Hwang HW, et al. Proc Natl Acad Sci U S A. 2009;106(9):3384. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dangi-Garimella S, Yun J, Eves EM, et al. The EMBO Journal. 2009;28(4):347. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viswanathan SR, Powers JT, Einhorn W, et al. Nature Genetics. 2009 doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gidekel S, Pizov G, Bergman Y, et al. Cancer Cell. 2003;4(5):361. doi: 10.1016/s1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 23.Korkola JE, Houldsworth J, Chadalavada RS, et al. Cancer Research. 2006;66(2):820. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
- 24.Cheng L. Cancer. 2004;101(9):2006. doi: 10.1002/cncr.20566. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, et al. PLoS ONE. 2008;3(3):e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ancelin K, Lange UC, Hajkova P, et al. Nature Cell Biology. 2006;8(6):623. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- 27.Garraway LA, Widlund HR, Rubin MA, et al. Nature. 2005;436(7047):117. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 28.Shivdasani RA, Mayer EL, Orkin SH. Nature. 1995;373(6513):432. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Getz G, Miska EA, et al. Nature. 2005;435(7043):834. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 30.Keller GM. Curr Opin Cell Biol. 1995;7(6):862. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 31.Kyba M, Perlingeiro RC, Daley GQ. Cell. 2002;109(1):29. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 32.West JA, Park IH, Daley GQ, et al. Nat Protoc. 2006;1(4):2026. doi: 10.1038/nprot.2006.303. [DOI] [PubMed] [Google Scholar]
- 33.Beard C, Hochedlinger K, Plath K, et al. Genesis. 2006;44(1):23. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 34.Koshimizu U, Watanabe M, Nakatsuji N. Dev Biol. 1995;168(2):683. doi: 10.1006/dbio.1995.1113. [DOI] [PubMed] [Google Scholar]
- 35.Ginsburg M, Snow MH, McLaren A. Development. 1990;110(2):521. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- 36.Stoger R, Kubicka P, Liu CG, et al. Cell. 1993;73(1):61. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 37.Yoon BJ, Herman H, Sikora A, et al. Nat Genet. 2002;30(1):92. doi: 10.1038/ng795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mager J, Montgomery ND, de Villena FP, et al. Nat Genet. 2003;33(4):502. doi: 10.1038/ng1125. [DOI] [PubMed] [Google Scholar]
- 39.Lucifero D, Mertineit C, Clarke HJ, et al. Genomics. 2002;79(4):530. doi: 10.1006/geno.2002.6732. [DOI] [PubMed] [Google Scholar]
- 40.Zaehres H, Daley GQ. Methods in Enzymology. 2006;420:49. doi: 10.1016/S0076-6879(06)20004-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.