Abstract
Palindromic regions are unstable and susceptible to deletion in prokaryotes and eukaryotes possibly due to stalled or slow replication. In the human genome, they also appear to become partially or completely deleted, while two palindromic AT-rich repeats (PATRR) contribute to known recurrent constitutional translocations. To explore the mechanism that causes the development of palindrome instabilities in humans, we compared the incidence of de novo translocations and deletions at PATRRs in human cells. Using a highly sensitive PCR assay that can detect single molecules, de novo deletions were detected neither in human somatic cells nor in sperm. However, deletions were detected at low frequency in cultured cell lines. Inhibition of DNA replication by administration of siRNA against the DNA polymerase alpha 1 (POLA1) gene or introduction of POLA inhibitors increased the frequency. This is in contrast to PATRR-mediated translocations that were never detected in similar conditions but were observed frequently in human sperm samples. Further deletions were found to take place during both leading- and lagging-strand synthesis. Our data suggest that stalled or slow replication induces deletions within PATRRs, but that other mechanisms might contribute to PATRR-mediated recurrent translocations in humans.
INTRODUCTION
Palindromic regions, or inverted repeats, in the genome are acknowledged to pose a threat to genome stability inducing deletions, translocations and duplications. Good examples of unstable repeats in humans are palindromic AT-rich repeats (PATRRs). PATRRs were first identified at the breakpoints of the constitutional t(11;22)(q23;q11), the only recurrent non-Robertsonian translocations in humans (1,2). Although balanced t(11;22) carriers have no clinical symptoms, they occasionally manifest infertility or recurrent spontaneous abortions. They are often identified after the birth of unbalanced offspring that present with a congenital malformation syndrome, Emanuel syndrome (supernumerary der(22)t(11;22) syndrome; MIM no. 609029) (3). Analysis of numerous unrelated t(11;22) cases reveal that the breakpoints are located within the 400–600 bp PATRRs on 11q23 and 22q11 (PATRR11 and PATRR22) (4). The center of the palindrome has been suggested to be susceptible to double-strand breaks (DSBs) inducing illegitimate chromosomal rearrangement (4–8). Indeed, de novo t(11;22)s were frequently identified in the sperm of normal healthy males (9). Further, PATRR22 has been known to be a hotspot for translocation breakpoints (10,11). Identification of PATRR-like sequences at the translocation breakpoints on other chromosome 22 translocation partner chromosomes supports the conclusion that palindrome-mediated chromosomal translocation appears to be one of the universal pathways for gross chromosomal rearrangements in humans (12–16).
The mechanism of genomic instability in palindromic regions has been extensively studied in Escherichia coli, Saccharomyces cerevisiae and mouse. In E. coli, a palindromic region is either partially or completely deleted (17,18). This genetic instability is because of secondary structure formed by intra-strand base-pairing and is generally related to DNA replication. Stalling of DNA synthesis or slow replication was observed in palindromic regions (18). Slow progression of the replication fork increases the likelihood of forming secondary structures at long tracts of single-stranded DNA residing in the lagging strand template. Such secondary structures may create an obstacle to fork progression leading to replication slippage, or may be a target for a structure-specific nuclease that leads to deletion or recombination (18,19). In S. cerevisiae, palindromic sequences induce meiotic or mitotic recombination by creating hotspots for DSBs (20,21). Palindromic regions induce deletions or chromosomal rearrangements more frequently in yeast that are deficient in DNA polymerase activity (22,23). In mammals, analysis of transgenic mice has demonstrated frequent deletions or insertions at such artificially created palindromic regions, indicative of substantial meiotic and mitotic instability (24–26).
Genomic instability induced by DNA palindromes in such model organisms often manifest as deletions between direct repeats located at either side of the palindromic center. Likewise, the PATRRs found in humans also appear to be susceptible to deletion including the center of symmetry, which is evidenced by the presence of size polymorphism among individuals (5,27,28). This means that deletions of the PATRR actually occurred in the course of human genome evolution, and the mechanism underlying this type of genomic instability might be consistent with presently accepted findings in other model organisms. However, since the incidence of de novo deletions might be rare, based on the limited variation in deletion polymorphisms, we have only limited information to verify this hypothesis. In contrast, the relatively high frequency of de novo translocations between PATRRs, often detectable in sperm from normal healthy males, should provide some clues regarding the mechanisms involved in translocation formation. De novo translocations could only be detected in sperm, and not in other somatic cells, suggesting that initiation of a PATRR-mediated translocation is likely to be independent of DNA replication unlike palindrome instabilities in other organisms (9). The finding that the frequency of de novo translocations does not appear to be affected by the increasing age of sperm donors also supports this hypothesis (29). Our recent comparative analysis of deletion endpoint sequences with those of translocation breakpoints indicates that replication slippage underlies the mechanism leading to deletions within the palindrome. However, a different mechanism appears to be responsible for translocations, but this hypothesis is still speculative (30).
To explore the mechanism leading to PATRR-mediated deletions and translocations in this study, we modulated DNA replication in human cells and examined the incidence of de novo deletions and translocations. We established a single molecule detection system for de novo deletions, and directly compared the frequency of de novo deletions with that of translocations under similar conditions.
RESULTS
Detection of de novo deletions within PATRR11
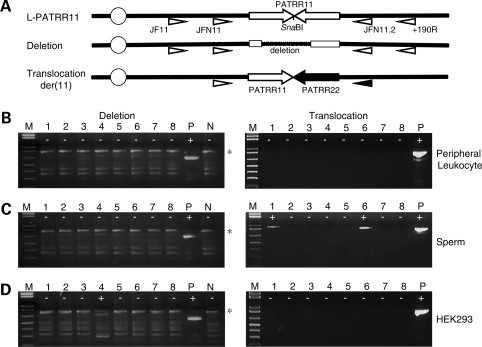
As we have previously reported, PATRR11 is polymorphic. The most common allele is a 450 bp PATRR11 (L-PATRR11) that comprises a nearly perfect palindrome (5). PCR detection of infrequent de novo deletions in template DNA that includes a vast excess of intact L-PATRR is very difficult since the primer set for deletion detection inevitably amplifies the intact PATRR11. We observed that there is a recognition site for the restriction enzyme SnaBI at the center of the L-PATRR11. This site should be missing in a deleted PATRR11 because the majority of deletions include the center. To prevent amplification of intact PATRR11s, we digested genomic DNA prior to performing the PCR for deletion detection. In fact, this procedure cannot completely eliminate the intact PATRR11, since the AT-rich nature of the PATRR11 promotes denaturation of template DNA leading to partial digestion at the central SnaBI site. To further increase the sensitivity, we re-digested the PCR products with SnaBI and performed nested PCR using an internal primer pair (Fig. 1A).
Figure 1.
Detection of de novo deletions and translocations by polymerase chain reaction (PCR). (A) Location of PCR primers. Arrows indicate each arm of the PATRR11 (white arrows) and the PATRR22 (black arrows). Deletion of the PATRR11 was detected by PCR amplification with primers (white triangles) designed on the region flanking to the PATRR11 after digestion of the template DNA with SnaBI. Translocations can be detected using one of the primers flanking the PATRR11 paired with one of the primers flanking the PATRR22 (black triangle). The centromere of chromosome 11 is represented as a circle. (B) Results of deletion-specific nested PCR (left panels) and translocation-specific PCR (right panels) of peripheral blood leukocyte DNA. (C) PCR results of sperm DNA. (D) PCR results of HEK293 cell lines. The largest bands correspond to the PCR products derived from an intact L-PATRR11 (775 bp, asterisk). Background bands observed in all of the lanes are partially or completely denatured molecules of the same PCR product. ‘Plus’ indicates positive PCR reactions, while ‘minus’ indicates negative reactions. M, size markers (1 kb ladder plus); P, positive controls; N, negative controls.
As a preliminary experiment, genomic DNA from an individual carrying a short PATRR11 allele (S-PATRR11) that lacks the central SnaBI site was mixed with various amounts of DNA from a typical L-PATRR11 homozygote to produce a dilution series. We performed nested polymerase chain reaction (PCR) using this serially diluted genomic DNA as template. The results demonstrate that amplification of a single molecule of deletion target sequence could be achieved (Supplementary Material, Fig. S1). The results suggest that this PCR system allows us to detect a single molecule of a deleted version of the PATRR11 in the PCR template of human genomic DNA from an L-PATRR11 homozygote.
Analysis of human samples
We first examined peripheral blood leukocyte DNA obtained from four human males homozygous for the L-PATRR11 allele. Among individuals with various PATRR11 genotypes, they should produce the highest frequency of de novo t(11;22) translocations in sperm a (28). The leukocyte DNAs did not yield any de novo deletion-specific PCR products (Fig. 1B). Translocation-specific PCR did not detect any PCR products in these leukocyte DNAs, either. As was expected, sperm samples from these four individuals yielded translocation-specific PCR products at a high frequency, whereas no de novo deletions were observed in the sperm DNAs (Fig. 1C).
Next, genomic DNAs from four human somatic cell lines (HEK293, HeLa, HepG2 and THP-1) cultured in standard conditions were also examined with the similar strategy. Genotype analyses revealed that all of the cell lines were homozygotes for the L-PATRR11. Highly sensitive PCR detected PCR products derived from de novo deletions in the genomic DNAs from these cell lines (Fig. 1D). Presence of both positive and negative PCRs in a single sample and size heterogeneity of the products provided evidence that the deletions arose de novo. Sequence analysis of the products allowed us to confirm that all of the PCR products were derived from the L-PATRR11s of the cell lines and that all were deleted within the PATRR11. We also confirmed that the deletion endpoints were different among multiple individual deletion products suggesting that each product originated from an independent deletion event. The frequency of de novo deletion was not high and was similar among the cell lines (Table 1). In clear contrast, no translocation-specific PCR products were observed in DNAs from these cell lines (Fig. 1D).
Table 1.
Frequency of the de novo deletions of the PATRR11 among cell lines
| Cell lines | Genotype | Polymerase chain reaction | Frequency |
|---|---|---|---|
| HEK293 | L-PATRR11 homozygotea | 3/40 | 2.4 × 10−6 |
| HeLa | L-PATRR11 homozygote | 3/40 | 2.4 × 10−6 |
| HepG2 | L-PATRR11 homozygote | 4/40 | 3.2 × 10−6 |
| THP-1 | L-PATRR11 homozygote | 3/40 | 2.4 × 10−6 |
aThis cell line is hypotriploid.
Inhibition of DNA polymerase by siRNA induces deletions of the PATRR, but not translocations
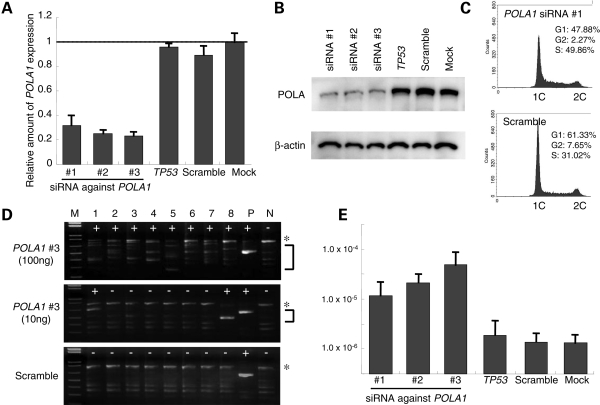
To examine the possible involvement of DNA replication in the mechanism of PATRR-mediated genomic instability in humans, we tested whether modulation of DNA replication affects the rate of PATRR-mediated deletion or translocation in human cell lines. We transfected siRNA against the DNA polymerase alpha 1 (POLA1) gene into human HEK293 cells. We used three different siRNAs against the human POLA1 gene. Samples were obtained at 72 h after transfection. Successful inhibition of POLA1 gene products was achieved by all three siRNAs, which was confirmed by examination of both RNA and protein levels (Fig. 2A and B). We examined the effect of the siRNAs on DNA replication by measuring DNA content. In cells transfected with the siRNA against the POLA1 gene, the height of the peak indicating G1 phase was decreased and the area under the curve indicating S phase was accordingly increased, suggesting that the number of cells in DNA replication is increased by slowed DNA replication (Fig. 2C).
Figure 2.
Partial inhibition of DNA replication by siRNA against the POLA1 gene. (A) Quantitative reverse transcriptase–polymerase chain reaction (RT–PCR) for the POLA1 gene transcript. Vertical axes indicate expression levels relative to GAPDH gene. (B) Western blot using antibodies against the POLA protein (upper panel). The membranes were reprobed with an anti-β-actin antibody (lower panel). (C) Histogram showing the DNA content. Horizontal axes indicate fluorescence intensity of propidium iodide, while vertical axes indicate the number of the cells. Estimated proportions of cells in each phase of the cell cycle are indicated on the upper right corner. (D) Results of PCR for de novo deletions. The asterisks indicate the PCR products derived from an intact L-PATRR11, while PCR products derived from de novo deletions are indicated by the bracket. ‘Plus’ indicates positive PCR reactions, while ‘minus’ indicates negative reactions. M, size markers (1 kb ladder plus); P, positive controls; N, negative controls. (E) Frequency of the de novo deletions. Vertical axis indicates the frequency expressed on a logarithmic scale. All the experiments were repeated three times. Error bars indicate the standard deviation.
We examined genomic DNA for de novo deletions using high-sensitivity PCR. The background deletion frequency of the HEK293 cells (2.4 × 10−6) is so small that the possibility that cells at the initiation of culture (7–10 × 104) would contain those with a deletion is rare. Even if it were to happen, we could still distinguish an inherent deletion from a de novo deletion based on the size of PCR products that should be uniform (Supplementary Material, Fig. S2). In siRNA-treated samples, de novo PCR products were generated in majority of reactions when 100 ng of template DNA was used for each reaction. The PCR products varied in size and were shorter than the product that was derived from the original L-PATRR (Fig. 2D). When 10 ng of DNA was used as template, the number of positive PCR reactions decreased, but the amount of positive PCR products was increased.
The rate of deletion induction for each of the siRNAs against the POLA1 gene was similar (Fig. 2E). Similar experiments were performed using other unrelated siRNAs, resulting in no increase of the de novo deletion frequency (Fig. 2E). We also performed similar experiments using another human cell line, HeLa cells, and the siRNAs induced a similar frequency of de novo deletions. As a negative control, we designed another PCR reaction at the non-palindromic region flanking the PATRR11 that also includes a SnaBI site, but we did not detect any deletions, suggesting that induction of de novo deletions by inhibition of the POLA1 gene is locus-specific (data not shown). Together, the slowed DNA replication induced by siRNA against the POLA1 gene increased the incidence of de novo deletions of the L-PATRR11 in human cell lines.
To examine whether modulation of DNA replication similarly induces PATRR-mediated translocations, we performed t(11;22)-specific PCR using the same genomic DNA samples obtained after the transfection with siRNA of the POLA1 gene. In contrast to the de novo deletions, no PCR product was detected in these samples (data not shown).
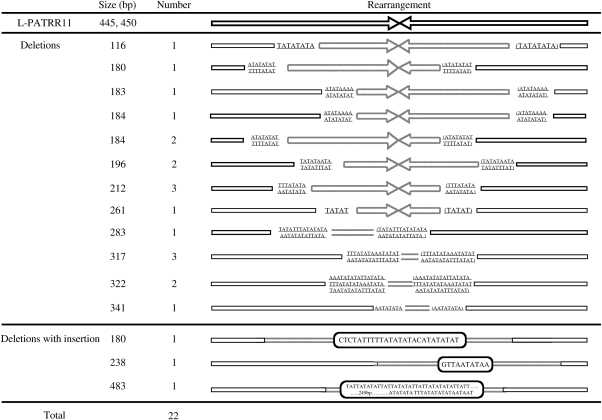
We analyzed sequence of 24 de novo deletions derived from HEK293 cells carrying a 445 bp and a 450 bp L-PATRR11 (GenBank accession nos AB334268 and AB235178, respectively). All of the deletions were asymmetric deletions that included the palindromic center of the PATRR11. The sizes of the deletions were greater than 100 bp. A considerable number of identical nucleotides (5–48 nt) were found at the breakpoints (Fig. 3), suggesting that the PATRRs utilized microhomology for deletion formation. Some of them were found to carry AT-rich insertions of unknown origin at the deletion junction. These properties agreed with those of polymorphic PATRR11 alleles and not with those of de novo t(11;22) translocations (30).
Figure 3.
Analysis of breakpoints of de novo deletions. Arrows indicate each arm of the PATRR11. Upper panel represents simple deletion. Dotted line depicts deleted regions. The nucleotides on the arrows indicate identical nucleotides at both of the proximal and distal breakpoints within the PATRR11. The nucleotides at the distal breakpoints are also shown in brackets. Lower panel represents deletions with insertion. The nucleotides in the boxes are the inserted sequences of unknown origin. Two deletions are not listed here since proximal breakpoints are located at AT-rich region just upstream of the PATRRs.
Administration of DNA replication inhibitors also induces deletion of the PATRR, but not translocation
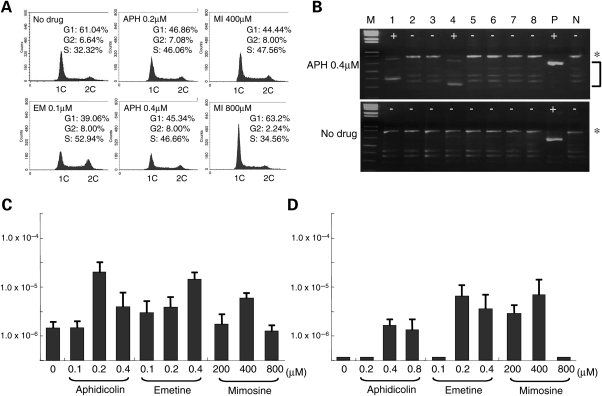
To confirm that inhibiting DNA replication induces deletions within the PATRR, we treated the human cell lines with aphidicolin, a known inhibitor of DNA polymerase alpha and delta. We empirically determined the optimal concentration of aphidicolin that slowed DNA replication without apparently affecting cell growth as 0.2–0.4 µm (Fig. 4A). Twenty-four hours in culture with this low dose of aphidicolin induced de novo deletions (Fig. 4B and C).
Figure 4.
Partial inhibition of DNA replication by administration of inhibitors of DNA replication. (A) Histogram showing the DNA content. Horizontal axes indicate fluorescence intensity of propidium iodide, while vertical axes indicate the number of cells. Estimated proportions of cells in each phase of the cell cycle are indicated on the upper right corner. APH, aphidicolin; EM, emetine; MI, mimosine. (B) Results of PCR for de novo deletion. Asterisks indicate the PCR products derived from an intact L-PATRR11, while PCR products derived from de novo deletion are indicated by a bracket. ‘Plus’ indicates positive PCR reactions, while ‘minus’ indicates negative reactions. M, size markers (1 kb ladder plus); P, positive controls; N, negative controls. (C) Frequency of the de novo deletions in HEK293 cells. (D) Frequency of the de novo deletions in PHA-stimulated peripheral lymphocytes. Vertical axes indicate the frequency expressed on a logarithmic scale. All the experiments were repeated three times. Error bars indicate the standard deviation.
We used other replication inhibitors to further examine the involvement of DNA replication in the induction of deletions at the palindromic region. Emetine, an inhibitor of protein synthesis, also has the potential to inhibit DNA replication. While aphidicolin inhibits both leading- and lagging-strand synthesis, emetine preferentially inhibits lagging-strand synthesis (31). Like aphidicolin, emetine induced de novo deletions at a concentration that induced slow DNA replication without apparently affecting cell growth (<0.4 µm) (Fig. 4A and C).
Mimosine is known as an inhibitor of the initiation of replication origins, but induces a slowed transit through S phase at a lower concentration (32). As was expected, mimosine also induced de novo deletions in a dose-dependent manner. Whereas a high concentration (>800 µm) induced G1 arrest and did not induce any de novo deletions (Fig. 4A and C), a low concentration of mimosine induced de novo deletions at a frequency similar to that of the other drugs, In contrast, we did not detect any t(11;22)-specific PCR products in any of these conditions (data not shown).
To exclude the possibility that this slow replication-dependent induction of de novo deletions is limited to specific cell lines, we performed similar experiments using PHA-stimulated peripheral blood lymphocytes. A standard 72-h culture with PHA did not induce any de novo deletions, but an additional 24 h in culture with aphidicolin, emetine or mimosine induced de novo deletions (Fig. 4D). This suggests that similar mechanisms work not only in cell lines but also in primary cultured cells. The frequency was slightly lower than that which was observed in cell lines, possibly because only a small subset of cells entered the cell cycle and underwent DNA replication in PHA-stimulated peripheral blood leukocytes. In clear contrast, we did not detect any t(11;22)-specific PCR products in PHA-stimulated peripheral blood lymphocytes under these conditions (data not shown).
Deletion of the PATRR is induced either on the leading or lagging strand
The fact that emetine induced de novo deletions supports the idea that deletion at the palindromic DNA induced by slowed DNA replication occurs during lagging-strand synthesis since emetine preferentially inhibits Okazaki fragment synthesis (31). To examine whether or not slowed DNA replication during leading-strand synthesis also induces deletions within the PATRR, we used the topoisomerase inhibitor, camptothecin, which inhibits progression of the replication fork by blocking topoisomerase I activity. We hypothesized that if the slowed POLA trails a more rapidly progressing helicase complex, the resulting long single-stranded DNA segment in the leading strand template might allow a secondary structure to form at the palindromic region leading to replication slippage. Inhibition of the progression of the helicase complex by inhibition of topoisomerase I, which precedes the helicase, will abolish the uncoupling of the polymerase and helicase and subsequent de novo deletion formation.
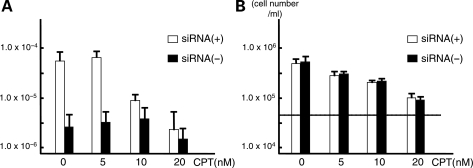
For this purpose, after transfection of siRNA against the POLA1 gene, we added various concentrations of camptothecin to the culture medium. We empirically determined the concentration that does not induce cell growth arrest (<30 nm). We collected the samples at 72 h after transfection. De novo deletion products were induced in cells transfected with siRNA, but addition of camptothecin decreased the de novo deletion frequency dramatically without drastic inhibition of cell growth (at 10 nm) (Fig. 5A and B). The frequency of de novo deletions was further decreased at 20 nm, but at this concentration the frequency of deletions in the absence of siRNA also decreased, mirroring the cell growth inhibition (Fig. 5A and B). These results indicate that addition of camptothecin partially inhibited the production of de novo deletions induced by slowed replication, suggesting that a subset of de novo deletions was generated at leading-strand synthesis (Fig. 6A).
Figure 5.
Induction of de novo deletion is abolished by addition of topoisomerase inhibitor. (A) Frequency of the de novo deletions. Horizontal axis indicates concentration of camptothecin, while vertical axis indicates the frequency of de novo deletions expressed on a logarithmic scale. Similar experiments were performed with or without transfection of siRNA against the POLA1 gene (white and black bars, respectively). (B) Cell numbers at 72 h after administration of camptothecin. Dotted line indicates initial number of cells (5 × 104/ml). The experiments were repeated three times. Error bars indicate the standard deviation.
Figure 6.
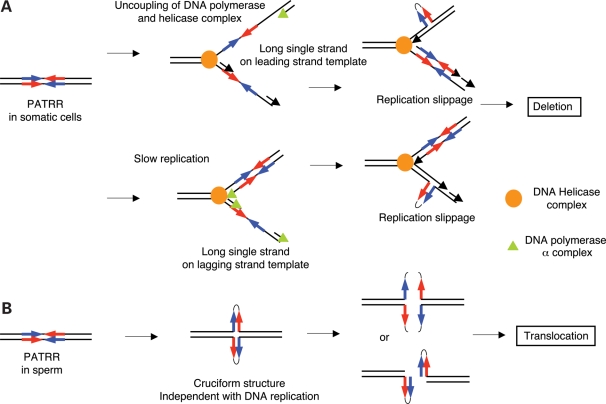
Mechanism of palindromic instabilities in humans. (A) Slow DNA polymerase-induced deletion of the PATRR. Inhibition of DNA polymerase by siRNA or aphidicolin uncouples the helicase complex and DNA polymerase complex. Long single-strand DNA in the leading strand template facilitates a hairpin formation at the palindromic region that may induce deletion by slippage (upper panel). Slow replication also promotes long single-strand DNA in the lagging strand template that may lead to secondary structure formation at the palindromic region (lower panel). (B) PATRR-mediated translocation. The mechanism which dictates that translocation may be driven by DNA replication-independent cruciform structure.
DISCUSSION
It has been challenging to detect de novo deletions of the PATRR by PCR similar to the translocation-detection system. A highly sensitive PCR system for translocation detection at the single molecule level is useful based on the fact that two primers that are located on different chromosomes cannot amplify PCR products in the normal situation, but can yield specific PCR products only in the presence of the translocation. However, a primer set for deletion detection can amplify PCR products derived from de novo deletions as well as those from the non-deleted PATRR, which constitutes the majority of the DNA in the template and inhibits amplification from a single molecule of a de novo deletion. In this study, we successfully established a highly sensitive detection system for deletion detection at a single molecule level. Using this system, we, for the first time, have detected de novo deletions of the PATRR in cultured human cells. Despite an anticipated bias owing to inability to detect larger deletion, this system is a useful method to monitor de novo deletion frequencies and compare them among various experimental conditions.
Since the PATRR has multiple sizes of short asymmetric variants, it has been anecdotal that the PATRR may undergo deletion in the course of human genome evolution (28). The basis of this hypothesis is that sequence analysis found all of the minor alleles of the PATRR11 to be derivatives of the typical L-PATRR11 (28,30). Under-representation of Alu-inverted repeats in human genome databases relative to direct repeats lends support to this hypothesis (33). Indeed, we detected de novo deletions of the PATRR, whose sequence properties support this hypothesis. The frequency was very rare as was predicted by the limited variation of size polymorphisms relative to the heterogeneity of translocation breakpoints (28,29). Thus, most of the polymorphic PATRR variants are likely transmitted from a small number of ancestral founders.
Although de novo deletion was rarely observed in cells cultured in standard conditions, the deletion frequency was remarkably increased in cells with slowed DNA replication. It is easy to imagine that replication of the palindromic region can be slowed or even stalled because of intra-strand base-pairing, which might lead to the formation of a hairpin structure. Perturbation of replication fork progression by inhibition of POLA would facilitate hairpin formation. Replication slippage is the major pathway for producing a deletion, which results from the formation of a free end at the stalled DNA followed by homology-dependent repair. Another important pathway is endonuclease cleavage of DNA secondary structure (18). The RAD50/MRE11 nuclease complex is known to cleave hairpin DNA that is formed in the replication fork (34).
Most of the examples of genomic instability originating from non-B DNAs have been explained by these replication-dependent phenomena. Some of the fragile sites in human chromosomes are suggestive of the involvement of DNA secondary structure as contributing to genomic instability. FRA3B manifests late replication and is sensitive to further replication delay by inhibitors of DNA strand synthesis leading to the induction of fragility (35,36). FRA16D, composed of highly AT-rich tandem repeats, is susceptible to DNA breaks during replication fork progression, and its instability is exacerbated by replication inhibitors (37). Trinucleotide repeat expansions are believed to arise by the formation of secondary structure in a lagging strand of the replication fork and are induced by replication inhibitors (38). Further, aphidicolin-induced replication stress leads to de novo deletions and duplications of tens to thousands of kilobases in human cells (39). All these data are consistent with our current findings.
It is formally accepted that slow progression of the replication fork increases the chance of forming secondary structures at long tracts of single-stranded DNA segments in the lagging strand template (17). Deletion is mainly mediated by slippage of the lagging strand because of this relative single-strandedness of the lagging strand template in the replication fork (Fig. 6A, lower panel). However, our data demonstrate that the palindromic deletions also occur on the leading strand synthesized under conditions when DNA replication is partially inhibited. Slowed DNA polymerase possibly uncouples the replicative helicase complex from the polymerase complex leading to long tracts of single-stranded DNA in the leading strand template (Fig. 6A, upper panel) (40). Since preference for deletion formation in lagging-strand synthesis is mainly based on findings observed in E. coli, the current observations might be specific to mammalian DNA. This deserves further investigation by differential inhibition of DNA polymerase delta and epsilon using siRNA.
Regarding translocation, slow DNA replication has been shown to induce translocations in yeast (23). However, we did not find any de novo translocations between the PATRRs in the presence of slowed DNA replication, where de novo deletion within the PATRR occurs. This suggests that the mechanism that leads to PATRR-mediated translocation in humans is totally different from the mechanism that drives deletions in humans, and also from what was found in yeast (30). The fact that polymorphism of the PATRR affects the frequency of de novo translocation implicates the involvement of potential secondary structure in PATRR-mediated translocations in humans (28,41,42). Recent experimental data suggest that palindromic sequence forms DNA hairpins, not cruciform structure, during DNA replication, which is responsible for replication-related palindromic instability (43). On the other hand, we established a plasmid-based translocation model using non-replicating plasmids and, in fact, only cruciform-extruding plasmids induce translocation-like rearrangements (44). Indeed, the observation of de novo translocations only in sperm (9) as well as the absence of an age-dependent increase in the translocation frequency (29) also appears to be inconsistent with the involvement of DNA replication in the process. Thus, it is conceivable that PATRR-mediated rearrangement is generated by genomic instability via a replication-independent DNA cruciform conformation (Fig. 6B).
MATERIALS AND METHODS
Assessment of de novo deletion and translocation by polymerase chain reaction
Genomic DNA was extracted using PUREGENE (Gentra) according to the manufacturer's protocol. Nested PCR was used for the detection of de novo deletions. PCR primers for amplification of the PATRR11 were designed as described previously with minor modification (45). Five micrograms of genomic DNA was digested with SnaBI for 16 h at 25°C. The conditions for the first PCR reaction were 40 cycles of 10 s at 98°C and 1 min at 60°C. After purification using columns (QIAGEN), the PCR products were again digested with SnaBI followed by column purification. The second PCR was performed using internal primers with the following condition: 35 cycles of 10 s at 98°C and 1 min at 60°C.
We amplified multiple batches of 100 ng of genomic DNA, each containing 33 000 haploids (n). The frequency of de novo deletion events was calculated as follows. We counted the number of positive PCR reactions per total reactions (p). The translocation frequency (q) was calculated on the basis that the probability of a positive PCR reaction corresponds to a total sum of a binomial series of the deletion frequency calculated using the equation: p = 1−(1 − q)n as described previously (9).
Translocation-specific PCR was performed using appropriate primer pair to amplify the der(11) junction fragment. One primer was located at the proximal region flanking the PATRR11 and the other at the distal region flanking the PATRR22. The PCR condition was 40 cycles of 10 s at 98°C and 5 min at 60°C. All frequency data were calculated based on the number of positive PCR reactions out of 40.
siRNA depletion
Human somatic cell lines (HEK293 and HeLa) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum on 10 cm dishes. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2. Logarithmically growing cells from the same initial plate were split into control and test aliquots. A total of 7–10 × 104 cells was cultured until 20% visual confluence and then subjected to transfection.
Three oligo sets (Stealth Select RNA, Invitrogen) were used for siRNA depletion of the POLA1 gene in HEK293 or HeLa cells; POLA-HSS108205 (sense: AAUAUUCUCAGUUCCACAUGUAGGG), POLA-HSS108206 (sense: UGGACAAGUCUACAGCUUUAUCUGC) and POLA-HSS108207 (sense: UAGAAUGUCACCUAGCAGACCAUCC). Another set of siRNA oligos against p53 as well as scrambled oligos were used as negative controls. The cells were harvested 72 h after transfection with the aid of Lipofectamine 2000 diluted in Opti-MEM (Invitrogen). Transfection efficiency measured by transfection of fluorescent oligos using FACScan (BD Bioscience) was greater than 95% in all experiments.
Total RNA was extracted using RNeasy (Qiagen). dT-primed cDNA was synthesized with the aid of Superscript III (Invitrogen). Quantitative RT-PCR for the POLA1 gene was performed with the TaqMan GEX primer/probe (POLA; Hs00213524, GAPDH; Hs99999905) using the ABI7300 Real Time PCR System (Applied Biosystem). For the TP53 gene, the LUX primer set (Invitrogen) was used for quantification of the expression level. The western blot was performed using goat polyclonal anti-POLA antibodies (sc-5921, Santa Cruz) and anti-β-actin antibody (Sigma–Aldrich).
DNA content was assessed by staining with propidium iodide (PI). Cells were lysed with 0.15% Triton X-100 in phosphate-buffered saline (PBS), treated with RNaseA (5 mg/ml) and then stained with 35 µg/ml of PI, followed by analysis on FACScan using CellQuest and ModFitLT software (BD).
Treatment with DNA replication inhibitors
Aphidicolin (Calbiochem) was dissolved in dimethyl sulfoxide (DMSO) to prepare a 1 mm solution. Emetine (Sigma) was dissolved in PBS to prepare a 1 mm solution. Mimosine (Sigma) was dissolved in 0.4 N NaOH and then pH adjusted to 8.4 prior to dilution with PBS (pH 8.4) to prepare a 10 mm solution. Logarithmically growing HEK293 or HeLa cells at 30–40% visual confluence were treated with various concentrations of these drugs for 24 h prior to harvest. We empirically determined the optimal concentration for slowed DNA replication. To partially inhibit DNA synthesis, we set the drug concentration for the experiments lower than the concentration that induced growth inhibition of the cell lines. Similar experiments were also performed on peripheral blood lymphocytes obtained from an L-PATRR11 homozygote. The various concentrations of drugs were added to lymphocytes cultured for 24 h starting at 72 h after stimulation with 10 µg/ml of PHA-P (Wako).
Camptothecin (Sigma) was dissolved in DMSO at 1 mg/ml (2.87 mm). Optimal concentration for slowed DNA replication was also empirically determined so as not to induce growth arrest. Cells (5.0 × 104/ml) with or without transfection of siRNA against the POLA1 gene were treated with camptothecin (0–20 nm) for 72 h prior to harvest.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by a grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to H.K.), and by a grant from the National Institutes of Health, USA (CA39926 to B.S.E.) and funds from the Charles E. H. Upham endowed chair (to B.S.E.).
Conflict of Interest statement. We declare no conflicts of interest.
REFERENCES
- 1.Kurahashi H., Inagaki H., Ohye T., Kogo H., Kato T., Emanuel B.S. Palindrome-mediated chromosomal translocations in humans. DNA Repair (Amst) 2006;b 5:1136–1145. doi: 10.1016/j.dnarep.2006.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurahashi H., Inagaki H., Ohye T., Kogo H., Kato T., Emanuel B.S. Chromosomal translocations mediated by palindromic DNA. Cell Cycle. 2006;5:1297–1303. doi: 10.4161/cc.5.12.2809. [DOI] [PubMed] [Google Scholar]
- 3.Zackai E.H., Emanuel B.S. Site-specific reciprocal translocation, t(11;22) (q23;q11), in several unrelated families with 3:1 meiotic disjunction. Am. J. Med. Genet. 1980;7:507–521. doi: 10.1002/ajmg.1320070412. [DOI] [PubMed] [Google Scholar]
- 4.Kurahashi H., Shaikh T.H., Hu P., Roe B.A., Emanuel B.S., Budarf M.F. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum. Mol. Genet. 2000;a 9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 5.Kurahashi H., Emanuel B.S. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 2001;a 10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- 6.Kurahashi H., Inagaki H., Hosoba E., Kato T., Ohye T., Kogo H., Emanuel B.S. Molecular cloning of a translocation breakpoint hotspot in 22q11. Genome Res. 2007;17:461–469. doi: 10.1101/gr.5769507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edelmann L., Spiteri E., Koren K., Pulijaal V., Bialer M.G., Shanske A., Goldberg R., Morrow B.E. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am. J. Hum. Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tapia-Paez I., Kost-Alimova M., Hu P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P. The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum. Genet. 2001;109:167–177. doi: 10.1007/s004390100560. [DOI] [PubMed] [Google Scholar]
- 9.Kurahashi H., Emanuel B.S. Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat. Genet. 2001;b 29:139–140. doi: 10.1038/ng1001-139. [DOI] [PubMed] [Google Scholar]
- 10.Budarf M.L., Eckman B., Michaud D., McDonald T., Gavigan S., Buetow K.H., Tatsumura Y., Liu Z., Hilliard C., Driscoll D., et al. Regional localization of over 300 loci on human chromosome 22 using a somatic cell hybrid mapping panel. Genomics. 1996;35:275–288. doi: 10.1006/geno.1996.0358. [DOI] [PubMed] [Google Scholar]
- 11.Spiteri E., Babcock M., Kashork C.D., Wakui K., Gogineni S., Lewis D.A., Williams K.M., Minoshima S., Sasaki T., Shimizu N., et al. Frequent translocations occur between low copy repeats on chromosome 22q11.2 (LCR22s) and telomeric bands of partner chromosomes. Hum. Mol. Genet. 2003;12:1823–1837. doi: 10.1093/hmg/ddg203. [DOI] [PubMed] [Google Scholar]
- 12.Kehrer-Sawatzki H., Haussler J., Krone W., Bode H., Jenne D.E., Mehnert K.U., Tummers U., Assum G. The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum. Genet. 1997;99:237–247. doi: 10.1007/s004390050346. [DOI] [PubMed] [Google Scholar]
- 13.Kurahashi H., Shaikh T.H., Takata M., Toda T., Emanuel B.S. The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am. J. Hum. Genet. 2003;72:733–738. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nimmakayalu M.A., Gotter A.L., Shaikh T.H., Emanuel B.S. A novel sequence-based approach to localize translocation breakpoints identifies the molecular basis of a t(4;22) Hum. Mol. Genet. 2003;12:2817–2825. doi: 10.1093/hmg/ddg301. [DOI] [PubMed] [Google Scholar]
- 15.Gotter A.L., Shaikh T.H., Budarf M.L., Rhodes C.H., Emanuel B.S. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum. Mol. Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gotter A.L., Nimmakayalu M.A., Jalali G.R., Hacker A.M., Vorstman J., Conforto Duffy D., Medne L., Emanuel B.S. A palindrome-driven complex rearrangement of 22q11.2 and 8q24.1 elucidated using novel technologies. Genome Res. 2007;17:470–481. doi: 10.1101/gr.6130907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trinh T.Q., Sinden R.R. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature. 1991;352:544–547. doi: 10.1038/352544a0. [DOI] [PubMed] [Google Scholar]
- 18.Leach D.R. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 19.Bzymek M., Lovett S.T. Evidence for two mechanisms of palindrome-stimulated deletion in Escherichia coli: single-strand annealing and replication slipped mispairing. Genetics. 2001;158:527–540. doi: 10.1093/genetics/158.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nag D.K., White M.A., Petes T.D. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989;340:318–320. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- 21.Gordenin D.A., Lobachev K.S., Degtyareva N.P., Malkova A.L., Perkins E., Resnick M.A. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol. Cell Biol. 1993;13:5315–5322. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruskin B., Fink G.R. Mutations in POL1 increase the mitotic instability of tandem inverted repeats in Saccharomyces cerevisiae. Genetics. 1993;134:43–56. doi: 10.1093/genetics/134.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemoine F.J., Degtyareva N.P., Lobachev K., Petes T.D. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 24.Collick A., Drew J., Penberth J., Bois P., Luckett J., Scaerou F., Jeffreys A., Reik W. Instability of long inverted repeats within mouse transgenes. EMBO J. 1996;15:1163–1171. [PMC free article] [PubMed] [Google Scholar]
- 25.Akgun E., Zahn J., Baumes S., Brown G., Liang F., Romanienko P.J., Lewis S., Jasin M. Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol. 1997;17:5559–5570. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunningham L.A., Cote A.G., Cam-Ozdemir C., Lewis S.M. Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol. Cell. Biol. 2003;23:8740–8750. doi: 10.1128/MCB.23.23.8740-8750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inagaki H., Ohye T., Kogo H., Yamada K., Kowa H., Shaikh T.H., Emanuel B.S., Kurahashi H. Palindromic AT-rich repeat in NF1 gene is hypervariable in human and evolutionally conserved among primates. Hum. Mutat. 2005;26:332–342. doi: 10.1002/humu.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato T., Inagaki H., Yamada K., Kogo H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H. Genetic variation affects de novo translocation frequency. Science. 2006;311:971. doi: 10.1126/science.1121452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato T., Yamada K., Inagaki H., Kogo H., Ohye T., Emanuel B.S., Kurahashi H. Age has no effect on de novo constitutional t(11;22) translocation frequency in sperm. Fertil. Steril. 2007;88:1446–1448. doi: 10.1016/j.fertnstert.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato T., Inagaki H., Kogo H., Ohye T., Yamada K., Emanuel B.S., Kurahashi H. Two different forms of palindrome resolution in the human genome: deletion or translocation. Hum. Mol. Genet. 2008;17:1184–1191. doi: 10.1093/hmg/ddn008. [DOI] [PubMed] [Google Scholar]
- 31.Burhans W.C., Vassilev L.T., Wu J., Sogo J.M., Nallaseth F.S., DePamphilis M.L. Emetine allows identification of origins of mammalian DNA replication by imbalanced DNA synthesis, not through conservative nucleosome segregation. EMBO J. 1991;10:4351–4360. doi: 10.1002/j.1460-2075.1991.tb05013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krude T. Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp. Cell Res. 1999;247:148–159. doi: 10.1006/excr.1998.4342. [DOI] [PubMed] [Google Scholar]
- 33.Stenger J.E., Lobachev K.S., Gordenin D., Darden T.A., Jurka J., Resnick M.A. Biased distribution of inverted and direct Alus in the human genome: implications for insertion, exclusion, and genome stability. Genome Res. 2001;11:12–27. doi: 10.1101/gr.158801. [DOI] [PubMed] [Google Scholar]
- 34.Lobachev K.S., Gordenin D.A., Resnick M.A. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 35.Le Beau M.M., Rassool F.V., Neilly M.E., Espinosa R., III, Glover T.W., Smith D.I., McKeithan T.W. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Hum. Mol. Genet. 1998;7:755–761. doi: 10.1093/hmg/7.4.755. [DOI] [PubMed] [Google Scholar]
- 36.Durkin S.G., Ragland R.L., Arlt M.F., Mulle J.G., Warren S.T., Glover T.W. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc. Natl Acad. Sci. USA. 2008;105:246–251. doi: 10.1073/pnas.0708097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H., Freudenreich C.H. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol. Cell. 2007;27:367–379. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z., Lau R., Marcadier J.L., Chitayat D., Pearson C.E. Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells. Am. J. Hum. Genet. 2003;73:1092–1105. doi: 10.1086/379523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arlt M.F., Mulle J.G., Schaibley V.M., Ragland R.L., Durkin S.G., Warren S.T., Glover T.W. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am. J. Hum. Genet. 2009;84:339–350. doi: 10.1016/j.ajhg.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cortez D. Unwind and slow down: checkpoint activation by helicase and polymerase uncoupling. Genes Dev. 2005;19:1007–1012. doi: 10.1101/gad.1316905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurahashi H., Inagaki H., Yamada K., Ohye T., Taniguchi M., Emanuel B.S., Toda T. Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J. Biol. Chem. 2004;279:35377–35383. doi: 10.1074/jbc.M400354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kogo H., Inagaki H., Ohye T., Kato T., Emanuel B.S., Kurahashi H. Cruciform extrusion propensity of human translocation-mediating palindromic AT-rich repeats. Nucleic Acids Res. 2007;35:1198–1208. doi: 10.1093/nar/gkm036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voineagu I., Narayanan V., Lobachev K.S., Mirkin S.M. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl Acad. Sci. USA. 2008;105:9936–9941. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inagaki H., Ohye T., Kogo H., Kato T., Hasbaira B., Taniguchi M., Shaikh T.H., Emanuel B.S., Kurahashi H. Chromosomal instability mediated by non-B DNA: cruciform conformation and not DNA sequence is responsible for recurrent translocation in humans. Genome Res. 2009;19:191–198. doi: 10.1101/gr.079244.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurahashi H., Shaikh T.H., Zackai E.H., Celle L., Driscoll D.A., Budarf M.L., Emanuel B.S. Tightly clustered 11q23 and 22q11 breakpoints permit PCR-based detection of the recurrent constitutional t(11;22) Am. J. Hum. Genet. 2000;b 67:763–768. doi: 10.1086/303054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.